Abstract
Planar cell polarity (PCP) signaling polarizes epithelial cells within the plane of an epithelium. Core PCP signaling components adopt asymmetric subcellular localizations within cells to both polarize and coordinate polarity between cells. Achieving subcellular asymmetry requires additional effectors, including some mediating post‐translational modifications of core components. Identification of such proteins is challenging due to pleiotropy. We used mass spectrometry‐based proximity labeling proteomics to identify such regulators in the Drosophila wing. We identified the catalytic subunit of protein phosphatase1, Pp1‐87B, and show that it regulates core protein polarization. Pp1‐87B interacts with the core protein Van Gogh and at least one serine/threonine kinase, Dco/CKIε, that is known to regulate PCP. Pp1‐87B modulates Van Gogh subcellular localization and directs its dephosphorylation in vivo. PNUTS, a Pp1 regulatory subunit, also modulates PCP. While the direct substrate(s) of Pp1‐87B in control of PCP is not known, our data support the model that cycling between phosphorylated and unphosphorylated forms of one or more core PCP components may regulate acquisition of asymmetry. Finally, our screen serves as a resource for identifying additional regulators of PCP signaling.
Keywords: APEX proximity labeling, planar cell polarity, Pp1‐87B protein phosphatase, Van Gogh
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Development
Mass spectrometry‐based proximity labeling proteomics identifies regulators of PCP signaling in the Drosophila wing. Among those, the catalytic subunit of protein phosphatase1, Pp1‐87B, regulates core protein polarization.
Introduction
Planar cell polarity (PCP) signaling controls the polarization of cells, typically within the plane of an epithelium, orienting asymmetric cellular structures, cell divisions, and cell migration. Much mechanistic understanding of PCP signaling has and will likely continue to derive from work using Drosophila as a model system. In flies, PCP signaling controls the orientation of hairs on the adult cuticle, chirality and orientation of ommatidia in the eye, orientation of cell divisions, and related processes in other tissues (Butler & Wallingford, 2017; Harrison et al, 2020). While much has been learned from studies in flies, medically important developmental defects and physiological processes in vertebrates are also under control of PCP signaling. Defects in the core PCP mechanism result in a range of developmental anomalies and diseases including open neural tube defects (reviewed in Simons & Mlodzik, 2008; Copp & Greene, 2010), conotruncal heart defects (Garriock et al, 2005; Phillips et al, 2005, 2007; Henderson et al, 2006; Gibbs et al, 2016), deafness (reviewed in Lu & Sipe, 2016; Montcouquiol & Kelley, 2020), situs inversus, and heterotaxy (reviewed in Santos & Reiter, 2010). PCP is also believed to participate in both early and late stages of cancer progression (reviewed in Zhan et al, 2017; VanderVorst et al, 2019) and during wound healing (Lee & Adler, 2004; Caddy et al, 2010). PCP polarizes skin and hair (Guo et al, 2004; Wang et al, 2006a, 2006b; Devenport & Fuchs, 2008) and the ependyma (Guirao et al, 2010; Hirota et al, 2010; Tissir et al, 2010; Ohata et al, 2014). Much of our knowledge of PCP signaling derived from the Drosophila model translates to studies of PCP in vertebrate model systems, thus informing the pathogenesis of these conditions.
The dedicated components of a core signaling module that acts to generate and amplify molecular asymmetry, and to coordinate the direction of polarization between neighboring cells, have been recognized for some time. Six known proteins in the core signaling module are the serpentine protein Frizzled (Fz) (Vinson & Adler, 1987; Vinson et al, 1989), the seven‐pass atypical cadherin Flamingo (Fmi; a.k.a. Starry night) (Chae et al, 1999; Usui et al, 1999), the four‐pass protein Van Gogh (Vang; a.k.a. Strabismus) (Taylor et al, 1998; Wolff & Rubin, 1998), and the cytosolic/peripheral membrane proteins Disheveled (Dsh) (Klingensmith et al, 1994; Thiesen et al, 1994), Diego (Dgo) (Feiguin et al, 2001), and the PET/Lim domain protein Prickle (Pk) (Gubb et al, 1999), which adopt asymmetric subcellular localizations that predict the morphological polarity pattern such as hairs in the fly wing (reviewed in Zallen, 2007; Butler & Wallingford, 2017). These proteins communicate at cell boundaries, recruiting one group to the distal side of cells, and the other to the proximal side, through the function of an incompletely understood feedback mechanism, thereby aligning the polarity of adjacent cells. The direction of core module polarization is proposed to be responsive to several kinds of signals tied to the tissue axes, including expression gradients, diffusible ligands, and mechanical forces (reviewed in Eaton & Julicher, 2011; Butler & Wallingford, 2017).
A variety of post‐translational modifications to the core PCP components have been recognized as regulators of polarization, and include serine–threonine and tyrosine phosphorylation, farnesylation, and ubiquitinylation (reviewed in Harrison et al, 2020). Furthermore, processes including oligomerization, internalization, and regulated transport are thought to play key roles in regulating polarization. Understanding these events is critical to understanding the PCP mechanism, yet a challenge has been to identify the proteins that carry them out, as many are broadly pleiotropic and are therefore not easily recognized in genetic screens. We report here the results of an APEX‐based proximity labeling screen. APEX, an active peroxidase, biotinylates proteins within a 5–50 nm radius of the probe in living cells (Bosch et al, 2021). This approach has been used several times successfully in living Drosophila tissues (Chen et al, 2015; Mannix et al, 2019; Li et al, 2020; Liu et al, 2020). By fusing APEX to the core PCP protein Vang, we labeled proteins likely to interact with the core PCP proteins in living Drosophila wing discs. In a secondary RNAi screen, many of these proteins can be shown upon knockdown to produce either or both of two typical PCP polarity phenotypes: misoriented wing hairs or a multiple wing hair phenotype.
Because several serine/threonine kinases have been suggested to regulate core PCP function, from our candidate list, we chose to further characterize Pp1‐87B, which encodes the catalytic subunit of protein phosphatase 1 (Pp1). By appropriately timed knockdown, we show that Pp1‐87B activity is necessary for properly oriented wing hairs, and for controlling the amount and subcellular localization of two core PCP proteins, Vang and Pk. Pp1‐87B knockdown genetically interacts with core PCP mutants, and with Dco/CKIε, the Drosophila and vertebrate homologs of a S/T kinase thought to regulate PCP in both Drosophila and vertebrates. We find that Pp1‐87B, either directly or indirectly, induces dephosphorylation of Vang in vivo. Furthermore, we identify PNUTS as a possible regulatory subunit of Pp1 that may provide specificity for PCP. Furthermore, our potential PCP interactome will serve as a resource for identification of other regulators of PCP signaling.
Results
Proximity labeling screen
To identify proteins participating in PCP that are not readily discovered in genetic screens, we fused APEX, along with myc and V5 tags, to the N‐terminus of Vang and expressed this under control of D174‐GAL4 or 71B‐GAL4 that each express throughout the wing. APEX::Vang expression in a vang mutant background showed normal planar polarity, verifying functionality of the engineered protein (Fig EV1). Streptavidin staining of third‐instar wing discs fixed after labeling detected biotinylation primarily at apicolateral junctions, substantially co‐localizing with Pk, similar to localization of native Vang. Furthermore, both Pk and Fmi were biotinylated by APEX::Vang in vivo (Fig EV2). Detection of Pk and Fmi, both known to be present in proximal PCP complexes, gave us confidence that APEX::Vang could effectively label the proximal PCP proteome. We note, however, that during polarization, complexes of opposite orientation are thought to interact, and indeed Vang has been shown to physically Interact with each of the other core proteins (Seifert & Mlodzik, 2007; Goodrich & Strutt, 2011; Adler, 2012) so that we expect to also label proteins associated with the distal complex.
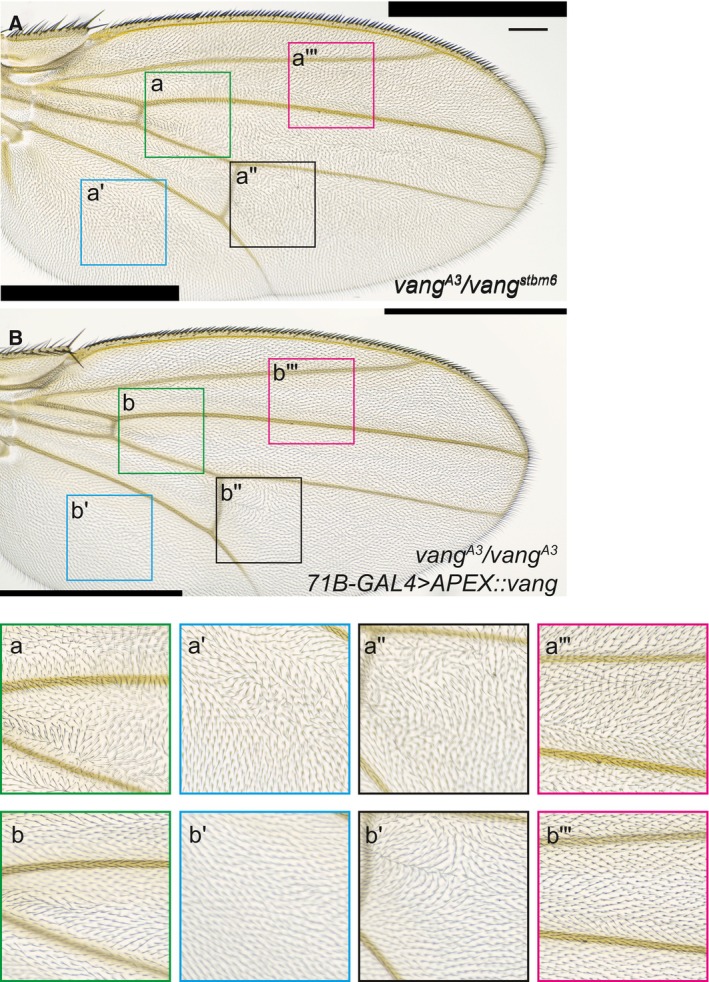
Figure EV1. Rescue of the vang stbm6 mutant phenotype by APEX::Vang expression.
-
AA vang (vang A3 /vang stbm6 ) mutant wing grown at 25° shows the stereotypical PCP polarity phenotype.
-
BA vang mutant wing ubiquitously expressing APEX::Vang (UAS‐Apex‐vang, vang A3 /vang A3 ; 71B GAL4/+) grown at 25° shows almost complete rescue.
-
a–a‴, b–b‴Magnified images of the boxed areas show nearly complete to complete rescue, with the exception of one area distal to the posterior cross‐vein (b″) that typically showed disturbed polarity. This is likely an overexpression phenotype, as growth at 29° made this polarity disturbance worse.
Data information: Scale bar = 100 μm.
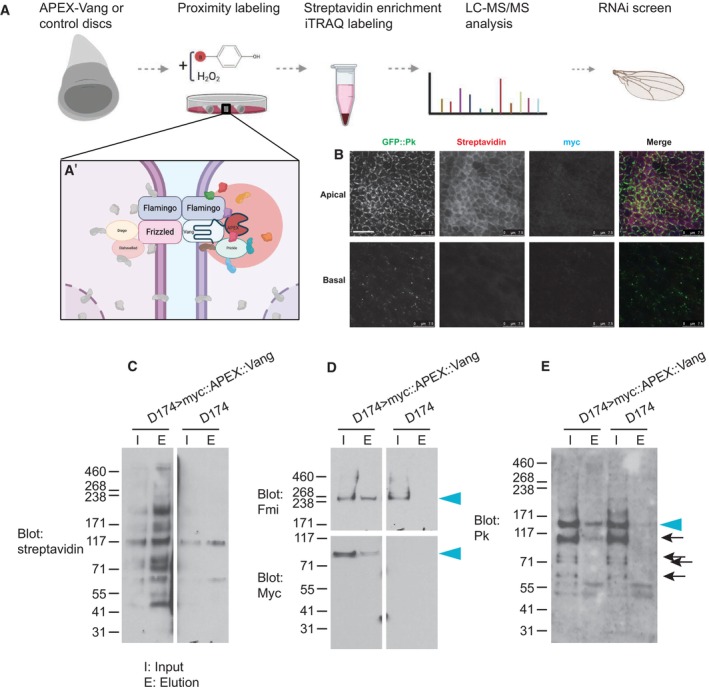
Figure EV2. Schematic and validation of the APEX‐labeling protocol.
-
A, A′Workflow for proximity labeling. (A′) Labeling is expected to biotinylate proteins in close proximity to the APEX::Vang probe.
-
BLabeled disc showing GFP::Pk, streptavidin detecting biotinylated proteins, Myc detecting Myc::APEX::Vang.
-
CInput and biotin enriched elution of APEX::Vang and control (D174) total protein.
-
DInput and elution probed for Fmi. The filter was cut and the upper portion probed for Fmi and the lower portion probed for Myc. Fmi and Myc::APEX::Vang are detected in the APEX::Vang but not the control elution (arrowheads).
-
EInput and elution probed for Pk. Pk is detected in the APEX::Vang but not the control elution (arrowhead). Degradation products of Pk are present in this blot (arrows).
Data information: Scale bar = 7.5 μm.
Since planar polarization is known to be underway in third instar (Sagner et al, 2012), we probed the PCP proteome in third‐instar wing discs. Four pools, two carrying D174‐GAL4 and UAS‐APEX::Vang (specific), and two carrying only D174‐GAL4 to be used for subtraction of background biotinylation (background), were collected and incubated with biotin phenol in S2 medium. Biotinylated proteins from lysates were enriched on streptavidin beads, released from beads by on‐bead trypsin digestion, and labeled with iTRAQ (isobaric tags for relative and absolute quantification) four‐plex reagents followed by liquid chromatography–mass spectrometry (LC–MS/MS). The relative level of a given protein identified by at least two unique peptides in each sample (2,028 in total; Dataset EV1) was obtained, and for each sample, the proteins in each background sample were used to generate the iTRAQ ratios. In total, four sets of iTRAQ ratios were generated (APEX::Vang replicate 1/control replicate 1; APEX::Vang replicate 1/control replicate 2; APEX::Vang replicate 2/control replicate 1; and APEX::Vang replicate 2/control replicate 2). A false‐positive rate (FPR) cutoff was calculated based on the distribution of positive and negative control genes. The genes known and assumed positive or thought to be functional regulators of, or otherwise associated with, PCP are positive controls while those known to localize in compartments such as the nucleus where PCP is inactive are negative controls (Rhee et al, 2013). At the 10% FPR threshold, 123 proteins scored in three or four datasets and are reported as the high‐confidence candidate PCP interactome, and an additional total of 520 proteins scored in two of four datasets and were considered a low‐confidence interactome (Dataset EV1). As expected, the high‐priority candidate interactome included several known and likely PCP‐related components. Many of the identified proteins have known relationships to other proteins, as determined using COMPLEAT (protein COMPLex Enrichment Analysis Tool; http://www.flyrnai.org/compleat/), and when their related proteins were also identified, this is also shown. We prioritized the high‐confidence interactome list for a secondary RNAi screen, and to this list, we added proteins that scored below this cutoff, but that we deemed to be good candidates either because they co‐complex with the highest scoring proteins or based on the nature of the gene product.
Using available RNAi lines from the TRiP and VDRC collections, we then performed a secondary RNAi screen for the candidate protein‐coding genes to test for abnormal adult wing hair polarity phenotypes characteristic of known PCP gene mutants. We initially drove RNAi expression with ptc‐Gal4 in the presence of Dicer‐2, and for lines that produced lethality, we re‐screened with clonal GAL4 expression, and in some cases, hh‐GAL4, ci‐GAL4, or ap‐GAL4. We note that efficiency of somatic expression varies with the different VALIUM vectors in the TRiP collection, so negative results, particularly for weaker expressors, may be false negatives (see Materials and Methods).
Dataset EV2 summarizes all of the proteins that scored positive in the RNAi screen for adult wing hair PCP phenotypes, specifically wing hair orientation defects most characteristic of core PCP mutants, and multiple wing hair phenotypes more characteristic of PCP effector mutants. Phenotypes were subjectively scored on a scale of 0–3 (no phenotype to strong phenotype), keeping in mind that a large contributor to strength is likely to be the specific properties of each individual RNAi line. This list comprises 95 proteins and includes 40 of the 123 high‐priority candidates from the APEX screen. A complete summary of the RNAi screen, including information about which RNAi lines were used, is shown in Dataset EV3. The data have been submitted to RSVP database, which is the resource for tracking and sharing the phenotype data of RNAi transgenic reagents (Perkins et al, 2015) (https://www.flyrnai.org/cgi‐bin/RSVP_search.pl).
Identification of Pp1‐87B as a regulator of PCP signaling
Among the high‐priority screened proteins, we identified the protein phosphatase 1 catalytic subunit Pp1‐87B. Three RNAi lines (BM32414, BM67911, and VDRC35024) driven by ptc‐gal4 produced considerable lethality, but escapers showed subtle wing hair orientation defects (Appendix Fig S1). The two Bloomington lines, BM32414 and BM67911, target independent sequences and by qRT–PCR suppress Pp1‐87B mRNA by roughly half. Furthermore, expression of the Pp1‐87B inhibitor I‐2 expressed under ptc‐gal4 control phenocopies that of the three RNAi lines, demonstrating that the observed polarity phenotypes are attributable to Pp1‐87B‐associated on‐target effects (Appendix Fig S2). Making RNAi‐expressing clones, we observed some swirls indicative of polarity defects. However, in pupal wings, a fluorescent clone marker showed dispersed signal, suggesting that some or all of the observed clonal phenotypes might result from the expected Pp1‐87B‐dependent cell lethality and resulting scars. Nine of the 122 proteins were co‐complexed with Pp1‐87B (Dataset EV1; http://www.flyrnai.org/compleat/), suggesting possible relationships between Pp1‐87B and other potential regulators of PCP. We note, however, that many substrates are expected, as Pp1‐87B is thought to be responsible for 80% of the phosphatase activity in Drosophila (Dombradi et al, 1990).
To further evaluate a possible role for Pp1‐87B in PCP control, we circumvented the lethality and other phenotypes associated with Pp1‐87B knockdown in the ptc domain by using a temperature‐sensitive allele of GAL80 to restrict loss of function to the early pupal period during which wing hair polarity is established. Using RNAi line (32414), white prepupae were raised at the restrictive temperature (18°) before shifting to the permissive temperature (29°) at the equivalent of either 12 h APF or 20 h APF (Fig 1). Adult wings consistently showed abnormal hair orientation in the ptc domain both distal and proximal to the anterior cross‐vein when shifted to the restrictive temperature at 12 h and much less frequently at 20 h APF. Similarly, when expressed throughout the wing using actin‐GAL4, polarity perturbation is seen in other regions of the wing (Fig EV3). Thus, appropriately modulated loss of function produced consistent PCP hair orientation phenotypes.
Figure 1. Pp1‐87B knockdown in the ptc domain disrupts planar polarization.
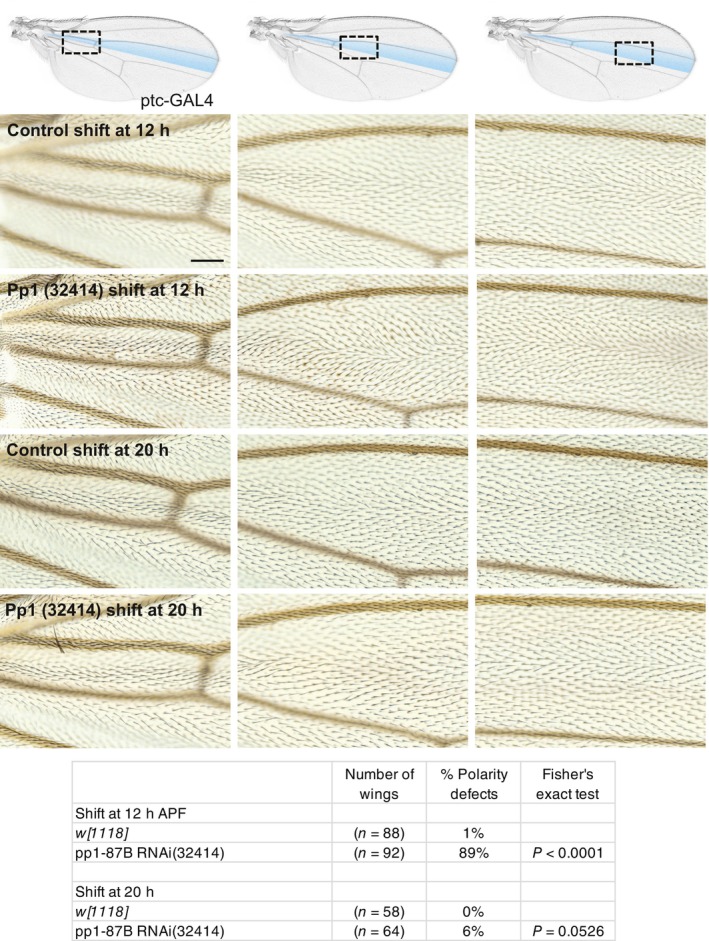
Control (w 1118 ) or Pp1‐87B (TRiP RNAi stock 32414) knockdown in the ptc domain at either 12 or 20 h APF. Three regions of the ptc domain proximal or distal to the anterior cross‐vein (acv) are shown. Knockdown by temperature shift to the permissive temperature (18°–29°) at the equivalent of 12 h APF but not at 20 h APF disrupted normal distal polarity, more prominently proximally than distally. The table indicates the percentage of wings that show a polarity perturbation. Scale bar = 50 μm.
Data information: The number of wings and percentages of each genotype are shown. Differences were compared using Fisher's exact test.
Source data are available online for this figure.
Figure EV3. ActinGAL4 driving Pp1‐87B RNAi throughout the wing induces polarity defects outside the ptc domain that are suppressed by simultaneous knockdown of Dco.
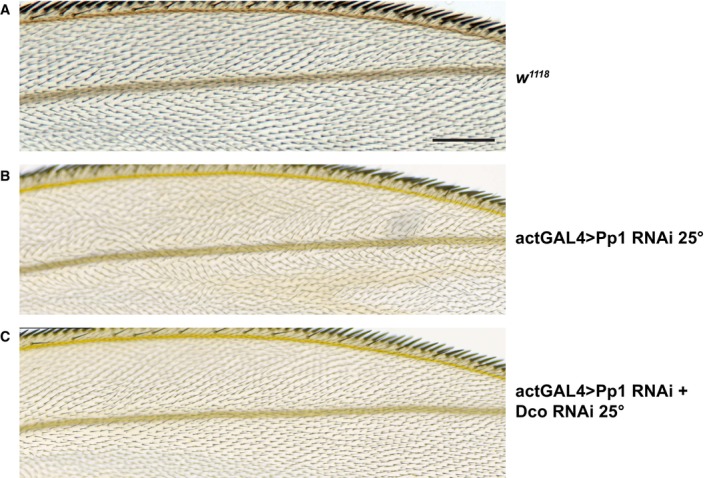
- Control (w 1118 ) wing showing a region near the anterior wing margin.
- Pp1‐87B (TRiP RNAi stock 32414) plus wRNAi knockdown raised at 25°C shows polarity disturbance in this region.
- Simultaneous expression of Dco RNAi and Pp1‐87B (TRiP RNAi stock 32414) raised at 25°C suppresses the polarity defect. > 10 wings for each of (A–C) were examined.
Data information: Scale bar = 100 μm.
Pp1 regulates polarization and localization of Vang
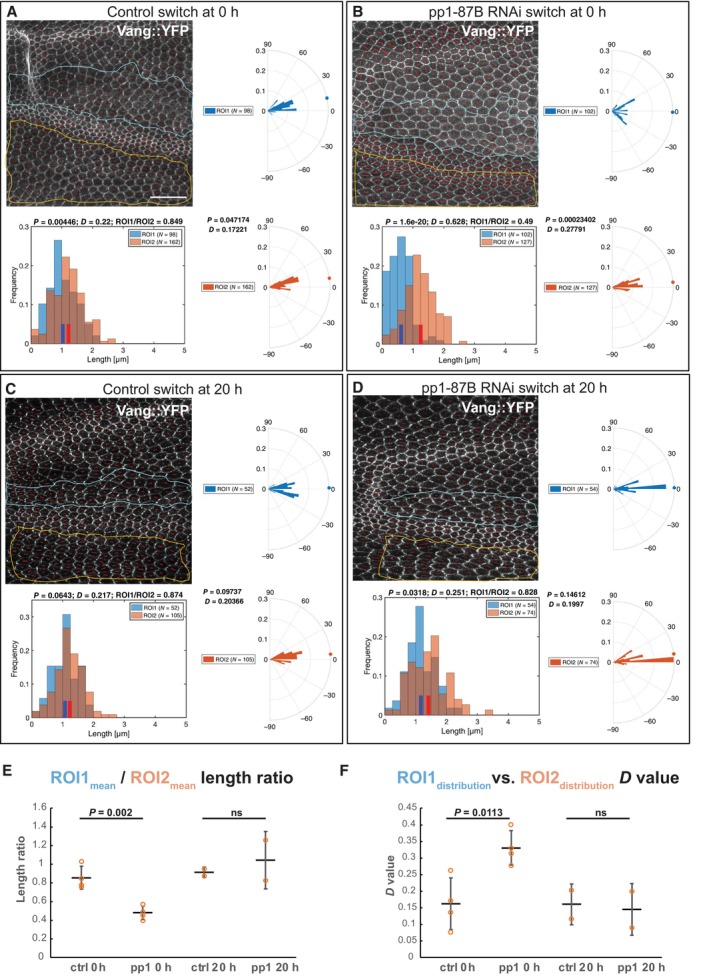
Proper subcellular localization of core PCP proteins correlates with, and is almost certainly responsible for, their ability to direct polarization. If Pp1‐87B affects wing hair polarity through the core PCP signaling machinery, it would be expected to alter the characteristic asymmetric subcellular localization of the core proteins to proximal or distal sides of the cell. To examine this, we again used temperature shifts (18°–29°) to knock down Pp1‐87B activity in the ptc domain of pupal wings expressing Vang::YFP beginning at 0 or 20 h APF, and harvested at the equivalent of 28 h APF. Vang::YFP within the ptc domain appeared less polarized than a control region just posterior to the fourth vein when shifted at 0 h but not at 20 h APF (Fig 2A–F). To quantify this result, the Fiji plug‐in “TissueMiner” (Aigouy et al, 2010; Etournay et al, 2016) was used to define and extract nematics where the length represents the degree of polarization and the angle represents the orientation of polarization in each cell. Using a custom MATLAB script (see Materials and Methods) to extract the lengths and compare means, we found that the degree of polarization in the ptc domain where Pp1‐87B activity is reduced is significantly less compared to the control region outside the ptc domain when knockdown is initiated at 0 h but not at 20 h APF. We similarly compared orientation of polarization by comparing angle distributions and found a significantly more dispersed orientation distribution in the Pp1‐87B knockdown domain compared to the control domain when knockdown is initiated at 0 h but not at 20 h APF. Pp1‐87B activity is therefore required before 20 h APF for complete polarization of Vang, and by proxy, the other core PCP signaling proteins.
Figure 2. Early but not late knockdown of Pp1‐87B in the ptc domain disrupts polarization of Vang subcellular localization.
-
A–DKnockdown (TRiP RNAi stock 32414) by temperature shift (18°–29°) was effected in wings expressing Vang::EYFP at the equivalent of 0 or 20 h APF and pupal wings were harvested at the equivalent of 28 h APF. The degree of polarization and orientation of polarization of Vang::EYFP was compared for cells in a portion of the ptc domain, defined by mRFP expression (ROI1), and an adjacent domain posterior to the L4 vein where no knockdown occurred (ROI2). Polarization of individual cells was determined using the Fiji plug‐in “TissueMiner” (Aigouy et al, 2010; Etournay et al, 2016) to determine nematics (red overlay lines), and both the length and orientation of each nematic were extracted using a custom MATLAB script (see Materials and Methods). Nematic lengths are plotted in histograms and orientations in rose plots. Individual examples are shown in (A–D). N indicates the number of cells included in the region of interest (ROI).
-
ERatios of mean nematic length, indicating degree of polarization in the ptc domain compared to the posterior domain, for four wings each shifted at 0 h APF and two each at 20 h APF are plotted. Shift at 0 h but not 20 h produced a significant decrease in mean length ratio (degree of polarization) (P = 0.002, Student's t‐test).
-
FDistributions of polarity orientation for the ptc domain and the posterior domain were compared for the same four wings each shifted at 0 h APF and two each at 20 h APF using the Kolmogorov–Smirnov test where D close to 0 indicates identical and D closer to 1 indicates different distributions. D‐values for shift at 0 h but not 20 h APF are significantly greater (P = 0.011, Student's t‐test), confirming the visual impression that orientations are more dispersed in the ptc domain compared to the posterior domain.
Data information: Scale bar = 20 μm.
In (A–D), nematic lengths are plotted in histograms and orientations in rose plots for the associated image and ROI. Distributions were compared using the Kolmogorov–Smirnov test where D close to 0 indicates identical and D closer to 1 indicates different distributions. In (E and F), means (E) and distributions (F) from multiple samples are plotted, and means are compared by Student's t‐test. The number of replicates (n) is shown as individual points in the plots in (E) and (F). Error bars indicate standard deviation.
Source data are available online for this figure.
In similar wings (18°–30° at 0 h APF), we also noticed that apicolateral junctional accumulation of Vang::YFP was increased where Pp1‐87B activity was reduced, and excess Vang accumulated along the lateral and basal membranes in these cells as well (Fig 3A). This does not appear to result from gross changes to cell and tissue architecture. A similar result is observed in third‐instar wing discs shifted to the permissive temperature 48 h prior to examination (Fig EV4). We examined the other core PCP proteins for subcellular localization in pupal wings and found that Pk, like Vang, accumulates apicolaterally and laterally, and is also at higher levels throughout the cytoplasm (Fig 3A and B). However, Fz, Dsh, and Fmi apical–basal localizations are minimally affected, if at all (Fig 3B and C). We conclude that in addition to perturbing polarization, Pp1‐87B knockdown regulates the quantity of the proximal proteins Vang and Pk that can accumulate both apicolaterally and basally.
Figure 3. Excess junctional Vang and cytoplasmic Pk accumulation upon Pp1‐87B knockdown.
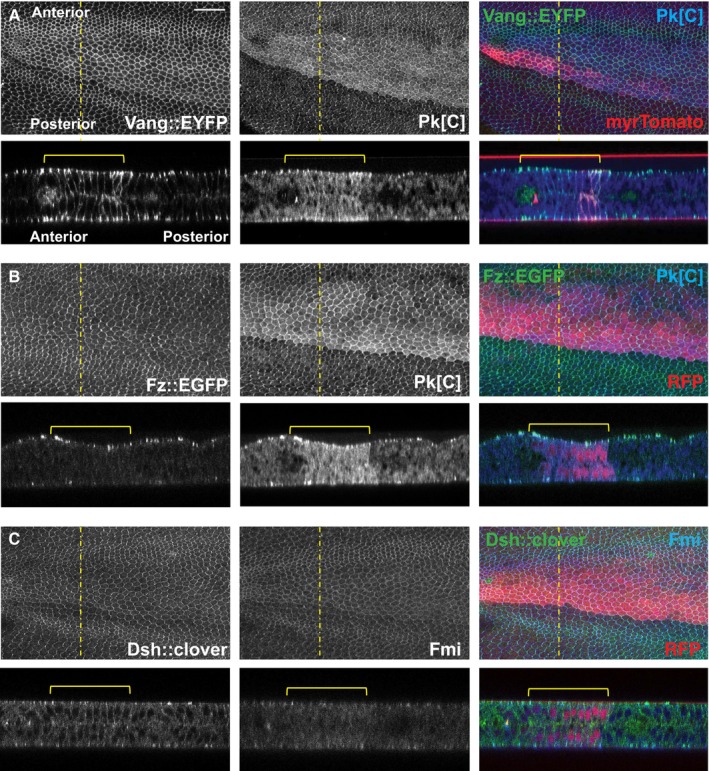
-
A–CPupal wings with Pp1‐87B knockdown in the ptc domain (TRiP RNAi stock 32414) by temperature shift (18°–30° at 0 h APF) as in Fig 2 were examined for Vang::EYFP (A), Pk (A and B) (antibody staining), Fz::EGFP (B), Dsh::clover (C), and Fmi (C) (antibody staining) in the X–Y and Y–Z planes. Excess Vang and Pk are observed in apicolateral junctions and along lateral membranes where Pp1‐87B is knocked down, whereas Fz, Dsh, and Fmi are not appreciably affected. The ptc domain is marked by myr‐Tomato or RFP as indicated. At least 10 wings of each genotype were examined. Scale bar = 50 μm.
Source data are available online for this figure.
Figure EV4. Vang accumulates at apical junctions and basolaterally in regions of a third‐instar wing disc where Pp1‐87B is knocked down.
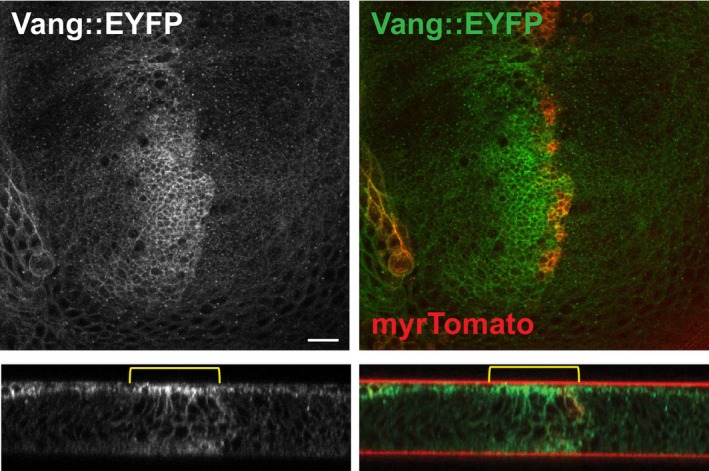
XY and Y‐Z planes are shown. myrTomato labels the ptc domain where Pp1‐87B (TRiP 32414) RNAi is expressed. Scale bar = 10 μm.
Pp1‐87B genetically interacts and colocalizes with core PCP components
If Pp1‐87B regulates the function of one or more core PCP proteins, either directly or indirectly, it might interact genetically with them. We produced a weak Pp1‐87B RNAi phenotype in the ptc domain by shifting flies from the restrictive to a partially permissive temperature (18° to 25°) at 0 h APF. We found that this phenotype was substantially enhanced by heterozygosity for vang, although not for fz (Fig 4). While the negative result for fz is not informative, enhancement by vang/+ provides additional evidence that Pp1‐87B regulates core PCP function. Consistent with this possibility, HA‐tagged Pp1‐87B expressed in the ptc domain is seen at apicolateral cell junctions where core PCP proteins localize as well as throughout the cytoplasm (Appendix Fig S3). Expression of Pp1‐87B in the ptc domain subtly perturbed polarity, if at all, and it did not significantly modify Vang polarization or apical–basal localization (Appendix Fig S4).
Figure 4. Pp1‐87B knockdown genetically interacts with vang .
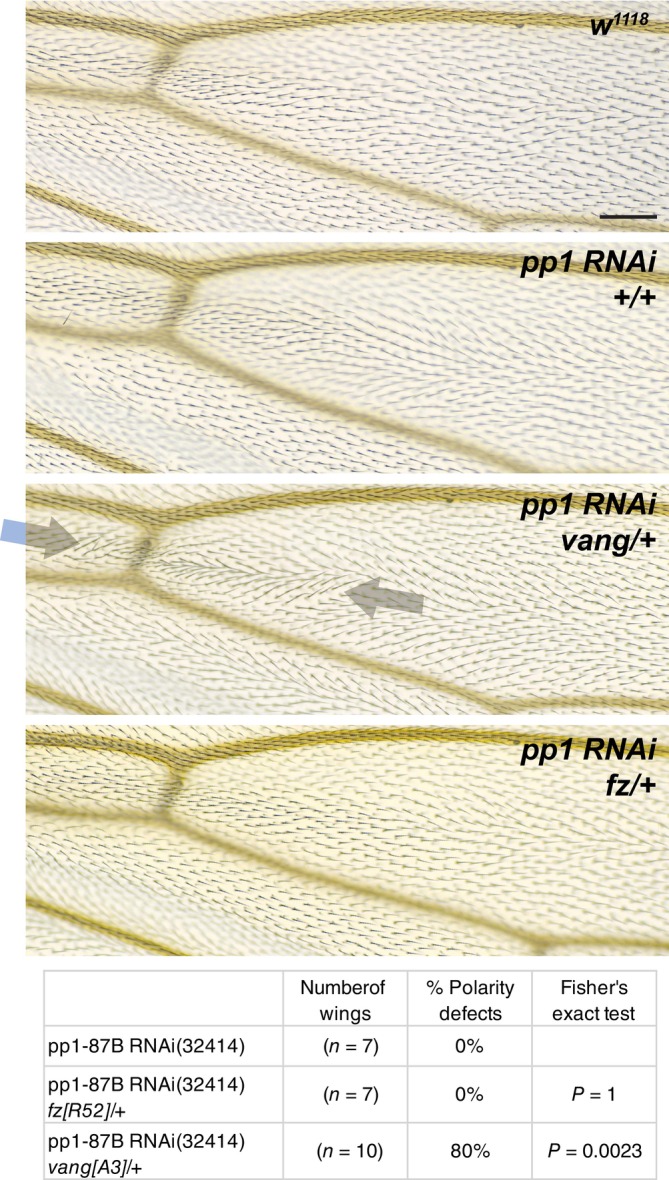
A weak Pp1‐87B RNAi knockdown phenotype in the ptc domain was produced by temperature shift (18°–25°) at 0 h APF. The aberrant polarity was enhanced by heterozygosity for vang A3 but not fz R52 . For consistency, the analysis was limited to the dorsal side of the wing. The table indicates the percentage of wings that show a polarity perturbation. The data in Figs 4 and 8 are from a contemporaneous experiment, and the same w 1118 wing is shown in each Figure. Scale bar = 50 μm.
Data information: The number of wings and percentages of each genotype are shown. Differences were compared using Fisher's exact test.
Source data are available online for this figure.
Pp1‐87B genetically interacts with Dco kinase
A number of serine/threonine kinases have been associated with PCP signaling, including PAR‐1, aPKC, CKIε/Dco, CK1δ, CK1γ/Gilgamesh, Ror2, Nek, Nemo, Drok, and Mink1 (Winter et al, 2001; Djiane et al, 2005; Ossipova et al, 2005; Klein et al, 2006; Strutt et al, 2006, 2019; Gao et al, 2011; Daulat et al, 2012; Gault et al, 2012; Schertel et al, 2013; Kelly et al, 2016; Weber & Mlodzik, 2017; Yang et al, 2017; Collu et al, 2018). Dco‐dependent phosphorylation of both the core proteins Dsh and Vang has been demonstrated, although evidence that their phosphorylation by Dco is direct is not definitive. We therefore asked if Dco might interact with Pp1‐87B to regulate PCP signaling. Knockdown of Pp1‐87B in the ptc domain (18°–29°) at 12 h APF induced a strong polarity phenotype in adult wings (Fig 1), while knockdown at 0 h APF (18°–31°) did not yield viable adults. Dco knockdown in the ptc domain under these conditions did not perturb polarity, but when we knocked down Dco together with Pp1‐87B (0 h APF, 18°–31°), the lethality induced by Pp1‐87B knockdown was strongly suppressed and the adult wing hairs showed only a subtle polarity defect (Fig 5A). Furthermore, simultaneous Dco knockdown partially suppressed the apical–basal mislocalization of Vang (Fig 5B and C). To rule out the possibility that Dco suppressed the Pp1‐87B lethality and polarity phenotype by modulating ptc expression, we did a similar experiment substituting actin‐GAL4 for ptc‐GAL4 and saw a similar result (Fig EV3). Together, these results suggest that Dco and Pp1‐87B intersect in their regulation of PCP.
Figure 5. dco knockdown suppresses the Pp1‐87B knockdown phenotype.
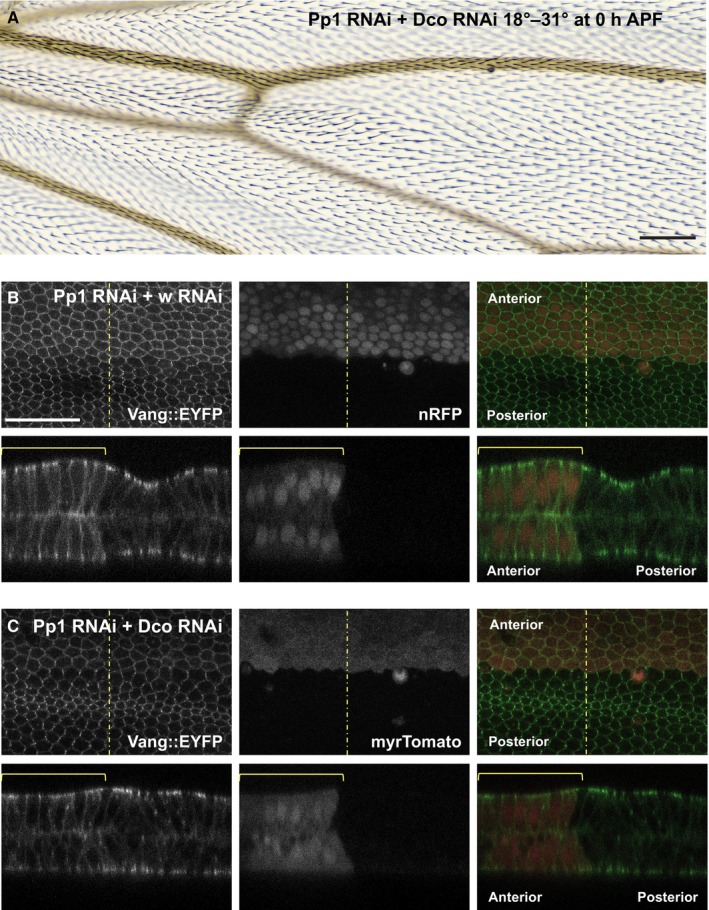
-
APp1‐87B RNAi (TRiP RNAi stock 32414 plus wRNAi) knockdown at 0 h APF produces lethality, but knockdown of Dco together with knockdown of Pp1‐87B (TRiP RNAi stock 32414 plus dco RNAi) rescues viability and produces only a weak PCP phenotype.
-
B, CVang::EYFP accumulates laterally and basally in 28 h APF pupal wings when Pp1‐87B RNAi is knocked down together with white (wRNAi; B) at 0 h APF as in (A); co‐expression of wRNAi somewhat weakens the effect compared to Pp1‐87B RNAi alone (Fig 1), likely due to GAL4 dilution. Knockdown of dco (dcoRNAi; C) together with Pp1‐87B RNAi partially suppresses lateral and basal Vang::EYFP accumulation relative to the control. At least 10 wings of each genotype were examined.
Data information: Scale bars = 50 μm.
Source data are available online for this figure.
Pp1‐87B modifies phosphorylation state of Vang in vivo
Our results thus far indicate that Pp1‐87B regulates core PCP polarization (Fig 2) and genetically interacts with vang but not fz (Fig 4), and with Dco, a kinase thought to phosphorylate Vang (Fig 5). Furthermore, the subcellular localization of Vang is modified by knockdown of Pp1‐87B (Fig 3). We therefore sought to determine whether Pp1‐87B might modify the phosphorylation status of Vang in vivo. Extracts from third‐instar wing discs expressing Pp1‐87B RNAi (32414 or 67911) or overexpressing Pp1‐87B, each under the control of 71B‐GAL4, were probed by Western blot (Fig 6). Control extract treated with lambda protein phosphatase showed a modest shift toward faster mobility, consistent with dephosphorylation of Vang. We detected a similar though perhaps more subtle shift in the Pp1‐87B overexpression condition. We could not reliably detect a shift toward hyperphosphorylation upon Pp1‐87B knockdown. Similarly, no shift was detectable with Dco knockdown, as has been reported by others, although such a shift was detected in transfection experiments (Kelly et al, 2016). These results are consistent with Vang being largely in a phosphorylated state in vivo, and with Vang being a substrate for Pp1‐87B phosphatase activity in vivo, although it is also possible that Pp1‐87B indirectly dephosphorylates Vang.
Figure 6. Pp1‐87B overexpression reduces phosphorylation of Vang.
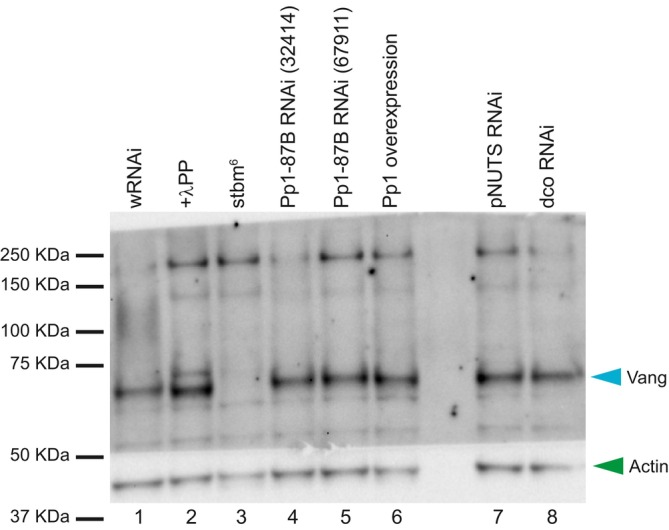
Vang runs as a single band (lane 1), and with λ‐protein phosphatase treatment runs faster, showing that it is phosphorylated in vivo (lane 2). We were unable to detect a shift toward hyperphosphorylation of Vang from wing discs expressing either of two Pp1‐87B RNAi lines throughout the wing under 71B‐GAL4 control (lanes 4 and 5), but Vang from wing discs broadly overexpressing Pp1::HA, Vang migrates slightly faster, suggesting less phosphorylation. We did not detect a shift from wing discs expressing pNUTS or dco RNAi. All extracts are from third‐instar wing discs. Actin was probed as a loading control. Two biological replicates were performed for this experiment.
Source data are available online for this figure.
PNUTS is a potential subunit of the Pp1 holoenzyme in PCP signaling
Pp1 functions as a holoenzyme, deriving substrate specificity by partnering with specific non‐catalytic subunits to find and act on its substrates in various contexts (Peti et al, 2013; Bertolotti, 2018). In vertebrates, known non‐catalytic subunits comprise a large family of structurally diverse proteins; in Drosophila, a modest number of non‐catalytic subunits has been identified (Bennett et al, 2006). Several known subunits were identified in our mass spec results, although they scored below our significance threshold: we were unable to produce a convincing polarity phenotype upon knockdown of ankyrin‐repeat, SH3‐domain, and proline‐rich region containing protein (ASPP), myosin binding subunit (Mbs), or Mars with any available RNAi line or driver we tested. Knockdown of Nebbish (Neb) produced a weak multiple wing hair phenotype, but at most a marginal hair orientation defect. We then examined several other known Pp1‐87B‐interacting proteins. Knockdown of phosphatase 1 nuclear‐targeting subunit (PNUTS) (Ciurciu et al, 2013) in the ptc domain induced a clear polarity phenotype proximal to the anterior cross‐vein (acv), where hypomorphic dco allelic combinations and two originally identified antimorphic Vang alleles have been shown to disrupt polarity (Taylor et al, 1998; Strutt et al, 2006) (Figs 7 and 8). This proximal phenotype differed in pattern on the dorsal and ventral sides of the wing, and notably, neither disruption extended substantially distal to the acv, where we see a robust Pp1‐87B knockdown phenotype. We were, however, able to detect a modest disruption to the polarity nematic distal to the acv (Fig EV5). This was not accompanied by a detectable change in apical or basal Vang::GFP accumulation (Appendix Fig S4).
Figure 7. PNUTS knockdown interacts with core PCP genes vang, fz, and dsh .
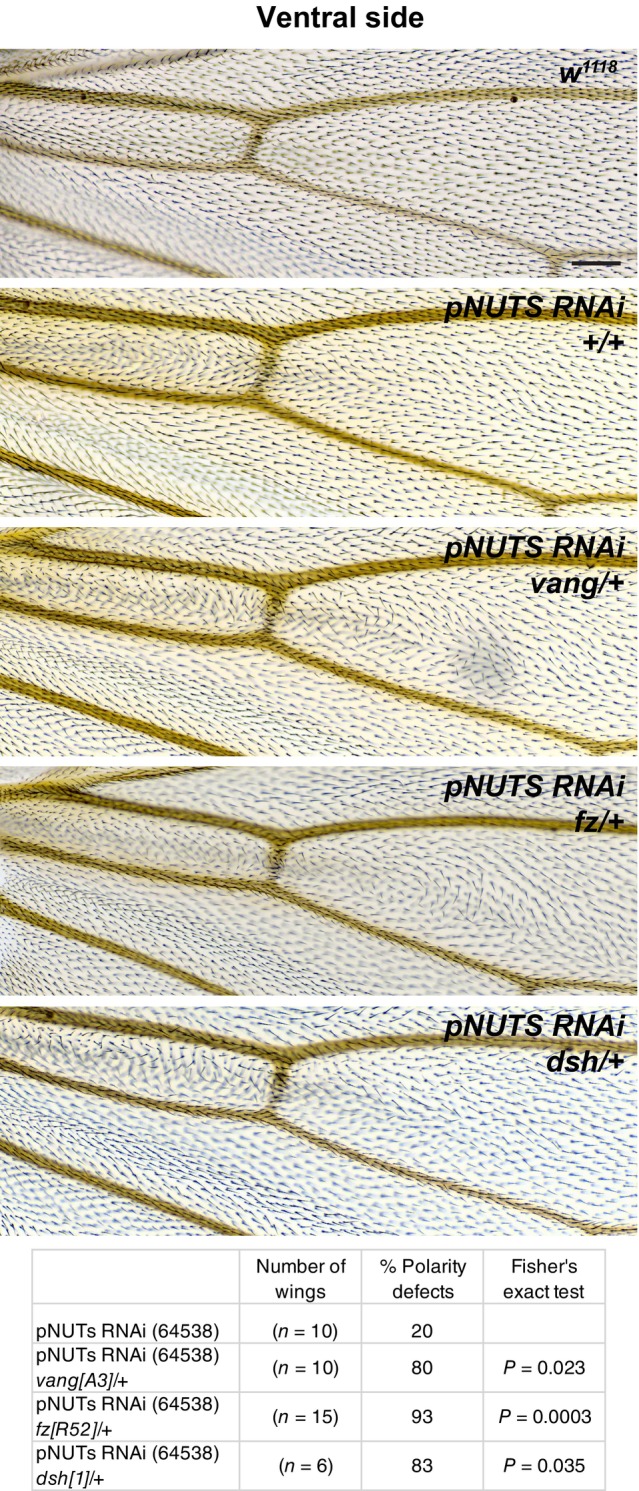
Because PNUTS knockdown (TRiP RNAi stock 64538) in the ptc domain produces substantially different patterns on the dorsal and ventral sides, we restricted interaction analyses to a single side, in this case, ventral. The weaker ventral phenotype is enhanced by heterozygosity for vang A3 , fz R52 , and dsh 1 . The table indicates the percentage of wings that show a polarity perturbation. Scale bar = 50 μm.
Data information: The number of wings and percentages of each genotype are shown. Differences were compared using Fisher's exact test.
Source data are available online for this figure.
Figure 8. PNUTS knockdown interacts with dco knockdown.
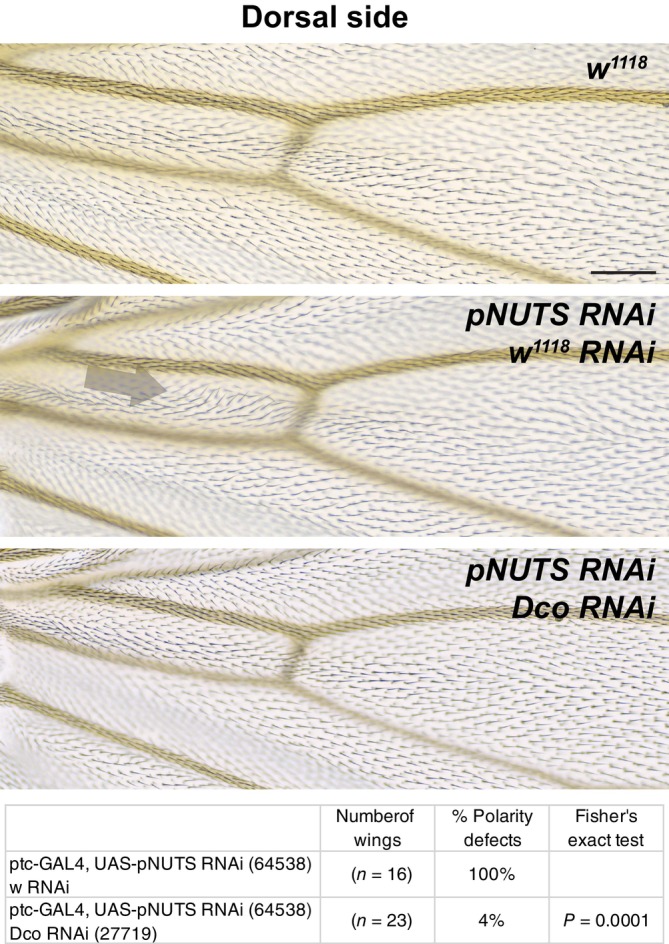
The dorsal PNUTS knockdown (TRiP RNAi stock 64538) phenotype in the ptc domain (arrow) was suppressed by simultaneous dco knockdown. Because PNUTS knockdown in the ptc domain produces substantially different patterns on the dorsal and ventral sides, we restricted interaction analyses to a single side, in this Case, dorsal. The table indicates the percentage of wings that show a polarity perturbation. The data in Figs 4 and 8 are from a contemporaneous experiment, and the same w 1118 wing is shown in each Figure. Scale bar = 50 μm.
Data information: The number of wings and percentages of each genotype are shown. Differences were compared using Fisher's exact test.
Source data are available online for this figure.
Figure EV5. Knockdown of PNUTS in the ptc domain disrupts polarization of Vang subcellular localization.
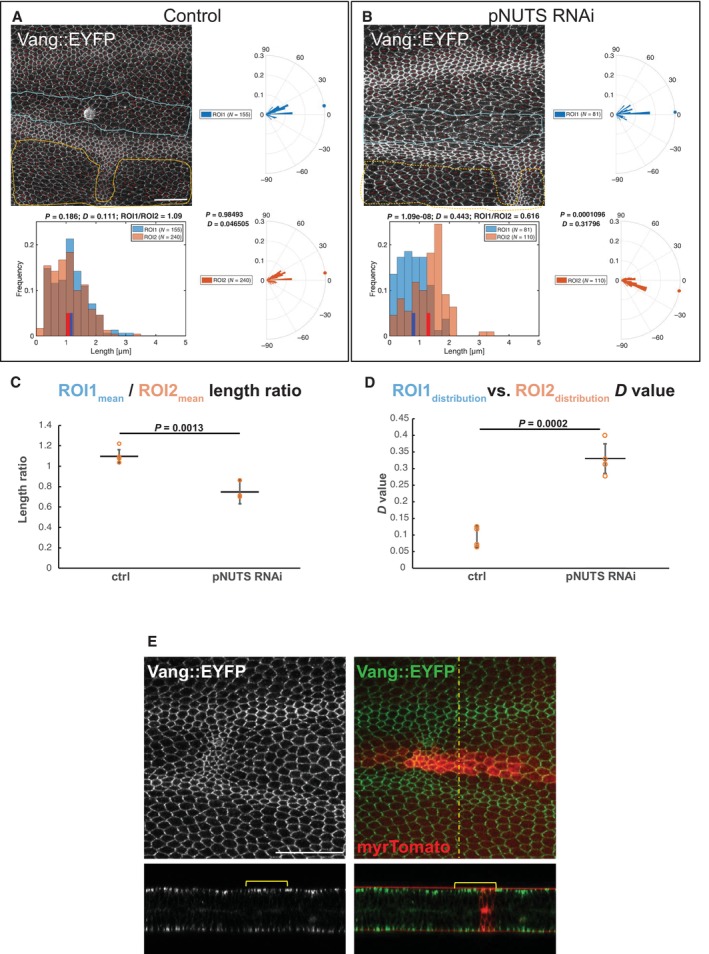
-
A, BKnockdown (TRiP RNAi stock 64538) was driven by ptc‐GAL4 in wings expressing Vang::EYFP, and pupal wings were harvested at 28 h APF. The degree of polarization and orientation of polarization of Vang::EYFP were compared for cells in the ptc domain, defined by mRFP expression (ROI1), and an adjacent domain posterior to the L4 vein where no knockdown occurred (ROI2). Polarization of individual cells was determined using the Fiji plug‐in “Tissue Analyzer” to determine nematics (red overlay lines), and both the length and orientation of each nematic were extracted using a custom MATLAB script (see Materials and Methods). Nematic lengths are plotted in histograms and orientations in rose plots. Individual examples are shown in (A, B). N indicates the number of cells included in the region of interest (ROI).
-
CRatios of mean nematic length, indicating degree of polarization in the ptc domain compared to the posterior domain, for four control and four PNUTS knockdown wings are plotted. PNUTS knockdown produced a significant decrease in mean length ratio (degree of polarization) (P = 0.0013, Student's t‐test).
-
DDistributions of polarity orientation for the ptc domain and the posterior domain were compared for the same four wings using the Kolmogorov–Smirnov test where D close to 0 indicates identical and D closer to 1 indicates different distributions. D‐values for PNUTS knockdown are significantly greater (P = 0.0002, Student's t‐test), confirming the visual impression that orientations are more dispersed in the ptc domain compared to the posterior domain.
-
EPNUTS knockdown does not cause excess accumulation or relocalization of Vang. Despite the diminution of polarization of Vang::EYFP upon PNUTS knockdown (Appendix Fig S4), Vang::EYFP does not accumulate to excess in apicolateral junctions or on the lateral membranes as was observed with knockdown of Pp1‐87B (Fig 3). Scale bars = 50 μm.
Data information: In (A, B), nematic lengths are plotted in histograms and orientations in rose plots for the associated image and ROI. Distributions were compared using the Kolmogorov–Smirnov test where D close to 0 indicates identical and D closer to 1 indicates different distributions. In (E and F), means (C) and distributions (D) from multiple samples are plotted, and means are compared by Student's t‐test. The number of replicates (n) is shown as individual points in the plots in (E) and (F). Error bars indicate standard deviation.
The PNUTS polarity phenotype was enhanced by heterozygosity of vang, fz, and to a lesser extent, dsh, and the enhanced polarity disruption in many instances extended distal to the position of the acv (Fig 7). Like Pp1‐87B, the PNUTS knockdown phenotype was suppressed by simultaneous Dco knockdown (Fig 8). Due to the different dorsal and ventral polarity patterns on PNUTS inhibition, we restricted each of these interaction analyses to a single side. No shift in Vang phosphorylation was detected on pNUTS knockdown, although this was not expected given no shift could be detected with Pp1‐87B knockdown (Fig 6). Thus, reduction in PNUTS results in many, although not all, of the alterations seen with reduction in Pp1‐87B, except that they are shifted toward a more proximal position in the wing.
Discussion
While the dedicated components of the core PCP mechanism have been known for some time, regulators that facilitate their activity, and which are likely to have pleiotropic effects, have been more challenging to identify in genetic screens. We employed an APEX‐based proximity labeling approach coupled with iTRAQ and LC–MS/MS to identify such regulators. Tagging Vang with APEX led to the identification of 123 high‐probability hits; these, plus selected lower probability hits, were secondarily screened by RNAi, and 91 showed either misorientation of wing hair polarity or multiple wing hair phenotypes (or both). The true‐positive rate may be higher given the variable efficiency of RNAi lines. Our screen therefore provides a resource for exploring previously uncharacterized regulators of PCP signaling.
Among the positive hits, we chose to further characterize the role of the S/T phosphatase catalytic subunit Pp1‐87B. Previously, Widerborst (wdb), a regulatory subunit of PP2A, was identified as a regulator of PCP, although its precise role is poorly defined (Hannus et al, 2002). Numerous post‐translational modifications of core PCP components have been proposed to control core PCP signaling, including phosphorylation of Drosophila and vertebrate homologs of Fmi, Fz, Vang, Pk, and Dsh (reviewed in Harrison et al, 2020). Of these, phosphorylation of Dsh and Vang has been most thoroughly investigated.
A phosphorylation‐dependent gel shift of Dsh is associated with PCP signaling (Axelrod, 2001; Shimada et al, 2001). CKIε/Dco, a S/T kinase, is required for PCP signaling in flies and vertebrates, and CKIε has been found to bind to and phosphorylate vertebrate Disheveled proteins (Dvl) in numerous cell culture and in vitro assays (Peter et al, 1990; Sakanaka et al, 1999; Kishida et al, 2001; McKay et al, 2001; Gao et al, 2002; Hino et al, 2003; Cong et al, 2004). Drosophila Dco is similarly able to phosphorylate Dsh in vitro, and mutation of S236, in a consensus motif predicted to be phosphorylated by Dco, blocks that ability (Klein et al, 2006; Strutt et al, 2006). Surprisingly, however, in S2 cells, a kinase‐dead CKIε induces Dsh phosphorylation as efficiently as does the functional kinase, suggesting that Dco, via a kinase‐independent mechanism, stimulates one or more other kinases to phosphorylate Dsh (Klein et al, 2006). In a cell culture model, MS mapping of Dsh phosphorylation identified many Fz‐dependent and ‐independent S/T phosphorylations (although surprisingly, not S236) (Yanfeng et al, 2011). However, none of these, either alone or in several combinations, were shown to be functionally important for PCP signaling in an in vivo rescue assay, although substantial redundancy cannot be ruled out. Furthermore, mutation of the of S/T target cluster in the C‐terminus did not disrupt rescue activity, and indeed the C‐terminus of Dsh, which accounts for the bulk of the phosphorylation‐dependent gel shift, is not required for rescue of PCP signaling. Thus, despite the extensive S/T phosphorylation of Dsh, these attempts failed to define functional significance for Dsh phosphorylation. In contrast, two subsequent observations provide at least weak evidence for functional significance: a modest genetic interaction between overexpressed Dco and dsh was observed, and a phosphomutant including S236 and seven adjacent S/T residues that failed to significantly disrupt polarity nonetheless showed a subtle decrease in Fmi asymmetry (Strutt et al, 2006, 2019). In sum, while Dsh S/T phosphorylation correlates with membrane localization, functional significance of this phosphorylation remains equivocal.
Of note, the MS analysis identified tyrosine phosphorylation (Y473) that was shown to be mediated by the Abl kinase and to have a relatively PCP‐specific requirement, as a Y473F mutant behaves much like the PCP‐specific dsh 1 allele (Singh et al, 2010; Yanfeng et al, 2011). Tyrosine phosphorylations have been proposed to regulate additional aspects of PCP signaling as well (Guo et al, 2013; Kelly et al, 2016; Humphries et al, 2023).
Drosophila and vertebrate Vang phosphorylation appears to be mediated by CKIε/Dco, likely directly (Gao et al, 2011; Kelly et al, 2016; Yang et al, 2017). Two clusters of S/T phosphorylation sites were identified in Vang, and their mutation disrupts PCP in flies and vertebrates (Gao et al, 2011; Ossipova et al, 2015; Kelly et al, 2016; Yang et al, 2017; Strutt et al, 2019). Because mutation to either phosphomutant or phosphomimetic residues disrupts PCP, an appealing explanation is that cycling between phosphorylated and unphosphorylated forms may be required (Strutt et al, 2019). Based on measurements of stability and localization asymmetry, it was proposed that phosphorylation destabilizes Vang in puncta and that cycling between phosphorylated and unphosphorylated forms is required for the mutual inhibition of oppositely oriented complexes that generates asymmetry (Butler & Wallingford, 2017; Strutt et al, 2019). This model is supported by the observation that phosphorylation of Vang is cell autonomously stimulated by Fz, and cell‐autonomous interaction between Fz and Vang complexes is hypothesized to destabilize one or the other (Tree et al, 2002; Amonlirdviman et al, 2005; Kelly et al, 2016). Consistent with this idea, Pk was proposed to stabilize Vang by inhibiting its phosphorylation (Strutt et al, 2019), but alternately, or perhaps in addition, Pk was proposed to destabilize Vang by promoting its internalization (Cho et al, 2015).
While Dco may participate in cycling Vang between phosphorylated and less phosphorylated states to regulate PCP, a corresponding phosphatase activity has not been reported. Here, we have provided evidence that the S/T phosphatase catalytic subunit Pp1‐87B might participate in such a role. Appropriately timed reduction in Pp1‐87B expression produces a polarity phenotype consistent with this function as assessed by wing hair polarity and subcellular localization of core PCP proteins and by genetic interaction with Vang. Antagonistic interaction between Pp1‐87B and Dco to modulate PCP further suggests that this kinase and Pp1‐87B oppositely regulate a common process. Vang accumulates and is mislocalized upon Pp1‐87B knockdown, consistent with its localization in puncta being destabilized by Fz‐dependent phosphorylation and stabilized by dephosphorylation.
Biochemical evidence for these activities is incomplete. Prior studies of Vang phosphorylation showed that upon co‐transfection into S2 cells with Dco, but not a kinase‐dead Dco, a shift reflecting presumptive phosphorylation was detected (Kelly et al, 2016). However, larval lysates from dco 2 or dco 3 hypomorphic mutants (Kelly et al, 2016) or Dco knockdown (Fig 6) did not show an appreciable shift. This may be due to remaining activity of the hypomorphic alleles, incomplete knockdown, or redundant activity of another kinase in vivo. It is also likely that the contribution of Dco‐dependent phosphorylation is a modest fraction of the total phosphorylation of Vang in vivo, making it difficult to detect. Treatment with the dual serine/threonine and tyrosine‐specific lambda phosphatase reveals significant phosphorylation of Vang in vivo, and tyrosine phosphorylation of Vang has also been demonstrated (Humphries et al, 2023). The subtle shift in phosphorylation we detect with Pp1‐87B overexpression, and lack of shift upon knockdown, are consistent with the possibility that Pp1‐87B phosphatase activity, in conjunction with pertinent regulatory components, acts on a limited set of Vang phosphorylation sites in the context of PCP signaling in vivo. Interestingly, pk mutants show a shift in Vang toward hyperphosphorylation, suggesting that Pk antagonizes Vang phosphorylation (Strutt et al, 2019), and the magnitude of this effect raises the possibility that it does more that impact phosphorylation of the sites potentially regulated by Dco and Pp1‐87B.
In sum, our data suggest that Pp1‐87B regulates the phosphorylation state of Vang, although we have not determined whether this action is direct or indirect. As Pp1‐87B is thought to be the most abundant and active S/T phosphatase (Dombradi et al, 1990), it would be unsurprising if it had multiple targets in the core PCP pathway. Cycling of Vang between phosphorylated and dephosphorylated states to facilitate mutual exclusion is expected to be contextually regulated: appropriately localized activity of Dco and the corresponding phosphatase Pp1 may fulfill this function.
That Vang accumulates and is mislocalized upon reduction in Pp1 activity is not simply predicted or explained by current understanding. Reduction in phosphorylated Vang's residence time in puncta with oppositely oriented complexes (Strutt et al, 2019) does not necessarily imply that the protein is itself destabilized, and indeed if excess Vang is expelled from such puncta, it may accumulate ectopically in cells. Similarly, excess phosphorylated Vang, perhaps having diminished residency in large puncta, might exit those large puncta and seed the formation of new, small puncta at apicolateral junctions. Furthermore, action of Pp1 on substrates other than Vang or its regulators might contribute to the phenomenon. The fate of this phosphorylated Vang population will require further study.
The substrate specificity of protein phosphatases is controlled by non‐catalytic subunits. While the catalytic subunits of Pp1 alone are highly promiscuous, in cells, they exist almost exclusively as subunits of a holoenzyme in which other subunits provide a high degree of substrate specificity (Brandt et al, 1974; Bertolotti, 2018). Hundreds of human regulatory subunits exist, and at least 40 putative regulatory subunits of perhaps many more in Drosophila have been identified (Bennett et al, 2006; Hendrickx et al, 2009; Bertolotti, 2018). Although no known Pp1 regulatory subunit reached significance in our screen, we identified a PCP phenotype with PNUTS knockdown, and interactions between PNUTS and other PCP components are quite similar to those of Pp1. Given the more limited territory in the wing in which PNUTS appears to act, contributions of other regulatory subunits are likely also involved.
Our data suggest a role for PNUTS in regulating PCP. Since the most dramatic effects of Pp1 are observed on Vang, one might speculate that PNUTS targets Pp1 to Vang. However, unlike Pp1, PNUTS knockdown does not regulate Vang accumulation or apical basal localization. Furthermore, PNUTS was previously shown to target Pp1 activity to RNA polymerase II (Ciurciu et al, 2013), raising the alternate possibility that PNUTS indirectly regulates PCP through broader changes in transcription. Furthermore, ribosome composition‐dependent translation of Vangl2 and Fz3 in differentiating human embryonic stem cells has recently been recognized, suggesting yet another potential target point (Genuth et al, 2022). Additional studies are required to determine how Pp1, PNUTS, and/or one or more other regulatory subunits are involved in regulating the core PCP machinery.
Materials and Methods
APEX::Vang flies
Vang cDNA from fly larvae was cloned into a Gateway pENTR vector and sequence verified. The Vang cDNA was recombined into a pUAST Gateway destination vector with 3xMyc‐APEX fused to the N‐terminus of Vang. Transgenic flies with site‐specific insertion of UAS‐3xMyc‐APEX‐Vang were made by injection of the plasmid together with PhiC31 integrase and targeted for insertion into the attP40 site on the second chromosome.
APEX labeling and sample preparation for Western blot and proteomic analyses
Third‐instar wing discs were biotinylated and prepared in batches for Western blots and mass spectrometry as follows. Dissected wing discs were collected into cold‐labeling buffer (PBS + 500 mM biotin phenol + 2 mM probenecid) on ice. Aliquots were then transferred to room‐temperature‐labeling buffer for 30 min. Labeling buffer was removed and 1 mM hydrogen peroxide in PBS was added to activate biotinylation for 1 min. The reaction was quenched immediately by adding quenching medium to a final concentration of 10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox in PBS. The tissue was washed in quench medium three times, then PBS two times. The discs were removed into cold PBS with protease inhibitors (Roche) and transferred to a 0.6 ml Eppendorf tube. An equal volume of 2× RIPA was added, tissue was hand ground, and then spun at 16,873 g for 10 min at 4°C. Supernatant was transferred to a new tube, immersed into liquid nitrogen and stored at −80°C. Protein concentration was determined using a BCA kit (Pierce).
Streptavidin bead binding
Streptavidin magnetic beads (Pierce; amount with sufficient binding capacity for protein amount) were washed with PBS, then with RIPA. Protein in RIPA was added to the washed beads and allowed to bind for 1 h at room temperature. Beads were then washed in RIPA three times. For Western blotting, 60 μl of 1× SDS–PAGE buffer was added to the beads and heated for 5 min at 95°C. The elution buffer E1 was removed from the beads.
Western blotting for Fmi and Pk
Samples were loaded on a Tris‐acetate 3–8% gel and HiMark prestained protein standard was loaded as size standards. The gel was run for 2 h and then proteins were transferred to a PVDF membrane. The membrane was blocked for 1 h in 5% dry milk in 50 mM Tris, 150 mM sodium chloride (blocking buffer) for Fmi, Myc, and Pk, or in Pierce protein free block for streptavidin. Membranes for Fmi, Myc, and Pk were probed with antibodies overnight at 4°C or with streptavidin as below for 1 h. Membranes were then washed, ECL applied, and then exposed to X‐ray film.
Fmi and Myc
Twenty microliter (13 μg) of input was loaded for each genotype/lane and the same volume of E1 (20 μl) was also loaded.
After transfer, the membrane was cut horizontally at 117 kDa.
The upper portion was incubated with Fmi antibody and the lower portion with Myc antibody.
Primary: anti‐Flamingo #74‐c (DSHB) 1:40 in 5% blocking buffer.
Secondary: donkey anti‐mouse HRP 1:5,000 in 5% blocking buffer.
Primary: anti‐Myc (source) 1:5,000 in 5% blocking buffer.
Secondary: donkey anti‐rabbit HRP 1:5,000 in 5% blocking buffer.
For Fmi, Western Bright ECL K12045‐D20‐sensitive HRP substrate for chemiluminescent Western blots was used. For myc, Supersignal West Pico chemiluminescent substrate was used.
Pk
Fifteen microliter (8 μg) of input was loaded for each genotype/lane and the same volume of E1 (15 μl) was also loaded.
Primary: Guinea pig anti‐pk (RRID:AB_2941929; Olofsson et al, 2014) 1:2,000 in 5% blocking buffer.
Secondary: Donkey anti‐guinea pig HRP 1:10,000 in 5% blocking buffer.
Proteins were visualized with Supersignal West Pico ECL.
Streptavidin
Twelve microliter (1.5 μg) of input was loaded for each genotype/lane and the same volume of E1 (12 μl) was also loaded.
After blocking for 1 h with Pierce protein‐free block, the membrane was probed with streptavidin horse radish peroxidase (SA10001) 1:50,000 in Pierce block for 1 h at RT.
Proteins were visualized with Supersignal West Pico ECL.
Samples for mass spectrometry
Two independent samples were generated for both genotypes, D174gal4 and D174gal4>APEX::myc::vang. Batches of biotinylated discs were prepared as above and numbered sequentially on the day of dissection and labeling. Even‐numbered and odd‐numbered tubes were pooled separately for each genotype. The final pooled volumes were approximately 1,800 μl. The total protein concentration of each sample was measured by BCA. Each sample was then bound to washed streptavidin beads as above. The beads were washed in PBS and sent for mass spec.
Mass spectrometry
Two milligram total input protein of each replicate was enriched for biotinylated proteins using streptavidin beads and analyzed by high‐resolution quantitative LC‐ MS/MS. On‐bead digestion was subsequently performed by incubating the beads in trypsin solution (80 μl 1 mM DTT and 5 μg/ml trypsin in 2 M urea in 50 mM Tris, pH 8) overnight to retrieve peptides of biotinylated proteins from the beads for the following analysis. The supernatant was removed and reduced (4 mM DTT), alkylated (10 mM iodoacetamide) for 30 min each, and digested with 0.5 μg of trypsin overnight (37°C). The resulting digested peptides were desalted and reconstituted in 30 μl iTRAQ reconstitution buffer. Four‐plex iTRAQ labeling was conducted per the manufacturer's instructions (SCIEX). Briefly, iTRAQ labels were reconstituted with ethanol to a final volume of 145 μl, followed by individual labeling at room temperature for 1 h by adding 140 μl iTRAQ reagent to the samples. Labels were used as follows: 114 for APEX::myc::vang, 115 for control, 116 for APEX::myc::vang, and 117 for control replicate B. Label incorporation was evaluated on an Orbitrap before quenching with 100 mM (final) Tris for 10 min at room temperature.
Peptides were separated over a 260‐min gradient using a heated PicoFrit (New Objective) column (50C) packed with 24 cm of 1.9 μm C18 material (Dr. Maisch) and run on a Q Exactive MS in data‐dependent mode where the top 12 most abundant precursors were selected for MS/MS prior to being placed on an exclusion list. Data were searched with Spectrum Mill (Agilent) using the UniProt Drosophila database. A fixed modification of carbamidomethylation of cysteine and variable modifications of N‐terminal protein acetylation, oxidation of methionine, and four‐plex iTRAQ labels were searched. The precursor mass tolerance and MS/MS tolerance were set to 20 ppm. The peptide and protein false discovery rates were set to 0.01.
Proteome analysis and prioritization of hits
Of 2,418 proteins identified, 2,064 proteins were identified by > 1 unique peptide and were selected for further analysis. Identified peptides with UniProt accession numbers were mapped to FlyBase gene identifiers using an ID mapping tool at UniProt that correspond to 2,028 unique annotated Drosophila genes (FlyBase release 6.10). All of the genes identified by iTRAQ along with their annotation are listed in Dataset EV1.
The log2 ratios of each replicate of experimental sample (D174‐GAL4, UAS‐APEX::Vang; replicates 114 and 116) to each replicate of the control sample (D174‐GAL4; replicates 115 and 117) (ratios: 114/115, 116/117, 116/115, and 116/117) were calculated and normalized to the median value of each channel, respectively.
To determine the significance cutoff of the iTRAQ ratio for each dataset, proteins were cross‐referenced to positive‐ and negative‐control lists. The positive‐control list was assembled manually based on literature, while a list of transcription factors and metabolic genes with high‐confidence annotation (Hu et al, 2015) was used as negative control.
The false‐positive rate (FPR) is calculated for each iTRAQ ratio using the equation as described in the literature (Rhee et al, 2013; Chen et al, 2015), where the numerator is the conditional probability of finding a false‐positive protein in a particular iTRAQ ratio range and is calculated as the percentage of proteins on the negative‐control list over all proteins identified on negative‐control list, and the denominator is the percentage of proteins on the positive control list over all proteins identified on the positive‐control list. The FPR is the ratio of these two conditional probabilities and is illustrated in the plot in Appendix Fig S5 for each iTRAQ ratio. We selected as our cut‐off point for each replicate the iTRAQ ratio at which the FPR reaches 0.1. All proteins above this cut‐off are > 10 times more likely to be true positives than false positives.
The genes that were above the cutoff in three or four of the four datasets were selected as high‐confidence hits while genes above the cutoff in two datasets were considered as low‐confidence hits (Dataset EV3). Protein complex annotation from COMPLEAT (Vinayagam et al, 2013) (www.flyrnai.org/compleat/) was used to identify complexes associated among the genes identified by APEX.
RNAi screen
The RNAi screen was first performed with constitutive GAL4 drivers (one of the following: ptc‐GAL4, ci‐GAL4, hh‐GAL4, and ap‐GAL4; the majority were initially screened with ptc‐GAL4). Progeny were mostly raised at 29°C; a minority were raised at 25°C as indicated. Those producing lethality were secondarily crossed to hsFlp>CD2>GAL4 or Dsh‐clover2; hsFlp>CD2>GAL4, the third‐instar larvae were heat shocked at 37°C for 30 min to induce clones, then raised at 25°C. Adult wings were mounted and examined for polarity phenotypes as described below.
qPCR protocol
RNA was obtained from 20 to 30 late third‐instar wing imaginal discs that were dissected and transferred to ice‐cold PBS + 0.2% Triton X‐100 (PBS‐T). We used 71B‐Gal4/UAS‐w RNAi as control and 71B‐Gal4/UAS‐Pp1‐87b RNAi (32414) and 71B‐GAL4/UAS‐Pp1‐87b RNAi (67911) to quantify the efficiency of both Pp1 RNAi lines. Discs were spun down at 9,000 g for 3 min, after which PBS‐T was removed and discs were snap frozen in liquid nitrogen.
Wing disc total RNA was extracted using the Qiagen RNeasy Mini RNA extraction kit, following manufacturer's instructions. RNA was then retrotranscribed to cDNA using Thermo Scientific's SuperScript IV first‐strand synthesis kit, following the manufacturer's protocol and using oligo(dT) primers.
The obtained cDNA was then diluted 1/100 and qPCR was performed on an Applied Biosystems StepOne Real‐Time PCR machine, using the Applied Biosystems PowerUp SYBR Green master mix. To normalize expression levels, we used RpL32 as reference gene. The following primers were used for the PCR reactions:
RpL32 F: ATGCTAAGCTGTCGCACAAATG
RpL32 R: GTTCGATCCGTAACCGATGT
Pp1 F‐I: GATCCGGGGACTTTGCTTGAA
Pp1 R‐I: GAGGAACAGGTAATTCGATTCCG
Pp1 F‐II: GCAATCGCTGGAGACGATCT
Pp1 R‐II: TCAAATCGGGACTGAGACCAC.
Temperature shift protocols
Expression levels of GAL4 driven RNAi lines and other transgenes were controlled by raising flies at defined temperatures through defined developmental time periods. Specific shifts were performed as indicated for each experiment. Because progression through development is temperature dependent, for simplicity, all developmental times are reported as adjusted to the equivalent times at 25°.
Adult wing hair imaging
Adult wings were dissected and washed with 70% EtOH and mounted in DPX (Sigma) solution. All adult wings were imaged on a Nikon Eclipse E1000M equipped with a Spot Flex camera (Model15.2 64 MP).
Immunohistochemistry
Pupal wings were dissected at indicated developmental time points after puparium formation (APF). Pupae were removed from their pupal cases and fixed for 60–90 min in 4% paraformaldehyde in PBS at 4°C. Wings were then dissected and extracted from the cuticle, and washed two times in PBST (PBS with 0.1% Triton X‐100). After blocking for 1 h in 5% bovine serum albumin in PBST at 4°C, wings were incubated with primary antibodies overnight at 4°C in the blocking solution. Incubations with secondary antibodies were done for 90 min at room temperature in PBST. Washes in PBST were performed three times after primary and secondary antibody incubations, and incubations in phalloidin (1:200 dilution) in PBST were done for 15 min followed by washing at room temperature before mounting. Stained wings were mounted in 15 μl Vectashield mounting medium (Vector Laboratories). Wing discs were isolated from third‐instar larvae and submerged in fixation solution (4% paraformaldehyde in PBS at 4°) for 10 min. All other procedures were the same as for pupal wing immunohistochemistry. Primary antibodies were as follows: mouse monoclonal anti‐Fmi (1:200 dilution, DSHB), guinea pig polyclonal anti‐Pk[C] (1:800 dilution, Olofsson et al, 2014), and rat monoclonal anti‐HA (clone 3F10, 1:200 dilution, Roche). Secondary antibodies from Thermo Fisher Scientific were as follows: 488‐donkey anti‐mouse, 488‐goat anti‐guinea pig, 546‐donkey anti‐goat, 633‐goat anti‐guinea pig, 633‐goat anti‐rat, and 647‐donkey anti‐mouse. Alexa 635‐ and Alexa 350‐conjugated phalloidin were from Thermo Fisher Scientific.
Quantification of Vang::EYFP polarity
Twenty‐eight hours, APF pupal confocal wing images were stacked into a max‐projected image and oriented as proximal to the left, anterior at the top. Non‐overlapping polarity nematics were generated with the FIJI plugin “TissueMiner” (Aigouy et al, 2010; Etournay et al, 2016) and overlaid on these images in a distinct color from the background image (in this case red). A custom‐built MATLAB script allows the user to mark two regions of interests (ROIs). This script identifies the uniquely colored nematic lines within a given ROI by connected component analysis and outputs the , , , pixel coordinates of each nematic inside each ROI. The length is then defined as
where is the xy image resolution, and the angle is defined as
The MATLAB script (based on version R2021a with the Image Processing and Statistics and Machine Learning Toolboxes installed) has been deposited at https://github.com/SilasBoyeNissen/song2023.
Western blot for Vang phosphorylation
Wing discs from wandering third‐instar larvae were rapidly dissected and transferred into ice‐cold PBS. Collected tissue was spun for 3 min at 9,000 g and the pellet snap frozen in liquid N2. A total of 20–30 discs of each genotype were collected. To prepare extracts, 50 μl of lysis buffer with (all samples except λ‐phosphatase treatment) or without phosphatase inhibitors (sample for λ‐phosphatase treatment) was added to thawed samples and the tissue lysed by multiple passages through a 27½ G needle and incubated on ice for 30 min. Samples were then spun at 20,000 g for 15 min at 4°C, the supernatant collected, and protein concentration measured by Bradford.
Lysis buffers
| Stock | Lysis buffer (w/ PP inhibitors) | Lysis buffer (w/o PP inhibitors) |
|---|---|---|
| 1 M HEPES, pH 7.5 | 20 mM HEPES pH 7.5 | 20 mM HEPES pH 7.5 |
| 4 M NaCl | 100 mM NaCl | 100 mM NaCl |
| 50 mM EDTA | 1 mM EDTA | 1 mM EDTA |
| Glycerol | 10% Glycerol | 10% Glycerol |
| 200 mM Na3O4V | 1 mM Na3O4V | — |
| 500 mM NaF | 1 mM NaF | — |
| NP‐40 | 1% v/v NP‐40 | 1% v/v NP‐40 |
| 2 M MgCl2 | 1 mM MgCl2 | 1 mM MgCl2 |
| 1,000× LPB/AAE | 1× LPB/AAE | 1× LPB/AAE |
1,000× LPB is 10 mg/ml leupeptin, 10 mg/ml pepstatin A, and 10 mg/ml bestatin dissolved in DMSO.
1,000× AAE is 1 M AEBSF (4‐(2‐aminoethyl)benzenesulfonyl fluoride hydrochloride), 800 μm aprotinin, and 15 mm E‐64 dissolved in DMSO.
For λ‐phosphatase treatment, 2 μl of λ‐protein phosphatase (NEB P0753S) was added and incubated at 37°C for 30 min and then placed on ice. Twenty microgram total protein per lane was mixed with 4× LDS (Invitrogen) 200 mM DTT to a final of 1× LDS 50 mM DTT and incubated at 50°C for 15 min.
Samples were loaded and run in a 7% Tris‐acetate gel (Invitrogen) in tris‐acetate buffer on ice at 45 V. Transfer to a preactivated PVDF membrane was done in 10% MeOH Towbin buffer at 4°C for 70 min at 400 mA constant intensity.
The membrane was cut at approximately 50 KDa. The low‐molecular‐weight portion was blocked in 10% Seablock diluted v/v in TBST (Tris‐buffered saline with 0.1% Tween® 20 detergent) for 30 min, and the piece of membrane with larger‐molecular‐weight proteins in 5% milk diluted w/v in TBST.
Membranes were incubated overnight in primary antibody o/n at 4°C: rabbit ⍺‐Vang (Rawls & Wolff, 2003) diluted 1:1,000 in 5% milk in TBST or rabbit ⍺‐actin (Sigma) diluted 1:2,000 in 10% Seablock in TBST followed by 3× wash in TBST for 10 min each. Membranes were developed in ECL reagent and imaged.
Genotypes for flies in each figure
Figure 1
(Pp1 knockdown): Ptc‐GAL4, UAS‐GFP/+; Tub‐GAL80ts/UAS‐Pp1‐87b RNAi (32414)
(Control): Ptc‐GAL4, UAS‐GFP/+; Tub‐GAL80ts/+
Figure 2
(B, D): Ptc‐GAL4/Tub‐GAL80ts; ActP‐Vang::EYFP, UAS‐myrTomato/UAS‐Pp1‐87b RNAi (32414)
(A, C): Ptc‐GAL4/Tub‐GAL80ts; ActP‐Vang::EYFP, UAS‐myrTomato/+
Figure 3
(Vang and Pk localization): ptc‐GAL4/tubP‐GAL80ts; actP‐vang::EYFP UAS‐myrTomato/UAS‐pp1RNAi
(Fz and Pk localization): ptc‐GAL4 UAS‐RFP/tubP‐GAL80ts; armP‐fz::EGFP/UAS‐pp1RNAi
(Dsh and Fmi localization): ptc‐GAL4 UAS‐RFP/tubP‐GAL80ts; pCas‐dsh::Clover/UAS‐pp1RNAi
Figure 4
(control)
w 1118
GAL80ts/ptc‐GAL4; UAS‐Pp1‐87B RNAi (32414)
GAL80ts/ptc‐GAL4; UAS‐Pp1‐87B RNAi (32414)/fz R52
vang A3 /ptc‐GAL4; UAS‐Pp1‐87B RNAi (32414)/tub‐Gal80ts
Figure 5
ptc‐Gal4/tub‐GAL80ts; UAS‐White RNAi/actP‐vang‐eYFP, UAS‐pp1 RNAi (32414)
ptc‐Gal4/Tub‐GAL80ts; UAS‐Dco RNAi (27719)/actP‐vang‐YFP, UAS‐Pp1‐87B RNAi (32414)
Figure 6
Lane 1: w 1118 ;; 71B‐GAL4/UAS‐white RNAi
Lane 2: w 1118 + λ‐protein phosphatase
Lane 3: w 1118 ; stbm 6
Lane 4: w 1118 ;; 71B‐GAL4/UAS‐Pp1‐87b RNAi (32414)
Lane 5: w 1118 ;; 71B‐GAL4/UAS‐Pp1‐87b RNAi (67911)
Lane 6: w 1118 ;; 71B‐GAL4/UAS‐Pp1::HA
Lane 7: w 1118 ;; 71B‐GAL4/UAS‐pNUTS RNAi (64538)
Lane 8: w 1118 ;; 71B‐GAL4/UAS‐dco RNAi (27719)
Figure 7
w 1118
ptc‐Gal4, UAS‐PNUTS RNAi (64538)
ptc‐Gal4, UAS‐PNUTS RNAi (64538)/vang A3
ptc‐Gal4, UAS‐PNUTS RNAi (64538); fz R52
dsh 1 /+; ptc‐Gal4, UAS‐PNUTS RNAi (64538)
Figure 8
w 1118
ptc‐GAL4, UAS‐PNUTS RNAi (64538); UAS‐white RNAi
ptc‐GAL4, UAS‐PNUTS RNAi (64538); UAS‐dco RNAi (27719)
Figure EV1
(A): vang A3 /vang stbm6
(B): UAS‐Apex‐vang, vang A3 /vang A3 ; 71B‐GAL4/+
Figure EV2
D174‐GAL4; UAS‐Myc::APEX::Vang/Cyo
D174‐GAL4
Figure EV3
(A): w 1118
(B): Tub‐GAL80ts/act‐GAL4; act‐vang‐EYFP, UAS‐Pp1 RNAi/UAS‐white RNAi
(C): Tub‐GAL80ts/act‐GAL4; act‐vang‐EYFP, UAS‐Pp1 RNAi/UAS‐dco RNAi
Figure EV4
ptc‐GAL4/tubP‐GAL80ts; actP‐vang::EYFP UAS‐myrTomato/UAS‐pp1RNAi
Figure EV5
(A): Ptc‐GAL4/+; ActP‐Vang::EYFP, UAS‐myrTomato/+
(B): Ptc‐GAL4/UAS‐PNUTS RNAi (64358); ActP‐Vang::EYFP, UAS‐myrTomato/+
Drosophila stocks not maintained in public repositories are available upon request.
Author contributions
Jeffrey D Axelrod: Conceptualization; supervision; funding acquisition; project administration; writing – review and editing. Norbert Perrimon: Conceptualization; funding acquisition; writing – review and editing. Song Song: Conceptualization; formal analysis; investigation; methodology; writing – original draft; writing – review and editing. Bomsoo Cho: Conceptualization; investigation. Alexis T Weiner: Formal analysis; investigation; writing – review and editing. Silas Boye Nissen: Formal analysis; investigation; methodology. Kaye Suyama: Investigation; methodology. Yanhui Hu: Conceptualization; formal analysis; writing – review and editing. Li He: Investigation. Tanya Svinkina: Investigation. Namrata D Udeshi: Data curation; investigation; writing – review and editing. Steven A Carr: Funding acquisition. Irene Ojeda Naharros: Investigation. Pablo Sanchez Bosch: Investigation.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
This work was funded by NIH R35GM131914 (JDA). NP is an investigator of the Howard Hughes Medical Institute (HHMI). We acknowledge support from NIH Transformative R01 grant 5R01DK121409 (SAC and NP). SBN was supported by the Novo Nordisk Foundation (grant awards NNF20OC0059462 and NNF21CC0073729) and the Stanford Bio‐X Program.
EMBO reports (2023) 24: e56997
Data availability
RNAi screen results: RSVP database (https://www.flyrnai.org/cgi‐bin/RSVP_search.pl). Polarity measurement computer scripts: GitHub (https://github.com/SilasBoyeNissen/song2023).
References
- Adler PN (2012) The frizzled/stan pathway and planar cell polarity in the Drosophila wing. Curr Top Dev Biol 101: 1–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigouy B, Farhadifar R, Staple DB, Sagner A, Roper JC, Julicher F, Eaton S (2010) Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila . Cell 142: 773–786 [DOI] [PubMed] [Google Scholar]
- Amonlirdviman K, Khare NA, Tree DR, Chen WS, Axelrod JD, Tomlin CJ (2005) Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science 307: 423–426 [DOI] [PubMed] [Google Scholar]
- Axelrod JD (2001) Unipolar membrane association of Dishevelled mediates Frizzled planar cell polarity signaling. Genes Dev 15: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D, Lyulcheva E, Alphey L, Hawcroft G (2006) Towards a comprehensive analysis of the protein phosphatase 1 interactome in Drosophila . J Mol Biol 364: 196–212 [DOI] [PubMed] [Google Scholar]
- Bertolotti A (2018) The split protein phosphatase system. Biochem J 475: 3707–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch JA, Chen CL, Perrimon N (2021) Proximity‐dependent labeling methods for proteomic profiling in living cells: an update. Wiley Interdiscip Rev Dev Biol 10: e392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt H, Killilea SD, Lee EY (1974) Activation of phosphorylase phosphatase by a novel procedure: evidence for a regulatory mechanism involving the release of a catalytic subunit from enxyme‐inhibitor complex(es) of higher molecular weight. Biochem Biophys Res Commun 61: 598–604 [DOI] [PubMed] [Google Scholar]
- Butler MT, Wallingford JB (2017) Planar cell polarity in development and disease. Nat Rev Mol Cell Biol 18: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddy J, Wilanowski T, Darido C, Dworkin S, Ting SB, Zhao Q, Rank G, Auden A, Srivastava S, Papenfuss TA et al (2010) Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev Cell 19: 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J, Kim MJ, Goo JH, Collier S, Gubb D, Charlton J, Adler PN, Park WJ (1999) The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 126: 5421–5429 [DOI] [PubMed] [Google Scholar]
- Chen CL, Hu Y, Udeshi ND, Lau TY, Wirtz‐Peitz F, He L, Ting AY, Carr SA, Perrimon N (2015) Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci USA 112: 12093–12098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B, Pierre‐Louis G, Sagner A, Eaton S, Axelrod JD (2015) Clustering and negative feedback by endocytosis in planar cell polarity signaling is modulated by ubiquitinylation of prickle. PLoS Genet 11: e1005259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurciu A, Duncalf L, Jonchere V, Lansdale N, Vasieva O, Glenday P, Rudenko A, Vissi E, Cobbe N, Alphey L et al (2013) PNUTS/PP1 regulates RNAPII‐mediated gene expression and is necessary for developmental growth. PLoS Genet 9: e1003885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collu GM, Jenny A, Gaengel K, Mirkovic I, Chin ML, Weber U, Smith MJ, Mlodzik M (2018) Prickle is phosphorylated by Nemo and targeted for degradation to maintain Prickle/Spiny‐legs isoform balance during planar cell polarity establishment. PLoS Genet 14: e1007391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H (2004) Casein kinase Iepsilon modulates the signaling specificities of dishevelled. Mol Cell Biol 24: 2000–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Greene ND (2010) Genetics and development of neural tube defects. J Pathol 220: 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulat AM, Luu O, Sing A, Zhang L, Wrana JL, McNeill H, Winklbauer R, Angers S (2012) Mink1 regulates beta‐catenin‐independent Wnt signaling via Prickle phosphorylation. Mol Cell Biol 32: 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D, Fuchs E (2008) Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol 10: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane A, Yogev S, Mlodzik M (2005) The apical determinants aPKC and dPatj regulate Frizzled‐dependent planar cell polarity in the Drosophila eye. Cell 121: 621–631 [DOI] [PubMed] [Google Scholar]
- Dombradi V, Axton JM, Barker HM, Cohen PT (1990) Protein phosphatase 1 activity in Drosophila mutants with abnormalities in mitosis and chromosome condensation. FEBS Lett 275: 39–43 [DOI] [PubMed] [Google Scholar]
- Eaton S, Julicher F (2011) Cell flow and tissue polarity patterns. Curr Opin Genet Dev 21: 747–752 [DOI] [PubMed] [Google Scholar]
- Etournay R, Merkel M, Popovic M, Brandl H, Dye NA, Aigouy B, Salbreux G, Eaton S, Julicher F (2016) TissueMiner: a multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. Elife 5: e14334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Hannus M, Mlodzik M, Eaton S (2001) The ankyrin repeat protein Diego mediates Frizzled‐dependent planar polarization. Dev Cell 1: 93–101 [DOI] [PubMed] [Google Scholar]
- Gao ZH, Seeling JM, Hill V, Yochum A, Virshup DM (2002) Casein kinase I phosphorylates and destabilizes the beta‐catenin degradation complex. Proc Natl Acad Sci USA 99: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y et al (2011) Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell 20: 163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock RJ, D'Agostino SL, Pilcher KC, Krieg PA (2005) Wnt11‐R, a protein closely related to mammalian Wnt11, is required for heart morphogenesis in Xenopus . Dev Biol 279: 179–192 [DOI] [PubMed] [Google Scholar]
- Gault WJ, Olguin P, Weber U, Mlodzik M (2012) Drosophila CK1‐gamma, gilgamesh, controls PCP‐mediated morphogenesis through regulation of vesicle trafficking. J Cell Biol 196: 605–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth NR, Shi Z, Kunimoto K, Hung V, Xu AF, Kerr CH, Tiu GC, Oses‐Prieto JA, Salomon‐Shulman REA, Axelrod JD et al (2022) A stem cell roadmap of ribosome heterogeneity reveals a function for RPL10A in mesoderm production. Nat Commun 13: 5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs BC, Damerla RR, Vladar EK, Chatterjee B, Wan Y, Liu X, Cui C, Gabriel GC, Zahid M, Yagi H et al (2016) Prickle1 mutation causes planar cell polarity and directional cell migration defects associated with cardiac outflow tract anomalies and other structural birth defects. Biol Open 5: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D (2011) Principles of planar polarity in animal development. Development 138: 1877–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb D, Green C, Huen D, Coulson D, Johnson G, Tree D, Collier S, Roote J (1999) The balance between isoforms of the prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes Dev 13: 2315–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi JM, Strehl L, Hirota Y, Desoeuvre A, Boutin C, Han YG et al (2010) Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat Cell Biol 12: 341–350 [DOI] [PubMed] [Google Scholar]
- Guo N, Hawkins C, Nathans J (2004) Frizzled6 controls hair patterning in mice. Proc Natl Acad Sci USA 101: 9277–9281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zanetti G, Schekman R (2013) A novel GTP‐binding protein‐adaptor protein complex responsible for export of Vangl2 from the trans Golgi network. Elife 2: e00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannus M, Feiguin F, Heisenberg CP, Eaton S (2002) Planar cell polarization requires Widerborst, a B′ regulatory subunit of protein phosphatase 2A. Development 129: 3493–3503 [DOI] [PubMed] [Google Scholar]
- Harrison C, Shao H, Strutt H, Strutt D (2020) Molecular mechanisms mediating asymmetric subcellular localisation of the core planar polarity pathway proteins. Biochem Soc Trans 48: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DJ, Phillips HM, Chaudhry B (2006) Vang‐like 2 and noncanonical Wnt signaling in outflow tract development. Trends Cardiovasc Med 16: 38–45 [DOI] [PubMed] [Google Scholar]
- Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M (2009) Docking motif‐guided mapping of the interactome of protein phosphatase‐1. Chem Biol 16: 365–371 [DOI] [PubMed] [Google Scholar]
- Hino S, Michiue T, Asashima M, Kikuchi A (2003) Casein kinase I epsilon enhances the binding of Dvl‐1 to Frat‐1 and is essential for Wnt‐3a‐induced accumulation of beta‐catenin. J Biol Chem 278: 14066–14073 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Meunier A, Huang S, Shimozawa T, Yamada O, Kida YS, Inoue M, Ito T, Kato H, Sakaguchi M et al (2010) Planar polarity of multiciliated ependymal cells involves the anterior migration of basal bodies regulated by non‐muscle myosin II. Development 137: 3037–3046 [DOI] [PubMed] [Google Scholar]
- Hu Y, Comjean A, Perkins LA, Perrimon N, Mohr SE (2015) GLAD: an online database of gene list annotation for Drosophila . J Genomics 3: 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AC, Molina‐Pelayo C, Sil P, Hazelett CC, Devenport D, Mlodzik M (2023) A Van Gogh/Vangl tyrosine phosphorylation switch regulates its interaction with core Planar Cell Polarity factors Prickle and Dishevelled. PLoS Genet 19: e1010849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LK, Wu J, Yanfeng WA, Mlodzik M (2016) Frizzled‐induced Van Gogh phosphorylation by CK1epsilon promotes asymmetric localization of core PCP factors in Drosophila . Cell Rep 16: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida M, Hino S, Michiue T, Yamamoto H, Kishida S, Fukui A, Asashima M, Kikuchi A (2001) Synergistic activation of the Wnt signaling pathway by Dvl and casein kinase Iepsilon. J Biol Chem 276: 33147–33155 [DOI] [PubMed] [Google Scholar]
- Klein TJ, Jenny A, Djiane A, Mlodzik M (2006) CKIepsilon/discs overgrown promotes both Wnt‐Fz/beta‐catenin and Fz/PCP signaling in Drosophila . Curr Biol 16: 1337–1343 [DOI] [PubMed] [Google Scholar]
- Klingensmith J, Nusse R, Perrimon N (1994) The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev 8: 118–130 [DOI] [PubMed] [Google Scholar]
- Lee H, Adler PN (2004) The grainy head transcription factor is essential for the function of the frizzled pathway in the Drosophila wing. Mech Dev 121: 37–49 [DOI] [PubMed] [Google Scholar]
- Li J, Han S, Li H, Udeshi ND, Svinkina T, Mani DR, Xu C, Guajardo R, Xie Q, Li T et al (2020) Cell‐surface proteomic profiling in the fly brain uncovers wiring regulators. Cell 180: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Papa A, Katchman AN, Zakharov SI, Roybal D, Hennessey JA, Kushner J, Yang L, Chen BX, Kushnir A et al (2020) Mechanism of adrenergic CaV1.2 stimulation revealed by proximity proteomics. Nature 577: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sipe CW (2016) Developmental regulation of planar cell polarity and hair‐bundle morphogenesis in auditory hair cells: lessons from human and mouse genetics. Wiley Interdiscip Rev Dev Biol 5: 85–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannix KM, Starble RM, Kaufman RS, Cooley L (2019) Proximity labeling reveals novel interactomes in live Drosophila tissue. Development 146: dev176644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RM, Peters JM, Graff JM (2001) The casein kinase I family in Wnt signaling. Dev Biol 235: 388–396 [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Kelley MW (2020) Development and patterning of the cochlea: from convergent extension to planar polarity. Cold Spring Harb Perspect Med 10: a033266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata S, Nakatani J, Herranz‐Perez V, Cheng J, Belinson H, Inubushi T, Snider WD, Garcia‐Verdugo JM, Wynshaw‐Boris A, Alvarez‐Buylla A (2014) Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron 83: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson J, Sharp KA, Matis M, Cho B, Axelrod JD (2014) Prickle/spiny‐legs isoforms control the polarity of the apical microtubule network in planar cell polarity. Development 141: 2866–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Dhawan S, Sokol S, Green JB (2005) Distinct PAR‐1 proteins function in different branches of Wnt signaling during vertebrate development. Dev Cell 8: 829–841 [DOI] [PubMed] [Google Scholar]
- Ossipova O, Kim K, Sokol SY (2015) Planar polarization of Vangl2 in the vertebrate neural plate is controlled by Wnt and Myosin II signaling. Biol Open 4: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang‐Zhou D, Flockhart I, Binari R, Shim HS et al (2015) The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA (1990) In vitro disassembly of the nuclear lamina and M phase‐specific phosphorylation of lamins by cdc2 kinase. Cell 61: 591–602 [DOI] [PubMed] [Google Scholar]
- Peti W, Nairn AC, Page R (2013) Structural basis for protein phosphatase 1 regulation and specificity. FEBS J 280: 596–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HM, Murdoch JN, Chaudhry B, Copp AJ, Henderson DJ (2005) Vangl2 acts via RhoA signaling to regulate polarized cell movements during development of the proximal outflow tract. Circ Res 96: 292–299 [DOI] [PubMed] [Google Scholar]
- Phillips HM, Rhee HJ, Murdoch JN, Hildreth V, Peat JD, Anderson RH, Copp AJ, Chaudhry B, Henderson DJ (2007) Disruption of planar cell polarity signaling results in congenital heart defects and cardiomyopathy attributable to early cardiomyocyte disorganization. Circ Res 101: 137–145 [DOI] [PubMed] [Google Scholar]
- Rawls AS, Wolff T (2003) Strabismus requires Flamingo and Prickle function to regulate tissue polarity in the Drosophila eye. Development 130: 1877–1887 [DOI] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, Merkel M, Aigouy B, Gaebel J, Brankatschk M, Julicher F, Eaton S (2012) Establishment of global patterns of planar polarity during growth of the Drosophila wing epithelium. Curr Biol 22: 1296–1301 [DOI] [PubMed] [Google Scholar]
- Sakanaka C, Leong P, Xu L, Harrison SD, Williams LT (1999) Casein kinase Iepsilon in the Wnt pathway: regulation of beta‐catenin function. Proc Natl Acad Sci USA 96: 12548–12552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N, Reiter JF (2010) Tilting at nodal windmills: planar cell polarity positions cilia to tell left from right. Dev Cell 19: 5–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C, Huang D, Bjorklund M, Bischof J, Yin D, Li R, Wu Y, Zeng R, Wu J, Taipale J et al (2013) Systematic screening of a Drosophila ORF library in vivo uncovers Wnt/Wg pathway components. Dev Cell 25: 207–219 [DOI] [PubMed] [Google Scholar]
- Seifert JR, Mlodzik M (2007) Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet 8: 126–138 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Usui T, Yanagawa S, Takeichi M, Uemura T (2001) Asymmetric colocalization of Flamingo, a seven‐pass transmembrane cadherin, and Dishevelled in planar cell polarization. Curr Biol 11: 859–863 [DOI] [PubMed] [Google Scholar]
- Simons M, Mlodzik M (2008) Planar cell polarity signaling: from fly development to human disease. Annu Rev Genet 42: 517–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Yanfeng WA, Grumolato L, Aaronson SA, Mlodzik M (2010) Abelson family kinases regulate Frizzled planar cell polarity signaling via Dsh phosphorylation. Genes Dev 24: 2157–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Price MA, Strutt D (2006) Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila . Curr Biol 16: 1329–1336 [DOI] [PubMed] [Google Scholar]
- Strutt H, Gamage J, Strutt D (2019) Reciprocal action of Casein Kinase Iepsilon on core planar polarity proteins regulates clustering and asymmetric localisation. Elife 8: e45107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Abramova N, Charlton J, Adler PN (1998) Van Gogh: a new Drosophila tissue polarity gene. Genetics 150: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H, Purcell J, Bennett M, Kansagara D, Syed A, Marsh JL (1994) Dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development 120: 347–360 [DOI] [PubMed] [Google Scholar]
- Tissir F, Qu Y, Montcouquiol M, Zhou L, Komatsu K, Shi D, Fujimori T, Labeau J, Tyteca D, Courtoy P et al (2010) Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat Neurosci 13: 700–707 [DOI] [PubMed] [Google Scholar]
- Tree DR, Ma D, Axelrod JD (2002) A three‐tiered mechanism for regulation of planar cell polarity. Semin Cell Dev Biol 13: 217–224 [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T (1999) Flamingo, a seven‐pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98: 585–595 [DOI] [PubMed] [Google Scholar]
- VanderVorst K, Dreyer CA, Konopelski SE, Lee H, Ho HH, Carraway KL 3rd (2019) Wnt/PCP signaling contribution to carcinoma collective cell migration and metastasis. Cancer Res 79: 1719–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayagam A, Hu Y, Kulkarni M, Roesel C, Sopko R, Mohr SE, Perrimon N (2013) Protein complex‐based analysis framework for high‐throughput data sets. Sci Signal 6: rs5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson CR, Adler PN (1987) Directional non‐cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila . Nature 329: 549–551 [DOI] [PubMed] [Google Scholar]
- Vinson CR, Conover S, Adler PN (1989) A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature 338: 263–264 [DOI] [PubMed] [Google Scholar]
- Wang Y, Badea T, Nathans J (2006a) Order from disorder: self‐organization in mammalian hair patterning. Proc Natl Acad Sci USA 103: 19800–19805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo N, Nathans J (2006b) The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner‐ear sensory hair cells. J Neurosci 26: 2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber U, Mlodzik M (2017) APC/C(Fzr/Cdh1)‐dependent regulation of planar cell polarity establishment via Nek2 kinase acting on dishevelled. Dev Cell 40: 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, Luo L (2001) Drosophila Rho‐associated kinase (Drok) links Frizzled‐mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105: 81–91 [DOI] [PubMed] [Google Scholar]
- Wolff T, Rubin GM (1998) Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila . Development 125: 1149–1159 [DOI] [PubMed] [Google Scholar]
- Yanfeng WA, Berhane H, Mola M, Singh J, Jenny A, Mlodzik M (2011) Functional dissection of phosphorylation of disheveled in Drosophila . Dev Biol 360: 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Garrett L, Feng D, Elliott G, Liu X, Wang N, Wong YM, Choi NT, Yang Y, Gao B (2017) Wnt‐induced Vangl2 phosphorylation is dose‐dependently required for planar cell polarity in mammalian development. Cell Res 27: 1466–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA (2007) Planar polarity and tissue morphogenesis. Cell 129: 1051–1063 [DOI] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M (2017) Wnt signaling in cancer. Oncogene 36: 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]


