Abstract
Worldwide, Urinary Tract Infections (UTIs) are an important health problem with many cases reported annually, women being the most affected. UTIs are relevant because they can become a recurrent condition, associated with different factors that contribute to the chronicity of the disease (cUTI). cUTI can be classified as persistent (peUTI) when the causative agent is the same each time the infection occurs or as reinfection (reUTI) when the associated microorganism is different. The purpose of this work was to characterize Escherichia coli isolates obtained in two prospective studies of patients with cUTI, to define which of them corresponded to peUTI and which to reUTI. A total of 394 isolates of E. coli were analyzed by agglutination with specific sera, antimicrobial susceptibility by diffusion disc test, and the phylogroups and presence of genes associated with virulence by PCR assays. Additionally, in some characterized strains adherence, invasiveness, and biofilm formation were analyzed by in vitro assays. The results showed that the peUTI strains belonged mainly to the classical UPEC serogroups (O25, O75, O6), were included in the B2 phylogroup, carried a great number of virulence genes, and were adherent, invasive, and biofilm-forming. Meanwhile, reUTI strains showed great diversity of serogroups, belonged mainly in the A phylogroup, and carried fewer virulence genes. Both peUTI and reUTI strains showed extensively drug-resistant (XDR) and multidrug-resistant (MDR) profiles in the antimicrobial susceptibility test. In conclusion, it appears that peUTIs are caused principally by classical UPEC strains, while reUTIs are caused by strains that appear to be a part of the common E. coli intestinal biota. Moreover, although both peUTI and reUTI strains presented different serotypes and phylogroups, their antimicrobial resistance profile (XDR and MDR) was similar, confirming the importance of regulating prophylactic treatments and seeking alternatives for the treatment and control of cUTI. Finally, it was possible to establish the features of the E. coli strains responsible for peUTI and reUTI which could be helpful to develop a fast diagnostic methodology.
Keywords: uropathogenic Escherichia coli, persistent UTI, reinfection UTI, urovirulence genes, drug-resistance
1. Introduction
With the development of antibiotic therapy in the mid-20th century, the clinical impact and devastating effect of infectious diseases was controlled. However, the inappropriate use of antimicrobials in the treatment of these diseases and their use in areas such as agribusiness and others, has been the cause of a significant increase of antimicrobial resistant bacteria, causing global population to currently face a major public health problem (1). Urinary Tract Infections (UTIs) represent a major health problem worldwide (2), predominantly more common in women and, although, UTIs occur at any stage of life, their highest incidence occurs between the ages of 15 and 24 years-old and in the post-menopausal period (3). The higher frequency of UTI in women has been linked to the proximity between the anus and the urethra, a situation that facilitates the contamination of the latter by feces (2, 4). Another relevant aspect of UTIs is their high rate of recurrence, thus becoming a chronic condition and affecting the quality of life due to the condition itself and the emotional and psychological effect it entails. In addition to the negative effects, their frequency with which cUTIs occur has an impact on the patient’s economy, either because of the medical costs or the need to be absent from work (5, 6). cUTIs are characterized by recurrent infections, which in turn are defined as persistent infections (peUTI), when the responsible microorganism is always the same; or reinfection (reUTI), in cases where the microorganism is different each time a new UTI occurs. To explain the etiology of both models it is pointed out that in peUTI, after the initial infection the pathogen establishes itself and resides within the epithelium of the urinary tract, avoiding the immune response of the host and protecting itself from the effect of antimicrobials (7). While the diversity of bacteria related to reUTI may be explained by the lack of genetic information that allows bacteria to maintain a stable form within the epithelium of the urinary tract, and therefore they can be eliminated more easily. However, depending on the patient’s susceptibility, they can be infected again through fecal contamination of the urethra by common intestinal microorganisms, or by some other mechanisms related to the patient’s poor hygienic habits (4). The treatment for both peUTI and reUTI is the prescription of antimicrobials, whose constant use alters the normal microbiota of the bladder, vagina, and gastrointestinal tract, favoring the selection and proliferation of multidrug-resistant strains. Escherichia coli is the main bacterial agent associated with UTI and is responsible for 70–95% of community-acquired infections and approximately 50% of nosocomial UTI cases (8–10). E. coli is a bacterium with a genome that presents great plasticity, a fact that has favored its great diversity formed by commensal clones that naturally colonize the intestine and contribute to the proper functioning of the host organism. However, in the evolutionary process, E. coli has generated pathogenic clones (pathotypes) that can cause intestinal diseases called diarrheogenic (DEC), and others more related to infections outside the intestine, the case of UTIs, named extraintestinal E. coli (ExTEC). The phenotypic diversity of E. coli is related to the composition of the different somatic (O) and flagellar (H) antigens expressed by the bacterium; both react to specific sera allowing to classify strains in different serogroups (O antigen only) and serotypes (both O and H antigens). By this method, some E. coli strains associated with UTIs have been designated as classic uropathogenic E. coli (UPEC) (11, 12). In the laboratory, we have the complete sera scheme to perform the antigenic characterization of E. coli, which allows us to know the serogroup and serotype of the bacterium and thus define the variety that causes the different types of UTIs. Previously, in two prospective studies conducted by our research group, urine samples from patients with cUTI were collected and analyzed monthly, followed up for a period of seven to 18 months, varying depending on the patient. From each positive urine culture, 10 colonies were selected, identified using biochemical tests, and serotyped; the strains were frozen for preservation and subsequent characterization (13, 14). The aim of this study was to perform the phenotypic and genotypic characterization of some of the E. coli strains previously isolated from the already mentioned studies, with the interest of analyzing the behavior of the isolates obtained from patients with peUTI and reUTI and thus propose strategies for the diagnosis, treatment, and potential control of cUTI.
2. Materials and methods
2.1. Bacteria
The study included 394 Escherichia coli isolates obtained from 131 urine samples from 39 adults and 17 children who participated in the two previously mentioned prospective studies of cUTI (13, 14).
2.2. Serotyping of Escherichia coli isolates
Escherichia coli strains were serotyped in a previous work (13, 14). In brief, the agglutination assays were performed using 96-well microtiter plates and rabbit antisera against O1 to O187 somatic (O) antigens and 53 flagellar (H) antigens prepared in rabbits (SERUNAM, registered trademark in Mexico, number 323,158/2015) using the method described by Orskov and Orskov (15).
2.3. Antimicrobial susceptibility
To assess antimicrobial susceptibility, the disk diffusion method was performed following the protocol described by Clinical and Laboratory Standards Institute (CLSI) (16). Susceptibility to 34 antimicrobials (Oxoid, United Kingdom) was assessed for 125 and 269 isolates from peUTI and reUTI, respectively. Discs impregned with ampicillin (AMP) 10 μg, piperacillin (PRL) 100 μg, carbenicillin (CAR) 100 μg, mecilanam (MEL) 10 μg, amox-clavulanic acid (AMC) 20–10 μg, piperacillin-tazobactam (TZP) 100–10 μg, cefazolin (KZ) 30 μg, cephalothin (KF) 30 μg, cefamandol (MA) 30 μg, cefepime (FEP) 30 μg, cefoperazone (CFP) 75 μg, cefoxitin (FOX) 30 μg, ceftriaxone (CRO) 30 μg, ceftazidime (CAZ) 30 μg, furoxime (CXM) 30 μg, meropenem (MEM) 10 μg, nitrofurantoin (F) 300 μg, aztreonam (ATM) 30 μg, gentamicin (CN) 10 μg, amikacin (KA) 30 μg, kanamycin (K)30 μg, tobramycin (TOB) 10 μg, streptomycin (S) 10 μg, tetracycline (TE) 30 μg, ciprofloxacin (CIP) 5 μg, norfloxacin (NOR) 10 μg, trimethoprim-sulfamethoxazole (SXT) 1.5/23.75 μg, nalidixic acid (NA) 30 μg, sulfonamides (S3) 250/300 μg, trimetroprim (W) 5 μg, chloramphenicol (C) 30 μg, fosfomycin (FOS) 200 μg, fosfomycin trometramol (FOT) 200 μg were used. The results were interpreted considering the diameter of the inhibition halo, categorized as susceptible (S), intermediate (I) and resistant (R) according to the CLSI (16) criteria, the E. coli ATCC 25922 reference strain was used as a control. Isolates classified as intermediate were re-classified as resistant, multidrug-resistant (MDR) strains were defined as: resistant to ≥1 agent in ≥3 antibiotic categories, extensively drug-resistant (XDR): resistant to ≥1 agent in all but <2 categories and pandrug-resistant (PDR) when resistant to all antimicrobial agents used (17). For each isolate, a resistance score was calculated, defined as the number of antibiotics to which that strain was resistant among the 34 antibiotics tested (18).
2.4. PCR assays
2.4.1. DNA extraction
DNA extraction was performed according to the manufacturing instructions of InstaGene Matrix kit (Bio-Rad, United States).
2.4.2. Phylogenetic analysis
Using the multiplex PCR method described by Clermont et al. (19), primers for chuA, yejA, and TspE4.C2 were used to identify the phylogenetic group of the different E. coli isolates (A, B1, B2, and D).
2.4.3. Virulence genes
Multiplex PCR (endpoint) was used to analyze the presence of 15 virulence genes associated with adhesion proteins (fimH, papA, papC), toxin production (sat), iron uptake (feoB, ireA, irp-2, sitA, iutA, fyuA), capsule synthesis (kpsMT-K1), pathogenicity island I (malX), and the enzyme associated with the degradation of antimicrobial peptides (ompT). Simplex assays were performed for sat, papA, ompT, iroND, malX, and duplex assays for the combinations (1) ibeA, iutA; (2) feoB, sitA; (3) fimH, ireA; (4) irp-2, kpsMTII and (5) fluA and fluB. The sequences of the primers are listed in Supplementary Table S1, PCR assays were performed under the following conditions: final volume 10 μL, reaction mixture consisting of 1.0 μL DNA, 0.4 μL (10 μM) of each primer, 5 μL (2X) PCR Master Mix (Thermo Scientific. United States). Amplification was performed using a MiniAmp thermal cyclerTM (Applied Biosystems., United States) according to the following conditions: denaturation step for 2 min (95°C); amplification of 30 cycles for 30 s (95°C), annealing temperature for 30 s, 1 min at 72°C and a final extension step for 7 min at 72°C (Supplementary Table S1). PCR products were analyzed by electrophoretic run using agarose gels (1.2%), stained with ethidium bromide (0.01%), and were visualized using a Cleaver Scientific TTD model OmniDoc Gel Documentation System (United Kingdom) ultraviolet light transilluminator. A virulence score was calculated for each isolate, defined as the number of virulence genes present in the strain with respect to the 15 genes analyzed (18).
2.5. In vitro assays
Some of the studied E. coli strains were selected to evaluate their in vitro properties in terms of adherence, invasiveness, and biofilm formation capacity to subsequently identify if there was a correlation between these properties and the cUTI type.
2.5.1. Adherence to cells
For this assay, 72 strains isolated from 37 urine cultures belonging to 12 patients with reUTI and 7 with peUTI who presented a positive urine culture for >3 months were selected. The presence of genes associated with fimbrial adhesins was the selection criterion for the E. coli strains used in this assay. The procedure described by Cravioto et al. (20) was used with some modifications. Briefly, 1 mL (2.5×105) of HEp-2 cells in suspension (ATCC CCL-23) was plated in a 24-well tissue culture microplate (Costar® United States) containing sterile 13 mm slides (Nunc Brand Products® United States) and Minimum Essential Medium (MEM) (Invitrogen. United States), supplemented with 10% Fetal Bovine Serum (FBS) (EquiLab, Canada). Cells were incubated for 24 h at 37° C in a 3% CO2 atmosphere, upon reaching 90% confluence the medium was removed and washed three times with sterile Phosphate-Buffered Saline (PBS) (1x), the bacterial inoculum was prepared from a previous culture (grown overnight) by adjusting the suspension to a concentration of 3.0 × 108 CFU/mL in MEM without FBS and antibiotic, and supplemented with 100 μL of 10% D-mannose, 1 mL of this was added to each well. The plates were incubated for 3 h at 37°C in 3% CO2 atmosphere, at the end of this time the medium with bacteria was removed and wells were washed twice with 1x PBS, the cell monolayer was fixed with methanol for 10 min and finally stained with 1% Giemsa. The number of adherent bacteria was counted independently by two persons using a microscope (100X), analyzing at least 15 fields of each preparation. The result was expressed as the average number of adherent bacteria per cell per duplicate assay. The following strains were used to evaluate adherence phenotypes: E. coli E2348/69 (localized adherence), E. coli 87,125 (diffuse adherence), E. coli 49,766 (aggregative adherence) and a non-adherent E. coli strain HB101.
2.5.2. Cell invasion
In this assay, 54 strains isolated from 27 urine cultures from 7 peUTI and 10 reUTI patients who had a positive urine culture for more than 3 consecutive months were tested. Shigella boydii 21639 and E. coli 1124 were used as positive controls and E. coli HB101 as a negative control. For invasion assays, the procedure described by Elsinghorst (21) with modifications was used. Briefly, HEp-2 cells (ATCC CCL-23) were cultured and infected following the same conditions described in the adherence assay. In this assay a first incubation of 3 h was performed, the medium was removed from the wells and washed three times with sterile PBS to eliminate the bacteria that failed to invade, 1 mL of MEM with Gentamicin (100 mg/mL) and lysozyme (300 mg/mL) was added, the plates were incubated again for 3 h at 37°C (22). At the end of the new incubation period the culture was washed twice with 1x PBS and the culture was fixed with methanol for 15 min and stained with 1% Giemsa for 20 min. The preparations were observed under the microscope and the cells with intracellular bacteria were counted. A bacterial quantification test was performed, briefly the culture medium was removed and 500 μL of Triton (0.1%) was added to each well for 15 min at room temperature to break the HEp-2 cells. The suspension obtained was subjected to serial ten-fold dilutions and plate count was performed using the drop-plate technique (10 μL) on MacConckey agar. The assay was repeated in two independent experiments, and the data analyzed was the average of invasive bacteria.
2.5.3. Biofilms
The biofilm formation capacity of 80 UPEC strains, 68 selected from 10 patients with peUTI and 12 from 3 patients with reUTI, all with a follow-up of more than 3 months, and with the presence of adherence-related genes, was analyzed. The method described by Christensen et al. (23) was used with minor modifications. Briefly, bacteria were inoculated in Luria Bertani broth (LB) and incubated at 37°C with constant shaking (200 rpm) until a 1 McFarland turbidity standard suspension (corresponding to 3×108 CFU/mL) of each isolate was reached. 50 μL volume of the previous suspension was placed in duplicate in 24-well plates (COSTAR, United States), 950 μL of MEM was added to each well and incubated at 37°C for 24, 48 and 72 h. The E. coli CFT073 strain was used as a positive control and E. coli HB101 as a negative control; as a blank, a well was used only with MEM without bacteria; the process was carried out under the same conditions used for the wells with bacteria. After each incubation time, the medium was removed from each well, washed twice with sterile water and dried at room temperature for at least 30 min. To each well, one milliliter of crystal violet solution (1%) was added and incubated for 20 min at room temperature. The dye was removed, and the wells were washed twice with distilled water. Once dry, one milliliter of 96% ethanol was added to each well and mixed using a micropipette, biofilm formation was defined by measuring O.D. at 570 nm using a microplate reader (Spectronic GenesysTM 2). All measurements were performed in two independent experiments, biofilm formation was classified as strong biofilm formers (4DOc < DO), moderate (2DOc < DO≤4DOc), weak (DOc<DO≤DO≤2DOc), or non-biofilm formers (DO≤DOc) (24). Strains that showed biofilm production were analyzed to identify the presence of the fluA (primer F: 5′-aggcaggaggaactgccagt-3′ and R: 5′-taaatgagggtgggtgcccgtgcc-3′) and fluB (primer F: 5′-cagccggatctgcc-3′ and R: 5′-actctggtgtttctggctgtt-3′) genes, alleles of the Ag43 antigen, following the protocol described by Zalewska-Piatek et al. (25) and Danese et al. (26).
2.6. Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8 software (GraphPad Sotfware, San Diego, CA, United States). The prevalence of virulence genes and antibiotic resistance patterns were compared between reUTI and peUTI by Fisher’s exact test. Virulence and resistance scores between groups (classical vs. Non-classical UPEC; Phylogroups; reUTI vs. peUTI) were compared by performing Student’s t-test. Additionally, linear regression was performed and Pearson’s correlation between virulence and resistance scores shown by the strains was calculated (18). For all analyses, a value of p < 0.05 was considered statistically significant.
3. Results
3.1. Serogroups and serotypes of the analyzed Escherichia coli strains
The serology of the 394 strains corresponded to 44 serogroups and 76 serotypes, 185 (47%) of the strains were included in 10 of the classical UPEC serogroups, 134 (34%) belonged to 34 different non-UPEC serogroups, 48 (12%) presented rough phenotype (R), and 28 (7%) of the strains did not agglutinate (ND, non-determined) with any of the 187 sera used (Table 1). The correlation between the type of UTI and serogroups, it was found that strains with classical UPEC serogroups belonged to patients with peUTI (p < 0.05). On the other hand, patients with reUTI, the strains belonged preferentially to non-UPEC serogroups or were not identified (p < 0.05). A more specific analysis showed that 52.5% of the strains belonged to the classical UPEC serogroups O25, O75, O8, O6, O1, and non-UPEC O9, O11, and O14 (Table 1). The serotype analysis in peUTI patients with a follow-up >7 months showed that O25: H4, O75: NM, O6: H1, and O9: NM were consistently isolated (p < 0.05). With respect to patients with reUTI the serotypes O8: NM, O8: H9, O1: H7, and O25: H4 were the most frequently identified. It was interesting to note that strains of serotype O25: H4 were isolated from both types of infections, peUTI and reUTI.
Table 1.
Escherichia coli serogroups in patients with peUTI and reUTI.
| Serogroup (No. of strains) | |||
|---|---|---|---|
| reUTI, n = 269 | peUTI, n = 125 | Total, n = 394 | |
| Classical UPEC | O25(21),O8(15),O1(10),O6(8),O16(6),O4(3),O21(3),O22(3),O75(3) | O25(69),O75(36),O6(6) | 184 (47%) |
| Non-Classical UPEC | O11(14),O14(12),O28ab(6),O32(6),O73(6),O174(6),O57(5),O76(4),O178(4),O12(3),O19(3),O45(3),O84(3),O96(3),O101(3),O109(3),O120(3),O147(3),O148(3),O153(3),O154(3),O170(3),boy8(3),O17(2),O23(2),O152(2),O49766(2),O18ac(1),O20(1),O49(1),O102(1),O168(1) | O9 (11) | 134 (34%) |
| Non-defined | OR(45),OND(28) | OR (3) | 76 (19%) |
3.2. Antimicrobial susceptibility
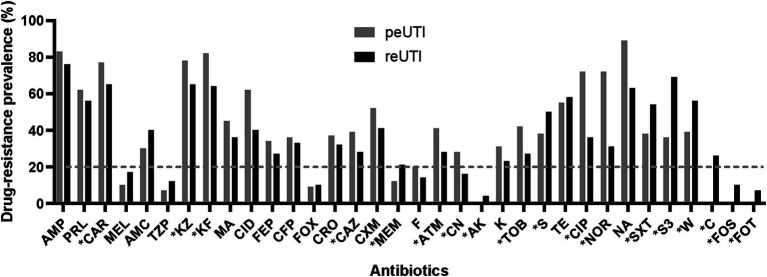
A relevant aspect in UTI is the poor response to antimicrobial treatment in a significant number of patients. The sensitivity results showed that 378 (96%) strains showed low resistance to mecilinam, piperacillin/tazobactam, cefoxitin, meropenem, amikacin, nitrofurantoin, chloramphenicol, fosfomycin, fosfomycin/trometramol. However, a different profile with high resistance to the penicillin family (ampicillin, piperacillin, carbenicillin), some cephalosporins (cefazolin, cephalotin) and nalidixic acid was identified in most strains (Figure 1). Comparative analysis showed significant higher resistance (p < 0.05) for carbenicillin, cefazolin, cephalotin, ceftazidime, aztreonam, gentamicin, tobramycin, ciprofloxacin, and norflozacin in peUTI strains and for meropenem, amikacin, streptomycin, trimethoprim-sulfamethoxazole, sulfonamides, trimetroprim, chloramphenicol, fosfomycin, and fosfomycin trometramol in reUTI isolates (Figure 1). Classification by resistance type showed that 27% (102/378) of the isolates were included in the XDR group and 59% (223/378) in MDR, and 14% (53/378) were considered still sensitive strains. A Drug-resistance score was used considering the total number of antimicrobials to which each isolated presented resistance (18). In this regard, UPEC strains from reUTI and peUTI did not show significantly different antibiotic resistance (p > 0.05) (Figure 2). However, strains isolated from children with peUTI were more resistant than isolates from children with reUTI (p < 0.05) and the same was observed for peUTI (p < 0.05) and reUTI (p < 0.05) in adults.
Figure 1.
Drug-resistance prevalence among UPEC isolates from patients with reUTI and peUTI. Ampicillin (AMP), piperacillin (PRL), carbenicillin (CAR), mecilanam (MEL), amox-clavulanic acid (AMC), piperacillin-tazobactam (TZP), cefazolin (KZ), cephalothin (KF), cefamandol (MA), cefepime (FEP), cefoperazone (CFP), cefoxitin (FOX), ceftriaxone (CRO), ceftazidime (CAZ), furoxime (CXM), meropenem (MEM), nitrofurantoin (F), aztreonam (ATM), gentamicin (CN), amikacin (KA), kanamycin (K), tobramycin (TOB), streptomycin (S), tetracycline (TE), ciprofloxacin (CIP), norfloxacin (NOR), trimethoprim-sulfamethoxazole (SXT), nalidixic acid (NA), sulfonamides (S3), trimetroprim (W), chloramphenicol (C), fosfomycin (FOS), fosfomycin trometramol (FOT). The dotted line marks the 20% resistance threshold. Fisher’s exact test was performed, *p < 0.05.
Figure 2.
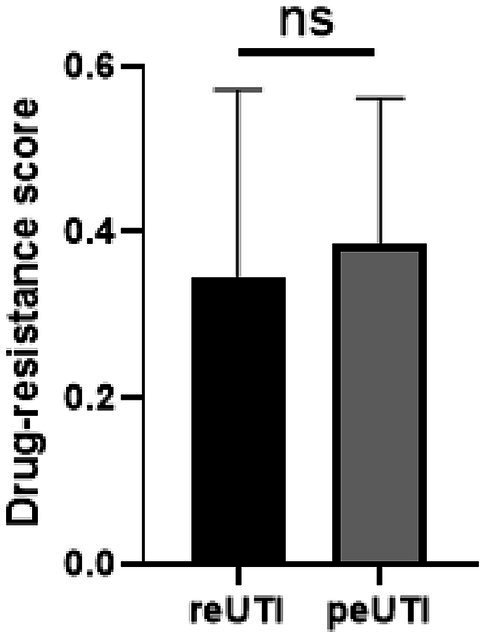
Distribution of the drug-resistance score in UPEC isolates from patients with reUTI and peUTI. The bars within each box plot show median values. The box covers the 25th percentile to the 75th percentile of the data. Bars above and below the box show 1.5 times the inter-quartile range. Whiskers show minimum and maximum values. Student’s t-test was performed, ns, statistically non-significant and *p < 0.05.
3.3. Phylogenetic diversity
Phylogenetic analysis of the E. coli strains showed that 96.4% (380/394) were included in one of the 4 phylogroups evaluated and only in 3.6% (14/394) of the isolates was not possible to define the group. Forty-four percent of the strains corresponded to phylogroup B2, 27% to A, 14.8% to D and 14% to B1; when performing a probability analysis to know the relationship of the phylogroup between patients with peUTI versus reUTI it was found that B2 was the most common in strains from patients with persistence (p < 0.0001) and A in isolates from patients with reinfections (Table 2).
Table 2.
Escherichia coli phylogroups prevalence in UPEC from peUTI and reUTI.
| Phylogroup | peUTI, n = 117 | reUTI, n = 263 | Total n = 380 | p-value |
|---|---|---|---|---|
| A | 0 | 104 (39.5%) | 104 (27%) | – |
| B1 | 0 | 53 (20.2%) | 53 (14%) | – |
| B2 | 114 (97.4%) | 52 (19.8%) | 166 (44%) | <0.0001 |
| D | 3 (2.6%) | 54 (20.5%) | 57 (14.8%) | – |
Fisher’s exact test was performed, only statistically significant p-values are shown.
3.4. Virulence genes
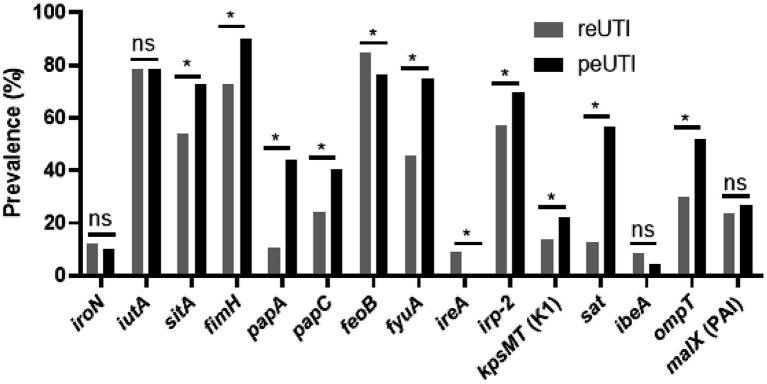
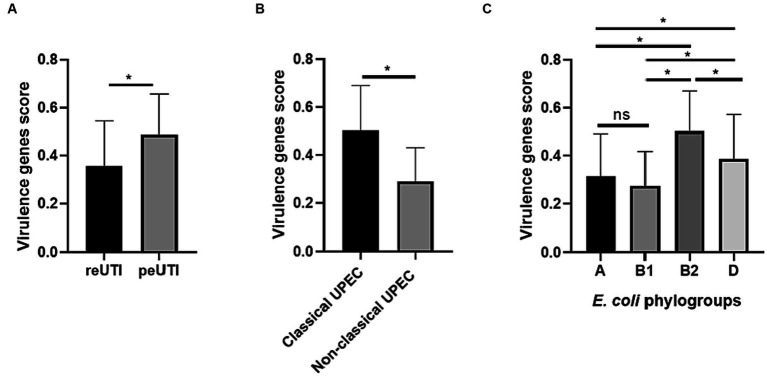
The assay to identify virulence genes (Figure 3), showed the presence of 6 genes (iutA, sitA, fimH, feoB, fyuA, and irp-2) in more than 50% of the strains analyzed. fimH, papA, papC (adhesins), sitA, fyuA, irp-2 (iron scavengers), kpsMT, and ompT (protectins) and sat (self-transported cytotoxin) were identified more frequently in strains isolated from peUTI patients (p < 0.05). On the other hand, feoB and ireA were observed more frequently in patients with reUTI (p < 0.05). To define whether the presence of the genes favors the persistence of the bacteria carrying them, virulence score analysis was used. This analysis showed that UPEC strains isolated from patients with peUTI presented a higher number of virulence genes (p < 0.0001), compared to strains from reUTI (Figure 4A); particularly, classical UPEC strains showed a higher virulence score regardless of the type of cUTI (p < 0.0001) (Figure 4B). Additionally, it was observed that strains from group B2 contained a higher number of the virulence genes analyzed, than strains included in other phylogroups (B2 vs. A, p < 0.0001; B2 vs. B1, p < 0.005; B2 vs. D, p < 0.05) (Figure 4C). UPEC strains from peUTI and reUTI showed a significant correlation (Pearson’s coefficient = 0.2171; R2 = 0.04, p < 0.0001) between virulence and antibiotic resistance scores.
Figure 3.
Virulence traits in UPEC isolates from patients with peUTI or reUTI. Fisher’s exact test was performed, ns, statistically non-significant and *p < 0.05.
Figure 4.
Escherichia coli isolates from complicated UTI. (A) Virulence genes score in strains from reUTI and peUTI, (B) Classical and Non-Classical UPEC groups, and (C) E. coli phylogroups. Student’s t-test was performed, ns, statistically non-significant and *p < 0.05.
3.5. In vitro assays
To corroborate whether the presence of certain genes was involved in the virulence of the strains and in the variety of cUTI (peUTI or reUTI) presented by the patients, in vitro tests were implemented to analyze the adherence, invasiveness, and biofilm formation of some of the bacteria included in the study.
3.5.1. Adherence to cells
Adherence capacity was evaluated in 72 strains from 6 patients with peUTI and 3 with reUTI, who were followed up for >3 months. The assay reported 20 (27.7%) adherence to cells with higher prevalence in isolates from patients with peUTI 70% (14/20). The correlation between adherence and serotype showed that 80% (16/20) had the classic UPEC serotypes O25: H4 (10), O75:NM (4), O6:H1 (2) and 4 the serotype O11:H25 (non-UPEC). The analysis of genes identified in the adherent strains reported the presence of fimH (38%), papA (36%) and papC (31%). As previously mentioned, fimH and papC were mainly identified in isolates from peUTI patients (p < 0.05).
3.5.2. In vitro invasiveness
To evaluate cell invasiveness, 54 strains were selected from peUTI and patients with reUTI. The assay showed that 37% of the strains were invasive, with a higher prevalence of 12/20 (60%) in isolates from patients with peUTI. Association analysis between invasiveness and serotype showed that 16 (80%) of the isolates belonged to the classical UPEC serotypes [O25:H4 (10), O6:H1 (4), O4:H5 (2)] and 4 (20%) the non-UPEC serotypes [O28ab:H4 (2), O45:HND (2)]. The quantitative invasiveness assay showed that the number of invasive bacteria isolated from peUTI was higher (2.8×103 CFU/mL) than those isolated from patients with reUTI (p < 0.05). E. coli O25:H4 strains were the most invasive (1.2×103-7×103 CFU/mL) reporting a logarithm higher than that obtained in Shigella boydii 21,639 (5×102 CFU/mL) and E. coli 1,124 (3×102 CFU/mL) strains used as controls.
3.5.3. Biofilm formation
Eighty strains isolated from 13 patients with cUTI were selected to evaluate biofilm formation, 29 were from 5 patients with peUTI and 51 from 8 with reUTI. The results reported a minimum absorbance of 0.2722 and a maximum of 0.6694, with a cut-off point of 0.339. With these data it was defined that 29/80 (36%) formed a biofilm classified as weak after 72 h of incubation, 12 strains were isolated from 3 patients with peUTI and 17 from 5 patients with reUTI, with no significant differences (p > 0.05). However, when performing the relationship between serotypes of the strains it was found that 17 (59%) belonged to serotypes O25:H4 (8), O4:H5 (5), O6:H1 (4) classic UPEC, and 11 (38%) to serotypes O11:H25 (2), O12:NM (4), O45:H (5) and one non-defined (ND), the statistical analysis between these classic and non-classic groups gives a significant value (p < 0.05). Ag43 is a membrane protein associated with biofilm formation identified in 26 E. coli strains, thus it was important to identify the flu alleles related to Ag43 expression. Gene analysis showed the presence of fluA and fluB in both UPEC-CFT073 and the 80 strains selected for biofilm formation, fluB was the most prevalent (83%) in strains that formed biofilms (p < 0.05) (Table 3).
Table 3.
Biofilm formation and genes profile in E. coli isolates from peUTI and reUTI.
| Gen | Biofilm producers, n = 29 | Non-biofilm producers, n = 51 | Total n = 80 |
|---|---|---|---|
| fluA (Ag43a) | 5 (17%) | 3 (6%) | 8 (10%) |
| fluB (Ag43b) | 22 (76%) | 8 (16%) | 30 (37%) |
| fluA/fluB | 2 (7%) | 1 (2%) | 3 (4%) |
| No genes | 0 | 39 (76%) | 39 (49%) |
4. Discussion
Escherichia coli strains are the most common bacteria isolated from urine of patients with acute and cUTI infections. In the cases where cUTI is caused by the same microorganism the infection is considered a peUTI, while in cases where the isolated microorganisms are different to those identified in the previous sample the infection is considered a reUTI. To define the characteristics of the E. coli that is causing the cUTI the isolate could be characterized by phenotypic (serological and biochemical typing) or genotypic (ribotyping, pulsed-field electrophoresis or MLST) methods (27–31). As previously mentioned, our work group has the complete sera scheme to perform the antigenic characterization of E. coli, which allows us to define the type of cUTI, i. e. peUTI or reUTI.
In two previous studies patients with cUTI were followed up for 7 to 18 months, where E. coli strains were isolated in approximately 70% of the samples (13, 14). Serotyping of E. coli isolates from some patients with cUTI, showed O25, O75, O6 serogroups (classic UPEC) in the different urine cultures evaluated, because the serogroup in each patient was the same always, the type of infection was classified as peUTI, same observations have been reported by other authors (29, 32, 33). However, it was interesting to observe in a patient with peUTI the identification of the O9 serogroup, not included in the classic UPEC pathotype and considered by other authors as enterohemorrhagic E. coli (34). This circumstance raises the possibility that gene transfer from UPEC to strains such as O9 is taking place in the intestine, favoring their survival in new environments such as the urinary tract (18, 35). When analyzing the data obtained during patient follow-up from it was found that initially the patient only presented reUTI, however, after several isolations of different E. coli serogroups O9 strains were identified and being isolated in the following urine cultures the infection changed to a peUTI. Furthermore, the genetic analysis of these O9 strains, showed the presence of genes linked to iron uptake (sitA, feoB, and irp2), which were also present in greater proportion in the classic UPEC strains (O25, O75, and O6). On the other hand, in reUTI cases a greater diversity of serogroups was identified, which in turn were different in each urine culture. Apparently different aspects may be related to cases of reUTI that favor infection by E. coli from the intestinal biota via cross-contamination (36). Although these patients can become free of infection when treated with antimicrobials, the predisposition to become infected favors a new strain to colonize them, causing the reinfection of the condition (37).
The phylogenetic distribution among the study strains was higher toward phylogroups B2 and A, this result contrasts with that reported by other authors, who mention that extraintestinal E. coli strains belong mainly to phylogroups B2 and D (18, 38). However, studies conducted in Russia, China, Iran, Portugal, Venezuela, and Mexico, reported similar results to those obtained in this study (13, 39–44). Phylogenetic variation in UPEC strains may be associated with host particularities such as the geographical area, climate and diet type in which they inhabit (45). Other predisposing factors such as anatomical alterations, metabolic diseases, immune status, and hygienic habits would allow gut commensal strains to become opportunistic pathogens (46). The results reported here confirmed that phylogroup B2 is particularly associated with peUTIs; in this regard, Thänert et al. (47) reported the presence of this type of strains preferentially associated with peUTIs, suggesting they might be colonizing the urinary tract, the intestine, or both habitats according to their ability for adaptation. Likewise, in a study on the virulence of strains belonging to phylogroup B2, using a murine model, it was reported that urine isolates are more virulent than those from feces (48). However, studies in France and Sweden evaluating the intestinal microbiota of children and adults, identified phylogroup B2 as carrying virulence genes associated with UPEC strains, which would corroborate that B2 phylogroup strains, despite being commensal, would be able to become extraintestinal pathogens (18, 49).
Virulence plays a very important role in the interaction with the host, in UPEC it has been documented that the number of virulence factors is proportional to the pathogenic potential and would facilitate the colonization of the urinary tract (50–52). In this study, it was observed that virulence-related genes of uropathogenic strains were more prevalent (mean virulence score: 0.501994 ± 0.1673) among strains from patients with peUTI than in those associated with reUTI. These genes could be associated with the ability of E. coli strains to resist in the urinary tract since it has been reported that the main genes associated with persistence are those related to adherence and iron uptake capacity (29, 53). Given the high prevalence of certain virulence factors in patients with peUTI, such as adhesins papA, papC, and fimH; those associated with iron uptake sitA, fyuA, irp-2, and Sat toxin (sat), whose cytotoxic and immunomodulatory effect has been associated with the survival and the potential generation of bloodstream infections and sepsis in producer strains (54), these could be used as a target for diagnosis or vaccine development, as proposed by Mobley and Alteri (55). On the other hand, it was observed that strains with classic UPEC serogroups and B2 and D phylogroups carried the highest number of virulence genes, similar observations documented by other authors (50, 56).
In the present work, employing immortalized HEp-2 larynx cell line, used as the gold standard to observe adherence patterns of diarrheagenic E. coli (57, 58), a low number of UPEC isolates showed diffusely adherent phenotype (59–61), although, in other works an adherence pattern of aggregative phenotype was reported (62). The ability of UPEC strains to adhere and invade bladder epithelial cells in the host has been related to the expression of different fimbrial adhesins (63). In this work, it was found adherent strains harboring fimH, papG, or papA, genes related to type I and type P adhesins, while the rest of the identified adherent strains could be associated with other adhesins such as type S, Curli and the aggregate-forming pili identified in hybrid UPEC/DAEC strains (63–66). In a previous study, we documented that E. coli strains could persist >6 months in the urinary tract of patients (14), a recent study by Hidad et al. (31) reported that E. coli strains were identified in their investigation for more than 1 year. In the present study, it was identified that 60% of the invasive strains were isolated from peUTI patients, associated with classical UPEC serogroups. Andersen et al. (67) reported that these UPEC strains can re-emerge from infected cells and invade adjacent cells.
Different studies indicate that cUTI (peUTI and reUTI) are associated with biofilm formation and antibiotic resistance (30, 68–70). In this study, weak in vitro biofilm formation was observed, where only 36% of the strains were positive. However, a relevant aspect to note is the presence of the b allele of Ag43, related to good biofilm formation (71), identified in 83% of the producing strains. Although the result contrasts with that reported by other authors who mentioned a high percentage of biofilm-forming UPEC strains (69, 72), it is likely that the results we obtained are related to the technique used, in which we perform energetic washes that could be detaching the biofilm.
Antibiotic prophylactic treatment is common in patients with cUTI (73), this favors the selection of uropathogenic resistant strains especially to those used as first choice (74). Although the CLSI only recommends sensitivity to five compounds (cefazolin, sulfonamides, trimethoprim, fosfomycin, and nitrofurantoin) for UPEC strains (16), given the conditions in our country due to the indiscriminate use of antibiotics and the fact that these are enterobacteria, it was decided to test for sensitivity to 32 antimicrobials. The results in this regard showed MDR in E. coli strains isolated from both reUTI and peUTI, mainly those belonging to the penicillin family, cephalosporins, sulfonamides and trimetroprim, a similar result reported in other countries (31, 75, 76). However, the E. coli from peUTI and reUTI demonstrated >80% susceptibility to certain antibiotics (mecilanam, piperacillin-tozobactam, cefoxitin, meropenem, nitrofurantoin, amikacin, chloramphenicol, fosfomycin, and fosfomycin-trometramol), which could be used empirically, with exceptions, according to the Infectious Diseases Society of America (IDSA) criteria (77). Relevantly, this MDR capability has also been described in E. coli strains from the intestine, the main reservoir of UPEC, and an explanation of how this resistance is transmitted between E. coli strains to potential UPEC (78). It is because of the rapid spread of MDR bacteria, that WHO mentioned E. coli as a critical priority pathogen for the development of new treatments (1).
Different authors have mentioned blueberries, D-mannose, probiotics or surgical treatments in patients with urinary tract complications as potential treatment alternatives (74). In two prospective studies conducted previously by our group bacterial lysates (autovaccines, i.e., immunostimulants) were used, and the remission of the clinical picture and control of the infection for periods ranging from 6 to more than 18 months was achieved (13, 14). However, improvement was only observed in patients with reUTI, which again confirms the presence of some other properties in the strains that cause peUTI. Some other alternatives for the treatment and control of cUTI should be developed, in the laboratory we are working with lysates prepared with the strains that were most frequently isolated in the prospective studies carried out. The preliminary results obtained in an animal model are encouraging; however, there is still a long way to go.
According to the results, the strains isolated from peUTI patients are professional bacteria in the generation of cUTI, with genetic and phenotypic characteristics of uropathogenic strains, however, the strains isolated from patients with reUTI correspond to strains of E. coli from the intestinal biota that have acquired part of the genetic information that allows them to reach the Urinary Tract and produce UTI but do not remain permanently. This is why they can be eliminated with antimicrobial treatment; however, due to factors related to the host, patients are reinfected but with a different microorganism each time, thus generating a reUTI.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
UH-C and CAE: study conception and design. UH-C, AN-O, MEC-B, JM-L, LMR-R, and AN-C: data collection. UH-C, REA-C, and CAE: draft manuscript preparation. UH-C, REA-C, and CAE: analysis and interpretation of results. All authors reviewed and approved the final version of the manuscript.
Acknowledgments
The authors wish to thank Facultad de Medicina, UNAM and Hospital Infantil de México Federico Gómez for the provided facilities. Specially, we are grateful to Lulú Estrada, for her ongoing administrative help, and Maribel Alvarado Cabello, Yolanda Pérez del Mazo, Gabriel Pérez Soto, and Luis León Alamilla, for their technical assistance in the laboratory.
Funding Statement
This research received funding through the Project PAPIIT-UNAM IN205322, federal funds HIM/2018/051 SSA 1554 and institutional support by the Research Division of Medicine Faculty UNAM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2023.1240392/full#supplementary-material
References
- 1.World Health Organization . (2017). Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Available at: https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12 (Accessed May 30, 2023).
- 2.Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. (2010) 7:653–60. doi: 10.1038/nrurol.2010.190 [DOI] [PubMed] [Google Scholar]
- 3.O’Brien VP, Hannan TJ, Schaeffer AJ, Hultgren SJ. Are you experienced? Understanding bladder innate immunity in the context of recurrent urinary tract infection. Curr Opin Infect Dis. (2015) 28:97–105. doi: 10.1097/QCO.0000000000000130, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Badr A, Al-Shaikh G. Recurrent urinary tract infections Management in Women. Sultan Qaboos Univ Med J. (2013) 13:359–67. doi: 10.12816/0003256, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigoryan L, Mulgirigama A, Powell M, Schmiemann G. The emotional impact of urinary tract infections in women: a qualitative analysis. BMC Womens Health. (2022) 22:182. doi: 10.1186/s12905-022-01757-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahumada-Cota RE, Olalde-Ramírez S, Hernández-Chiñas U, Acevedo-Monroy S, Eslava Campos CA. Infecciones del tracto urinario en México, un problema de salud pública. Revista Tendencias en Docencia e Investigación en Química. (2022) 8:728–34. [Google Scholar]
- 7.Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. (2001) 69:4572–9. doi: 10.1128/IAI.69.7.4572-4579.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Schembri MA, Hultgren SJ. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular Uropathogenic E. coli bladder infection. FEMS Microbiol Rev. (2012) 36:616–48. doi: 10.1111/j.1574-6976.2012.00339.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reitzer L, Zimmern P. Rapid growth and metabolism of Uropathogenic Escherichia coli in relation to urine composition. Clin Microbiol Rev. (2020) 33:e00101-19. doi: 10.1128/CMR.00101-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tullus K, Shaikh N. Urinary tract infections in children. Lancet. (2020) 395:1659–68. doi: 10.1016/S0140-6736(20)30676-0 [DOI] [PubMed] [Google Scholar]
- 11.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. (2010) 8:207–17. doi: 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 12.Paniagua-Contreras GL, Monroy-Pérez E, Rodríguez-Moctezuma JR, Domínguez-Trejo P, Vaca-Paniagua F, Vaca S. Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J Microbiol Immunol Infect. (2017) 50:478–85. doi: 10.1016/j.jmii.2015.08.005, PMID: [DOI] [PubMed] [Google Scholar]
- 13.Ahumada-Cota RE, Hernandez-Chiñas U, Milián-Suazo F, Chávez-Berrocal ME, Navarro-Ocaña A, Martínez-Gómez D, et al. Effect and analysis of bacterial lysates for the treatment of recurrent urinary tract infections in adults. Pathogens. (2020) 9:102. doi: 10.3390/pathogens9020102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández-Chiñas U, Chávez-Berrocal ME, Ahumada-Cota RE, Navarro-Ocaña A, Rocha-Ramírez LM, Pérez-del Mazo Y, et al. Prospective study in children with complicated urinary tract infection treated with autologous bacterial lysates. Microorganisms. (2021) 9:1811. doi: 10.3390/microorganisms9091811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orskov F, Orskov I. Escherichia coli serotyping and disease in man and animals. Can J Microbiol. (1992) 38:699–704. doi: 10.1139/m92-115 [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute (CLSI) . Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100, 31st ed. Wayne, PA: Clinical and Laboratory Standards Institute; (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x, PMID: [DOI] [PubMed] [Google Scholar]
- 18.Massot M, Daubié A-S, Clermont O, Jauréguy F, Couffignal C, Dahbi G, et al. Phylogenetic, virulence and antibiotic resistance characteristics of commensal strain populations of Escherichia coli from community subjects in the Paris area in 2010 and evolution over 30 years. Microbiol. (2016) 162:642–50. doi: 10.1099/mic.0.000242, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic Group. Appl Environ Microbiol. (2000) 66:4555–8. doi: 10.1128/AEM.66.10.4555-4558.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cravioto A, Tello A, Villafán H, Ruiz J, del Vedovo S, Neeser JR. Inhibition of localized adhesion of enteropathogenic Escherichia coli to HEp-2 cells by immunoglobulin and oligosaccharide fractions of human colostrum and breast milk. J Infect Dis. (1991) 163:1247–55. doi: 10.1093/infdis/163.6.1247, PMID: [DOI] [PubMed] [Google Scholar]
- 21.Elsinghorst EA. Measurement of invasion by gentamicin resistance. Methods Enzymol. (1994) 236:405–20. doi: 10.1016/0076-6879(94)36030-8 [DOI] [PubMed] [Google Scholar]
- 22.Aribam SD, Hirota J, Kusumoto M, Harada T, Shiraiwa K, Ogawa Y, et al. A rapid differentiation method for enteroinvasive Escherichia coli. J Microbiol Methods. (2014) 98:64–6. doi: 10.1016/j.mimet.2013.11.012 [DOI] [PubMed] [Google Scholar]
- 23.Christensen GD, Simpson WA, Anglen JO, Gainor BJ. Methods for evaluating attached bacteria and biofilms In: An YH, Friedman RJ, editors. Handbook of bacterial adhesion: principles, methods, and applications. Totowa, NJ: Humana Press; (2000). 213–33. [Google Scholar]
- 24.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. (2000) 40:175–9. doi: 10.1016/s0167-7012(00)00122-6 [DOI] [PubMed] [Google Scholar]
- 25.Zalewska-PiaTek B, Pia Tek R, Olszewski M, Kur J. Identification of antigen Ag43 in uropathogenic Escherichia coli Dr+ strains and defining its role in the pathogenesis of urinary tract infections. Microbiology. (2015) 161:1034–49. doi: 10.1099/mic.0.000072 [DOI] [PubMed] [Google Scholar]
- 26.Danese PN, Pratt LA, Dove SL, Kolter R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol. (2000) 37:424–32. doi: 10.1046/j.1365-2958.2000.02008.x [DOI] [PubMed] [Google Scholar]
- 27.Bergström T, Lincoln K, Orskov F, Orskov I, Winberg J. Studies of urinary tract infections in infancy and childhood. 8. Reinfection vs. relapse in recurrent urinary tract infections. Evaluation by means of identification of infecting organisms. J Pediatr. (1967) 71:13–20. doi: 10.1016/s0022-3476(67)80224-5, PMID: [DOI] [PubMed] [Google Scholar]
- 28.Brauner A, Jacobson SH, Kühn I. Urinary Escherichia coli causing recurrent infections--a prospective follow-up of biochemical phenotypes. Clin Nephrol. (1992) 38:318–23. PMID: [PubMed] [Google Scholar]
- 29.Johnson JR, O’Bryan TT, Delavari P, Kuskowski M, Stapleton A, Carlino U, et al. Clonal relationships and extended virulence genotypes among Escherichia coli isolates from women with a first or recurrent episode of cystitis. J Infect Dis. (2001) 183:1508–17. doi: 10.1086/320198 [DOI] [PubMed] [Google Scholar]
- 30.Soto SM, Smithson A, Horcajada JP, Martinez JA, Mensa JP, Vila J. Implication of biofilm formation in the persistence of urinary tract infection caused by uropathogenic Escherichia coli. Clin Microbiol Infect. (2006) 12:1034–6. doi: 10.1111/j.1469-0691.2006.01543.x, PMID: [DOI] [PubMed] [Google Scholar]
- 31.Hidad S, van der Putten B, van Houdt R, Schneeberger C, Kuil SD. Recurrent E. coli urinary tract infections in nursing homes: insight in sequence types and antibiotic resistance patterns. Antibiotics. (2022) 11:1638. doi: 10.3390/antibiotics11111638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vosti KL. A prospective, longitudinal study of the behavior of serologically classified isolates of Escherichia coli in women with recurrent urinary tract infections. J Infect. (2007) 55:8–18. doi: 10.1016/j.jinf.2007.01.006, PMID: [DOI] [PubMed] [Google Scholar]
- 33.Paniagua-Contreras GL, Monroy-Pérez E, Bautista A, Reyes R, Vicente A, Vaca-Paniagua F, et al. Multiple antibiotic resistances and virulence markers of uropathogenic Escherichia coli from Mexico. Pathog Glob Health. (2018) 112:415–20. doi: 10.1080/20477724.2018.1547542, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro A, van der Ploeg C, Rogé A, Licona-Moreno D, Delgado G, Morales-Espinosa R, et al. Diversity of potentially pathogenic Escherichia coli O104 and O9 serogroups isolated before 2011 from Fecal samples from children from different geographic regions. Microorganisms. (2021) 9:2227. doi: 10.3390/microorganisms9112227, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowrouzian FL, Clermont O, Edin M, Östblom A, Denamur E, Wold AE, et al. Escherichia coli B2 phylogenetic subgroups in the infant gut microbiota: predominance of Uropathogenic lineages in Swedish infants and enteropathogenic lineages in Pakistani infants. Appl Environ Microbiol. (2019) 85:e01681–19. doi: 10.1128/AEM.01681-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, et al. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci Transl Med. (2013) 5:184ra60. doi: 10.1126/scitranslmed.3005497, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooton TM. Clinical practice. Uncomplicated urinary tract infection. N Engl J Med. (2012) 366:1028–37. doi: 10.1056/NEJMcp1104429 [DOI] [PubMed] [Google Scholar]
- 38.Micenková L, Bosák J, Štaudová B, Kohoutová D, Čejková D, Woznicová V, et al. Microcin determinants are associated with B2 phylogroup of human fecal Escherichia coli isolates. Microbiology. (2016) 5:490–8. doi: 10.1002/mbo3.345, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grude N, Potaturkina-Nesterova NI, Jenkins A, Strand L, Nowrouzian FL, Nyhus J, et al. A comparison of phylogenetic group, virulence factors and antibiotic resistance in Russian and Norwegian isolates of Escherichia coli from urinary tract infection. Clin Microbiol Infect. (2007) 13:208–11. doi: 10.1111/j.1469-0691.2006.01584.x, PMID: [DOI] [PubMed] [Google Scholar]
- 40.Derakhshandeh A, Firouzi R, Motamedifar M, Motamedi Boroojeni A, Bahadori M, Arabshahi S, et al. Distribution of virulence genes and multiple drug-resistant patterns amongst different phylogenetic groups of uropathogenic Escherichia coli isolated from patients with urinary tract infection. Lett Appl Microbiol. (2015) 60:148–54. doi: 10.1111/lam.12349 [DOI] [PubMed] [Google Scholar]
- 41.Tong Y, Sun S, Chi Y. Virulence genotype and phylogenetic groups in relation to Chinese herb resistance among Escherichia coli from patients with acute pyelonephritis. Afr J Tradit Complement Altern Med. (2014) 11:234–8. doi: 10.4314/ajtcam.v11i3.33, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halaji M, Fayyazi A, Rajabnia M, Zare D, Pournajaf A, Ranjbar R. Phylogenetic group distribution of Uropathogenic Escherichia coli and related antimicrobial resistance pattern: a meta-analysis and systematic review. Front Cell Infect Microbiol. (2022) 12:790184. doi: 10.3389/fcimb.2022.790184, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millán Y, Hernández E, Millán B, Araque M. Distribución de grupos filogenéticos y factores de virulencia en cepas de Escherichia coli uropatógena productora de β-lactamasa CTX-M-15 aisladas de pacientes de la comunidad en Mérida. Venezuela Revista Argentina de Microbiología. (2014) 46:175–81. doi: 10.1016/S0325-7541(14)70069-0, PMID: [DOI] [PubMed] [Google Scholar]
- 44.Narciso A, Nunes F, Amores T, Lito L, Melo-Cristino J, Duarte A. Persistence of uropathogenic Escherichia coli strains in the host for long periods of time: relationship between phylogenetic groups and virulence factors. Eur J Clin Microbiol Infect Dis. (2012) 31:1211–7. doi: 10.1007/s10096-011-1431-7, PMID: [DOI] [PubMed] [Google Scholar]
- 45.Stoppe N, De C, Silva JS, Carlos C, MIZ S, Saraiva AM, et al. Worldwide phylogenetic Group patterns of Escherichia coli from commensal human and wastewater treatment plant isolates. Front Microbiol. (2017) 8. doi: 10.3389/fmicb.2017.02512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhuri RR, Henderson IR. The evolution of the Escherichia coli phylogeny. Infect Genet Evol. (2012) 12:214–26. doi: 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 47.Thänert R, Choi J, Reske K, Hink T, Thänert A, Ma W, et al. Persisting uropathogenic Escherichia coli lineages show signatures of niche-specific within-host adaptation mediated by mobile genetic elements. Cell Host Microbe. (2022) 30:1034–1047.e6. doi: 10.1016/j.chom.2022.04.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, et al. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol Biol Evol. (2007) 24:2373–84. doi: 10.1093/molbev/msm172, PMID: [DOI] [PubMed] [Google Scholar]
- 49.Nowrouzian FL, Wold AE, Adlerberth I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis. (2005) 191:1078–83. doi: 10.1086/427996 [DOI] [PubMed] [Google Scholar]
- 50.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. (1999) 67:546–53. doi: 10.1128/IAI.67.2.546-553.1999, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bien J, Sokolova O, Bozko P. Role of Uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. (2012) 2012:681473. doi: 10.1155/2012/681473, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terlizzi ME, Gribaudo G, Maffei ME. Uro pathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol. (2017) 8:1566. doi: 10.3389/fmicb.2017.01566, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd AL, Rasko DA, Mobley HLT. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. (2007) 189:3532–46. doi: 10.1128/JB.01744-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freire CA, Silva RM, Ruiz RC, Pimenta DC, Bryant JA, Henderson IR, et al. Secreted autotransporter toxin (sat) mediates innate immune system evasion. Front Immunol. (2022) 13:844878. doi: 10.3389/fimmu.2022.844878, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mobley HLT, Alteri CJ. Development of a vaccine against Escherichia coli urinary tract infections. Pathogens. (2015) 5:1. doi: 10.3390/pathogens5010001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dadi BR, Abebe T, Zhang L, Mihret A, Abebe W, Amogne W. Distribution of virulence genes and phylogenetics of uropathogenic Escherichia coli among urinary tract infection patients in Addis Ababa. Ethiopia BMC Infect Dis. (2020) 20:108. doi: 10.1186/s12879-020-4844-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scaletsky ICA, Fabbricotti SH, Silva SOC, Morais MB, Fagundes-Neto U. Hep-2–adherent Escherichia coli strains associated with acute infantile Diarrhea, São Paulo. Brazil Emerg Infect Dis. (2002) 8:855–8. doi: 10.3201/eid0808.010492, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miranda-Estrada LI, Ruíz-Rosas M, Molina-López J, Parra-Rojas I, González-Villalobos E, Castro-Alarcón N. Relationship between virulence factors, resistance to antibiotics and phylogenetic groups of uropathogenic Escherichia coli in two locations in Mexico. Enferm Infecc Microbiol Clin. (2017) 35:426–33. doi: 10.1016/j.eimc.2016.02.021, PMID: [DOI] [PubMed] [Google Scholar]
- 59.Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J. (1987) 6:829–31. doi: 10.1097/00006454-198709000-00008 [DOI] [PubMed] [Google Scholar]
- 60.Girón JA, Jones T, Millán-Velasco F, Castro-Muñoz E, Zárate L, Fry J, et al. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J Infect Dis. (1991) 163:507–13. doi: 10.1093/infdis/163.3.507, PMID: [DOI] [PubMed] [Google Scholar]
- 61.Mathewson JJ, Cravioto A. HEp-2 cell adherence as an assay for virulence among diarrheagenic Escherichia coli. J Infect Dis. (1989) 159:1057–60. doi: 10.1093/infdis/159.6.1057, PMID: [DOI] [PubMed] [Google Scholar]
- 62.Modgil V, Mahindroo J, Narayan C, Kalia M, Yousuf M, Shahi V, et al. Comparative analysis of virulence determinants, phylogroups, and antibiotic susceptibility patterns of typical versus atypical Enteroaggregative E. coli in India. PLoS Negl Trop Dis. (2020) 14:e0008769. doi: 10.1371/journal.pntd.0008769, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lüthje P, Brauner A. Virulence factors of uropathogenic E. Coli and their interaction with the host. Adv Microb Physiol. (2014) 65:337–72. doi: 10.1016/bs.ampbs.2014.08.006 [DOI] [PubMed] [Google Scholar]
- 64.Cordeiro MA, Werle CH, Milanez GP, Yano T. Curli fimbria: an Escherichia coli adhesin associated with human cystitis. Braz J Microbiol. (2016) 47:414–6. doi: 10.1016/j.bjm.2016.01.024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schüroff PA, Salvador FA, Abe CM, Wami HT, Carvalho E, Hernandes RT, et al. The aggregate-forming pili (AFP) mediates the aggregative adherence of a hybrid-pathogenic Escherichia coli (UPEC/EAEC) isolated from a urinary tract infection. Virulence. (2021) 12:3073–93. doi: 10.1080/21505594.2021.2007645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vega-Hernández R, Ochoa SA, Valle-Rios R, Jaimes-Ortega GA, Arellano-Galindo J, Aparicio-Ozores G, et al. Flagella, type I fimbriae and curli of Uropathogenic Escherichia coli promote the release of proinflammatory cytokines in a coculture system. Microorganisms. (2021) 9:2233. doi: 10.3390/microorganisms9112233, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andersen TE, Khandige S, Madelung M, Brewer J, Kolmos HJ, Møller-Jensen J. Escherichia coli uropathogenesis in vitro: invasion, cellular escape, and secondary infection analyzed in a human bladder cell infection model. Infect Immun. (2012) 80:1858–67. doi: 10.1128/IAI.06075-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falk KN, Satola SW, Chassagne F, Northington GM, Quave CL. Biofilm production by Uropathogens in postmenopausal women with recurrent and isolated urinary tract infection. Female Pelvic Med Reconstr Surg. (2022) 28:e127–32. doi: 10.1097/spv.0000000000001124, PMID: [DOI] [PubMed] [Google Scholar]
- 69.Zhao F, Yang H, Bi D, Khaledi A, Qiao M. A systematic review and meta-analysis of antibiotic resistance patterns, and the correlation between biofilm formation with virulence factors in uropathogenic E. coli isolated from urinary tract infections. Microb Pathog. (2020) 144:104196. doi: 10.1016/j.micpath.2020.104196, PMID: [DOI] [PubMed] [Google Scholar]
- 70.Nasrollahian S, Halaji M, Hosseini A, Teimourian M, Armaki MT, Rajabnia M, et al. Genetic diversity, carbapenem resistance genes, and biofilm formation in UPEC isolated from patients with catheter-associated urinary tract infection in north of Iran. Int J Clin Pract. (2022) 2022:9520362. doi: 10.1155/2022/9520362, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo J-M, Schembri MA. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun. (2007) 75:3233–44. doi: 10.1128/IAI.01952-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naziri Z, Kilegolan JA, Moezzi MS, Derakhshandeh A. Biofilm formation by uropathogenic Escherichia coli: a complicating factor for treatment and recurrence of urinary tract infections. J Hosp Infect. (2021) 117:9–16. doi: 10.1016/j.jhin.2021.08.017, PMID: [DOI] [PubMed] [Google Scholar]
- 73.Khan A, Jhaveri R, Seed PC, Arshad M. Update on associated risk factors, diagnosis, and Management of Recurrent Urinary Tract Infections in children. J Pediatric Infect Dis Soc. (2019) 8:152–9. doi: 10.1093/jpids/piy065, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Brien VP, Hannan TJ, Nielsen HV, Hultgren SJ. Drug and vaccine development for the treatment and prevention of urinary tract infections. Microbiol Spectr. (2016) 4:1–42. doi: 10.1128/microbiolspec.UTI-0013-2012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haaijman J, Stobberingh EE, van Buul LW, Hertogh CMPM, Horninge H. Urine cultures in a long-term care facility (LTCF): time for improvement. BMC Geriatr. (2018) 18:221. doi: 10.1186/s12877-018-0909-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hisano M, Bruschini H, Nicodemo AC, Gomes CM, Lucon M, Srougi M. The bacterial Spectrum and antimicrobial susceptibility in female recurrent urinary tract infection: how different they are from sporadic single episodes? Urology. (2015) 86:492–7. doi: 10.1016/j.urology.2015.05.033, PMID: [DOI] [PubMed] [Google Scholar]
- 77.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. (2011) 52:e103–20. doi: 10.1093/cid/ciq257, PMID: [DOI] [PubMed] [Google Scholar]
- 78.Mota R, Pinto M, Palmeira J, Gonçalves D, Ferreira H. Multidrug-resistant bacteria as intestinal colonizers and evolution of intestinal colonization in healthy university students in Portugal. Access Microbiol. (2020) 3:acmi000182. doi: 10.1099/acmi.0.000182, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.