Abstract
Mitochondrial diseases are a group of disorders defined by defects in oxidative phosphorylation caused by nuclear‐ or mitochondrial‐encoded gene mutations. A main cellular phenotype of mitochondrial disease mutations is redox imbalances and inflammatory signaling underlying pathogenic signatures of these patients. One method to rescue this cell death vulnerability is the inhibition of mitochondrial translation using tetracyclines. However, the mechanisms whereby tetracyclines promote cell survival are unknown. Here, we show that tetracyclines inhibit the mitochondrial ribosome and promote survival through suppression of endoplasmic reticulum (ER) stress. Tetracyclines increase mitochondrial levels of the mitoribosome quality control factor MALSU1 (Mitochondrial Assembly of Ribosomal Large Subunit 1) and promote its recruitment to the mitoribosome large subunit, where MALSU1 is necessary for tetracycline‐induced survival and suppression of ER stress. Glucose starvation induces ER stress to activate the unfolded protein response and IRE1α‐mediated cell death that is inhibited by tetracyclines. These studies establish a new interorganelle communication whereby inhibition of the mitoribosome signals to the ER to promote survival, implicating basic mechanisms of cell survival and treatment of mitochondrial diseases.
Keywords: IRE1α, MALSU1, mitochondrial disease, mitoribosome, tetracyclines
Subject Categories: Molecular Biology of Disease, Translation & Protein Quality
This study identifies a link between mitochondrial protein translation and endoplasmic reticulum stress, where activation of mitoribosome quality control suppresses unfolded protein responses. These results illustrate a novel interorganelle communication that adds insight into mechanisms of cell survival in normal physiology and disease.
Introduction
Mitochondrial diseases (MDs) are a rare, clinically heterogenous, and heritable class of diseases characterized by dysfunctional mitochondria and bioenergetic defects (Nunnari & Suomalainen, 2012; Gorman et al, 2016; Wallace, 2018). Mitochondrial function relies on proteins originating from both nuclear and mitochondrial genomes, and mutations in genes from either genome can result in MD (Nunnari & Suomalainen, 2012; Gorman et al, 2016; Wallace, 2018; Kummer & Ban, 2021; Bennett et al, 2022a, 2022b). The hallmark phenotypes of MD include bioenergetic defects resulting in cellular redox imbalance, inflammatory signaling, and tissue damage. MD manifests in tissues of high energetic (aerobic) demands and commonly results in neuromuscular deficiencies (Gorman et al, 2016). Specifically, mtDNA mutations in mitochondrial complex I (e.g., A3796G ND1) and tRNA leucine (e.g., A3243G tRNALeu(UUR)) constitute the most clinically prevalent MDs, presenting as Leber hereditary optic neuropathy (LHON) and mitochondrial encephalomyopathy with lactic acidosis and stroke‐like episodes (MELAS), respectively (Gorman et al, 2016). Although the genetic causes of MD are now well understood, pathological signaling mechanisms remain poorly defined, and treatment strategies are very limited.
We have shown using a high‐throughput chemical screen that antibiotic tetracyclines promote survival and fitness in models of MD (Perry et al, 2021). Antibiotics that inhibit the bacterial ribosome, like tetracyclines, also target the evolutionarily conserved mitochondrial ribosome (mitoribosome), constituting a novel treatment paradigm by modulating mitochondrial protein synthesis for MD therapy. Treatment of MD cells with tetracyclines rescues glucose‐deprivation induced cell death by reversing inflammatory gene expression and restoring redox homeostasis, most notably NADPH/NADP+ ratios, consistent with inhibition of p38 mitogen activated protein kinase (MAPK) (Perry et al, 2021). This is also consistent with and substantiated by known mechanisms of cell death in mitochondrial complex I disease (Balsa et al, 2020). Further, tetracyclines improved survival and fitness of NDUFS4−/− mice, where the neuromuscular decline accompanying this MD was delayed, and neuroinflammatory signaling was suppressed (Perry et al, 2021). Interestingly, pharmacological inhibition of the endoplasmic reticulum (ER)‐resident unfolded protein response (UPR) stress sensor inositol‐requiring enzyme 1 (IRE1α) or downstream p38 MAPK inflammatory response can rescue MDs from glucose deprivation (Soustek et al, 2018). Additionally, activation of protein kinase R (PKR)‐like endoplasmic reticulum kinase (PERK) rescues MD cells from nutrient deprivation (Balsa et al, 2019). These studies position the UPR as a key regulator of MD cell survival and death phenotypes under nutrient stress conditions, where protein misfolding and ER stress responses are exacerbated (Balsa et al, 2019; Huang et al, 2019; Carreras‐Sureda et al, 2022). The role of mitoribosomes in signaling to the ER, and how tetracyclines may modulate this response, is unknown, prompting the current study.
Originating from an endosymbiotic event between a eukaryotic progenitor and an α‐proteobacterium, mitochondria once functioned as independent bacterial organisms. Throughout evolution, mitochondria have maintained their own genome along with transcription and translation machinery (Ott et al, 2016; Kummer & Ban, 2021). Specifically, mitochondrial ribosomes (mitoribosomes) are responsible for the expression of mtDNA‐encoded protein subunits of the electron transport chain. Translation by the mitoribosome is analogous to the bacterial ribosome, with functionally conserved phases of translation initiation, elongation, and termination (Kummer & Ban, 2021). Accordingly, antibiotics that target the bacterial ribosome, like tetracyclines, are also known to target the mitoribosome (Wilson, 2014; Moullan et al, 2015). Specifically, tetracyclines target the bacterial ribosome by binding to 16 s rRNA, disrupting the accommodation of amino‐acyl tRNAs in the A‐site of the ribosome‐mRNA complex and thereby inhibiting elongation of nascent peptides (Wilson, 2014). Although not yet structurally determined, it can be reasoned that tetracyclines work in a similar fashion at the mitoribosome. Mitochondrial translation is initiated upon recognition and accommodation of formyl‐methionine tRNA, with subsequent amino‐acyl tRNAs recruited and translocated in a GTP‐dependent fashion, assisted by a series of mitochondrial elongation factors (Kummer & Ban, 2021). Disruption in elongation is accompanied by the recruitment of rescue factors to maintain mitoribosome integrity and translation fidelity. Elongational stalling induced by aberrant tRNA accommodation results in a “splitting” event, where the large and small mitoribosome subunits (mtLSU and mtSSU, respectively) are dissociated and bound by a series of quality control proteins (Desai et al, 2020; Kummer & Ban, 2021). First, a protein module composed of MALSU1, LOR8F8, and NDUFAB1 (MALSU1 module) binds to the mtLSU, sterically inhibiting reassociation of the mtSSU (Brown et al, 2017; Desai et al, 2020). Subsequently, a series of rescue factors (MTRES1 and mtRF‐R) bind and stabilize nascent peptide and P‐site tRNA (Desai et al, 2020). Mitochondrial translation initiation factor 3 (MTIF3) plays similar roles in dissociating, binding, and stabilizing the mtSSU (Luo et al, 2019; Khawaja et al, 2020; Itoh et al, 2022; Remes et al, 2023). These quality control proteins identify a means by which the cell can recycle native‐disrupted or inhibited mitoribosomes to bypass energetically costly rounds of ribosome biogenesis. The role of these proteins, and more generally of mitoribosomes, in signaling for cell survival or death has not yet been described, offering a starting point for the current work toward the investigation into the molecular mechanisms of tetracycline‐induced cell survival.
In the current work, we investigate signaling mechanisms that originate at the mitoribosome in response to tetracyclines in MD. We illustrate that tetracyclines mediate the recruitment of the mitochondrial quality control protein MALSU1 to the mtLSU, indicating that elongational stalling and splitting are initial responses to tetracycline treatment. Using CRISPR‐Cas9 technology, we identify that MALSU1 and MTIF3, but not other rescue factors, are required for tetracycline‐induced survival response in MD cells under glucose starvation. We further show that tetracyclines reverse cell‐death signaling in the ER through the attenuation of IRE1α oligomerization and inhibition of effector UPR function associated with all three arms of the UPR. MALSU1‐deficient MD cells sensitive to nutrient stress exhibit heightened sensitivity toward the activation of ER stress, which is not reversed by tetracyclines. This work highlights a novel signaling between the mitochondria and ER that can be leveraged for the treatment of MD.
Results
Tetracyclines require mitoribosome splitting and quality control protein MALSU1 to promote survival in ND1 complex I mutant cells
Mutations in mitochondrial genes result in increased sensitivity to nutrient deprivation, such that glucose deprivation can induce cell death. Specifically, cells cultured in equimolar concentrations of galactose in place of glucose become dependent on mitochondrial metabolism, resulting in cells harboring mitochondrial mutations being particularly sensitive to cell death (Soustek et al, 2018; Balsa et al, 2019, 2020; Perry et al, 2021). We previously showed that inhibition of mitochondrial translation by tetracyclines rescues mitochondrial disease models from cell death by a currently unknown mechanism (Perry et al, 2021). Here, utilizing mitochondrial disease complex I (A3796G ND1) mutant cybrid cells, we interrogate mitoribosome quality control pathways as potential mediators of tetracycline‐induced rescue under glucose deprivation.
Recent cryogenic electron microscopy studies in phosphodiesterase 12 (PDE12) mutant HEK293 cells indicate that mitoribosome elongational stalling induced by aberrant tRNA accommodation promotes mitoribosome splitting and recruitment of quality control factors (Desai et al, 2020). Doxycycline inhibits bacterial protein synthesis by binding to the bacterial SSU rRNA in the decoding region, stalling translation by disrupting the accommodation of tRNA (Wilson, 2014). This mechanism is similar to defective tRNA processing in PDE12 mutant cells (Desai et al, 2020). With this in mind, we sought to investigate mitoribosome elongational stalling and quality control factors as initial molecular signals in promoting doxycycline‐induced survival in ND1 mitochondrial mutant cells. ND1 mutations are commonly found in MD patients (Gorman et al, 2016), and we have previously used this cell model to validate genes and compounds, including tetracyclines, that rescue cell death under glucose‐deprivation conditions (Soustek et al, 2018; Balsa et al, 2019, 2020; Perry et al, 2021).
The mitochondrial translation cycle includes distinct stages of initiation, elongation, and termination, where conserved classes of proteins regulate progression between each phase (Kummer & Ban, 2021). To investigate if the general inhibition of mitochondrial translation was sufficient to promote survival in ND1 mutants, we compared the effects of mitoribosome‐targeting doxycycline with a distinct inhibitor, N‐formyl methionine mimetic actinonin, that stalls mitochondrial translation by inhibiting peptide deformylase (Richter et al, 2013, 2015) (PDF, Fig 1A), the enzyme responsible for the removal of N‐formyl groups on nascent peptides as they emerge from the ribosome exit tunnel prior to folding (Richter et al, 2013, 2015). It has also been suggested that actinonin inhibits mitochondrial translation indirectly through inhibition of the mitochondrial AAA‐protease (Richter et al, 2015), illustrating another non‐mitoribosome targeting mechanism. Nonetheless, the mechanism of inhibition of mitochondrial translation by actinonin is distinct to that of tetracyclines. Treatment with actinonin was unable to promote survival in ND1 cells at a wide range of concentrations (Fig 1A), suggesting that inhibition of mitochondrial translation is not necessarily sufficient for the promotion of MD cell survival during glucose starvation. We have also previously identified (Perry et al, 2021) that other mitoribosome‐targeting inhibitors pentamidine (Sun & Zhang, 2008) and retapamulin (Wilson, 2014; Meydan et al, 2019) promoted survival similar to doxycycline. Together, these results suggest that general suppression of mitochondrial protein synthesis is not sufficient to promote survival, rather, a signal must be initiated through direct inhibition of the mitoribosome. In an unbiased approach to identify factors involved in doxycycline rescue, we utilized the Dependency Map (DepMap) proteomics datasets, where repurposing screens have been performed to investigate the utility of current FDA approved small molecules, including doxycycline, for cancer treatment (Corsello et al, 2020) (Fig 1B). Here, positive correlation between the protein abundance and sensitivity to doxycycline identified proteins involved in cell death. Hence, we sought to identify proteins for which high expression was associated with reduced sensitivity to doxycycline, which we reasoned might be involved in promoting doxycycline‐mediated survival in models of MD (Perry et al, 2021). Gene‐ontology enrichment analysis identified high (negative) correlation with numerous mitochondrial protein subclasses including “mitochondrion” and “transit peptide,” and more specifically the mitoribosome including “translational elongation” and “large mitoribosome subunit” (Fig 1B). Interestingly, these studies identified survival correlation with mitoribosome quality control protein MALSU1, in line with potential elongational stalling and splitting mechanism described above. To note, MTRES1 and mtRF‐R did not correlate with doxycycline treatment. MRPL14, an accessory subunit of the mtLSU that MALSU1 binds during splitting (Brown et al, 2017; Desai et al, 2020), was also identified in this screen, further substantiating the importance of this binding module in surviving cancer cells and potentially in mitochondrial mutant cells in the context of doxycycline mechanisms of action (Fig 1B).
Figure 1. Tetracyclines require mitoribosome splitting and quality control protein MALSU1 to induce survival in ND1 mutant cells.
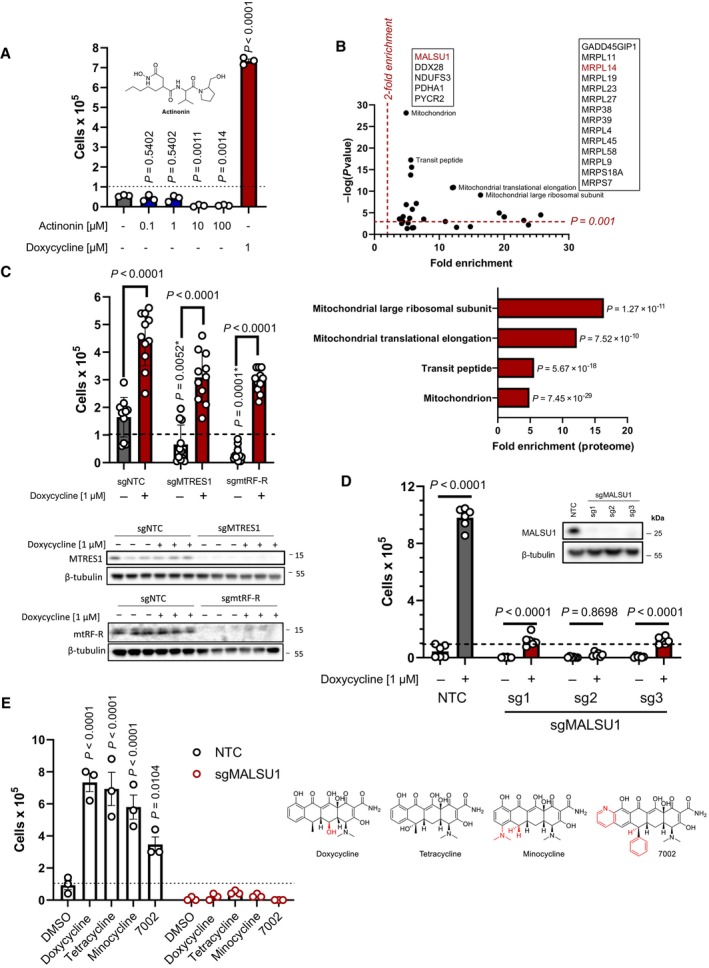
- ND1 cells treated with a range of concentrations (0.1, 1.0, 10, 100 μM) of peptide deformylase inhibitor actinonin do not promote survival under galactose conditions (error bars represent the average ± s.e.m. of n = 3 independent biological samples). Statistical significance (P < 0.05) between actinonin and untreated cultures was calculated using one‐way ANOVA, multiple comparisons.
- DepMap drug‐repurposing proteomics database indicates enrichment of mitochondrial and more specifically mitoribosome proteins in the context of doxycycline. Gene‐ontology enrichment was determined using DAVID pathway analysis, where significant proteins were determined (P < 0.001) and compared to entire proteome. Note enrichment of MALSU1 associated with surviving cells in the context of doxycycline. Statistical significance was determined as previously described (Huang da et al, 2009).
- Depletion (CRISPR‐Cas9) of mitochondrial quality control rescue factors MTRES1 and mtRF‐R did not reverse capacity of tetracyclines to rescue ND1 cells from galactose induced cell death (error bars represent the average ± s.e.m. of n = 11 biological replicates over N = 4 experiments). During survival experiments, dashed‐line represents ND1 seeding density, where cells were exposed to glucose deprivation (galactose supplementation) for 4 days. P‐values* with asterisk denote comparisons with DMSO treated NTC cells. Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC, sgMTRES1, and sgmtRF‐R cells was calculated using two‐way ANOVA, multiple comparisons.
- MALSU1 expression is required for tetracyclines induced survival (error bars represent the average ± s.e.m. of n = 6 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC and sgMALSU1 (sg1‐sg3 independent small‐guide RNA) was calculated using two‐way ANOVA, multiple comparisons.
- The broad class of tetracycline antibiotics rescue ND1 cells from nutrient deprivation depending on MALSU1. Doxycycline, tetracycline, minocycline, and 7002 were utilized as structurally diverse tetracycline analogs that cannot rescue ND1 mutant cells that lack MALSU1 (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistical significance (P < 0.05) between tetracycline analogs and untreated cells across NTC and sgMALSU1 was calculated using two‐way ANOVA, multiple comparisons. Chemical modifications of each respective analog from parental tetracycline template are highlighted in red.
Source data are available online for this figure.
To interrogate the role of the mitoribosome quality control pathway in facilitating tetracycline‐induced survival in mitochondrial ND1 mutant cells, we utilized CRISPR Cas9 gene editing to knock‐out (KO) respective proteins involved in this pathway. Elongational stalling results first in splitting of the large and small subunit where MALSU1, along with LOR8F8 and NDUFAB1 (MALSU1 module), binds and stabilizes the large subunit. Rescue factors MTRES1 and mtRF‐R then bind and stabilize the nascent peptide and tRNA. We first knocked out MTRES1, mtRF‐R, and MALSU1 to identify which, if any, proteins may be involved in doxycycline survival (Fig 1C and D). These studies illustrated that doxycycline was still able to rescue ND1 cell death in the absence of MTRES1 or mtRF‐R (Fig 1C), but survival was significantly reduced in the absence of MALSU1, where the recruitment of the entire MALSU1 module, including NDUFAB1 and LOR8F8, is expected to be compromised (Fig 1D). It was also noted that basal survival rates of ND1 cells in galactose were drastically reduced in MTRES1 and mtRF‐R KO conditions (Fig 1C), indicating that these cells rely on mitoribosome quality control under nutrient stress, but doxycycline specifically requires the MALSU1 module to promote survival (Fig 1D). We further investigated whether this mechanism can be applied to the general tetracycline class of antibiotics by testing additional analogs, including the parental tetracycline, along with synthetic analogs minocycline and 7002, a compound we previously illustrated to inhibit mitochondrial translation and promote survival in mitochondrial mutants (Perry et al, 2021) (Fig 1E). These studies illustrate that tetracycline antibiotics universally depend on MALSU1 to promote survival in ND1 cybrid cells (Fig 1E). Ectopic overexpression of MALSU1 in complex I mutant cells independent of tetracyclines was unable to promote survival of ND1 mutant cells under glucose deprivation, indicating that MALSU1 is required but not sufficient to promote survival. It also suggests that the survival signal requires the tetracycline‐inhibited mitoribosome for MALSU1 to protect against cell death (Fig EV1A–C).
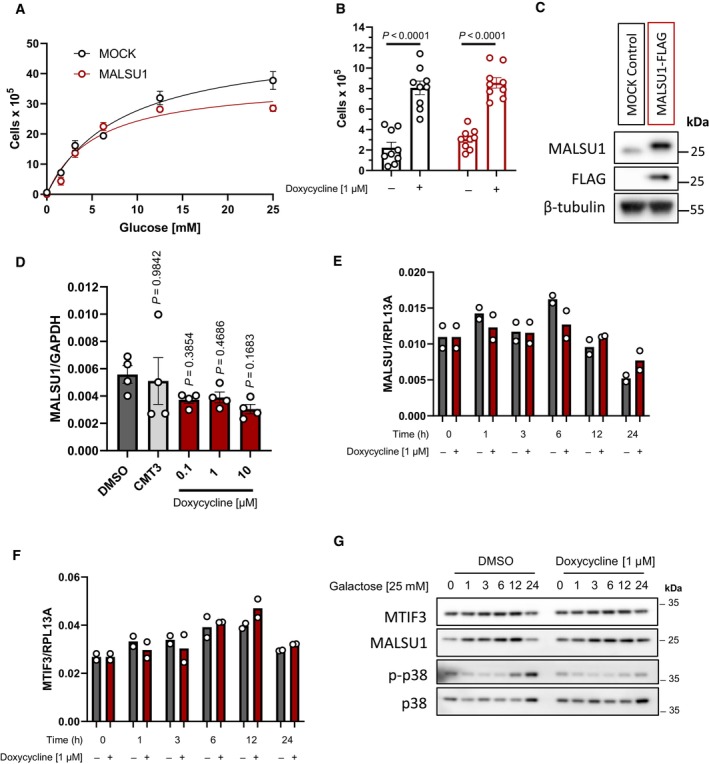
Figure EV1. Ectopic overexpression (OE) of MALSU1 does not protect ND1 mutant cybrid cells from nutrient deprivation. ND1 cells were transduced with lentiviral vector containing MALSU1 under the constitutive control of the CMV‐promotor.
-
AND1 control (MOCK) and MALSU1‐OE cells exposed to varying extent of glucose deprivation (96 h) follow similar cell death responses (error bars represent the average ± s.e.m. of n = 3 biological replicates per concentration).
-
BMALSU1‐OE ND1 cells do not synergize with doxycycline (1 μM) in promoting survival (96 h, error bars represent the average ± s.e.m. of n = 9 biological replicates across N = 3 independent experiments). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across MOCK and MALSU1‐OE cells was calculated using two‐way ANOVA, multiple comparisons.
-
CStable MALSU1‐flag tag expression in ND1 cells.
-
DQuantitative PCR of ND1 cells (galactose, 48 h) treated with varying concentrations of doxycycline or inactive tetracycline analog CMT3 indicate mRNA expression levels of MALSU1 are unchanged (error bars represent the average ± s.e.m. of n = 4 biological replicates). Statistical significance (P < 0.05) between doxycycline or CMT3 treated and untreated cells was calculated using one‐way ANOVA, multiple comparisons.
-
E, FTime‐course experiments indicate that doxycycline does not alter (E) MALSU1 or (F) MTIF3 transcript levels (n = 2 technical replicates).
-
GND1 cells exposed to glucose deprivation for indicated time periods with and without doxycycline indicate stability of MALSU1 by doxycycline at the 24 h time point. Note MTIF3 expression levels are unchanged. Note doxycycline suppresses the phosphorylation of p38 MAPK at later time points (12–24 h) as a control.
Mitoribosome dissociating factor MTIF3 is partially required for doxycycline to rescue cell death in mitochondrial disease mutant cells
Having observed that MALSU1 is required to promote survival in mitochondrial mutant cells by tetracycline, we investigated the requirement for other mitoribosome quality control factors. Mitochondrial initiation factor 3 (MTIF3) has been previously implicated in binding the mitoribosome small subunit during biogenesis (Khawaja et al, 2020; Itoh et al, 2022) and during translation to access polycistronic transcripts (Remes et al, 2023). Specifically, MTIF3 sterically blocks the premature association of large‐ and small subunits (Khawaja et al, 2020; Itoh et al, 2022), analogous to the role of MALSU1 at the large subunit (Brown et al, 2017; Desai et al, 2020). Further, MTIF3 has ribosome dissociative activity (Luo et al, 2019), qualifying it as a potential mediator of mitoribosome splitting following tetracycline treatment and downstream cell survival promoting effects. CRISPR‐Cas9 depletion of MTIF3 in ND1 cells resulted in a reduced ability of doxycycline to promote cell survival (Fig 2A and B), implicating this protein in potentiating signaling at the tetracycline‐inhibited mitoribosome, albeit to a lesser extent than MALSU1 (Fig 2B).
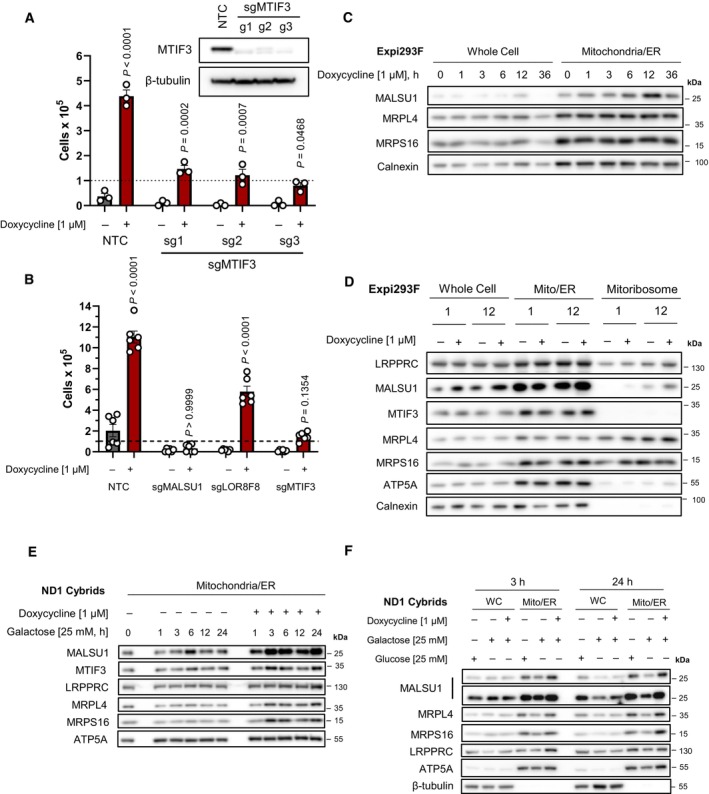
Figure 2. MTIF3 potentiates tetracycline‐induced cell survival in ND1 mutant cells.
-
ACRISPR‐Cas9 KO of MTIF3 in ND1 cybrid cells reduces doxycycline's ability to promote survival (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC and sgMTIF3 (sg1‐sg3) was calculated using two‐way ANOVA, multiple comparisons.
-
BCRISPR‐Cas9 KO of MALSU1 and MTIF3 drastically reduce the ability of tetracyclines to promote survival when compared to LOR8F8‐KO (error bars represent the average ± s.e.m. of n = 6 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC, sgMALSU1, sgLOR8F8, and sgMTIF3 cells was calculated using two‐way ANOVA, multiple comparisons.
-
CMALSU1 is recruited to the mitochondria at early time points. Crude mitochondria (mitochondria/ER) were isolated from Expi293F cells at varying time points following doxycycline treatment. Note maximum mitochondrial MALSU1 at the 12‐h time point.
-
DMitoribosome isolation in Expi293F cells indicates doxycycline promotes the accumulation of MALSU1 staring at 1 h, with drastic increases at the 12‐h time point. Note mitochondrial accumulation of MALSU1 at 12 h. MTIF3 expression, along with mitochondrial and mitoribosome accumulation were unchanged with doxycycline.
-
E, F(E) Mitochondria were isolated from ND1 mutant cybrid cells exposed to galactose with or without doxycycline for varying time points, with rapid mitochondrial accumulation of MALSU1 after 1 h, that maximizes from 3 to 24 h of doxycycline treatment. Note subtle increases in MTIF3, along with MRPL4 and MRPL6 at the 3‐h time points. (F) Matched glucose controls at the 3‐ and 24‐h time point illustrate a decrease in mitochondrial MALSU1, MRPL4, and MRPS16 upon galactose treatment that is stabilized with doxycycline. Note increased whole cell expression level of MALSU1 in doxycycline treated cultures at the 24‐h time point.
Source data are available online for this figure.
To further define MALSU1 as a key player in survival, we dissected the role of the MALSU1 module by knocking‐out LOR8F8. LOR8F8, a LYRM‐chaperone, binds to and recruits NDUFAB1 to MALSU1, constituting the mature MALSU1 module capable of binding the split mitoribosome (Brown et al, 2017). Similar to MALSU1 and MTIF3, LOR8F8‐KO‐sensitized cells to glucose deprivation (Fig 2B). However, doxycycline was able to efficiently rescue LOR8F8 deficient cells to a much greater extent than MALSU1 and MTIF3 KO cells (Fig 2B). The ability of doxycycline to promote survival in the LOR8F8‐KO cells indicates that MALSU1, but not other MALSU1‐module proteins, is necessary to promote survival.
We next sought to investigate the regulation of MALSU1 at the mitochondria and mitoribosome in response to tetracyclines. To study the recruitment of different quality control factors to the purified mitoribosome, we utilized HEK293 suspension cells (Expi293F), a model previously used to study mitoribosome elongational stalling and quality control (Desai et al, 2020). ND1 mutant cybrid cells grown in monolayer culture did not provide sufficient mitochondria to purify appreciable quantities of the mitoribosome, so Expi293F suspension cells were used as an alternate system to study changes in the molecular composition and architecture of the doxycycline‐inhibited mitoribosome. To first understand the kinetics of MALSU1 recruitment, we performed a time‐course experiment, isolating mitochondria from Expi293F cells at a series of different time points following doxycycline treatment (Fig 2C). Here, we found that levels of MALSU1 within mitochondria were highest at 12 h following doxycycline treatment (Fig 2C). Using these timepoints as a guide, we isolated mitoribosomes from Expi293F cells at early (1 h) and later (12 h) time points to assess MALSU1‐mitoribosome recruitment. We found that MALSU1 levels were detectable at the mitoribosome after 1 h, with much larger increases observed after 12 h of doxycycline treatment (Fig 2D). We also evaluated the regulation of MTIF3 in these experiments, observing that doxycycline did not significantly alter the amount of this protein present at the mitochondria or mitoribosome (Fig 2D), highlighting MALSU1 as a key mediator of signaling in response to elongational stalling. The necessity of MTIF3 for doxycycline‐induced survival, however, implicates this protein in potentiating the survival signal initiated by tetracycline treatment. Similar to Expi293F cells, the abundance of MALSU1 in mitochondrial fractions of ND1 mutant cells increased following doxycycline treatment, with accumulation starting at 1 h and maximizing from 3 to 24 h (Fig 2E). We also observed a notable increase in mitochondrial accumulation of MTIF3, along with mitoribosome proteins MRPL4 and MRPS16, potentially reflecting a stimulation of mitochondrial quality control or biogenesis in response to doxycycline treatment in the context of mitochondrial mutations and glucose starvation (Fig 2E). Interestingly, when compared to matched glucose controls, we observed that MALSU1 and MRPs are decreased upon glucose starvation and are stabilized by doxycycline, most notably at the 24‐h time point (Fig 2F). We observed concomitant increases in MALSU1 protein in the whole cell extract at 24 h without substantial changes in transcript levels (Figs 2F and EV1D–G), indicating likely increased mitochondrial stability of MALSU1 at the tetracycline‐inhibited mitoribosome. These results highlight a potential stabilization of MALSU1 at the mitochondria in response to tetracyclines at early timepoints.
Tetracyclines mediate MALSU1 recruitment to the mitoribosome large subunit
To investigate the mechanisms whereby MALSU1 is required for ND1 mutant cell survival under glucose starvation, we sought to further investigate doxycycline‐dependent regulation of MALSU1 recruitment to the mitoribosome. Rapid isolation of the mitoribosome, initially from Expi293F cells, was achieved through successive sucrose gradient centrifugations to obtain mitochondria, which were then solubilized in dodecyl‐β‐D‐maltoside (β‐DDM) and cardiolipin. This efficiently extracted mitoribosomes from the inner mitochondrial membrane for fractionation of the mtSSU, mtLSU, and monosomes (Desai et al, 2020).
Expi293F cells were treated with doxycycline for 48 h to induce mitoribosome stalling, after which mitoribosomes were isolated for analysis. Doxycycline‐treated mitochondria, and more drastically the mitoribosomes, exhibited increased MALSU1 abundance (Fig 3A). These results are consistent with recruitment of MALSU1 to the mitoribosome at earlier timepoints (Fig 2D) and may reflect increases in its mitochondrial import and/or stability in response to doxycycline. To investigate whether MALSU1 was enriched at the large subunit in doxycycline‐treated cultures, we sucrose‐gradient‐fractionated mitoribosomes to separate and analyze the location of MALSU1. Here, we found that MALSU1 was enriched at the mtLSU upon doxycycline treatment (Fig 3B). Additionally, we observed a shift in the sedimentation of the MALSU1‐bound mtLSU with doxycycline treatment, indicating a potential change in the size and/or molecular shape of the large subunit in this condition when compared to controls (Fig 3B). To validate these effects in ND1 cybrid cells, crude mitochondria were isolated, solubilized with β‐DDM and cardiolipin, and fractionated by sucrose gradients in a similar fashion as described above. Mitochondria from CRISPR‐Cas9 MALSU1‐depleted ND1 cells were analyzed in parallel to investigate the necessity of MALSU1 in shifting the sedimentation pattern of the doxycycline treated large subunit. As observed in Expi293F suspension cells, doxycycline treatment in ND1 cells resulted in an increase in mitochondrial MALSU1 (Fig 3C) that was enriched at the large subunit (Fig 3D and E). The shift in large subunit sedimentation was again observed with doxycycline treatment (Fig 3D), mimicking Expi293F cultures (Fig 3B). The sedimentation pattern of the large mitoribosome subunit was still altered in MALSU1‐KO cells (Fig 3F), indicating that doxycycline may split the large subunit independent of MALSU1, although MALSU1 binding to the large subunit is required for survival (Fig 1D and E).
Figure 3. Tetracyclines mediate MALSU1 recruitment to the mitoribosome large subunit.
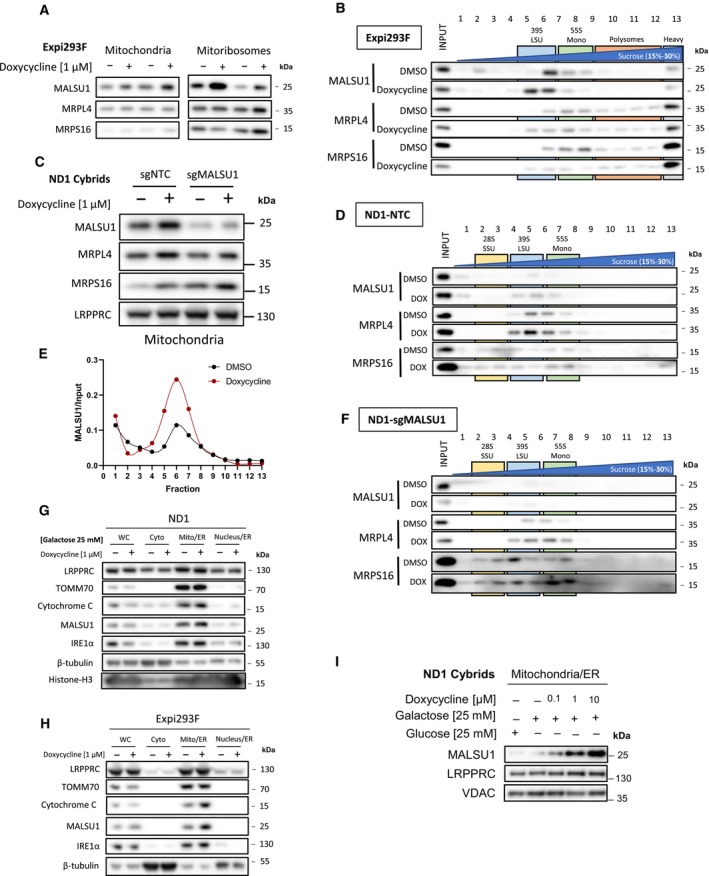
-
A, B(A) Isolated mitochondria and mitoribosomes from Expi293F exhibit enrichment of MALSU1 in doxycycline treated cultures, that is (B) associated with the large subunit of the mitoribosomes evidenced by sucrose gradient sedimentation (15–30% sucrose). Sedimentation of large and small subunit were assessed by MRPL4 and MRPS16, respectively. Duplicates of mitochondria and mitoribosomes in (A, n = 2 independent experiments) are the inputs for (B). Note shift in MALSU1 and MRPL4 upon doxycycline treatment.
-
CIsolation of crude mitochondria from doxycycline (1 μM) treated ND1 cells (non‐target control (NTC) and sgMALSU1). Note increased MALSU1 expression in doxycycline‐treated mitochondria.
-
DCrude mitochondria from ND1‐NTC cells were sucrose gradient (15–30%) sedimented and analyzed for MALSU1 as in (B). Note shift and enrichment of MALSU1 and MRPL4 with doxycycline treatment.
-
EQuantitation of MALSU1 elution profile in sucrose gradients of crude mitochondria from ND1 cells derived from (D), normalized to input signal of MALSU1.
-
FSucrose gradient (15–30%) sedimentation of MALSU1‐depleted ND1 cells with or without doxycycline treatment. Crude mitochondria from (C) are inputs for (D) and (F).
-
G, HCell fractionation of (G) ND1 and (H) Expi293F suspension cultures treated with doxycycline. Note enrichment of MALSU1 specifically in mitochondrial fractions and decreases in mitochondrial markers TOM70 and Cytochrome C at the whole cell level.
-
IND1 cells treated with increasing concentrations of doxycycline illustrate dose‐dependent increases in MALSU1 specifically at the mitochondria without changes in total mitochondria (VDAC, LRPPRC).
Source data are available online for this figure.
To further investigate the subcellular localization of MALSU1 in response to doxycycline, we fractionated ND1 (Fig 3G) and Expi293F (Fig 3H) cells to identify if MALSU1 is enriched in specific compartments in response to mitoribosome stalling. These experiments showed that MALSU1 is specifically (Fig 3G and H) and dose‐dependently (Fig 3I) enriched at the mitochondria of doxycycline‐treated cells, without changes in mitochondrial abundance of the matrix protein LRPPRC or the outer‐mitochondrial membrane protein TOM70. It has been previously reported that MALSU1 has a short half‐life (Morgenstern et al, 2021) and that its expression is lost in rho0 cells (Wanschers et al, 2012), highlighting a tightly regulated and mitochondria‐specific localization. Increased whole cell levels of MALSU1 upon doxycycline treatment at earlier timepoints (e.g., ND1 24 h and Expi293F 1 and 12 h, Figs 2D–F and EV1G) may reflect an accumulation of MALSU1 at the mitochondria due to its stabilization at the mitoribosome. MALSU1 expression is apparently unchanged at the whole cell level when compared to increases in mitochondrial fractions at later time points (48 h). In these conditions, this may suggest remodeling of the mitochondrial proteome in doxycycline treated cells, that when normalized to mitochondrial protein content reflects higher MALSU1 levels. This is consistent with decreases in mitochondrial markers TOM70 and cytochrome C (Fig 3G and H). These studies support the notion that MALSU1 binding to the mitoribosome large subunit drives tetracycline‐induced survival responses and shows a direct association between the extent of mitoribosome inhibition and MALSU1 recruitment.
Taken together, mitoribosome stalling caused by doxycycline results in mitoribosome splitting where doxycycline acts as a “splitting factor” and MALSU1 binds and stabilizes the large subunit to provide a pro‐survival signal. Based on its partial requirement in doxycycline‐induced cell survival (Fig 2A and B) and its reported ribosome dissociating activity (Luo et al, 2019), it is possible that MTIF3 cooperates in tetracycline‐induced ribosome splitting to promote MALSU1 binding.
Tetracyclines promote survival through MALSU1‐dependent suppression of ER stress
We have previously reported that pharmacological inhibition of ER stress sensor/transducer IRE1α promotes survival in mitochondrial complex I disease cells (Soustek et al, 2018), implicating ER stress in promoting death of cells with defective mitochondrial respiration. To determine whether doxycycline‐induced survival involves modulation of this pathway, we tested if doxycycline treatment reversed the activation of X‐box binding protein 1 short fragment (XBP1s), an ER‐stress‐induced transcription factor produced through translation of an alternatively spliced mRNA generated through the endoribonuclease activity of IRE1α (Huang et al, 2019; Carreras‐Sureda et al, 2022). A potent transcription factor, XBP1s, regulates the expression of a wide variety of ER‐homeostatic genes. Western blot analysis indicated that XBP1s is strongly and specifically activated in ND1 cells, but not WT cells, after 48 and 72 h under glucose restriction conditions (Fig 4A). Doxycycline treatment efficiently reduced XBP1s activation at 48 h and completely eliminated it at 72 h, consistent with improved cell survival and reduced PARP1 cleavage (Fig 4A). Interestingly, CMT3, a tetracycline analog that does not exhibit mitoribosome translation inhibition or rescue mitochondrial mutants (Perry et al, 2021), did not reverse the activation of XBP1s (Fig 4A). These results indicate that inhibition of mitochondrial translation by doxycycline results in the pro‐survival resolution of ER stress that is exacerbated in complex I mutants.
Figure 4. Tetracyclines promote survival through MALSU1‐dependent suppression of ER stress and IRE1α signaling.
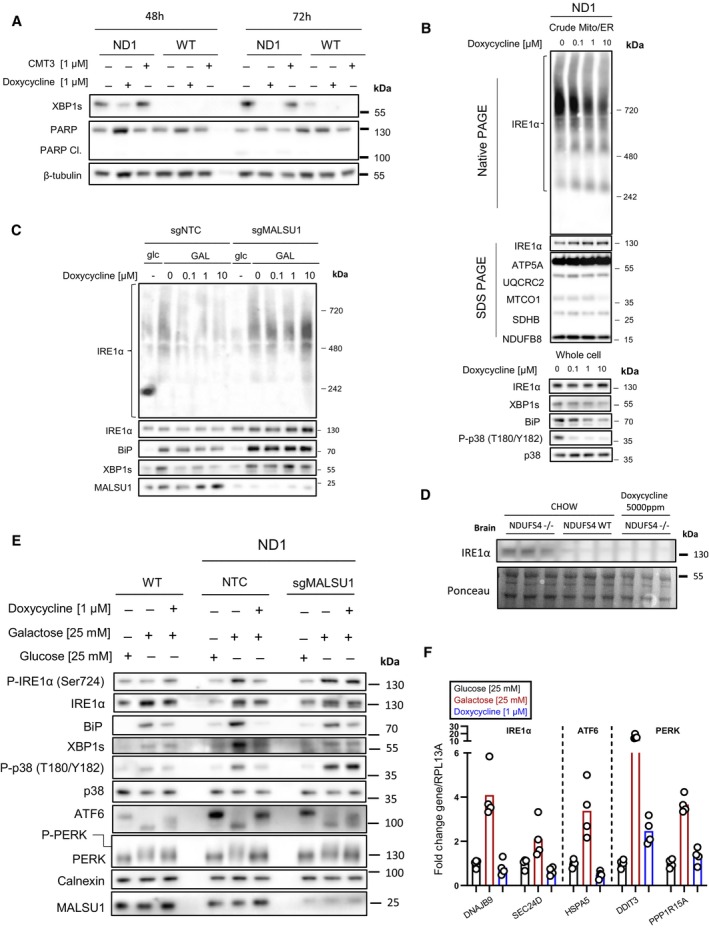
-
ADoxycycline (1 μM) reverses activation of IRE1α as evidenced by decrease in XBP1s in ND1 cells (galactose, 48 and 72 h). Inactive CMT3 tetracycline analog does not reverse XBP1s. Suppression of XBP1s at 72 h is associated with reversed PARP cleavage.
-
BBlue‐Native PAGE indicates dose‐dependent reductions in high‐order IRE1α oligomers in isolated crude mitochondria (Mitochondria‐ER). Doxycycline dose‐dependently inhibits activation of XBP1s, ER chaperone BiP, and phosphorylation of p38 MAPK in ND1 cells (galactose, 48 h).
-
CIRE1α oligomerization status in mitochondria‐ER fractions and effector UPR function in ND1 cells is MALSU1 dependent. Note increased oligomerization of IRE1α upon galactose treatment that it attenuated in doxycycline treated cultures (non‐target control, NTC), with reductions in BiP and XBP1s. IRE1α oligomerization status was not attenuated by doxycycline in ND1 cells lacking MALSU1, and consistent with BiP and XBP1s.
-
DIRE1α expression levels in the brains of NDUFS4−/− mice indicate increased expression when compared to WT and doxycycline‐supplemented counterparts. Results represent whole brains isolated from three independent mice under each indicated condition (n = 3 independent biological replicates).
-
E, FTetracyclines attenuate all three arms of the UPR. (E) Control (WT) and ND1 cybrids with (NTC) or without (sgMALSU1) MALSU1 were cultured in glucose, galactose (with or without doxycycline) for 48 h and analyzed for activation of IRE1α (phosphorylation), ATF6 (cleavage), and PERK (gel‐shift phosphorylation), along with expression levels of XBP1s and BiP, along with phosphorylation status of p38 MAPK. Note activation of IRE1α, PERK, and ATF6 in ND1 mutants that is suppressed with doxycycline in a MALSU1‐dependent fashion. (F) Genes downstream of all three arms of the UPR are activated with galactose, and suppressed with doxycycline. Genes from respective pathways include IRE1a‐XBP1s arm (DNAJB9, SEC24D), ATF6 arm (HSPA5), and PERK (DDIT3, PPP1R15A). qPCR data represents the average fold change expression levels from the average of n = 4 technical replicates across n = 2 biological samples.
Source data are available online for this figure.
Unfolded proteins in the ER bind to IRE1α on its luminal domain, promoting dimer/oligomerization and activation of the cytosolic kinase and RNAase domains, transducing ER stress (Huang et al, 2019). To further investigate doxycycline effects on IRE1α activation, we analyzed its oligomerization state using blue‐native polyacrylamide gel electrophoresis (BN‐PAGE, Fig 4B) as previously described (Sundaram et al, 2018). In whole cell extracts, we did not observe substantial changes in the oligomerization state with doxycycline treatment (Fig EV2A), inconsistent with results on XBP1s activation (Fig 4A). It has been previously reported that IRE1α can be enriched at mitochondrial ER contact sites (Takeda et al, 2019), a potential subcellular location wherein the oligomerization state is regulated. Consistent with this, we found IRE1α to be enriched in crude mitochondrial (Mito/ER) samples (Fig 3G and H). Hence, due to IRE1α enrichment with mitochondria, along with doxycycline mode of action at the mitoribosome, we isolated crude mitochondria containing ER fractions from ND1 cells and performed BN‐PAGE to analyze the IRE1α oligomerization status in the ER concentrated at the mitochondria. Here, we observed a marked dose‐dependent decrease in oligomerization state of IRE1α with doxycycline treatment without changes in total IRE1α levels or associated mitochondrial markers (Fig 4B). These results indicate that ND1 cell‐survival decisions through IRE1α are to some extent controlled at the mitochondria‐associated ER, and that a signal may be initiated at the mitochondrial ribosome upon elongational stalling with doxycycline treatment. These effects were also observed for additional markers of ER stress, including the ER lumen UPR chaperone BiP and downstream p38 MAPK phosphorylation, which were both reduced in a similar fashion to XBP1s (Fig 4B). In fact, we have previously illustrated that pharmacological inhibition of p38 (SB203580) rescues survival in ND1 mutant cells concomitant with metabolic changes similar to that of doxycycline treatment implicating MAPK signaling in the survival mechanism (Perry et al, 2021). Treatment of ND1 cells with IRE1α inhibitor 4μ8C reverses the galactose‐induced phosphorylation of p38 (Fig EV2B) similar to doxycycline or chemically distinct and active pentacycline‐based mitoribosome inhibitor 7002, but not the inactive CMT3 (Fig EV2C). This illustrates that MAPK activation is downstream of IRE1α and can be controlled by mitoribosome inhibition under these conditions.
Figure EV2. Inhibition of mitochondrial translation with tetracyclines inhibits IRE1α and MAPK signaling, dependent on MALSU1.
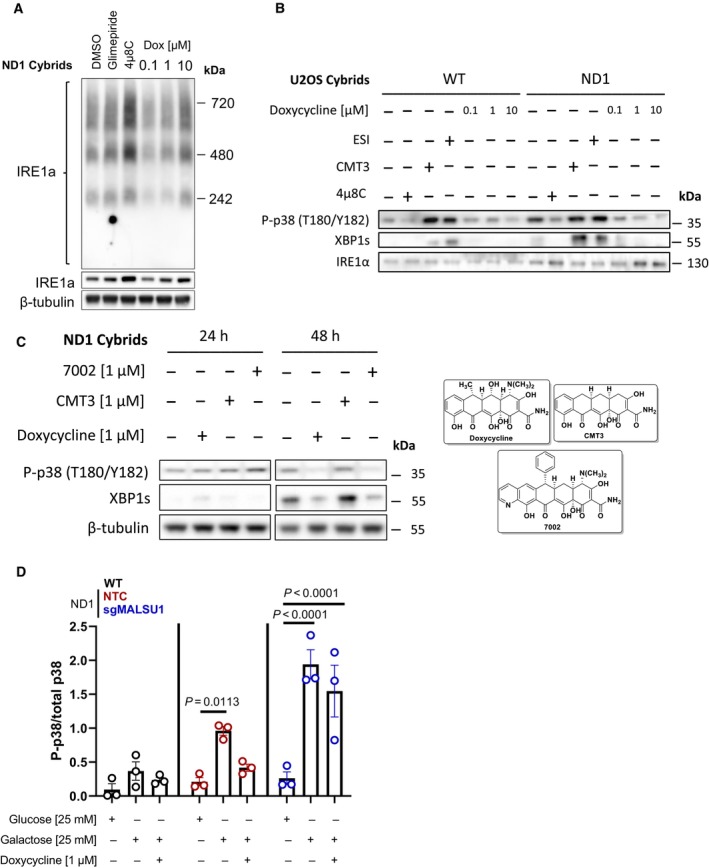
- Whole cell IRE1α oligomerization states under galactose conditions assessed by blue‐native PAGE. BN‐PAGE of IRE1α with whole cell extracts does not capture doxycycline regulation of high‐order oligomers when compared to ER concentrated at the mitochondria (Mito/ER extracts, Fig 4B). Glimepiride (25 μM) and IRE1α inhibitor 4μ8C (20 μM), compounds previously identified to rescue complex I mutants from nutrient deprivation, were used as controls. Note compensatory increase in IRE1α expression and oligomerization in 4μ8C treated cells.
- The activation of p38 MAPK is downstream of IRE1α and can be modulated by ERAD and mitochondrial translation. WT and ND1 cells were cultured in galactose media for 48 h and analyzed for the activation of MAPK and UPR signaling as evidenced by phosphorylation of p38 (P‐p38) and expression of XBP1s, respectively. Pharmacological inhibition of IRE1α with 4μ8C (20 μM) and p97 inhibitor ESI (20 μM) indicate P‐p38 and XBP1s are downstream of IRE1α and can be controlled by ERAD. CMT3 (1 μM) does not affect P‐p38 MAPK or XBP1s levels when compared to doxycycline (1 μM) indicating inhibition of mitochondrial translation, not off target effects of tetracyclines, modulates UPR and MAPK signaling.
- UPR and MAPK signaling are activated after 48 h of galactose stress in ND1 cybrids, where doxycycline (1 μM) and chemically distinct 7002 (1 μM), but not CMT3 (1 μM) attenuate these responses.
- Doxycycline suppresses p38 MAPK phosphorylation depending on MALSU1. Data represents the average ± s.e.m. p38 phosphorylation level normalized to total p38 using western blot densitometry (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistics across glucose, galactose, and doxycycline treated cultures in WT, ND1, and sgMALSU1 cells was calculated using two‐way ANOVA, multiple comparisons.
MALSU1, over other mitoribosome quality control proteins, is required for doxycycline‐induced survival (Fig 1D and E). To investigate if MALSU1 is required for doxycycline‐induced inactivation of IRE1α, we compared IRE1α oligomerization and XBP1s activation between doxycycline treated control and MALSU1‐KO mutant cells. In control and MALSU1 KO cells, we observe an increase in IRE1α oligomerization at mitochondrial contacts, along with XBP1s and BiP activation with galactose treatment (Fig 4C). However, unlike in control cells, IRE1α oligomerization, XBP1s cleavage, and BiP activation are not repressed in MALSU1 KO cells with doxycycline treatment, consistent with inability of doxycycline to inhibit phosphorylation of p38 MAPK (Fig EV2D) and promote survival of MALSU1 KO cells (Fig 4C). These studies show that MALSU1 must be present for doxycycline to modulate IRE1α‐mediated UPR activity in ND1 mitochondrial mutant cells.
NDUFS4−/− mice are a widely employed preclinical model of mitochondrial disease, largely characterized by neuromuscular decline (Kruse et al, 2008). Previously, we used this model to investigate the utility of doxycycline in vivo, where we found doxycycline supplemented mice exhibited improved survival and fitness when compared to controls (Perry et al, 2021). Specifically, doxycycline delayed the development of neurological defects in NDUFS4 KO brains. Thus, we investigated the status of IRE1α in the brains of NDUFS4 KO mice (Fig 4D). We found that NDUFS4 KO brains exhibited marked increased expression of IRE1α protein levels when compared to their control and doxycycline‐treated counterparts (Fig 4D). These results indicate that chronic treatment of doxycycline may attenuate the UPR in brains with mitochondrial dysfunction, suppressing IRE1α. Taken together, doxycycline‐induced survival signaling at elongating mitoribosmes requires MALSU1 and initiates cross‐talk between mitoribosome quality control and ER‐stress effectors, highlighting an important survival mechanism in mitochondrial disease mutants under nutrient stress.
Canonical regulation of IRE1α involves the interplay between inhibitory binding of BiP to monomeric IRE1α on the luminal domain and the activating recruitment of BiP to unfolded proteins in the ER lumen, releasing BiP from IRE1α to chaperone and maintain protein homeostasis (Huang et al, 2019). This release allows for dimerization of IRE1α, transducing initial pro‐survival UPR. If ER proteostasis is unable to be re‐established, unfolded proteins bind to the IRE1α luminal domain directly, causing oligomerization activating cell death responses (Huang et al, 2019). MARCH5, a mitochondrial outer membrane ubiquitin ligase, has recently been implicated in attenuating the oligomerization of IRE1α under prolonged ER stress (Takeda et al, 2019). With marked decreases in IRE1α oligomerization upon mitochondrial translation inhibition with doxycycline (Fig 4B and C), we sought to investigate if this regulation was mediated through MARCH5. CRISPR Cas9 KO of MARCH5‐sensitized ND1 cells to galactose‐induced ER stress with heightened levels of XBP1s and P‐p38, but this response could still be reversed by doxycycline treatment (Fig EV3A). Importantly, doxycycline still rescued MARCH5‐depleted ND1 cells from galactose‐induced cell death (Fig EV3B), suggesting that MARCH5 is not involved in the cross‐talk between the doxycycline inhibited mitochondrial ribosome and IRE1α activation at the ER.
Figure EV3. Outer‐mitochondrial membrane E3‐ubiquitin ligase MARCH5 is not required for tetracyclines pro‐survival mechanism in mitochondrial mutant cells.
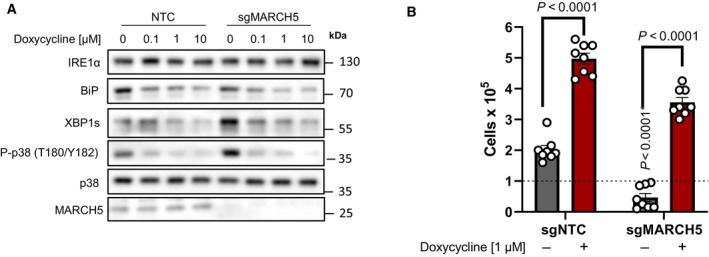
- Doxycycline dose‐dependently inhibits UPR and MAPK signaling in ND1 non‐target control (NTC) cells and cells depleted of MARCH5 under galactose conditions. Note basal levels of XBP1s and P‐p38 are increased in sgMARCH5 cells, indicating enhanced sensitivity to ER stress under these conditions.
- ND1 cybrids cultured in galactose conditions for 4 days can be rescued by doxycycline independent of MARCH5 expression status (error bars represent the average ± s.e.m. of n = 8 independent biological replicates across N = 3 independent experiments). Statistics of doxycycline treated and untreated cultures across NTC and sgMARCH5 cells were calculated using two‐way ANOVA, multiple comparisons.
Doxycycline treatment results in marked MALSU1‐dependent decreases in ER stress as evidenced by reduced IRE1α oligomerization and activation and downstream reductions in XBP1s, BiP, and phosphorylation of p38 MAPK (Figs 4A–C and EV2D). To further investigate tetracyclines effects on the UPR, we analyzed the activation of additional UPR sensors PERK and ATF6. Similar to IRE1α, PERK, and ATF6 are ER membrane proteins with luminal sensing and cytosolic signal‐transducing domains (Sundaram et al, 2018; Wiseman et al, 2022). Activation occurs following the accumulation of unfolded proteins in the lumen that out‐compete protein folding chaperones (Sundaram et al, 2018; Wiseman et al, 2022). ATF6 is proteolytically processed to translocate first to the golgi, and then to the nucleus where the cleaved product acts as a transcription factor, promoting the expression of ER‐stress responsive genes (e.g., BiP/HSPA5) (Wiseman et al, 2022). PERK is activated upon oligomerization and trans‐autophosphorylation, once active it phosphorylates eif2α, resulting in the specific translation of the transcription factor ATF4 and the subsequent induction of UPR genes (e.g., DDIT3, PPP1R15A) (Madhavan et al, 2022). Hence, we investigated the activity of all three UPR sensors upon mitochondrial mutation and glucose deprivation, how this activity is regulated by doxycycline, and whether the effects are MALSU1‐dependent (Fig 4E). Here, we observe that all three UPR sensors are strongly activated upon glucose deprivation in ND1 cells, as evidenced by increased IRE1α phosphorylation, ATF6 cleavage, and PERK phosphorylation, all of which could be suppressed by doxycycline (Fig 4E). Consistent with cell survival (Fig 1D and E) and IRE1α oligomerization (Fig 4C) data, MALSU1 expression was required for the doxycycline‐dependent resolution of UPR activation for all arms of the UPR, and these results can be expanded to downstream UPR effector XBP1s, phosphorylation of p38 MAPK, and luminal chaperone BiP (Fig 4E). Activation of each respective arm of the UPR can be linked to the expression of specific ER‐stress‐responsive genes (Wiseman et al, 2022) and can be monitored using qPCR. Consistent with the observed activation of IRE1α, ATF6, and PERK on the protein level, transcripts linked to each specific arm were found to be activated upon nutrient stress, and doxycycline attenuated this activation (Fig 4F). These results show that glucose deprivation results in activation of all arms of the ER UPR, and that doxycycline resolves this global ER stress in a manner dependent on the mitoribosome quality control protein MALSU1.
Mitochondrial ND1 mutant cells exhibit increased ER protein loading that is attenuated by tetracyclines
Intrigued by the ability of tetracyclines to suppress ER stress in mitochondrial mutants, we evaluated the contribution of different ER protein homeostasis pathways. ER stress and homeostasis are largely regulated by (i) ER‐associated protein degradation (ERAD), (ii) ER protein secretion, and (iii) ER protein translocation or insertion (Wiseman et al, 2022). To test if tetracyclines are resolving ER stress by activating ERAD, we utilized specific inhibitor of p97‐induced protein degradation eeyarestatin I (ESI, Fig 5A–C) (Wang et al, 2008, 2010). It was noted in these experiments that mitochondrial mutant cells were more dependent on ERAD for survival under glucose deprivation when compared to WT, as the potency of ESI was increased under galactose conditions in ND1 mutant cells (Fig 5A) to a greater extent than in WT cells (Fig 5B). Galactose‐induced phosphorylation of p38 MAPK and XBP1 splicing were increased in ND1 when compared to WT cells, exacerbated by ESI, and suppressed by IRE1α inhibition, illustrating that ERAD modulates MAPK and XBP1s UPR signaling (Fig EV2B). However, we observed that treatment with ESI did not reverse the ability of doxycycline to promote survival in mitochondrial mutant cells under glucose deprivation (Fig 5C), suggesting that doxycycline does not promote survival by enhancing ERAD.
Figure 5. Mitochondrial mutant cells exhibit increased ER protein loading that is attenuated by tetracyclines.
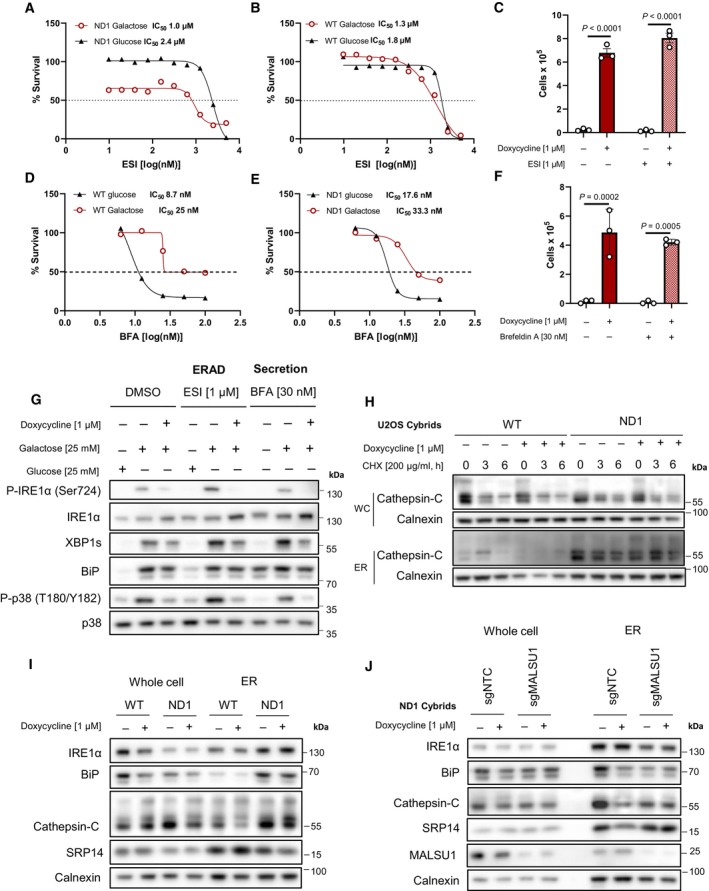
-
A, BDose‐dependent treatment with p97 specific ERAD inhibitor eeyarestatin I (ESI) shows (A) mitochondrial mutant cells are more dependent on ERAD when compared to (B) WT as indicated by enhanced potency (IC50) of ESI in ND1 cells under galactose‐stress conditions. (B) Note ESI potency is unchanged between glucose and galactose conditions in WT cells (IC50). Data points in (A) and (B) represent the average of n = 2 biological replicates.
-
CInhibition of ERAD with ESI does not reverse doxycycline‐induced survival (error bars represent the average ± s.e.m. of n = 3 independent biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells with or without ESI was calculated using two‐way ANOVA, multiple comparisons.
-
D, EDose–response curves of secretion inhibitor brefeldin A (BFA) in WT and ND1 cells under glucose and galactose conditions. Note cells are less sensitive to BFA under galactose conditions (IC50 = 25–33 nM) when compared to glucose (IC50 = 8–17 nM) in WT and ND1 cells, respectively. Data points represent the average of n = 2 biological replicates.
-
FInhibition of secretion with BFA does not reverse doxycycline‐induced survival (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells with or without BFA was calculated using two‐way ANOVA, multiple comparisons.
-
GInhibition of ERAD and secretion induce ER stress and the UPR that can be reversed by doxycycline, evidenced by IRE1α and p38 MAPK phosphorylation and XBP1s expression levels. Note enhanced galactose‐induced expression levels of XBP1s and basal levels of BiP that indicate ESI and BFA are on target.
-
HER protein cathepsin‐C loading is increased in isolated microsomes (ER) upon mitochondrial mutation, which is attenuated by doxycycline. Whole cell (WC) expression levels are unchanged between WT and ND1 cells. Cycloheximide treatment indicates comparable half‐lives between control and doxycycline treated cultures and isolated microsomes.
-
IMicrosomal cathepsin‐C levels that are increased upon ND1 mutation are attenuated by doxycycline, and associated with decreases in BiP and SRP14 in these fractions.
-
JDepletion of MALSU1 in ND1 cells reverses doxycycline regulation of BiP, cathepsin‐C, and SRP14 at the microsome.
Source data are available online for this figure.
To assess the effects of tetracyclines on ER‐protein secretion, we utilized the secretion inhibitor brefeldin A (BFA), which interferes with ER‐Golgi protein transport (Fujiwara et al, 1988; Orci et al, 1991). It was noted that WT and ND1 cells are less sensitive to BFA under galactose conditions, illustrating a potential decrease in secretory pathway when glucose is limiting (Fig 5D and E). Nonetheless, ND1 cells treated with BFA at IC50 value concentrations can still be rescued by doxycycline (Fig 5F). Consistent with these results, doxycycline can reverse the activation of the UPR in ND1 cells treated with ESI or BFA as evidenced by XBP1s and BiP protein levels, as well as IRE1α and p38 phosphorylation status (Fig 5G). Importantly, galactose‐induced XBP1s and basal expression levels of BiP are increased in ESI and BFA treated cells cultured in galactose, consistent with inhibition of ERAD and secretion, respectively (Fig 5G).
To assess the effects of tetracyclines on ER protein translocation, we probed tetracycline‐treated mitochondrial mutant cells and isolated microsomes (ER) for the accumulation of cathepsin‐C, a glycosylated and secreted protein that can be used as a marker for translocation in ER fractions (Ruiz‐Canada et al, 2009). Here, we found that microsomes derived from ND1 cells exhibit a substantial increase in cathepsin‐C when compared to those derived from WT cells, and doxycycline suppressed this accumulation (Fig 5H and I). Expression or stability of cathepsin‐C was not altered in whole cell extracts as indicated by cycloheximide chase experiments, and expression levels were comparable between WT and ND1 cells (Fig 5H and I) suggesting that ER protein loading is specifically increased upon mitochondrial mutation, consistent with increased BiP in the ER from ND1 cells that is reduced by tetracyclines (Fig 5I). Due to the necessity of MALSU1 in promoting survival of doxycycline‐treated cells, we investigated the effect of doxycycline on ER protein loading in mitochondrial mutant cells lacking MALSU1. We found that tetracyclines were unable to modulate cathepsin‐C microsome levels in MALSU1‐deficient cells (Fig 5J), further implicating this mitoribosome quality control response in the regulation of ER‐protein loading induced UPR. Interestingly, we noted a decrease in numerous ER markers in MALSU1‐depleted microsomes including IRE1α, BiP, cathepsin‐C, and calnexin, with a relative increase in SRP14 when compared to microsomes from control cells (Fig 5J). Altered microsome expression levels of ER proteins may potentially indicate a maladaptive response upon MALSU1 depletion that results in aberrant adaptive UPR function, in line with heightened p38 MAPK phosphorylation in MALSU1‐deficient cells (Figs 4E and EV2D). Nonetheless, in the absence of MALSU1, basal microsome levels of cathepsin‐C which are insensitive to doxycycline treatment are associated with increased sensitivity to glucose starvation (Fig 1D and E).
Endoplasmic reticulum‐protein translocation is initially regulated by the signal recognition particle (SRP), which binds to the ribosome nascent chain complex to promote co‐translational translocation into the ER lumen (Akopian et al, 2013; Voorhees & Hegde, 2015). We observed a marked decrease in SRP subunit SRP14 in doxycycline‐treated microsomes from ND1 cells (Fig 5I) that was not observed in the absence of MALSU1 (Fig 5J) further supporting a decrease in ER‐protein translocation in doxycycline‐treated cultures. Additionally, SRP14 is relatively more abundant in the microsomes of MALSU1‐depleted cells compared to NTC cells when normalized to other ER markers, which may suggest a maladaptive increase in associated ribosomes. Interestingly, doxycycline's ability to reduce ER stress is independent of global translation rates, evidenced by unchanged puromycin incorporation in galactose treated ND1 cells (Fig 6A). Increased protein insertion into the ER of mitochondrial mutant cells may thus explain the increased dependence of cells with ND1 mutation on ERAD (Fig 5A) and the enhanced sensitivity to ER stress as presented in Fig 4A. Further, we examined known mitochondrial stress response factor DELE1 in mediating doxycycline‐induced survival. Here, we show that ATF4 is not activated in cells lacking DELE1 when treated with doxycycline or other mitochondrial toxins including oligomycin (Fig 6B), and doxycycline can rescue cells independent of DELE1 and at concentrations that do not induce ATF4 (1 μM, Fig 6C). This data further supports the prospect of a novel signaling mechanism originating at the mitoribosome that communicates to the ER to promote survival. Thus, we present a model whereby inhibition of mitoribosome elongation with tetracyclines activates mitoribosome quality control and resolves ER stress and IRE1α signaling, potentially through modulation of the rate at which newly synthesized proteins are inserted into the ER (Fig 6D).
Figure 6. Tetracyclines promote survival independent of global translation rates or the mitochondrial integrated stress response factor DELE1.
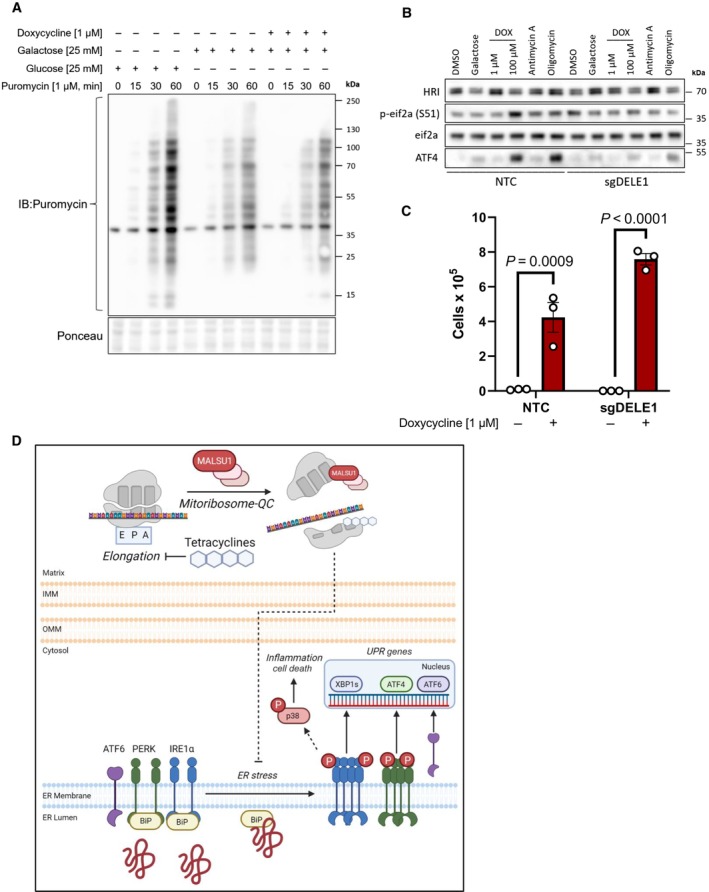
- Tetracyclines do not alter global translation rates as evidenced by puromycin incorporation assays. ND1 cells cultured for 48 h in glucose, galactose, and galactose supplemented with doxycycline (1 μM) and pulsed with puromycin to assay global translation rates. Note decrease in translation upon glucose starvation that is not further altered with doxycycline (representative of n = 2 independent experiments).
- Activation of the integrated stress response with mitochondrial DELE1 is not required for tetracyclines to promote survival in mitochondrial mutant cells. ND1 cybrid cells treated with mitochondrial targeting agents (6 h) activate the integrated stress response as indicated by increases in P‐eif2α and ATF4. Note changes in P‐eif2α and ATF4 seen in NTC cells are suppressed in cells lacking DELE1 (sgDELE1). Cells were cultured under glucose conditions except for where noted (galactose). Doxycycline (1 μM, 100 μM), antimycin A (40 nM), and Oligomycin (1.25 ng/μl).
- ND1 cells deprived of glucose for 7 days can be rescued by doxycycline (1 μM) independent of DELE1 expression status (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistics of doxycycline treated and untreated cultures across NTC and sgDELE1 cells were calculated using two‐way ANOVA, multiple comparisons.
- Proposed model of pro‐survival MALSU1‐dependent resolution of ER stress by tetracyclines through attenuation of ER stress and IRE1α signaling.
Source data are available online for this figure.
Discussion
Mitochondrial disease represents a clinically heterogenous and difficult to treat subset of metabolic diseases, where mechanisms of disease progression and pathological signaling are largely unknown (Nunnari & Suomalainen, 2012; Gorman et al, 2016; Wallace, 2018; Bennett et al, 2022b). Here, we present a model whereby inhibition of mitochondrial translation with tetracyclines splits the mitoribosome and recruits MALSU1, reducing lethal ER stress and IRE1α signaling (Fig 6D). Mechanisms of cross‐talk between the mitochondria and ER have been widely explored in normal physiology and disease processes including but not limited to autophagy, bioenergetics, calcium signaling, redox balance, protein import, and metabolite exchange (Carreras‐Sureda et al, 2022); aspects commonly accompanied by pathological transformation in MD. Here we show for the first time a functional cross‐talk between a mitoribosome associated protein and the unfolded protein response that expands our current knowledge and scope of interorganelle communication.
As the protein processing hub of the cell, the ER promotes the proper glycosylation, folding, membrane insertion, and delivery of proteins to their subcellular destination (Wiseman et al, 2022). Defects in these processing mechanics result in accumulation of misfolded proteins in the ER lumen, activating the UPR through a series of ER‐membrane stress sensors IRE1α, PERK, and ATF6 (Carreras‐Sureda et al, 2022; Wiseman et al, 2022). IRE1α, the most conserved UPR protein, contains an ER luminal domain that interacts with ER chaperones (namely BiP) and/or misfolded proteins and a cytosolic kinase/RNAase domain responsible for transducing luminal protein stress (Huang et al, 2019; Carreras‐Sureda et al, 2022; Wiseman et al, 2022). Heightened levels of misfolded proteins activate IRE1α through oligomerization and auto‐transphosphorylation, promoting RNAase activity to cleave XBP1 mRNA resulting in the expression of UPR effector XBP1s (Huang et al, 2019). XBP1s, a transcription factor and specific marker of IRE1α‐mediated UPR activity, localizes to the nucleus to promote the expression of genes involved in ER expansion, protein folding, ERAD, secretion, and inflammation. If ER stress remains unresolved, prolonged IRE1α signaling activates pathways that promote cell death (Huang et al, 2019). Our studies reveal that mitochondrial mutations sensitize cells to the ER stress‐induced activation of IRE1α and that doxycycline can suppress its activation along with other UPR sensors PERK and ATF6 in a MALSU1‐dependent fashion (Fig 4). Doxycycline was able to suppress ER stress chaperone BiP, inhibit the phosphorylation of p38 downstream of IRE1α, and suppress the activation of genes from all three arms of the UPR (Fig 4). These observations suggest that tetracyclines resolve global ER stress. Using pharmacological tools, enhanced ERAD or ER‐Golgi secretory processes were disqualified as potential mechanisms through which doxycycline exerts these effects, as doxycycline was still able to promote survival when these processes were inhibited (Fig 5). Mitochondrial mutant cells exhibited strong increases in the abundance of ER‐protein cathepsin‐C in isolated microsome fractions when compared to WT, and this was attenuated in doxycycline treated cells (Fig 5). Additionally, mitochondrial mutants were more dependent on ERAD for survival (Fig 5A), where maladaptive protein loading may require this mechanism of protein homeostasis for survival in stressed conditions. Together, these results suggest that the increases in phosphorylation of IRE1α and concomitant activation of XBP1s and p38 in mitochondrial mutants cultured under glucose deprivation may be due to increased basal protein loading in the ER. The pro‐survival effects of doxycycline may stem from its ability to attenuate ER protein translocation and accompanied ER stress, downstream of inhibition of the mitoribosome. Activated UPR and SRP‐mediated co‐translational translocation have been recently observed in patient fibroblasts with aberrant complex IV assembly (Sturm et al, 2023), substantiating our observations and highlighting a maladaptive response to mitochondrial mutations.
At ER‐mitochondrial contacts, IRE1α oligomerization and activity are regulated by the outer mitochondrial membrane E3‐ubiquitin ligase MARCH5 (Takeda et al, 2019). Reminiscent of tetracyclines, MARCH5‐mediated ubiquitylation of IRE1α attenuates high‐order oligomerization, controlling the extent of UPR and cell fate. We illustrate that tetracyclines promote survival in MD cells independent of MARCH5 (Fig EV3). Mitochondrial stress, including translational stalling, can be communicated to the cytosol through the integrated stress response (ISR) (Fessler et al, 2020; Guo et al, 2020). DELE1, a mitochondrial protein, is cleaved by protease OMA1 under mitochondrial stress conditions, resulting in localization of a cleaved DELE1 fragment to the cytosol where it binds to and activates the heme regulated kinase (HRI), which subsequently phosphorylates eIF2α (p‐eif2α) to activate the ISR and induce the expression of activating transcription factor 4 (ATF4) (Fessler et al, 2020; Guo et al, 2020). To test if DELE1 is involved in the doxycycline survival mechanisms, we used CRISPR‐Cas9 to KO DELE1 and assessed the ability of doxycycline to promote survival during glucose deprivation (Fig 6B and C). These studies illustrated that doxycycline at higher concentrations (100 μM) acutely induced p‐eif2α and ATF4 in a DELE1‐dependent fashion; however, deletion of DELE1 did not impair the ability of doxycycline to promote cell survival (Fig 6B and C). This is consistent with our previous findings (Perry et al, 2021) where knockdown of ATF4 with RNAi and inhibition of p‐eif2α with ISRIB did not prevent doxycycline from promoting survival, disqualifying ATF4 and most likely other upstream ISR‐transducers in the survival mechanism. Thus, we illustrate that tetracyclines promote survival independent of known mitochondrial proteins that are annotated to mediate stress responses: UPR targeting MARCH5 and ISR mediating DELE1, indicating a novel regulation of IRE1α originating at the mitochondria.
The maintenance of the mitochondrial proteome requires an intricate balance between the import of nuclear‐encoded proteins synthesized on cytosolic ribosomes, and the expression of mitochondrial‐encoded proteins from mitoribosomes (Kummer & Ban, 2021). Encoding genes for rRNA and tRNA, mtDNA also encodes 13 hydrophobic protein subunits of the electron transport chain complexes (Itoh et al, 2021; Kummer & Ban, 2021). Due to their hydrophobic nature, mitochondrial‐encoded nascent proteins are inserted cotranslationally into the inner mitochondrial membrane, where the mitoribosome LSU is tethered to insertase OXA1L, and specific chaperones direct and coordinate the assembly of mitochondrial‐encoded subunits with nuclear‐encoded subunits (Itoh et al, 2021). Elongational stalling of mitoribosomes during this process activates mitochondrial quality control factors (Desai et al, 2020) where we illustrate that doxycycline mediates a recruitment of MALSU1 to the mtLSU (Fig 3), which is required for the alleviation of ER stress and cell survival (Figs 1, 2, 4 and 5). In this regard, we claim that general suppression of mitochondrial translation is not sufficient to promote survival of mitochondrial mutants, but rather, the action of tetracyclines in splitting the mitoribosome is required for the survival mechanism. These claims are substantiated with the observation that a non‐mitoribosome‐targeting translation inhibitor, actinonin, did not promote survival (Fig 1A). These mechanistic insights can be expanded to other antibiotics that target tRNA accommodation phase of mitoribosome elongation including pentamidine (Sun & Zhang, 2008), or mtLSU peptidyl transferase targeting pleuromutilins (Wilson, 2014; Meydan et al, 2019); compounds we identified and validated to rescue mitochondrial mutant cell death. Our data indicates that MALSU1‐bound ribosomes constitute a signaling platform that communicates translation states in the mitochondria to the ER to resolve maladaptive increases in protein loading. Further, we illustrate that MTIF3 potentiates tetracycline‐induced survival (Fig 2). It is possible that MTIF3 works upstream of MALSU1, where its ability to dissociate mitoribosomes (Luo et al, 2019) and stabilize the mtSSU may further support the ability of tetracyclines to split the ribosome. There is recent evidence of coordinate regulation between mitochondrial and cytosolic ribosomes (Couvillion et al, 2016; Soto et al, 2022), and thus attenuation of mitochondrial translation by tetracyclines may induce a specific and currently unknown signal that reduces cytosolic protein loading in the ER which depends on specific components of the mitoribosome quality control. Defining this regulation between translation systems and its role in modulating compartmentalized protein homeostasis are important areas to focus future research efforts in light of the current work.
Materials and Methods
Cell lines, treatments, and culture conditions
ND1 and WT cybrid cells previously utilized by us (Perry et al, 2021) were generated by R. Vogel and J. Smeitink (Radboud University Medical Centre) using patient fibroblast mitochondria carrying either WT or the A3796G (ND1) mutation and were routinely validated for identity and contamination. Cybrid cells were cultured at 37°C and 5% CO2 in DMEM high glucose (25 mM, Gibco) medium supplemented with sodium pyruvate (1 mM), fetal bovine serum (10%), and penicillin–streptomycin (100 U/ml). For glucose deprivation experiments, DMEM no‐glucose (Gibco, 11966‐025) was supplemented with equimolar (25 mM) galactose, sodium pyruvate (1 mM), fetal bovine serum (10%), and penicillin–streptomycin (100 U/ml). Expi293F cells (Gibco) were cultured in suspension in Expi293 expression medium (A14351‐01) without antibiotics using the manufactures protocol. Cells were maintained at a density of 0.5–5 × 106 cells/ml, and cell viability (trypan blue) was assessed upon subculture to maintain viability > 95%.
Antibodies and reagents
Reagents used in this study include: Doxycycline hyclate (Sigma), tetracycline (Santa Cruz Biotechnology), minocycline (Santa Cruz Biotechnology), 7002, CMT‐3 (MedChem Express), actinonin (Sigma), eeyarestatin I (Sigma), 4μ8C (Sigma), n‐dodecyl β‐D‐maltoside (β‐DDM, Anatrace), cardiolipin (18:1, Avanti Polar Lipids), and β,γ‐methyleneguanosine 5′‐triphosphate sodium salt (GMPPCP, Sigma).
Antibodies used in this study include: C12orf65 (mtRF‐R, abcam, ab122448, 1:1,000), C6orf203 (MTRES1, abcam, ab151066, 1:1,000), C7orf30 (MALSU1, Proteintech, 22838‐1‐AP, 1:1,000), MRPL4 (Proteintech, 27484‐1‐AP, 1:1,000), MRPS16 (Proteintech, 16735‐1‐AP, 1:1,000), LRPPRC (Proteintech, 21175‐1‐AP, 1:1,000), TOM70 (Santa Cruz, sc390545, 1:1,000), IRE1α (Cell Signaling, 14C10, 1:1,000), β‐tubulin, Histone‐H3 (abcam, ab52946, 1:1,000), VDAC (Santa Cruz, sc390996, 1:1,000), XBP1s (Cell Signaling, D2C1F, 1:1,000), PARP (Cell Signaling, 46D11, 1:1,000), BiP (Cell Signaling, C50B12, 1:1,000), P‐p38 (T180/Y182, Cell Signaling, D3F9, 1:1,000), p38 (total, Cell Signaling, 1:1,000), pan – OXPHOS (ATP5A1, UQCRC2, MTCO1, SDHB, NDUFB8, abcam, STN‐19467, 1:500), Cathepsin‐C (R&D Systems, AF1071, 1:1,000), SRP14 (Proteintech, 11528‐1‐AP, 1:1,000), Calnexin (Cell Signaling, C5C9, 1:1,000), MARCH5 (abcam, ab185054, 1:1,000), HRI (EIF2AK1, abcam, 1:1,000), p‐eif2α (S51, Cell Signaling, 119A11, 1:1,000), eif2α (total, Cell Signaling, 1:1,000), ATF4 (Cell Signaling, D4B8, 1:1,000), MTIF3 (Proteintech, 14219‐1‐AP, 1:1,000), PERK (Cell Signaling, D11A8, 1:1,000), ATF6 (Cell Signaling, D4ZBV, 1:1,000), and Puromycin (Kerafast, 3RH11, 1:1,000).
Cell survival assays
Survival assays of mitochondrial mutant ND1 cybrid cells were carried out as previously described by us (Perry et al, 2021). Cells were seeded at 1.0 × 105 cells/well in 6‐well plates in glucose‐deprivation media. Doxycycline, actinonin, or eeyarestatin I (ESI) were added at indicated concentrations at the time of seeding. Cells were then incubated for 96 h at which time the surviving cells were counted via trypan‐blue exclusion method. ESI dose–response curves were generated using SRB by seeding ND1 or WT cells with or without glucose at varying concentrations. Cells were then incubated for 48 h after which media was aspirated and analyzed for cell survival via SRB as indicated above. Dose–response curves were generated, and IC50 values were calculated using GraphPad prism software and illustrate the concentration at which 50% of cells survive when compared to nontreated control cells in each respective media condition (glucose or galactose).
CRISPR‐Cas9 gene editing
CRISPR‐Cas9 gene editing experiments were performed using the GeCKO (Shalem et al, 2014) protocol. sgRNA guides were cloned into lentiCRISPR‐V2 plasmids containing the puromycin resistance vector (Addgene, 98290), transformed into Stbl3 competent E. coli cells, and purified using standard methods. HEK293T cells were then seeded in 6‐well dishes (6 × 105 cells/well) and reverse transfected using Polyfect (Qiagen, 301105) following the manufacturers protocol. Transfection was carried out using 880 ng of lentiCRISPR‐V2 with respective target‐ or nontargeting control sgRNA, 600 ng psPAX2 (Addgene, 12260), and 300 ng pMD2 (Addgene, 12259). Following 24 h transfection, media was replaced with DMEM and virus was generated for an additional 48 h. Lentiviral medium was then harvested, filtered (0.45 μm), and added with polybrene (conc, Cat. No.) onto ND1 cells that had been seeded and adhered to 6 well dishes (3 × 105 cells/well) overnight. Twenty‐four hours after transduction, cells were split into 10 cm dishes and puromycin (0.5 μg/ml) was added and cells were selected for 7–10 days before used for experiments. The following sgRNA sequences were used: mtRF‐R: 5′‐ACCTTTACAACGATGCCTGA‐3′. MTRES1: 5′‐ATGATGTTGTCCTGAAGACG‐3′. MALSU1: sg1 5′‐CAGGCCGCGGACAAAGTTTG‐3′; sg2 5′‐CCCACTAATGTGGCGCAGGG‐3′; sg3 5′‐GCGGAGGGGACGGTCAACGA‐3′. DELE1: 5′‐TGATAATAAAGGACCGCCTG‐3′. MARCH5: 5′‐CCAGGCCTGTCTACAACGCT‐3′. LOR8F8: 5′‐TACCCGCTTAACTGCCAGCG‐3′. MTIF3: sg1 5′‐GCAGAGTATCAGCTCATGAC‐3′ sg2 5′‐GCAATAGGGGACAACTGTGC‐3′ sg3 5′‐AGGCGAACCCCAAAACTGGT‐3′.
Dependency Map proteomics gene‐ontology analysis
Proteomic correlation of doxycycline was scored and ranked by P‐value (DepMap, PRISM repurposing screen; Corsello et al, 2020), where proteins enriched in cells surviving under doxycycline treatment were identified (P < 0.0001). Within this statistically significant subset of proteins, positive correlates were further analyzed for gene‐ontology (GO) enrichment against the entire proteome using DAVID functional annotation bioinformatics (https://david.ncifcrf.gov/) (Huang da et al, 2009). GO‐terms and their respective calculated fold enrichment were ranked and plotted. High fold enrichment represented a high number of proteins within a given GO‐term that were enriched in doxycycline‐treated cells.
Subcellular fractionations
Subcellular fractionations were carried out using differential centrifugation as previously described (Wieckowski et al, 2009). ND1 or WT cybrid cells were seeded in 2 × 15 cm dishes in glucose media and allowed to reach 80% confluence. Cells were then rinsed with PBS, and galactose media was then added with doxycycline at indicated concentrations and were incubated for 48 h. Cells were then harvested by scraping and pelleted at 300 × g for 5 min at 4°C. Cells were resuspended in PBS, pelleted, and then resuspended in 5 ml of cold isolation buffer (IB, 225 mM mannitol, 75 mM sucrose, 0.1 mM EGTA, 30 mM HEPES‐HCl pH 7.4). Cells were than dounce‐homogenized (~60 up‐down strokes) on ice and centrifuged at 1,000 × g at 4°C for 5 min. Supernatant containing mitochondria, ER (microsomes), lysosomes, and cytosol was then decanted into an additional 5 ml of IB and centrifuged at 10,000 × g at 4°C for 10 min to obtain crude mitochondria (mitochondria/ER) in the pellet. Supernatant containing cytosol, microsomes, and lysosomes was decanted and kept on ice, and mitochondria/ER fractions were resuspended in mitochondria resuspension buffer (MRB, 250 mM mannitol, 5 mM HEPES pH 7.4, and 0.5 mM EGTA). MRB was centrifuged at 10,000 × g at 4°C for 10 min, supernatant was aspirated, and crude mitochondria were snap frozen in liquid nitrogen and stored at −80°C. Lysosomes were then separated from microsomes and cytosol by centrifugation (20,000 × g at 4°C for 30 min), pelleted, and snap frozen. Microsomes in the supernatant were isolated by ultracentrifugation (100,000 × g for 60 min at 4°C) and resuspended in IB with protease inhibitors and snap frozen in liquid nitrogen and kept at −80°C. An aliquot of the resulting final supernatant containing cytosol was also snap frozen in liquid nitrogen and kept at −80°C.
Western blotting and blue‐native PAGE
SDS‐ and Native‐PAGE were carried out using standard protocols. For SDS‐PAGE, cells were resuspended in RIPA buffer (Cell Signaling, 9806) with protease and phosphatase inhibitors at 4°C, and protein content was determined using standard BCA colorimetric protocols (Pierce 23228). Microsomes in IB and crude mitochondria in MRB (from Subcellular fractionations) were directly quantified and solubilized with Laemmli SDS‐sample buffer (Bio‐Rad, 1610747). Ten to twenty microgram of protein from respective samples were run on 4–12% SDS‐PAGE gels (Invitrogen, NP0336BOX). Proteins were then transferred to PVDF membranes and probed with specific antibodies. Native‐PAGE analysis was carried out as previously described (Sundaram et al, 2018) with some modifications. Mitochondria/ER (100 μg) were solubilized in 50 μl of Native Page Buffer (Invitrogen, 2360376) supplemented with 1.5% digitonin and protease inhibitors. Cells were then incubated at 4°C for 10 min to complete lysis, and centrifuged at 18,500 × g for 20 min (4°C) to remove insoluble material. Supernatant was then carefully removed and resuspended in 0.5% G‐250 (Invitrogen, BN20041). Samples (10 μg) were then added to each well, and protein complexes were separated on 3–12% NativePAGE gels (Thermo Fisher Scientific, BN1003BOX). Proteins were transferred to PVDF membranes and probed with specific antibodies. Buffers used for NativePAGE include: NativePAGE Running Buffer (20×; Thermo Fisher Scientific, BN2001) and NativePAGE Cathode Buffer Additive (20×; Thermo Fisher Scientific, BN2002).
Isolation of mitoribosomes
Mitoribosomes were isolated following a procedure similar to previously described (Desai et al, 2020). Expi293F cells (750 ml, 6 × 106 cells/ml) treated with doxycycline (1 μM) or vehicle (DMSO) for 48 h were harvested by centrifugation (300 × g, 5 min, 4°C). Cells were resuspended in cold PBS and repelleted. Cells were then resuspended in mitochondrial isolation buffer (MIB, 50 mM HEPES‐KOH pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitors) at a ratio of 6 ml MIB per 1 g of pellet. Cells were left to swell at 4°C for 10 min, after which were brought to isotonic concentrations of sucrose and mannitol by adding calculated volumes of 4× sucrose mannitol buffer (SM4, 280 mM sucrose, 840 mM mannitol, 50 mM HEPES‐KOH pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitors). The resulting solution MIB + SM4 (MIBSM) will be used in following steps. Cells were then dounce homogenized (~60 up‐ down‐passes) and cellular debris and nuclei were centrifuged (800 × g, 4°C, 15 min). Supernatant‐1 was filtered and pellet was resuspended in fresh MIBSM for 15 passes. Resulting solution was again centrifuged and supernatant‐2 was combined through a cheesecloth with supernatant‐1. This solution was clarified by centrifugation a (1,000 × g at 4°C for 15 min) and resulting supernatant was collected. Crude mitochondrial pellet was then obtained by centrifugation (10,000 × g at 4°C for 15 min) and was resuspended in MIBSM buffer with DNAase (0.1 U/ml) and left rocking at 4°C for 30 min. Mitochondria were pelleted (10,000 × g at 4°C for 15 min), resuspended in 1 ml of SEM buffer (250 mM sucrose, 20 mM HEPES‐KOH pH 7.5, 1 mM EDTA), and loaded onto a discontinuous sucrose gradient (15–60%) constructed as previously described (Desai et al, 2020). Mitochondria were obtained, snap frozen in liquid nitrogen, and kept at −80°C.
To obtain mitoribosomes, mitochondria were thawed at 4°C and combined with equal parts of lysis buffer (25 mM HEPES‐KOH pH 7.4, 150 mM KCl, 50 mM MgOAc, 1.5% β‐DDM, 0.15 mg cardiolipin, 0.5 mM GMPPCP, 2 mM DTT, and protease inhibitors). GMPPCP was utilized to prevent dissociation of GTPases during the mitoribosome isolation and not included during live cell culture and doxycycline inhibition experiments. Combined solution was mixed and gently homogenized with a dounce homogenizer and was left to rock at 4°C for 30 min. Lysed material was centrifuged at 20,000 × g for 30 min to clarify, and supernatant containing mitoribosomes was carefully decanted. Lysis solution (2 ml) was added on top of a sucrose cushion (1 ml, 1 M sucrose (34%), 20 mM HEPES‐KOH pH 7.4, 100 mM KCl, 20 mM MgOAc, 0.6% β‐DDM, 0.06 mg/ml cardiolipin, 0.25 mM GMPPCP, 2 mM DTT) in SW 55 Ti tubes. Samples were centrifuged (231,550 × g at 4°C for 1 h) to obtain crude mitoribosome pool. Mitoribosomes were reconstituted in 100 μl of resuspension buffer (20 mM HEPES‐KOH pH 7.4, 100 mM KCl, 20 mM MgOAc, 0.3% β‐DDM, 0.03 mg/ml cardiolipin, 0.25 mm GMPPCP, 2 mM DTT) and loaded on top of a 13–30% linear sucrose gradient15 in SW 55 Ti tubes. Samples were centrifuged (213,626 × g at 4°C for 90 min), and entire volume was fractionated into 13 equal parts. Samples were snap frozen and kept for western blotting analysis. Inputs represent crude mitoribosome pool.
ND1 mitoribosomes were analyzed in a similar fashion, but crude mitochondria (see Subcellular fractionations) were directly lysed and subjected to linear sucrose gradient fractionation without purification of mitoribosome pool. Inputs represent crude mitochondria in these experiments.
Quantitative PCR
Cells were seeded (1.0 × 105 cells/well) in 6 well dishes and treated with the indicated compounds for 48 h under glucose starvation conditions. RNA was then isolated using Trizol (Invitrogen, 15596‐026) and a Zymo‐Spin Direct‐zol RNA Kit (Zymo Research, R2050) following the manufactures instructions. 0.5–1 μg of RNA was then used to reverse transcribe cDNA using a High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813) following. cDNA was then mixed with SYBR Green qPCR master mix (Applied Biosystems, 4309155) and amplified on a CFX 384 Real‐Time system (Bio‐Rad) for analysis. Primers used for this study include Human: MALSU1 F‐CTGCTCGCCCCACTAATG, R‐ACTTGGGACCAGTATGATCTGC. MTIF3. DNAJB9 F‐TCTTAGGTGTGCCAAAATCGG, R‐TGTCAGGGTGGTACTTCATGG. SEC24D F‐GTCAACAAGGTTACGTGGCTA, R‐TAGTGCCCATAATGAGGTGGA. HSPA5 F‐CATCACGCCGTCCTATGTCG, R‐CGTCAAAGACCGTGTTCTCG. DDIT3 F‐GGAAACAGAGTGGTCATTCCC, R‐CTGCTTGAGCCGTTCATTCTC. PPP1R15A F‐ATGATGGCATGTATGGTGAGC, R‐AACCTTGCAGTGTCCTTATCAG.
Cycloheximide chase experiments
ND1 or WT cybrid cells were seeded in glucose in 2 × 15 cm2 dishes per condition and allowed to reach 90% confluence. Cells were pulsed with cycloheximide (200 μg/ml) for 0, 3, and 6 h after which they were harvested and snap frozen in liquid nitrogen. Whole cells and microsomes were then isolated (see Subcellular fractionations) for western blot analysis.
Puromycin incorporation analysis of global cellular translation
ND1 cells were seeded in 6 well dishes cultured in glucose, galactose, or galactose supplemented with 1 μM doxycycline and were incubated for 48 h. Cells were then pulsed with puromycin (1 μM) for the indicated time periods and harvested in RIPA buffer for western blotting as described above. Whole‐cell puromycinylated peptides were probed for with an anti‐puromycin antibody.
Animal experiments
Animal studies were performed as previously reported by us (Perry et al, 2021). C57BL/6J NDUFS4−/− or WT mice supplemented with doxycycline or normal chow diets were euthanized at ~p55 and brains were immediately harvested and snap frozen in liquid nitrogen and kept at −80°C. Brain tissue was then pulverized with mortar and pestle until a fine powder was obtained. Tissue was then resuspended in RIPA buffer for western blot analysis.
Ethical considerations
Animal studies were performed in compliance with the Institutional Animal Care and Use Committee protocol approved by the Beth Israel Deaconess Medical Center Animal Facility.
Statistics
Replicates presented are derived from independent biological samples. Statistical analysis between two samples was done using student t‐test, and greater than two groups using one‐ or two‐way ANOVA with multiple comparisons where appropriate. Statistical significance was determined with P‐value < 0.05*, < 0.01**, < 0.001***, and < 0.0001**** as noted.
Author contributions
Conor T Ronayne: Conceptualization; formal analysis; supervision; funding acquisition; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing. Thomas D Jackson: Investigation; writing – review and editing. Christopher F Bennett: Funding acquisition; investigation; writing – review and editing. Elizabeth A Perry: Funding acquisition; investigation; writing – review and editing. Noa Kantorovic: Investigation; writing – review and editing. Pere Puigserver: Conceptualization; resources; formal analysis; supervision; funding acquisition; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank the entire Puigserver laboratory for helpful discussions and insights for the duration of the project. We also would like to extend our gratitude to the department of cancer biology at Dana Farber and department of cell biology at Harvard Medical School for their support. This work was funded by the NIH grants, R56 AG074527/AG/NIA and RO1 CA181217/CA/NCI (to PP), T32 CA236754‐02 (to CTR), F32 GM125243‐01A1 NIGMS (to CFB), and F30 DE028206‐01A1 NIDCR (to EAP).
EMBO reports (2023) 24: e57228
Data availability
New datasets were not generated as a result of the current study. Source data for this study has been included.
References
- Akopian D, Shen K, Zhang X, Shan SO (2013) Signal recognition particle: an essential protein‐targeting machine. Annu Rev Biochem 82: 693–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsa E, Soustek MS, Thomas A, Cogliati S, Garcia‐Poyatos C, Martin‐Garcia E, Jedrychowski M, Gygi SP, Enriquez JA, Puigserver P (2019) ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK‐eIF2alpha axis. Mol Cell 74: e876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsa E, Perry EA, Bennett CF, Jedrychowski M, Gygi SP, Doench JG, Puigserver P (2020) Defective NADPH production in mitochondrial disease complex I causes inflammation and cell death. Nat Commun 11: 2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Latorre‐Muro P, Puigserver P (2022a) Mechanisms of mitochondrial respiratory adaptation. Nat Rev Mol Cell Biol 23: 817–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Ronayne CT, Puigserver P (2022b) Targeting adaptive cellular responses to mitochondrial bioenergetic deficiencies in human disease. FEBS J 289: 6969–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Rathore S, Kimanius D, Aibara S, Bai XC, Rorbach J, Amunts A, Ramakrishnan V (2017) Structures of the human mitochondrial ribosome in native states of assembly. Nat Struct Mol Biol 24: 866–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras‐Sureda A, Kroemer G, Cardenas JC, Hetz C (2022) Balancing energy and protein homeostasis at ER‐mitochondria contact sites. Sci Signal 15: eabm7524 [DOI] [PubMed] [Google Scholar]
- Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG, Humeidi R, Peck D, Wu X, Tang AA et al (2020) Discovering the anti‐cancer potential of non‐oncology drugs by systematic viability profiling. Nat Cancer 1: 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS (2016) Synchronized mitochondrial and cytosolic translation programs. Nature 533: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Yang H, Chandrasekaran V, Kazi R, Minczuk M, Ramakrishnan V (2020) Elongational stalling activates mitoribosome‐associated quality control. Science 370: 1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer‐Bender MF, Hanf M, Philippou‐Massier J, Krebs S, Zischka H, Jae LT (2020) A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579: 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y (1988) Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem 263: 18545–18552 [PubMed] [Google Scholar]
- Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, Suomalainen A, Thorburn DR, Zeviani M, Turnbull DM (2016) Mitochondrial diseases. Nat Rev Dis Primers 2: 16080 [DOI] [PubMed] [Google Scholar]
- Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA, Kampmann M (2020) Mitochondrial stress is relayed to the cytosol by an OMA1‐DELE1‐HRI pathway. Nature 579: 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huang S, Xing Y, Liu Y (2019) Emerging roles for the ER stress sensor IRE1alpha in metabolic regulation and disease. J Biol Chem 294: 18726–18741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Andrell J, Choi A, Richter U, Maiti P, Best RB, Barrientos A, Battersby BJ, Amunts A (2021) Mechanism of membrane‐tethered mitochondrial protein synthesis. Science 371: 846–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Khawaja A, Laptev I, Cipullo M, Atanassov I, Sergiev P, Rorbach J, Amunts A (2022) Mechanism of mitoribosomal small subunit biogenesis and preinitiation. Nature 606: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaja A, Itoh Y, Remes C, Spahr H, Yukhnovets O, Hofig H, Amunts A, Rorbach J (2020) Distinct pre‐initiation steps in human mitochondrial translation. Nat Commun 11: 2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse SE, Watt WC, Marcinek DJ, Kapur RP, Schenkman KA, Palmiter RD (2008) Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab 7: 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer E, Ban N (2021) Mechanisms and regulation of protein synthesis in mitochondria. Nat Rev Mol Cell Biol 22: 307–325 [DOI] [PubMed] [Google Scholar]
- Luo Y, Su R, Wang Y, Xie W, Liu Z, Huang Y (2019) Schizosaccharomyces pombe Mti2 and Mti3 act in conjunction during mitochondrial translation initiation. FEBS J 286: 4542–4553 [DOI] [PubMed] [Google Scholar]
- Madhavan A, Kok BP, Rius B, Grandjean JMD, Alabi A, Albert V, Sukiasyan A, Powers ET, Galmozzi A, Saez E et al (2022) Pharmacologic IRE1/XBP1s activation promotes systemic adaptive remodeling in obesity. Nat Commun 13: 608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S, Marks J, Klepacki D, Sharma V, Baranov PV, Firth AE, Margus T, Kefi A, Vazquez‐Laslop N, Mankin AS (2019) Retapamulin‐assisted ribosome profiling reveals the alternative bacterial proteome. Mol Cell 74: 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M, Peikert CD, Lubbert P, Suppanz I, Klemm C, Alka O, Steiert C, Naumenko N, Schendzielorz A, Melchionda L et al (2021) Quantitative high‐confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab 33: 2464–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B et al (2015) Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep 10: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A (2012) Mitochondria: in sickness and in health. Cell 148: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Tagaya M, Amherdt M, Perrelet A, Donaldson JG, Lippincott‐Schwartz J, Klausner RD, Rothman JE (1991) Brefeldin A, a drug that blocks secretion, prevents the assembly of non‐clathrin‐coated buds on Golgi cisternae. Cell 64: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Ott M, Amunts A, Brown A (2016) Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem 85: 77–101 [DOI] [PubMed] [Google Scholar]
- Perry EA, Bennett CF, Luo C, Balsa E, Jedrychowski M, O'Malley KE, Latorre‐Muro P, Ladley RP, Reda K, Wright PM et al (2021) Tetracyclines promote survival and fitness in mitochondrial disease models. Nat Metab 3: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remes C, Khawaja A, Pearce SF, Dinan AM, Gopalakrishna S, Cipullo M, Kyriakidis V, Zhang J, Dopico XC, Yukhnovets O et al (2023) Translation initiation of leaderless and polycistronic transcripts in mammalian mitochondria. Nucleic Acids Res 51: 891–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myohanen M, Greco D, Cannino G, Jacobs HT, Lietzen N, Nyman TA, Battersby BJ (2013) A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol 23: 535–541 [DOI] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Suomi F, Battersby BJ (2015) Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness. J Cell Biol 211: 373–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Canada C, Kelleher DJ, Gilmore R (2009) Cotranslational and posttranslational N‐glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG et al (2014) Genome‐scale CRISPR‐Cas9 knockout screening in human cells. Science 343: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto I, Couvillion M, Hansen KG, McShane E, Moran JC, Barrientos A, Churchman LS (2022) Balanced mitochondrial and cytosolic translatomes underlie the biogenesis of human respiratory complexes. Genome Biol 23: 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustek MS, Balsa E, Barrow JJ, Jedrychowski M, Vogel R, Jan S, Gygi SP, Puigserver P (2018) Inhibition of the ER stress IRE1alpha inflammatory pathway protects against cell death in mitochondrial complex I mutant cells. Cell Death Dis 9: 658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G, Karan KR, Monzel AS, Santhanam B, Taivassalo T, Bris C, Ware SA, Cross M, Towheed A, Higgins‐Chen A et al (2023) OxPhos defects cause hypermetabolism and reduce lifespan in cells and in patients with mitochondrial diseases. Commun Biol 6: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Zhang Y (2008) Pentamidine binds to tRNA through non‐specific hydrophobic interactions and inhibits aminoacylation and translation. Nucleic Acids Res 36: 1654–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram A, Appathurai S, Plumb R, Mariappan M (2018) Dynamic changes in complexes of IRE1alpha, PERK, and ATF6alpha during endoplasmic reticulum stress. Mol Biol Cell 29: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Nagashima S, Shiiba I, Uda A, Tokuyama T, Ito N, Fukuda T, Matsushita N, Ishido S, Iwawaki T et al (2019) MITOL prevents ER stress‐induced apoptosis by IRE1alpha ubiquitylation at ER‐mitochondria contact sites. EMBO J 38: e100999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS (2015) Structures of the scanning and engaged states of the mammalian SRP‐ribosome complex. Elife 4: e07975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (2018) Mitochondrial genetic medicine. Nat Genet 50: 1642–1649 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y (2008) Inhibition of p97‐dependent protein degradation by Eeyarestatin I. J Biol Chem 283: 7445–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Shinkre BA, Lee JG, Weniger MA, Liu Y, Chen W, Wiestner A, Trenkle WC, Ye Y (2010) The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane‐binding domain and a p97/VCP inhibitory group. PLoS One 5: e15479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanschers BF, Szklarczyk R, Pajak A, van den Brand MA, Gloerich J, Rodenburg RJ, Lightowlers RN, Nijtmans LG, Huynen MA (2012) C7orf30 specifically associates with the large subunit of the mitochondrial ribosome and is involved in translation. Nucleic Acids Res 40: 4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P (2009) Isolation of mitochondria‐associated membranes and mitochondria from animal tissues and cells. Nat Protoc 4: 1582–1590 [DOI] [PubMed] [Google Scholar]
- Wilson DN (2014) Ribosome‐targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12: 35–48 [DOI] [PubMed] [Google Scholar]
- Wiseman RL, Mesgarzadeh JS, Hendershot LM (2022) Reshaping endoplasmic reticulum quality control through the unfolded protein response. Mol Cell 82: 1477–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]


