Abstract
The prevalence of allergic diseases, such as asthma, rhinitis, eczema, and sick building syndrome (SBS), has increased drastically in the past few decades. Current medications can only relieve the symptoms but not cure these diseases whose development is suggested to be greatly impacted by the indoor microbiome. However, no study comprehensively summarizes the progress and general rules in the field, impeding subsequent translational application. To close knowledge gaps between theoretical research and practical application, we conducted a comprehensive literature review to summarize the epidemiological, environmental, and molecular evidence of indoor microbiome studies. Epidemiological evidence shows that the potential protective indoor microorganisms for asthma are mainly from the phyla Actinobacteria and Proteobacteria, and the risk microorganisms are mainly from Bacilli, Clostridia, and Bacteroidia. Due to extremely high microbial diversity and geographic variation, different health-associated species/genera are detected in different regions. Compared with indoor microbial composition, indoor metabolites show more consistent associations with health, including microbial volatile organic compounds (MVOCs), lipopolysaccharides (LPS), indole derivatives, and flavonoids. Therefore, indoor metabolites could be a better indicator than indoor microbial taxa for environmental assessments and health outcome prediction. The interaction between the indoor microbiome and environmental characteristics (surrounding greenness, relative humidity, building confinement, and CO2 concentration) and immunology effects of indoor microorganisms (inflammatory cytokines and pattern recognition receptors) are briefly reviewed to provide new insights for disease prevention and treatment. Widely used tools in indoor microbiome studies are introduced to facilitate standard practice and the precise identification of health-related targets.
Keywords: Indoor microbial metabolite, Asthma, Rhinitis, Eczema, Sick building syndrome, Environmental characteristic
Graphical abstract
1. Introduction
The prevalence of allergic diseases, including asthma, rhinitis, eczema, and sick building syndrome (SBS), has increased drastically since the 1960s [1]. The number of asthma, rhinitis, and eczema patients is estimated to exceed 350 million, 1 billion, and 400 million in the 2010s, respectively, leading to billions of economic and medical costs per year [[2], [3], [4]]. The increasing trend is more pronounced in developing countries than in developed countries. In China, childhood asthma increased from 0.91% in 1999 to 6.8% in 2010, indicating a six-fold increase in 11 years [5]. The prevalence of self-reported allergic rhinitis rose from 11.1% in 2005 to 17.7% in 2011, increasing by 59.5% in six years [6]. In Shanghai, the prevalence of childhood eczema increased from 0.7% in 2000 to 8.3% in 2012, representing a 12-fold increase in 12 years [7]. The prevalence of mucosal and general symptoms of SBS in junior high schools was 22.7% and 20.4%, respectively, in Taiyuan, China [8].
Currently, there is no cure for these diseases. Corticosteroids are the primary medication for asthma and rhinitis, but they only serve as symptom relievers, not the cure. Therefore, new disease treatment or prevention strategies require further exploration. Epidemiological studies reported that many personal and environmental factors are associated with the onset and development of asthma, rhinitis, and eczema, including genetic predisposition, preterm delivery, allergen exposure, air pollution, environmental tobacco smoke, and dietary supplement [[9], [10], [11]]. Efforts have been made to translate these epidemiological progressions into practical prevention strategies. For example, the smoking ban in public places significantly reduced asthma-related hospitalization in Finland [12,13]. In Denmark, dietary supplementation of vitamin D3 and long-chain polyunsaturated fatty acid (fish oil) to pregnant mothers reduced the asthma risk for children [14,15].
Recent studies reported that the indoor microbiome played a critical role in the onset and development of allergic diseases, providing a new promising strategy for disease prevention. Modern humans spend 90% of their time indoors, and the percentage goes even higher during the COVID-19 pandemic [16,17]. Indoor occupants inhale millions of microorganisms and microbial metabolites per hour from this environment, significantly impacting their health [18,19]. The importance of indoor microbiome is well-demonstrated in the “farm effects”. The prevalence of asthma, rhinitis, and SBS is significantly lower in farm and rural regions than in urban regions [20,21]. Subsequent studies show that indoor microbiome diversity and composition is the primary factor shaping the “farm effect” [21,22]. Many studies reported asthma- and rhinitis-related indoor microorganisms in various indoor environments across different geographic regions [[23], [24], [25]]. Studies in molecular immunology also confirmed the role of indoor microorganisms in disease occurrence and development [26].
Although progress has been made, it is still challenging to translate theoretical advances into prevention strategies, and several issues need to be addressed. First, a general epidemiological pattern between the indoor microbiome and disease needs to be established. Many studies reported the health-related indoor microorganisms in different countries and cities. However, no general pattern was summarized from these surveys. It is, therefore, unclear whether some health-related indoor microorganisms are widely distributed or restricted in a local region. Also, it is unclear whether the protective or risk microorganisms are mainly restricted in specific phylogenetic lineage or widely distributed in the whole phylogenetic tree. Reviewing previous studies can establish a catalog of protective and risk microorganisms and metabolites, which is useful for environmental assessment and health outcome prediction. Second, environmental factors affecting indoor microbiome diversity, composition, and abundance have not been comprehensively summarized. Accurate identification of environmental characteristics can provide potential intervention strategies for allergic diseases. Environmental characteristics can thus be controlled at an appropriate level for the colonization of the beneficial microbiome indoors. Third, the molecular target and mechanism of the indoor microorganisms should be uncovered to provide candidate microorganisms and targets for disease treatment. Answering these questions will offer new insights and strategies for environmental assessment and intervention, health outcome prediction, and disease treatment, promoting translational application and benefiting human health.
To close knowledge gaps between indoor microbiome research and translational application, we conducted a comprehensive literature search and interpretation and summarized epidemiological, environmental, molecular, and bioinformatic progress in the indoor microbiome area. This review aims to summarize general rules to answer the four questions proposed above and inspire new insights into disease prevention and treatment strategies.
2. Epidemiological evidence of indoor microbiome and allergic diseases
A primary object of the indoor microbiome study is to characterize the major protective and risk microbial species and metabolites, facilitating environmental assessment and disease prevention. The following subsections summarize the epidemiology evidence on microbial diversity, composition, and metabolites for multiple allergic diseases, including asthma, rhinitis, dermatitis, and SBS.
2.1. Indoor microbial load and allergic diseases
Previous studies mainly used the cultivation approach or quantitative PCR (qPCR) to assess the overall load of indoor fungi and bacteria. The total fungal load was commonly reported as a risk factor for allergic diseases. Epidemic studies in schools reported that higher levels of indoor airborne mold were associated with rhinitis and asthma development in students [27,28] and nasal mucosal inflammation and inflammatory biomarkers in nasal lavage in teachers [29]. Home studies in adults reported that total airborne mold was associated with asthma and atopy [30,31]. Besides the airborne mold, the total fungi and mold species in dust in homes, schools, and daycare centers are also quantified [8,[32], [33], [34]]. The total fungal DNA level in indoor dust was consistently associated with airway inflammations in children in school studies, including asthma symptoms, lower lung function, and elevated fractional exhaled nitric oxide, a marker of allergic airway inflammation. The total fungal DNA level in dust was also associated with SBS symptoms, including ocular and nasal symptoms and tiredness [35]. Higher absolute concentrations of Aspergillus sp., Penicillium sp., and Aspergillus versicolor in school dust increase the risk of airway inflammation in students [36].
The health effect of the total bacterial load was not consistently concluded. The total airborne bacteria was observed to be greater in homes of asthmatic adults [30] but was protectively associated with nasal mucosal inflammation and level of an eosinophilic inflammatory marker in teachers in a school study [29].
2.2. Indoor microbial richness and allergic diseases
The effect of overall indoor microbial richness on asthma, rhinitis, and eczema is controversial. “Hygiene hypothesis” proposes that higher indoor microbial diversity protects against allergic diseases. The hypothesis was first proposed by Strachan [37]. He found significantly less allergic sensitization and allergic rhinitis (hay fever) in subjects with many siblings than in subjects with no siblings and suggested that children in large families were protected against allergic sensitization. The “hygiene hypothesis” was subsequently framed as the “diversity hypothesis,” which proposed that early childhood exposure to diverse microorganisms might protect against allergic diseases by contributing to the maturation of the immune system [[38], [39], [40]]. The “diversity hypothesis” has been tested in several cohort studies, and for comparative reasons, we only presented below a few studies using culture-independent sequencing approaches and the number of observed microbial operational taxonomic units (OTUs) to represent microbial richness.
Several birth or children cohort studies reported that a higher indoor microbial richness protected against asthma development. For example, Ege et al. collected the home dust from dairy farms and adjacent non-farm areas in Germany and Finland. They reported that indoor microbial richness in childhood was associated with a lower risk of asthma development [22]. Similarly, two birth-cohort studies in Finland reported that a high home bacterial richness in early life reduced the risk of asthma and allergic rhinitis [41,42]. However, several studies did not support the protective effects of high microbial richness. A study in the USA reported a positive association between household bacterial richness and asthma severity among children [43]. Another study in Finland and Germany reported that high bacterial diversity of home exposure decreased the sensitization to inhalant allergens in urban populations but increased the risk in rural populations [42]. Therefore, it is still not conclusive whether early childhood exposure to diverse indoor microorganisms could provide beneficial health effects to children.
The “diversity hypothesis” has also been tested in adult populations. A study in Taiyuan, China, reported that the indoor microbial richness in taxonomic class Clostridia was positively associated with asthma prevalence, and the richness in class Alphaproteobacteria, Actinobacteria, and Cyanobacteria was protectively (negatively) associated with asthma prevalence [44]. These results suggested that the richness in specific taxonomic classes rather than the whole microbial community is important for disease outcomes. However, it is unclear whether the pattern could be generalized to a broader population or whether exposure to these microbial classes in early childhood could have the same health effects on children as on adults. The timing of indoor microbial exposure seems to have a large effect on the onset of disease [20]. More studies are needed to clarify the question.
2.3. Indoor microbial taxa and asthma, rhinitis, and allergy
Characterizing health-associated indoor microbial taxa is important for environmental assessment and translational application. Traditional culture-dependent or low-throughput sequencing characterized a few beneficial and risk microorganisms for asthma and allergy, including Staphylococcus sciuri, Acinetobacter lwoffii, and mold species Aspergillus and Penicillium [26,[45], [46], [47], [48]].
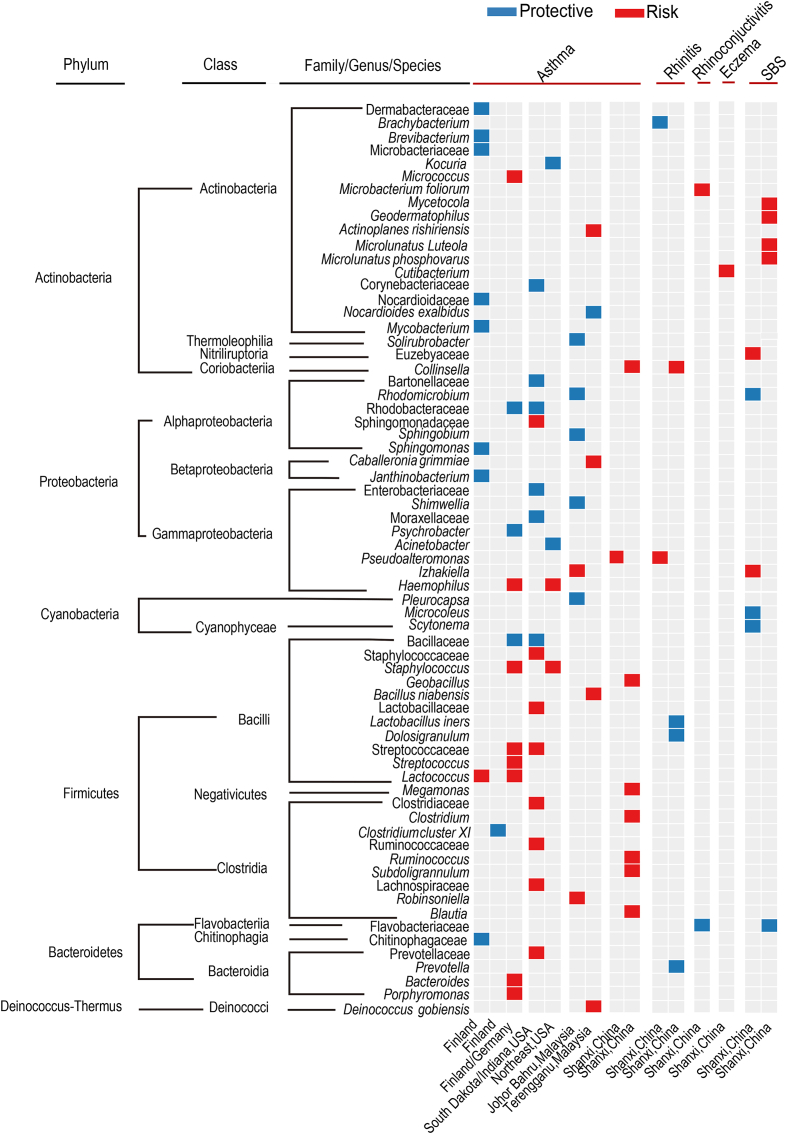
Culture-independent high-throughput sequencing techniques reported a much higher number of health-associated indoor microorganisms. The potential protective and risk health-associated bacterial and fungal species from representative studies are summarized in Fig. 1, Fig. 2. Half of the studies were conducted in the home environment, and a major issue was that the health-associated microorganisms were different among studies and showed strong geographic variation. A study in Germany reported that Acinetobacter, Lactobacillus, Neisseria, Staphylococcus, and Corynebacterium were inversely associated with asthma and allergies [49]. A multicenter case-control study in European adults reported that Clostridium cluster XI was protective for asthma [50]. Two studies in Finland reported different health-related microorganisms. One study showed that Sphingobacteria, Alphaproteobacteria, and Cyanobacteria decreased asthma risk while Streptococcaceae increased asthma risk [23]. Another study reported that the household Lactococcus was a risk factor for asthma, whereas an Actinomycetales cluster was a protective factor [41]. Studies in the USA also reported different health-associated microorganisms. A study in South Dakota and Indiana reported that Corynebacteriaceae, Bacillaceae, Rhodobacteraceae, and Moraxellaceae were negatively associated with asthma, whereas Prevotellaceae, Lactobacillaceae, Clostridiaceae, and Ruminococcaceae were positively associated with asthma [51]. A study in the northeast reported 202 positive and 171 negative associations between microbial taxa and asthma development [52]. Two studies in Connecticut/Massachusetts and California examined the household microbiome of children and reported that fungal Cryptococcus and Volutella were associated with asthma risk [43,53]. Therefore, the health-associated microorganisms showed strong geographic variation, even for studies conducted in the same country.
Fig. 1.
Indoor bacteria taxa associated with asthma, rhinitis, eczema, and SBS. Bacterial taxa positively and negatively associated with diseases and symptoms are coded with red and blue colors, respectively. The health-associated taxa were mainly identified by regression analysis between the abundance of taxa and health outcomes in these selected studies [21,23,25,41,44,[50], [51], [52],54,55,57,84].
Fig. 2.
Indoor fungi and protists associated with asthma and SBS. Fungal taxa positively and negatively associated with diseases and symptoms are coded with red and blue colors, respectively. The health-associated taxa were mainly identified by regression analysis between the abundance of taxa and health outcomes in these selected studies [25,43,53,57,84,96].
A similar variation has been observed in public or multi-occupant indoor environments. Solirubrobacter, Rhodomicrobium, Sphingobium, and Shimwellia were protective taxa for asthma in Johor Bahru, Malaysia [54], whereas Nocardioides exalbidus was protective taxa in Terengganu, Malaysia [55]. A comparative study in three centers of Malaysia reported that the potential protective bacteria were mainly from phylum Actinobacteria in Johor Bahru, Cyanobacteria in Terengganu, and Proteobacteria in Penang, indicating that different regions/cities have different health-related microorganisms [56]. Two studies in Shanxi, China, also reported different health-associated microorganisms. A high school study reported that Brachybacterium sp. P6-10-X1 was a potential protective bacterium for rhinitis, and Psuedoalteromonas was positively associated with asthma and rhinitis [57]. A preschool study reported that Collinsella and Cutibacterium were potential risk bacteria for rhinitis and eczema, and Prevotella, Lactobacillus iners, and Dolosigranulum were potential protective bacteria for rhinitis [25].
Efforts have been made to integrate diversified observations into a general pattern. Fu et al. reported an interesting pattern in university dormitories in Taiyuan, China, stating that the health-associated taxa were consistent at the higher taxonomic level, such as phylum and class levels [44]. In this study, the potential risk microbial taxa for asthma were from the class Bacilli and Clostridia, including Geobacillus, Ruminococcus, Blautia, Clostridium, Subdoligranulum, and Megamonas. The phenomenon is called functional coherence in high bacterial taxonomic rank, which is also observed in Fig. 1 that integrates results from multiple studies. The figure shows that microbial taxa from Firmicutes (Bacilli and Clostridia) and Bacteroidetes are mainly risk factors for asthma, whereas microbial taxa from Alphaproteobacteria and Actinobacteria are protective factors for asthma. Functional coherence has been reported in other ecological studies. For example, the ratio of bacterial phyla Firmicutes/Bacteroidetes and Proteobacteria are widely used markers to assess human gut dysbiosis [58,59]. Therefore, functional coherence presents an interesting hypothesis for indoor microorganisms and health. However, more studies are still needed to confirm the pattern since different observations were reported. For example, Kirjavainen et al. reported that the abundance of Bacteroidetes and Clostridia were protectively associated with asthma [23]. The study by Fu et al. also proposed that the source of the indoor microbiome impacts health outcomes: the Clostridia taxa, including Ruminococcus, Blautia, Clostridium, Subdoligranulum, and Megamonas, were present in high abundance in the human gut and thus might be derived from the human, whereas Alphaproteobacteria and Actinobacteria were mainly derived from the outdoor environment [44]. This claim also needs further verification.
2.4. Indoor microbial metabolites, cell wall compounds, and asthma, rhinitis, and allergies
Most indoor metabolite studies use low-throughput techniques to characterize specific metabolites, including microbial volatile organic compounds (MVOCs), mycotoxin, and microbial cell wall compounds.
MVOCs are risk factors for asthma and allergy. For example, Araki et al. assessed eight MVOCs in diffusive air samples in single-detached homes in six cities and reported that 1-Octen-3-OL concentration was associated with allergic rhinitis and rhinoconjunctivitis [60,61]. A case-control study by Choi et al. reported that a high level of 28 MVOCs and indoor absolute humidity was significantly associated with doctor-diagnosed asthma [62].
Mycotoxins are suggested to be risk factors for respiratory health. Laboratory experiments showed that mycotoxin exposure could modulate gene expression in the asthma pathways and increase inflammatory cytokines in murine models and human airway epithelial cells [63,64]. Also, surveys in Europe showed that settled dust derived from moisture-damaged rooms contained more mycotoxins than un-damaged rooms, posing potential adverse health effects [65,66]. However, the dose remains an important but controversial issue. Kirjavainen et al. surveyed 333 mycotoxins and found that none of the individual mycotoxins were associated with an increased risk of asthma [66]. It is still unclear whether mycotoxin level in the indoor environment is high enough to cause asthma and rhinitis.
Indoor studies also surveyed cell wall compounds of gram-negative bacteria (endotoxins/lipopolysaccharides [LPS]), gram-positive bacteria (muramic acid), and fungi (ergosterol). Most studies reported that the total endotoxin level in home dust was inversely related to asthma, rhinitis, and allergies [22,67]. Endotoxin exposure in early life could reduce subsequent airway responsiveness and sensitization in later life, a phenomenon called endotoxin/LPS tolerance. A meta-analysis study also supported the conclusion [68]. However, the role of specific LPS compounds seems different. Norback et al. reported that C10 3-OH was protectively associated with asthma, and C14 3-OH was positively associated with asthma [32]. No general rule was reported for muramic acid and ergosterol. Muramic acid was reported to be negatively associated with asthma in Austria, Switzerland, Germany, and China [[69], [70], [71]]. However, two studies in Europe reported a positive association between muramic acid and asthma [72,73]. Ergosterol was reported to be positively or negatively associated with doctor-diagnosed asthma and rhinitis [32,33]. β-1,3-glucan, a major constituent of fungal cell walls, was reported as positively, negatively, or null associated with asthma development [[74], [75], [76], [77]].
A recent study using high-throughput untargeted liquid chromatography-mass spectrometry (LC-MS) reported that indoor indole derivatives and flavonoids provide protective effects against asthma and rhinitis. The study profiled 2663 indoor metabolomics/chemicals in junior high school classrooms in three cities of Malaysia and reported that plant secondary metabolites flavonoids and microbial metabolites indole derivatives were protectively associated with asthma (Fig. 3), and synthetic chemicals, including pesticides, fragrance, and detergents, were positively associated with asthma [56]. The potential health-related microorganisms were different among the three centers, but the health-related metabolites/chemicals were largely consistent. The results were confirmed by a home study in Shanghai (unpublished data, Supplementary). The study surveyed homes of 149 children and found that indoles, keto acids (mainly derived from microorganisms), and flavonoids (mainly derived from plants) were protectively associated with asthma and rhinitis, and mycotoxins and synthetic chemicals, including herbicides, insecticides, and food additives were positively associated with asthma and rhinitis. Therefore, indoor metabolites/chemicals showed consistent health effects in studies with different geographic locations and populations, suggesting that it could be a better indicator than the indoor microbiome for environmental and health assessments.
Fig. 3.
The potential protective flavonoids and indole derivatives for asthma and rhinitis. Flavonoids and indole derivatives protectively associated with asthma and rhinitis in Malaysia and Shanghai are coded in blue color. The data were taken from a multicenter association analysis in Malaysia [56] and unpublished data in Shanghai.
2.5. Indoor microbial taxa and sick building syndrome
SBS is defined by World Health Organization (WHO) in 1983 as a group of non-specific symptoms that appear to be linked to time spent in a building, including dry or irritated eyes, runny or blocked nose, dry or sore throat, rashes or itching on the skin, headache, and tiredness [78]. Specifically, several symptoms of SBS are specific allergic symptoms, including itchy eyes, skin rashes, and runny nose, whereas several other symptoms of SBS are non-specific symptoms that may not relate to allergy, including fatigue and headache [79]. Previous epidemiology studies reported that indoor mold and total fungal DNA were risk factors for the new onset of SBS symptoms [8,[80], [81], [82], [83]]. By qPCR, a study in Malaysia reported that Aspergillus versicolor, Streptomyces, and total fungi were associated with SBS symptoms [34].
Two studies used high-throughput sequencing and regression model to disentangle specific microbial taxa associated with SBS symptoms. A study in junior high schools in Johor Bahru, Malaysia, reported that Rhodomicrobium from Alphaproteobacteria and Scytonema and Microcoleus from Cyanobacteria were negatively associated with ocular and throat symptoms and tiredness among students, and Izhakiella from Gammaproteobacteria and Euzebyaceae from Actinobacteria were positively associated with ocular and throat symptoms and tiredness [84]. Izhakiella has also been reported as a potential risk bacterium for asthma [54]. A health association study in urban and rural schools in Shanxi Province, China, reported that Actinobacteria increased SBS risks, including Geodermatophilus, Friedmanniella luteola, Microlunatus phosphovorus, and Mycetocola sp. [21].
Although the number of studies is limited, the association pattern between SBS and asthma seems different from that between SBS and allergy. Actinobacteria is mainly protectively associated with asthma but positively associated with SBS (Fig. 1), indicating different etiological mechanisms between the two diseases.
2.6. Indoor microbial metabolites, cell wall compounds, and SBS symptoms
Indoor mycotoxins and MVOCs are risk factors for SBS. Straus reported that macrocyclic trichothecene mycotoxins (MTM) released from Stachybotrys chartarum were associated with SBS symptoms in buildings [85]. Delmulle et al. reported that 16 mycotoxins were positively associated with SBS, including aflatoxins, roridin, nivalenol, deoxynivalenol, ochratoxin, T-2 toxin, verrucarin, verrucarol, neosolaniol, and sterigmatocystin [86]. Indoor MVOCs, including 2-pentanol, 2-hexanone, 2-pentylfuran, 3-methylfuran, and 1-Octen-3-OL, were reported to be positively associated with SBS symptoms [87,88]. Specifically, 1-Octen-3-OL and 3-methylfuran were associated with mucosal symptoms, such as nasal, throat, or ocular symptoms.
Similar to asthma, the total level of LPS from bacterial cell walls seems to be mainly protective. In two studies, the total LPS level in indoor dust was negatively associated with the incidence of SBS symptoms [8,33]. However, the role of specific LPS compounds is largely unclear. For example, C14 3-OH and C18 3-OH were positively associated with SBS symptoms in a Malaysian school study but negatively associated with SBS symptoms in a Chinese school study [8,33].
Other techniques are also used to infer the health effects of microbial metabolites. By shotgun metagenomics sequencing, Fu et al. reported that the lysine biosynthesis pathways and the relative abundance of biotin, folate, peptidoglycan, gamma-aminobutyric acid (GABA), short-chain fatty acids (SCFAs), and serotonin were protectively associated with SBS symptoms [21]. The protective effects of these metabolites were also reported in the human gut, indicating that indoor exposure could pose a similar health effect as food exposure [[89], [90], [91], [92], [93], [94]]. A limitation of the study is that the metabolic potentials were inferred from the abundance of functional genes; it was unclear whether these metabolites were produced. Thus, future metabolomics studies using mass spectrometry are needed to verify the results.
2.7. Indoor microbiome and respiratory infection
Besides allergic diseases, two studies explored associations between the indoor microbiome and respiratory infections. An interesting but surprising phenomenon is that, similar to asthma, potential protective bacteria for respiratory infections were mainly from taxonomic classes Actinobacteria, Cyanobacteria, and Alphaproteobacteria. A university dormitory study in China reported that the taxa richness (defined as the number of observed OTUs) in Actinobacteria and the abundance of two Actinobacteria genera (Micrococcus and an uncultured Actinobacterium) were protectively associated with respiratory infections. Potential risk genera were opportunistic pathogens mainly from Gammaproteobacteria, including Shinella, Buttiauxella, Haemophilus, Klebsiella, and Raoultella [95]. A junior school study in Malaysia reported 19 potential protective microbial genera for respiratory infections, mainly from Actinobacteria, Alphaproteobacteria, and Cyanobacteria [96]. The implication of this observation is still unclear.
2.8. Summary of health associations for indoor microbiome and metabolites
Studies reported different health-associated genera and species for asthma, rhinitis, and SBS. Several factors could explain the phenomenon. First, the number of environmental microbial species is extremely diverse (>1 trillion) [97]. Many environmental microorganisms could enter the indoor environment by air or other transmission routes. Two surveys in household dust and neonatal intensive care unit reported that >10,000 microbial species were presented in each room [98,99], much higher than in an individual gut (approximately 160 species) [100]. Second, the indoor microbiome shows strong geographic distribution [98,[101], [102], [103]]. Distant geographic regions have different microbial compositions and health-related microorganisms. The drastic indoor microbiome variation is partly due to the large geographic variation of soil and air microbiome. The relative abundance of soil bacterial, fungal, and archaeal taxa from sampling sites a few centimeters apart can vary considerably [104,105]. A continental-scale microbiome survey also revealed that geographic variation is the major factor shaping the air microbiome structure [106]. Third, many environmental characteristics affect the indoor microbiome, including relative humidity, outdoor pollutants, vegetation, and various indoor characteristics (discussed in detail in section 3). The differences in environmental characteristics further lead to microbial variation. Fourth, the various research strategies and tools are used among studies, which could be another reason for the inconsistent results (discussed in section 5). At higher taxonomic levels (phylum and class levels), the health-associated taxa seem to be mainly consistent (Fig. 1). For asthma, the potential protective taxa seem to be mainly from the phylum Actinobacteria and Proteobacteria. Potential risk taxa seem to be mainly from the phylum Firmicutes (class Clostridia and Bacilli) and Bacteroidetes. Also, the association pattern differs between SBS and asthma/rhinitis. Actinobacteria is mainly a protective factor for asthma and rhinitis but a risk factor for SBS. All these observations still need further verifications.
Compared with indoor microbial composition, indoor microbial metabolites show a more consistent pattern with health. For example, mycotoxins and MVOCs are frequently identified as risk factors for asthma, allergy, and SBS symptoms, and total LPS/endotoxin concentration has been largely characterized as a protective factor. A high-throughput untargeted indoor metabolites study also reported consistent health effects of indoles, flavonoids, and synthetic chemicals [56]. Thus, it is tempting to speculate that indoor metabolites could be a better indicator than the indoor microbiome for environment and health assessments. More studies with high-throughput techniques are needed to verify the hypothesis, including targeted or untargeted LC-MS. If the pattern holds true, a catalog of important protective and risk metabolites/chemicals should be established for environmental assessments. A comprehensive and accurate environmental assessment could indicate whether an indoor room is safe or not, providing valuable information for disease prevention.
3. Environmental characteristics and indoor microbiome: potential strategies for disease prevention
Many outdoor and indoor environmental characteristics affect the abundance of protective and risk microorganisms. Disentangling these effects could provide new insights into environmental intervention. Specific environmental characteristics can be used to regulate the abundance of health-related indoor microorganisms at an appropriate level to improve occupants’ health.
The indoor microbial community is influenced by various outdoor environmental characteristics, including latitude, relative humidity, precipitation, geographic characteristics, ambient outdoor pollutant, vegetation, and soil type (Fig. 4). Danko et al. conducted a global-scale metagenomic survey in 60 cities across the globe and reported that city-microbiome were dominated by terrestrial, aquatic taxa, and skin commensals. The global microbiome varied geographically, and each city had its unique microbiome signature [107]. An inter-continental microbial survey in hotel rooms reported that the structure of bacterial and fungal communities varied upon latitude, relative humidity, proximity to the sea, and visible mold indoors [103]. The relative abundance of Aspergillus was inversely correlated with latitude and accounted for 80% of the total fungi in tropical hotels. A study in traditional Japanese houses reported that the microbial concentration on indoor surfaces was positively correlated with the lowest annual relative humidity [108]. Urbanization also impacts indoor microbial composition. A higher percentage of built area, defined as areas covered by industrial, commercial, or residential buildings, is inversely correlated to indoor microbial diversity [109]. The indoor microbiome in a traditional farm differs from an industrialized farm and urban areas [23,51]. Greenness and vegetation around residential buildings impact bacterial and fungal diversity [110]. An intervention study in daycare centers reported that the plantation from forests increased the richness of Proteobacteria in soil and skin and decreased the relative abundance of pathogenic bacteria in the saliva and skin of children [111]. The ambient outdoor air pollutant is also an important factor influencing the diversity and composition of the indoor microbiome. The outdoor NO2, SO2, and PM10 concentrations were associated with the variation of the indoor microbial composition in schools in Malaysia and China [25,55].
Fig. 4.
Associations between environmental characteristics and indoor microbiome. The green and red lines indicate positive and negative associations between environmental characteristics and microorganisms, respectively. The black lines indicate significant associations with microbial composition.
Indoor characteristics and human activities can also affect the composition and diversity of the indoor microbiome (Fig. 4) [112]. Human occupants and pets contribute to the indoor microbiome through sheds and contacts. Lax et al. profiled microbial communities in the doorknob, kitchen counter, light switch, and floor over six weeks and observed a unique microbiome signature for each family [113]. When families moved in, their signature microbiome occupied the new home efficiently and diminished quickly a few days after they left. Keeping dogs and plants increased indoor bacterial diversity and was suggested to be beneficial for human health [44,114]. Indoor characteristics and lifestyles influence the microbial community. Increased building confinement and cleaning are associated with decreased microbial diversity, increased Gram-negative bacteria, and increased antimicrobial resistance genes [115]. Many studies reported that surface materials affect indoor microbiome composition. For example, two studies in Malaysia reported that the indoor microbial community also varies with the age of the building, quality of the interior, type of floor, and indoor surfaces [44,103]. A study in commercial aircraft cabins reported that dry-skin commensals were enriched on textile surfaces, and oily/moist-skin commensals were enriched on leather surfaces [116]. A hotel microbiome study reported that the old or worn furniture and building materials increased the abundance of Saccharopolyspora [103]. Also, microbial composition varied significantly among common building materials under humidity conditions [117]. Surface materials also affect the attachment or transmission of microorganisms, and porous surfaces harbor more microorganisms than smooth surfaces [[118], [119], [120]].
Besides the direct interaction between environmental characteristics and the indoor microbiome, there are more complex interactions between environmental characteristics, indoor microorganisms, and health. In these cases, the health effects of environmental characteristics could be mediated by indoor microorganisms. For instance, the adverse effects of house dust mite allergens on respiratory infection were partly mediated by increasing the abundance of adverse microbial taxa [96]. The potential health effects of outdoor NO2 on rhinitis respiratory infections were fully and inversely mediated by indoor microorganisms [25,96]. Fu et al. observed that the indoor CO2 concentration (representing crowdedness) was negatively associated with Actinoplanes rishiriensis, a potential risk bacteria for asthma, and the weight of vacuumed dust (representing cleanness) was positively associated with Nocardioides exalbidus, a potential protective bacteria for asthma [55]. Moreover, indoor mold or dampness may reduce the relative abundance of potential asthma-protective bacterial genera Rhodomicrobium [54] and increase the abundance of Aspergillus, Eupenidiella, and Exobasidium [103].
Overall, many environmental characteristics can affect indoor microbial diversity and composition. Some environmental characteristics are more feasible to manipulate, such as greenness and vegetation around residential buildings, indoor relative humidity, indoor surface type material, building confinement, and indoor CO2 concentration. For example, indoor Alphaproteobacteria can reduce asthma and rhinitis risks, and Alphaproteobacteria are mainly derived from the outdoor environment. Using biodiverse forest vegetation in daycare centers increased Alphaproteobacterial diversity on children’s skin [111], suggesting more contact with these protective microorganisms. Similar intervention strategies can also be tested in various indoor environments. Studies could also build experimental setups or chambers to manipulate multiple environmental characteristics and find the optimal combination. The optimal environmental characteristics could increase or maintain the homeostasis of beneficial indoor microorganisms while reducing the pathogenic and risk microorganisms along with virulent and antimicrobial resistance genes. Murine models can be used to test the health effects of environmental manipulation. The combined effects of environmental characteristics and indoor microorganisms could be an outstanding solution for allergic diseases.
4. Indoor microbiome and immunological modulation
Molecular immunological studies in the indoor microbiome can pinpoint key molecular markers and pathways for disease development and attenuation, providing valuable targets for disease treatment. Compared to extensive immunological studies in human gut microbiota, the number of indoor microorganisms is limited, but some progress has also been reported in the past few years.
Epigenetic modification is likely to play an important role between indoor microbiome/metabolite and allergic diseases. Environmental exposure impacts gene transcription through DNA methylation or histone methylation, acetylation, and phosphorylation [[121], [122], [123]]. The altered epigenetic status often regulates, up or down, the expression of transcription promoters involved in the activation of immunological pathways. It is strongly believed that the period from pregnancy to the first year of life is an optimal time window for epigenetic alterations related to environmental impact. Current studies mainly investigate the epigenetic mechanisms of food ingestion and gut/intestinal microbiome, and we expect to see more epigenetic studies in the indoor microbiome and metabolites soon.
The crosstalk between indoor microorganisms and inflammatory cytokines is important for disease development. Many studies applied indoor dust from healthy or asthma homes to cell lines and murine models to test the immunological effects. Rich and diverse indoor microbial exposure can train innate and adaptive immune systems by activating regulatory T (Treg) cells and reduce the production of inflammatory cytokines (tumor necrosis factor, TNF; interferon γ, IFNγ), IgE, and type 2 immune responses [20]. For example, the striking disparities in asthma prevalence in the Amish (low asthma) and Hutterite populations (high asthma) were due to the increasing neutrophils and decreasing eosinophils in peripheral blood in Amish children [51]. Distillation of house dust extract from Amish homes to the nasal cavity of laboratory mice reduced peripheral eosinophils, cytokines, airway hyperactivity, and airway inflammation. Similarly, an ex vivo mitogenic stimulation experiment revealed that house dust from healthy children reduced the expression of type 1 immunity-associated cytokines, including IFNγ, interleukin-1β (IL-1β), IL-6, IL-12, and immunoglobulin-like transcript 4 (ILT4) [23]. A European survey of early-life exposure reported that endotoxin levels from children’s mattresses were inversely correlated to TNF-α, IFNγ, IL-10, and IL-12, indicating that diverse bacteria exposure could reduce immune responses in children [67]. Protective indoor microorganisms A. lwoffii F78 and Lactococcus lactis G121, both isolated from farm cowsheds, induced TH1-polarizing cytokines and reduced airway inflammation [47,48]. Also, the indoor microbiome can change the abundance of gut microbiota. Exposure of house dust from low asthma homes to mice increased the Lactobacillus johnsonii, a beneficial microorganism in the gastrointestinal tract, and decreased CD11c+/CD11b+ and CD11c+/CD8+ cells and airway TH2 cytokine expression and airway inflammation [124]. Overall, current investigations propose that early-life exposure to protective farm/beneficial microorganisms can modulate the maturation of the immune system from type 2 to type 1 response and reduce the occurrence of allergic diseases [125].
Besides inflammatory cytokines, the role of pattern recognition receptors, including Toll-like receptors (TLRs) and NOD-like receptors (NLRs), is also explored. Conrad et al. isolated A. lwoffii F78 from the cowsheds in the traditional farm environment and tested its immunological effects in mice [46]. Intranasal distillation of A. lwoffii F78 protected against the development of asthma and airway inflammation in the mother and progeny by regulating the expression of TLR signaling. Stein et al. showed that the endosomal acidification and TLR8 receptor mediated the protective effects of L. lactis G121 [48]. Hagner et al. isolated Staphylococcus sciuri W620 from farm-derived dust [45]. Intranasally exposing the species to mice activated TLR2 and nucleotide-binding oligomerization domain 2 (NOD2) receptors and initiated the human monocyte-derived dendritic cell maturation to prevent airway inflammation.
Current studies suggest that a “good” indoor microbiome can be recognized by pattern recognition receptors, promote immune maturation, balance cytokine production, and reduce asthma and allergy. However, compared to extensive epidemiological studies, the number of molecular immunological studies is limited since most candidate species cannot be cultivated in laboratory conditions. New cultivation methods, such as coculture and cultivation by simulated environments, have been developed in recent years [126,127], which could expand the catalog of culturable microorganisms and facilitate a deeper understanding of health-related indoor microorganisms. Microorganism cultivation can also facilitate uncovering of the health effects of microbial metabolites. The key metabolites for a beneficial microorganism can be characterized by mass spectrometry and molecular immunology. With this progress, future studies could provide more therapeutic targets for disease prevention and treatment.
5. Practices and analyzing tools for indoor microbiome studies
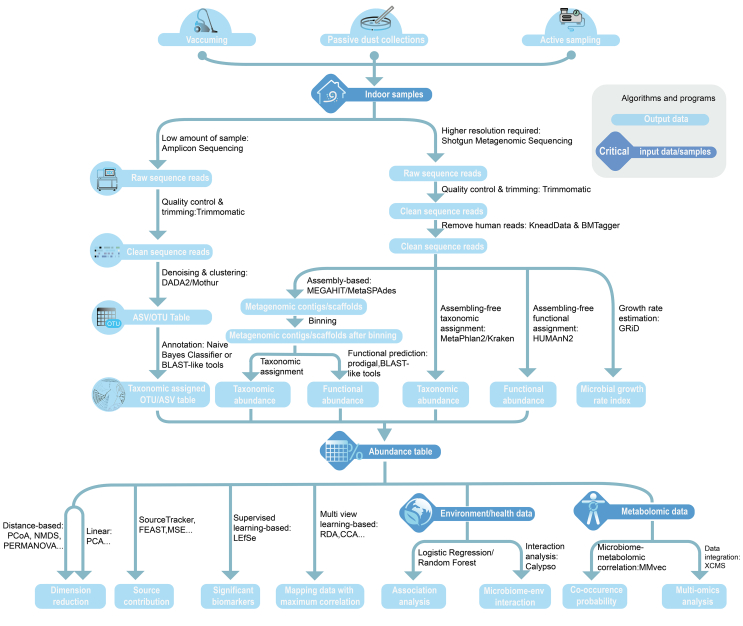
The indoor microbiome is a newly developed area. Many tools and programs have been developed for microbial sampling, data analysis, and health associations [128,129]. As we discussed in the previous section, various research strategies and tools are used among studies, which could be an important reason for the inconsistent results in epidemiological studies. Therefore, widely used tools are introduced in this section to facilitate the standard practice in this research area (Fig. 5).
Fig. 5.
Tools and workflow of indoor microbiome data analysis. Algorithms and programs are shown on lines, critical input data/samples are shown in diamond symbols, and output data are shown in rounded rectangle symbols. ASV, amplicon sequence variants; OTU, operational taxonomic unit; PCA, principal component analysis; RDA, redundancy analysis; CCA, canonical correspondence analysis; PERMANOVA, permutational multivariate analysis of variance.
Several strategies are used to sample indoor microorganisms, including vacuuming, passive dust collectors, and active sampling by air pumps [130,131]. The strengths and limitations of each sampling strategy are summarized in Table 1. Different sampling strategies may impact the analysis and interpretation. The microbial diversity in floor vacuum dust is generally higher than in air dust from passive and active sampling. For example, Yamamoto et al. reported that the fungal richness in vacuumed floor dust was 40% higher than in active sampled air dust [132]. A study on crawling infants reported that the bacterial richness in vacuumed floor dust was higher than in active sampled air dust at the breathing levels of infants and adults [133]. The best practice is to choose an appropriate sampling strategy matching a specific scientific question. For example, vacuum sampling can capture long-term indoor microbial exposure, and thus may be optimal for studying allergic diseases for which it is of great importance. In contrast, active sampling for air dust may be an optimal option for studies aiming to characterize short-term or transient microbial exposure. Multiple sampling strategies can be considered if the aim is to characterize comprehensive indoor microbial exposure.
Table 1.
Strengths and limitations of indoor dust sampling strategies.
| Dust sampling strategies | Sampling details | Strengths | Limitations |
|---|---|---|---|
| Vacuum dust sampling | Collect indoor dust from the floor, mattress, chairs, tables, curtains, bookshelves, and other upper surface dust with a vacuum cleaner |
|
|
| Passive dust sampling by collectors | Settled airborne dust sampling by passive dust collectors such as petri dish, TefTex material (a polytetrafluoroethylene fiber sampling cloth), and electrostatic dustfall collectors (EDCs) |
|
|
| Active airborne dust sampling | Active airborne dust sampling by an air pump or BioSampler |
|
|
Amplicon and shotgun metagenomics sequencing is the most widely used culture-independent sequencing strategy, and their strengths and limitations are summarized in Table 2 [[134], [135], [136]]. A major limitation of amplicon sequencing is functional inference. Bioinformatics tools, such as PICRUSt2 [137], have been developed to predict functional potentials based on taxonomic information, but the approach is inaccurate and relies on reference genomes [138]. Shotgun metagenomic sequencing is a standard approach to characterizing the functional potentials of the indoor microbiome. It can produce much more data than amplicon sequencing (2–20 Gb for shotgun metagenomics versus 5–50 Mb for amplicon sequencing). However, extensive computational resources and time are needed, and cloud computation or supercomputer computation could, therefore, be a potential solution to reduce computational time.
Table 2.
Strengths and limitations of sequencing strategy in the indoor microbiome study.
| Sequencing strategy | Sequencing platform | Sequencing statistics | Strengths | Limitations |
|---|---|---|---|---|
| Amplicon sequencing | Illumina MiSeq, NextSeq | Read length: 2 × 100 bp–2 × 250 bp Sequencing depth: 30,000–100,000 reads |
|
|
| PacBio Sequel, Sequel II, Nanopore MinION | Read length: >1500 bp Sequencing depth: 4000–10,000 reads |
|
1. More expansive than second-generation amplicon sequencing 2. Low sequencing depth and output (limit in detecting low-frequency variants) 3. No accurate functional inference |
|
| Shotgun metagenomics sequencing | Illumina HiSeq, NovaSeq | Read length: 2 × 100 bp–2 × 250 bp Sequencing depth: 20–100 million reads |
|
|
The first step of bioinformatic analysis is to check, trim, and process raw sequence data. It is important for indoor shotgun metagenome studies, as up to 80% of the sequenced reads can be derived from human skin sheddings [116]. The human data can be removed by KneadData and BMTagger. For amplicon data, 16S rRNA genes from chloroplast and mitochondria should also be removed [139]. Afterward, amplicon sequences are clustered and annotated by DADA2 and Deblur [140,141]. The exact sequences are searched against amplicon reference databases, such as Silva, Greengenes, UNITE, and NCBI [[142], [143], [144]]. For metagenomics, the data can be processed first by assembly based tools, such as metaSPAdes or MEGAHIT [145,146], or assembly free tools, such as MetaPhlAn or HUMAnN2 [147,148]. The taxonomic and functional profiles are annotated by public databases, such as NCBI, KEGG, EggNOG, or GO [149,150]. OTU or amplicon sequence variants (ASV) tables containing the number of microbial features are produced after annotation, which are multidimensional structured tables with thousands of rows and columns challenging for human interpretation. Thus, dimensional reduction statistics are needed to present the data in a human-readable output. Bray-Curtis and UniFrac distance are widely used methods to compute microbial community differences [151], and the latter approach integrates phylogenetic information in the distance estimation. Ordination is a common approach to visualizing microbial community and functional variation. Common ordination strategies include unconstrained ordination, including principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS), and constrained ordination, including redundancy analysis (RDA) and canonical correspondence analysis (CCA). Permutational multivariate analysis of variance (PERMANOVA/Adonis) and analysis of similarities (ANOSIM) can provide statistical support for the distance and ordination analysis [152,153].
QIIME and QIIME2 are the most widely used platforms for microbiome data analysis and visualization [154,155]. QIIME supports a wide range of microbial community analysis, including raw data processing, microbial taxonomic profiling, diversity and richness estimation, core microorganism identification, network analysis, habitat inference, and visualization. Third-party plugins in QIIME2 can perform sequence and taxonomic assignment, time-series analysis, machine learning, and multi-omics analysis, including metagenomics, metatranscriptomics, metaproteomics, and metabolomics [155,156]. Besides the two platforms, community efforts have developed an R-based platform for microbiome data exploration and visualization [157].
Many studies used linear or logistic regression models in Stata, SPSS, and R to investigate the relationship between the indoor microbiome and health outcomes [43,54,57,158]. One issue is that a few hundred to thousand microbial taxa need to be tested, which may produce false-positive results, a phenomenon called the multiple comparison problem [159]. Multiple comparison correction methods have been applied, such as Bonferroni and Benjamini-Hochberg procedures, but these methods can be overly conservative and remove true-positive candidates. A new program DS-FDR uses discrete false-discovery rate to increase the detection power and sensitivity of multiple comparison correction [160]. LEfSe and metagenomeSeq are other widely used tools to characterize featured microorganisms and functional genes between two or more sample groups [161,162]. The tools are less stringent than regression and report more candidate microbial taxa and functional genes/pathways. A machine learning anchor-based method was also developed to identify potential protective and risk microbial taxa and clusters [23]. The approach is used to model the indoor microbiome from the farm environment as a training dataset and predict the asthma risks in urban homes.
Multi-omic and multidimensional data analysis is a challenging and promising new research area with only a few tools available. XCMS is an online platform that automatically integrates metabolomic, transcriptomic, and proteomic data onto metabolic pathways [163]. Microbe-metabolite vectors (mmvec) is a newly developed neural network program to infer the co-occurrence between microbial taxa and metabolites [164]. The program uses neural networks to calculate the probability that a metabolic molecule is related to a specific microorganism. SourceTracker and FEAST are tools to analyze the compositional structure of microbiome data and identify the potential sources by Bayesian and expectation-maximization algorithms [165,166]. Calypso is a user-friendly web server to mine and visualize microbiome-environment interaction [167]. SparseMCMM is an R function that disentangles causal mediation effects within the high-dimensional data [168].
Many other tools are also widely used in indoor microbiome studies. GRiD is a computational tool to estimate microbial growth rate (replication rate) in shotgun metagenomics data with ultra-low coverage (0.2×) [169], precisely fitting for the low reads coverage in indoor microbiome studies. PathoFact is a newly developed pipeline to calculate the abundance of virulence factors, bacterial toxins, and antimicrobial resistance genes in metagenomic data [170], estimating the microbial pathogenic potentials in the indoor environment. Microbiome search engine (MSE) searches query microbiome samples against a public database with 346,877 microbiome samples and reports a list of highly similar microbiome samples [171,172]. However, due to the low taxonomic resolution of 16S rRNA, MSE could produce high similarity scores among samples from different habitats, and thus the results should be treated with caution [173].
Besides computational tools, many online databases are useful for indoor microbiome and metabolomics data analysis and interpretation, including Comprehensive Antibiotic Resistance Database (CARD), Virulence Factor Database (VFDB), Toxin and Toxin Target Database (T3DB), World Urban Database (WUDAPT), Global Burden of Disease Database (GHDx), PubChem, and METLIN [2,[174], [175], [176], [177], [178], [179], [180], [181]]
Many algorithms and tools have been developed in microbiome data analysis. However, there are still limitations and gaps in the high-level analysis. Disentangling multi-omic and multidimensional data is still challenging. For example, current studies use correlation or machine learning approaches to infer the association or co-occurrence probability between each microorganism and metabolite, which only considers the abundance of the two subjects. Other factors, such as overall microbial composition/diversity and environmental characteristics, can impact the interactions and should be integrated into the statistical models. Mediation analysis is a common approach to disentangle the interactions among environmental characteristics, the indoor microbiome, and health. However, the correct application of mediation analysis requires several assumptions, including the correct specification of causal order and direction, which can be problematic in multilevel and multivariate mediation models [182]. Thus, new computational tools or modeling are needed to identify critical health-related and therapeutical targets for allergic diseases.
6. Conclusion and future perspectives
In this review, we summarized major progress in the indoor microbiome field in the past few years. First, a general rule of the indoor microbiome and metabolites has been summarized from epidemiological studies. Different health-related microorganisms have been reported in different studies due to the extremely high diversity of indoor microorganisms and large geographic variation. Thus, it is challenging to establish a universal catalog of protective and risk microorganisms for environmental assessment and health outcome prediction. Indoor microbial metabolites show a more consistent pattern with health, which could be a better indicator than the indoor microbiome for environment and health assessments. Second, the interaction between the indoor microbiome and environmental characteristics and immunology effects of indoor microorganisms is briefly reviewed. Third, widely used tools in indoor microbiome studies are introduced to facilitate standard practice in indoor microbiome research.
The current preventive strategies are mostly regulations of individual behaviors and mainly for childhood asthma, including smoking bans and nutrient supplementation during pregnancy. Smoking bans and legislation in public indoor environments have reduced hospital admission rates for childhood asthma [183,184]. For pregnant women with a family history of asthma and allergies, supplementing Vitamin D3 and fish oil during pregnancy reduces the risk of wheeze and respiratory tract infections at pre-school age, but not asthma, eczema, and allergic sensitization [15]. Indoor microbiome research can provide more intervention strategies for disease control and prevention (Fig. 6). The most extensively studied microbial intervention strategy are prebiotics and probiotics. However, the preventive effects of these microorganisms and microbial substrates on allergic diseases are still not conclusive. Some studies showed that there was not enough evidence to support the protective role of probiotics in allergic rhinitis, while other studies showed probiotics had a preventive role in allergic rhinitis [185,186]. Indoor and environmental microbiome surveys can provide a broader catalog of beneficial microorganisms and explore a better combination of microorganisms and substrates. Besides oral supplementation, these microorganisms and microbial substrates may have the potential to be used in the indoor environment. For example, the aerosol spray with beneficial microorganisms and metabolites can be installed in the mechanical ventilation or air conditioning system, reducing the risk of allergic diseases. Also, environmental characteristics, such as greenness and vegetation, relative humidity, indoor surface material, building confinement, and CO2 concentration, could be set at an optimal level to maintain “a healthy indoor microbiome” and promote the health of occupants. In addition, an indoor microbiome/metabolites index for environmental assessment and health outcome prediction can be established to predict whether an indoor space is healthy or risky for occupants. These new disease prevention and treatment options have outstanding application potential to improve human life quality.
Fig. 6.
Future directions and perspectives in the indoor microbiome field. Comprehensive research will facilitate translational application.
Author contributions
Xi Fu: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. Zheyuan Ou: Data curation, Methodology, Visualization. Yu Sun: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors have declared no conflicts of interests.
Acknowledgments
This work was supported by the Natural Science Foundation of Guangdong Province (2020A1515010845 and 2021A1515010492) and the Science and Technology Program of Guangzhou (202102080362).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eehl.2022.09.002.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Eder W., Ege M.J., von Mutius E. The asthma epidemic. N. Engl. J. Med. 2006;355(21):2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Stanaway J.D., Afshin A., Gakidou E., Lim S.S., Abate D., Abate K.H., et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1923–1994. doi: 10.1016/S0140-6736(18)32225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savouré M., Bousquet J., Jaakkola J.J.K., Jaakkola M.S., Jacquemin B., Nadif R. Worldwide prevalence of rhinitis in adults: a review of definitions and temporal evolution. Clin. Transl. Allergy. 2022;12(3) doi: 10.1002/clt2.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanifin J.M., Reed M.L. A population-based survey of eczema prevalence in the United States. Dermatitis : contact, atopic, occupational. Drug. 2007;18(2):82–91. doi: 10.2310/6620.2007.06034. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Li B., Huang C., Yang X., Qian H., Deng Q., et al. Ten cities cross-sectional questionnaire survey of children asthma and other allergies in China. Chin. Sci. Bull. 2013;58(34):4182–4189. [Google Scholar]
- 6.Wang X.D., Zheng M., Lou H.F., Wang C.S., Zhang Y., Bo M.Y., et al. An increased prevalence of self-reported allergic rhinitis in major Chinese cities from 2005 to 2011. Allergy. 2016;71(8):1170–1180. doi: 10.1111/all.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu F., Yan S., Li F., Cai M., Chai W., Wu M., et al. Prevalence of childhood atopic dermatitis: an urban and rural community-based study in Shanghai, China. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X., Zhao Z., Nordquist T., Larsson L., Sebastian A., Norback D. A longitudinal study of sick building syndrome among pupils in relation to microbial components in dust in schools in China. Sci. Total Environ. 2011;409(24):5253–5259. doi: 10.1016/j.scitotenv.2011.08.059. [DOI] [PubMed] [Google Scholar]
- 9.Castro-Rodriguez J.A., Forno E., Rodriguez-Martinez C.E., Celedon J.C. Risk and protective factors for childhood asthma: what is the evidence? J. Allergy Clin. Immunol. Pract. 2016;4(6):1111–1122. doi: 10.1016/j.jaip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunlop J., Matsui E., Sharma H.P. Allergic rhinitis: environmental determinants. Immunol. Allergy Clin. 2016;36(2):367–377. doi: 10.1016/j.iac.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Hellings P.W., Klimek L., Cingi C., Agache I., Akdis C., Bachert C., et al. Non-allergic rhinitis: position paper of the European academy of allergy and clinical immunology. Allergy. 2017;72(11):1657–1665. doi: 10.1111/all.13200. [DOI] [PubMed] [Google Scholar]
- 12.Burki T.K. Asthma control: learning from Finland’s success. Lancet Respir. Med. 2019;7(3):207–208. doi: 10.1016/S2213-2600(19)30030-X. [DOI] [PubMed] [Google Scholar]
- 13.Haahtela T., Valovirta E., Bousquet J., Mäkelä M. The Finnish allergy programme 2008-2018 works. Eur. Respir. J. 2017;49(6) doi: 10.1183/13993003.00470-2017. [DOI] [PubMed] [Google Scholar]
- 14.Chawes B.L., Bønnelykke K., Stokholm J., Vissing N.H., Bjarnadóttir E., Schoos A.M., et al. Effect of vitamin D3 supplementation during pregnancy on risk of persistent wheeze in the offspring: a randomized clinical trial. JAMA. 2016;315(4):353–361. doi: 10.1001/jama.2015.18318. [DOI] [PubMed] [Google Scholar]
- 15.Bisgaard H., Stokholm J., Chawes B.L., Vissing N.H., Bjarnadóttir E., Schoos A.M., et al. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in offspring. N. Engl. J. Med. 2016;375(26):2530–2539. doi: 10.1056/NEJMoa1503734. [DOI] [PubMed] [Google Scholar]
- 16.Klepeis N.E., Nelson W.C., Ott W.R., Robinson J.P., Tsang A.M., Switzer P., et al. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. J. Expo. Anal. Environ. Epidemiol. 2001;11(3):231–252. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- 17.Weerakoon S.M., Jetelina K.K., Knell G. Longer time spent at home during COVID-19 pandemic is associated with binge drinking among US adults. Am. J. Drug Alcohol Abuse. 2021;47(1):98–106. doi: 10.1080/00952990.2020.1832508. [DOI] [PubMed] [Google Scholar]
- 18.Oh H.J., Ma Y., Kim J. Human inhalation exposure to aerosol and health effect: aerosol monitoring and modelling regional deposited doses. Int. J. Environ. Res. Publ. Health. 2020;17(6) doi: 10.3390/ijerph17061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian J., Hospodsky D., Yamamoto N., Nazaroff W.W., Peccia J. Size-resolved emission rates of airborne bacteria and fungi in an occupied classroom. Indoor Air. 2012;22(4):339–351. doi: 10.1111/j.1600-0668.2012.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Mutius E., Vercelli D. Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol. 2010;10:861. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 21.Fu X., Ou Z., Zhang M., Meng Y., Li Y., Chen Q., et al. Classroom microbiome, functional pathways and sick-building syndrome (SBS) in urban and rural schools - potential roles of indoor microbial amino acids and vitamin metabolites. Sci. Total Environ. 2021;795 doi: 10.1016/j.scitotenv.2021.148879. [DOI] [PubMed] [Google Scholar]
- 22.Ege M.J., Mayer M., Normand A.C., Genuneit J., Cookson W.O., Braun-Fahrlander C., et al. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 2011;364(8):701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 23.Kirjavainen P.V., Karvonen A.M., Adams R.I., Täubel M., Roponen M., Tuoresmäki P., et al. Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat. Med. 2019;25(7):1089–1095. doi: 10.1038/s41591-019-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dannemiller K.C., Gent J.F., Leaderer B.P., Peccia J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air. 2016;26(2):179–192. doi: 10.1111/ina.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y., Meng Y., Ou Z., Li Y., Zhang M., Chen Y., et al. Indoor microbiome, air pollutants and asthma, rhinitis and eczema in preschool children–a repeated cross-sectional study. Environ. Int. 2022;161 doi: 10.1016/j.envint.2022.107137. [DOI] [PubMed] [Google Scholar]
- 26.Renz H., Skevaki C. Early life microbial exposures and allergy risks: opportunities for prevention. Nat. Rev. Immunol. 2021;21(3):177–191. doi: 10.1038/s41577-020-00420-y. [DOI] [PubMed] [Google Scholar]
- 27.Simoni M., Cai G.H., Norback D., Annesi-Maesano I., Lavaud F., Sigsgaard T., et al. Total viable molds and fungal DNA in classrooms and association with respiratory health and pulmonary function of European schoolchildren. Pediatr. Allergy Immunol. 2011;22(8):843–852. doi: 10.1111/j.1399-3038.2011.01208.x. official publication of the European Society of Pediatric Allergy and Immunology. [DOI] [PubMed] [Google Scholar]
- 28.Smedje G., Norbäck D. Incidence of asthma diagnosis and self-reported allergy in relation to the school environment--a four-year follow-up study in schoolchildren. Int. J. Tubercul. Lung Dis.: the official journal of the International Union against Tuberculosis and Lung Disease. 2001;5(11):1059–1066. [PubMed] [Google Scholar]
- 29.Norbäck D., Wålinder R., Wieslander G., Smedje G., Erwall C., Venge P. Indoor air pollutants in schools: nasal patency and biomarkers in nasal lavage. Allergy. 2000;55(2):163–170. doi: 10.1034/j.1398-9995.2000.00353.x. [DOI] [PubMed] [Google Scholar]
- 30.Björnsson E., Norbäck D., Janson C., Widström J., Palmgren U., Ström G., et al. Asthmatic symptoms and indoor levels of micro-organisms and house dust mites. Clin. Exp. Allergy: J British Society for Allergy and Clinical Immunology. 1995;25(5):423–431. doi: 10.1111/j.1365-2222.1995.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 31.Matheson M.C., Abramson M.J., Dharmage S.C., Forbes A.B., Raven J.M., Thien F.C.K., et al. Changes in indoor allergen and fungal levels predict changes in asthma activity among young adults. Clin. Exp. Allergy. 2005;35(7):907–913. doi: 10.1111/j.1365-2222.2005.02272.x. [DOI] [PubMed] [Google Scholar]
- 32.Norbäck D., Markowicz P., Cai G.-H., Hashim Z., Ali F., Zheng Y.-W., et al. Endotoxin, ergosterol, fungal DNA and allergens in dust from schools in Johor Bahru, Malaysia- associations with asthma and respiratory infections in pupils. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0088303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norbäck D., Hashim J.H., Markowicz P., Cai G.-H., Hashim Z., Ali F., et al. Endotoxin, ergosterol, muramic acid and fungal DNA in dust from schools in Johor Bahru, Malaysia — associations with rhinitis and sick building syndrome (SBS) in junior high school students. Sci. Total Environ. 2016;545-546:95–103. doi: 10.1016/j.scitotenv.2015.12.072. [DOI] [PubMed] [Google Scholar]
- 34.Norback D., Hashim J.H., Cai G.H., Hashim Z., Ali F., Bloom E., et al. Rhinitis, ocular, throat and dermal symptoms, headache and tiredness among students in schools from Johor Bahru, Malaysia: associations with fungal DNA and mycotoxins in classroom dust. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0147996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norback D. An update on sick building syndrome. Curr. Opin. Allergy Clin. Immunol. 2009;9(1):55–59. doi: 10.1097/ACI.0b013e32831f8f08. [DOI] [PubMed] [Google Scholar]
- 36.Norbäck D., Cai G.H., Kreft I., Lampa E., Wieslander G. Fungal DNA in dust in Swedish day care centres: associations with respiratory symptoms, fractional exhaled nitrogen oxide (FeNO) and C-reactive protein (CRP) in serum among day care centre staff. Int. Arch. Occup. Environ. Health. 2016;89(2):331–340. doi: 10.1007/s00420-015-1076-4. [DOI] [PubMed] [Google Scholar]
- 37.Strachan D.P. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garn H., Potaczek D.P., Pfefferle P.I. The hygiene hypothesis and new perspectives-current challenges meeting an old postulate. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.637087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Mutius E. The "hygiene hypothesis" and the lessons learnt from farm studies. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.635522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfefferle P.I., Keber C.U., Cohen R.M., Garn H. The hygiene hypothesis - learning from but not living in the past. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.635935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karvonen A.M., Kirjavainen P.V., Taubel M., Jayaprakash B., Adams R.I., Sordillo J.E., et al. Indoor bacterial microbiota and development of asthma by 10.5 years of age. J. Allergy Clin. Immunol. 2019;144(5):1402–1410. doi: 10.1016/j.jaci.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 42.Hyytiäinen H., Kirjavainen P.V., Täubel M., Tuoresmäki P., Casas L., Heinrich J., et al. Microbial diversity in homes and the risk of allergic rhinitis and inhalant atopy in two European birth cohorts. Environ. Res. 2021 doi: 10.1016/j.envres.2021.110835. [DOI] [PubMed] [Google Scholar]
- 43.Dannemiller K.C., Gent J.F., Leaderer B.P., Peccia J. Indoor microbial communities: influence on asthma severity in atopic and nonatopic children. J. Allergy Clin. Immunol. 2016;138(1):76–83. doi: 10.1016/j.jaci.2015.11.027. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu X., Li Y., Meng Y., Yuan Q., Zhang Z., Wen H., et al. Derived habitats of indoor microbes are associated with asthma symptoms in Chinese university dormitories. Environ. Res. 2021;194 doi: 10.1016/j.envres.2020.110501. [DOI] [PubMed] [Google Scholar]
- 45.Hagner S., Harb H., Zhao M., Stein K., Holst O., Ege M.J., et al. Farm-derived Gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy. 2013;68(3):322–329. doi: 10.1111/all.12094. [DOI] [PubMed] [Google Scholar]
- 46.Conrad M.L., Ferstl R., Teich R., Brand S., Blümer N., Yildirim A.Ö., et al. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J. Exp. Med. 2009;206(13):2869–2877. doi: 10.1084/jem.20090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Debarry J., Garn H., Hanuszkiewicz A., Dickgreber N., Blümer N., von Mutius E., et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J. Allergy Clin. Immunol. 2007;119(6):1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 48.Stein K., Brand S., Jenckel A., Sigmund A., Chen Z.J., Kirschning C.J., et al. Endosomal recognition of Lactococcus lactis G121 and its RNA by dendritic cells is key to its allergy-protective effects. J. Allergy Clin. Immunol. 2017;139(2):667–678. doi: 10.1016/j.jaci.2016.06.018. e5. [DOI] [PubMed] [Google Scholar]
- 49.Ege M.J., Mayer M., Schwaiger K., Mattes J., Pershagen G., van Hage M., et al. Environmental bacteria and childhood asthma. Allergy. 2012;67(12):1565–1571. doi: 10.1111/all.12028. [DOI] [PubMed] [Google Scholar]
- 50.Pekkanen J., Valkonen M., Taubel M., Tischer C., Leppanen H., Karkkainen P.M., et al. Indoor bacteria and asthma in adults: a multicentre case-control study within ECRHS II. Eur. Respir. J. 2018;51(2) doi: 10.1183/13993003.01241-2017. [DOI] [PubMed] [Google Scholar]
- 51.Stein M.M., Hrusch C.L., Gozdz J., Igartua C., Pivniouk V., Murray S.E., et al. Innate immunity and asthma risk in amish and hutterite farm children. N. Engl. J. Med. 2016;375(5):411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connor G.T., Lynch S.V., Bloomberg G.R., Kattan M., Wood R.A., Gergen P.J., et al. Early-life home environment and risk of asthma among inner-city children. J. Allergy Clin. Immunol. 2018;141(4):1468–1475. doi: 10.1016/j.jaci.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dannemiller K.C., Mendell M.J., Macher J.M., Kumagai K., Bradman A., Holland N., et al. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air. 2014;24(3):236–247. doi: 10.1111/ina.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu X., Norbäck D., Yuan Q., Li Y., Zhu X., Hashim J.H., et al. Indoor microbiome, environmental characteristics and asthma among junior high school students in Johor Bahru, Malaysia. Environ. Int. 2020;138 doi: 10.1016/j.envint.2020.105664. [DOI] [PubMed] [Google Scholar]
- 55.Fu X., Meng Y., Li Y., Zhu X., Yuan Q., Ma’pol A., et al. Air Quality, Atmosphere & Health; 2021. Associations between Species-Level Indoor Microbiome, Environmental Characteristics, and Asthma in Junior High Schools of Terengganu, Malaysia. [Google Scholar]
- 56.Sun Y., Zhang M., Ou Z., Meng Y., Chen Y., Lin R., et al. Indoor microbiome, microbial and plant metabolites, chemical compounds and asthma symptoms in junior high school students: a multicentre association study in Malaysia. Eur. Respir. J. 2022 doi: 10.1183/13993003.00260-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu X., Ou Z., Zhang M., Meng Y., Li Y., Wen J., et al. Indoor bacterial, fungal and viral species and functional genes in urban and rural schools in Shanxi Province, China–association with asthma, rhinitis and rhinoconjunctivitis in high school students. Microbiome. 2021;9(1):138. doi: 10.1186/s40168-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019;12(4):843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 59.Magne F., Gotteland M., Gauthier L., Zazueta A., Pesoa S., Navarrete P., et al. The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12(5):1474. doi: 10.3390/nu12051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Araki A., Kanazawa A., Kawai T., Eitaki Y., Morimoto K., Nakayama K., et al. The relationship between exposure to microbial volatile organic compound and allergy prevalence in single-family homes. Sci. Total Environ. 2012;423:18–26. doi: 10.1016/j.scitotenv.2012.02.026. [DOI] [PubMed] [Google Scholar]
- 61.Araki A., Kawai T., Eitaki Y., Kanazawa A., Morimoto K., Nakayama K., et al. Prevalence of asthma, atopic dermatitis, and rhinitis and MVOC exposure in single family homes-A survey in 6 cities of Japan. Epidemiology. 2011;22(1):S40–S41. [Google Scholar]
- 62.Choi H., Schmidbauer N., Bornehag C.G. Volatile organic compounds of possible microbial origin and their risks on childhood asthma and allergies within damp homes. Environ. Int. 2017;98:143–151. doi: 10.1016/j.envint.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 63.Miller J.D., McMullin D.R. Fungal secondary metabolites as harmful indoor air contaminants: 10 years on. Appl. Microbiol. Biotechnol. 2014;98(24):9953–9966. doi: 10.1007/s00253-014-6178-5. [DOI] [PubMed] [Google Scholar]
- 64.Ferreira Lopes S., Vacher G., Ciarlo E., Savova-Bianchi D., Roger T., Niculita-Hirzel H. Primary and immortalized human respiratory cells display different patterns of cytotoxicity and cytokine release upon exposure to deoxynivalenol, nivalenol and fusarenon-X. Toxins. 2017;9(11):337. doi: 10.3390/toxins9110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peitzsch M., Sulyok M., Täubel M., Vishwanath V., Krop E., Borràs-Santos A., et al. Microbial secondary metabolites in school buildings inspected for moisture damage in Finland, The Netherlands and Spain. J. Environ. Monit. 2012;14(8):2044–2053. doi: 10.1039/c2em30195d. [DOI] [PubMed] [Google Scholar]
- 66.Kirjavainen P.V., Taubel M., Karvonen A.M., Sulyok M., Tiittanen P., Krska R., et al. Microbial secondary metabolites in homes in association with moisture damage and asthma. Indoor Air. 2016;26(3):448–456. doi: 10.1111/ina.12213. [DOI] [PubMed] [Google Scholar]
- 67.Braun-Fahrländer C., Riedler J., Herz U., Eder W., Waser M., Grize L., et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N. Engl. J. Med. 2002;347(12):869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 68.Williams L.K., Ownby D.R., Maliarik M.J., Johnson C.C. The role of endotoxin and its receptors in allergic disease. Ann. Allergy Asthma Immunol. 2005;94(3):323–332. doi: 10.1016/S1081-1206(10)60983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Strien R.T., Engel R., Holst O., Bufe A., Eder W., Waser M., et al. Microbial exposure of rural school children, as assessed by levels of N-acetyl-muramic acid in mattress dust, and its association with respiratory health. J. Allergy Clin. Immunol. 2004;113(5):860–867. doi: 10.1016/j.jaci.2004.01.783. [DOI] [PubMed] [Google Scholar]
- 70.Zhao Z., Sebastian A., Larsson L., Wang Z., Zhang Z., Norbäck D. Asthmatic symptoms among pupils in relation to microbial dust exposure in schools in Taiyuan, China. Pediatr. Allergy Immunol. 2008;19(5):455–465. doi: 10.1111/j.1399-3038.2007.00664.x. official publication of the European Society of Pediatric Allergy and Immunology. [DOI] [PubMed] [Google Scholar]
- 71.Valkonen M., Taubel M., Pekkanen J., Tischer C., Rintala H., Zock J.P., et al. Microbial characteristics in homes of asthmatic and non-asthmatic adults in the ECRHS cohort. Indoor Air. 2018;28(1):16–27. doi: 10.1111/ina.12427. [DOI] [PubMed] [Google Scholar]
- 72.Karvonen A.M., Hyvarinen A., Rintala H., Korppi M., Taubel M., Doekes G., et al. Quantity and diversity of environmental microbial exposure and development of asthma: a birth cohort study. Allergy. 2014;69(8):1092–1101. doi: 10.1111/all.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tischer C., Zock J.P., Valkonen M., Doekes G., Guerra S., Heederik D., et al. Predictors of microbial agents in dust and respiratory health in the Ecrhs. BMC Pulm. Med. 2015;15:48. doi: 10.1186/s12890-015-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z., Reponen T., Hershey G.K. Fungal exposure and asthma: IgE and non-IgE-mediated mechanisms. Curr. Allergy Asthma Rep. 2016;16(12):86. doi: 10.1007/s11882-016-0667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi H., Byrne S., Larsen L.S., Sigsgaard T., Thorne P.S., Larsson L., et al. Residential culturable fungi, (1-3, 1-6)-β-d-glucan, and ergosterol concentrations in dust are not associated with asthma, rhinitis, or eczema diagnoses in children. Indoor Air. 2014;24(2):158–170. doi: 10.1111/ina.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maheswaran D., Zeng Y., Chan-Yeung M., Scott J., Osornio-Vargas A., Becker A.B., et al. Exposure to Beta-(1,3)-D-glucan in house dust at age 7-10 is associated with airway hyperresponsiveness and atopic asthma by age 11-14. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0098878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iossifova Y.Y., Reponen T., Bernstein D.I., Levin L., Kalra H., Campo P., et al. House dust (1-3)-beta-D-glucan and wheezing in infants. Allergy. 2007;62(5):504–513. doi: 10.1111/j.1398-9995.2007.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.WHO. World Health Organization Indoor air pollutants: exposure and health effects. EURO Rep. Stud. 1983;78:1–42. [Google Scholar]
- 79.Laumbach R.J., Kipen H.M. Bioaerosols and sick building syndrome: particles, inflammation, and allergy. Curr. Opin. Allergy Clin. Immunol. 2005;5(2):135–139. doi: 10.1097/01.all.0000162305.05105.d0. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X., Sahlberg B., Wieslander G., Janson C., Gislason T., Norback D. Dampness and moulds in workplace buildings: associations with incidence and remission of sick building syndrome (SBS) and biomarkers of inflammation in a 10 year follow-up study. Sci. Total Environ. 2012;430:75–81. doi: 10.1016/j.scitotenv.2012.04.040. [DOI] [PubMed] [Google Scholar]
- 81.Sahlberg B., Norback D., Wieslander G., Gislason T., Janson C. Onset of mucosal, dermal, and general symptoms in relation to biomarkers and exposures in the dwelling: a cohort study from 1992 to 2002. Indoor Air. 2012;22(4):331–338. doi: 10.1111/j.1600-0668.2012.00766.x. [DOI] [PubMed] [Google Scholar]
- 82.Zhao G., Yin G., Inamdar A.A., Luo J., Zhang N., Yang I., et al. Volatile organic compounds emitted by filamentous fungi isolated from flooded homes after Hurricane Sandy show toxicity in a Drosophila bioassay. Indoor Air. 2017;27(3):518–528. doi: 10.1111/ina.12350. [DOI] [PubMed] [Google Scholar]
- 83.Scheel C.M., Rosing W.C., Farone A.L. Possible sources of sick building syndrome in a Tennessee middle school. Archives of environmental health. 2001;56(5):413–417. doi: 10.1080/00039890109604476. [DOI] [PubMed] [Google Scholar]
- 84.Fu X., Norbäck D., Yuan Q., Li Y., Zhu X., Hashim J.H., et al. Association between indoor microbiome exposure and sick building syndrome (SBS) in junior high schools of Johor Bahru, Malaysia. Sci. Total Environ. 2021;753 doi: 10.1016/j.scitotenv.2020.141904. [DOI] [PubMed] [Google Scholar]
- 85.Straus D.C. Molds, mycotoxins, and sick building syndrome. Toxicol. Ind. Health. 2009;25(9–10):617–635. doi: 10.1177/0748233709348287. [DOI] [PubMed] [Google Scholar]
- 86.Delmulle B., De Saeger S., Adams A., De Kimpe N., Van Peteghem C. Development of a liquid chromatography/tandem mass spectrometry method for the simultaneous determination of 16 mycotoxins on cellulose filters and in fungal cultures. Rapid Commun. Mass Spectrom. : RCM (Rapid Commun. Mass Spectrom.) 2006;20(5):771–776. doi: 10.1002/rcm.2373. [DOI] [PubMed] [Google Scholar]
- 87.Araki A., Kawai T., Eitaki Y., Kanazawa A., Morimoto K., Nakayama K., et al. Relationship between selected indoor volatile organic compounds, so-called microbial VOC, and the prevalence of mucous membrane symptoms in single family homes. Sci. Total Environ. 2010;408(10):2208–2215. doi: 10.1016/j.scitotenv.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 88.Sahlberg B., Gunnbjörnsdottir M., Soon A., Jogi R., Gislason T., Wieslander G., et al. Airborne molds and bacteria, microbial volatile organic compounds (MVOC), plasticizers and formaldehyde in dwellings in three North European cities in relation to sick building syndrome (SBS) Sci. Total Environ. 2013;444:433–440. doi: 10.1016/j.scitotenv.2012.10.114. [DOI] [PubMed] [Google Scholar]
- 89.Theiler A., Bärnthaler T., Platzer W., Richtig G., Peinhaupt M., Rittchen S., et al. Butyrate ameliorates allergic airway inflammation by limiting eosinophil trafficking and survival. J. Allergy Clin. Immunol. 2019;144(3):764–776. doi: 10.1016/j.jaci.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 90.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silva Y.P., Bernardi A., Frozza R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020;11(25) doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Siucinska E. Γ-Aminobutyric acid in adult brain: an update. Behav. Brain Res. 2019;376 doi: 10.1016/j.bbr.2019.112224. [DOI] [PubMed] [Google Scholar]
- 94.Wolf A.J., Underhill D.M. Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 2018;18(4):243–254. doi: 10.1038/nri.2017.136. [DOI] [PubMed] [Google Scholar]
- 95.Fu X., Li Y., Meng Y., Yuan Q., Zhang Z., Norbäck D., et al. Associations between respiratory infections and bacterial microbiome in student dormitories in Northern China. Indoor Air. 2020;30(5):816–826. doi: 10.1111/ina.12677. [DOI] [PubMed] [Google Scholar]
- 96.Fu X., Yuan Q., Zhu X., Li Y., Meng Y., Hashim J.H., et al. Associations between the indoor microbiome, environmental characteristics and respiratory infections in junior high school students of Johor Bahru, Malaysia. Environmental science Processes & impacts. 2021;23(8):1171–1181. doi: 10.1039/d1em00115a. [DOI] [PubMed] [Google Scholar]
- 97.Locey K.J., Lennon J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA. 2016;113(21):5970. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barberán A., Dunn Robert R., Reich Brian J., Pacifici K., Laber Eric B., Menninger Holly L., et al. The ecology of microscopic life in household dust. Proc. Biol. Sci. 2015;282(1814) doi: 10.1098/rspb.2015.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brooks B., Firek B.A., Miller C.S., Sharon I., Thomas B.C., Baker R., et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2014;2(1):1. doi: 10.1186/2049-2618-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barberán A., Ladau J., Leff J.W., Pollard K.S., Menninger H.L., Dunn R.R., et al. Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl. Acad. Sci. U.S.A. 2015;112(18):5756–5761. doi: 10.1073/pnas.1420815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Amend A.S., Seifert K.A., Samson R., Bruns T.D. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. U. S. A. 2010;107(31):13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fu X., Li Y., Yuan Q., Cai G.H., Deng Y., Zhang X., et al. Continental-scale microbiome study reveals different environmental characteristics determining microbial richness, composition, and quantity in hotel rooms. mSystems. 2020;5(3) doi: 10.1128/mSystems.00119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017;15:579. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 105.O’Brien S.L., Gibbons S.M., Owens S.M., Hampton-Marcell J., Johnston E.R., Jastrow J.D., et al. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 2016;18(6):2039–2051. doi: 10.1111/1462-2920.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leung M.H.Y., Tong X., Bøifot K.O., Bezdan D., Butler D.J., Danko D.C., et al. Characterization of the public transit air microbiome and resistome reveals geographical specificity. Microbiome. 2021;9(1):112. doi: 10.1186/s40168-021-01044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Danko D., Bezdan D., Afshin E.E., Ahsanuddin S., Bhattacharya C., Butler D.J., et al. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell. 2021;184(13):3376–3393. doi: 10.1016/j.cell.2021.05.002. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kokubo M., Fujiyoshi S., Ogura D., Nakajima M., Fujieda A., Noda J., et al. Relationship between the microbiome and indoor temperature/humidity in a traditional Japanese house with a thatched roof in Kyoto, Japan. Diversity. 2021;13(10) [Google Scholar]
- 109.Parajuli A., Gronroos M., Siter N., Puhakka R., Vari H.K., Roslund M.I., et al. Urbanization reduces transfer of diverse environmental microbiota indoors. Front. Microbiol. 2018;9:84. doi: 10.3389/fmicb.2018.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dockx Y., Taubel M., Bijnens E.M., Witters K., Valkonen M., Jayaprakash B., et al. Residential green space can shape the indoor microbial environment. Environ. Res. 2021;201 doi: 10.1016/j.envres.2021.111543. [DOI] [PubMed] [Google Scholar]
- 111.Roslund M.I., Puhakka R., Nurminen N., Oikarinen S., Siter N., Gronroos M., et al. Long-term biodiversity intervention shapes health-associated commensal microbiota among urban day-care children. Environ. Int. 2021;157 doi: 10.1016/j.envint.2021.106811. [DOI] [PubMed] [Google Scholar]
- 112.Leung M.H., Lee P.K. The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: a review. Microbiome. 2016;4(1):21. doi: 10.1186/s40168-016-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lax S., Smith D.P., Hampton-Marcell J., Owens S.M., Handley K.M., Scott N.M., et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mäki J.M., Kirjavainen P.V., Täubel M., Piippo-Savolainen E., Backman K., Hyvärinen A., et al. Associations between dog keeping and indoor dust microbiota. Sci. Rep. 2021;11(1):5341. doi: 10.1038/s41598-021-84790-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mahnert A., Moissl-Eichinger C., Zojer M., Bogumil D., Mizrahi I., Rattei T., et al. Man-made microbial resistances in built environments. Nat. Commun. 2019;10(1):968. doi: 10.1038/s41467-019-08864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun Y., Fu X., Li Y., Yuan Q., Ou Z., Lindgren T., et al. Shotgun metagenomics of dust microbiome from flight deck and cabin in civil aviation aircraft. Indoor Air. 2020;30(6):1199–1212. doi: 10.1111/ina.12707. [DOI] [PubMed] [Google Scholar]
- 117.Lax S., Cardona C., Zhao D., Winton V.J., Goodney G., Gao P., et al. Microbial and metabolic succession on common building materials under high humidity conditions. Nat. Commun. 2019;10(1):1767. doi: 10.1038/s41467-019-09764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao B., Dewald C., Hennig M., Bossert J., Bauer M., Pletz M.W., et al. Microorganisms @ materials surfaces in aircraft: potential risks for public health? - a systematic review. Trav. Med. Infect. Dis. 2019;28:6–14. doi: 10.1016/j.tmaid.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 119.Vaglenov K. Auburn University; 2014. Survival and Transmission of Selected Pathogens on Airplane Cabin Surfaces and Selection of Phages Specific for campylobacter Jejuni. [Google Scholar]
- 120.Sze-To G.N., Yang Y., Kwan J.K., Yu S.C., Chao C.Y. Effects of surface material, ventilation, and human behavior on indirect contact transmission risk of respiratory infection. Risk Anal.: an official publication of the Society for Risk Analysis. 2014;34(5):818–830. doi: 10.1111/risa.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Acevedo N., Alashkar Alhamwe B., Caraballo L., Ding M., Ferrante A., Garn H., et al. Perinatal and early-life nutrition, epigenetics, and allergy. Nutrients. 2021;13(3) doi: 10.3390/nu13030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Potaczek D.P., Alashkar Alhamwe B., Miethe S., Garn H. In: Allergic Diseases – from Basic Mechanisms to Comprehensive Management and Prevention. Traidl-Hoffmann C., Zuberbier T., Werfel T., editors. Springer International Publishing; Cham: 2022. Epigenetic mechanisms in allergy development and prevention; pp. 331–357. [Google Scholar]
- 123.Potaczek D., Harb H., Michel S., Alhamwe B.A., Renz H., Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017;9 doi: 10.2217/epi-2016-0162. [DOI] [PubMed] [Google Scholar]
- 124.Fujimura K.E., Demoor T., Rauch M., Faruqi A.A., Jang S., Johnson C.C., et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111(2):805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.von Mutius E., Smits H.H. Primary prevention of asthma: from risk and protective factors to targeted strategies for prevention. Lancet. 2020;396(10254):854–866. doi: 10.1016/S0140-6736(20)31861-4. [DOI] [PubMed] [Google Scholar]
- 126.Martiny A.C. High proportions of bacteria are culturable across major biomes. ISME J. 2019;13(8):2125–2128. doi: 10.1038/s41396-019-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stewart E.J. Growing unculturable bacteria. J. Bacteriol. 2012;194(16):4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Knight R., Vrbanac A., Taylor B.C., Aksenov A., Callewaert C., Debelius J., et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 129.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017;35(9):833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 130.Frankel M., Timm M., Hansen E.W., Madsen A.M. Comparison of sampling methods for the assessment of indoor microbial exposure. Indoor Air. 2012;22(5):405–414. doi: 10.1111/j.1600-0668.2012.00770.x. [DOI] [PubMed] [Google Scholar]
- 131.Adams R.I., Tian Y., Taylor J.W., Bruns T.D., Hyvärinen A., Täubel M. Passive dust collectors for assessing airborne microbial material. Microbiome. 2015;3(1):46. doi: 10.1186/s40168-015-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yamamoto N., Hospodsky D., Dannemiller K.C., Nazaroff W.W., Peccia J. Indoor emissions as a primary source of airborne allergenic fungal particles in classrooms. Environ. Sci. Technol. 2015;49(8):5098–5106. doi: 10.1021/es506165z. [DOI] [PubMed] [Google Scholar]
- 133.Hyytiäinen H.K., Jayaprakash B., Kirjavainen P.V., Saari S.E., Holopainen R., Keskinen J., et al. Crawling-induced floor dust resuspension affects the microbiota of the infant breathing zone. Microbiome. 2018;6(1):25. doi: 10.1186/s40168-018-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnson J.S., Spakowicz D.J., Hong B.-Y., Petersen L.M., Demkowicz P., Chen L., et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019;10(1):5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wenger A.M., Peluso P., Rowell W.J., Chang P.-C., Hall R.J., Concepcion G.T., et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019;37(10):1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Callahan B.J., Wong J., Heiner C., Oh S., Theriot C.M., Gulati A.S., et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47(18):e103. doi: 10.1093/nar/gkz569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Douglas G.M., Maffei V.J., Zaneveld J., Yurgel S.N., Brown J.R., Taylor C.M., et al. bioRxiv; 2019. PICRUSt2: An Improved and Extensible Approach for Metagenome Inference. [Google Scholar]
- 138.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hanshew A.S., Mason C.J., Raffa K.F., Currie C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods. 2013;95(2):149–155. doi: 10.1016/j.mimet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2(2):e00191. doi: 10.1128/mSystems.00191-16. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Quast C., Pruesse E., Gerken J., Peplies J., Yarza P., Yilmaz P., et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nilsson R.H., Larsson K.H., Taylor A.F.S., Bengtsson-Palme J., Jeppesen T.S., Schigel D., et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47(D1):D259–D264. doi: 10.1093/nar/gky1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li D., Luo R., Liu C.M., Leung C.M., Ting H.F., Sadakane K., et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 146.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Franzosa E.A., McIver L.J., Rahnavard G., Thompson L.R., Schirmer M., Weingart G., et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods. 2018;15(11):962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Beghini F., McIver L.J., Blanco-Miguez A., Dubois L., Asnicar F., Maharjan S., et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021:10. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.The Gene Ontology C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McArdle B.H., Anderson M.J. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82(1):290–297. [Google Scholar]
- 153.Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18(1):117–143. [Google Scholar]
- 154.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bokulich N.A., Dillon M.R., Zhang Y., Rideout J.R., Bolyen E., Li H., et al. q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems. 2018;3(6):e00219. doi: 10.1128/mSystems.00219-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shetty S.A., Lahti L. Microbiome data science. J. Biosci. 2019;44(5):115. [PubMed] [Google Scholar]
- 158.Dannemiller K.C., Lang-Yona N., Yamamoto N., Rudich Y., Peccia J. Combining real-time PCR and next-generation DNA sequencing to provide quantitative comparisons of fungal aerosol populations. Atmos. Environ. 2014;84:113–121. [Google Scholar]
- 159.Noble W.S. How does multiple testing correction work? Nat. Biotechnol. 2009;27(12):1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang L., Amir A., Morton J.T., Heller R., Arias-Castro E., Knight R. Discrete false-discovery rate improves identification of differentially abundant microbes. mSystems. 2017;2(6):e00092. doi: 10.1128/mSystems.00092-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60–R. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10(12):1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huan T., Forsberg E.M., Rinehart D., Johnson C.H., Ivanisevic J., Benton H.P., et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods. 2017;14:461. doi: 10.1038/nmeth.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Morton J.T., Marotz C., Washburne A., Silverman J., Zaramela L.S., Edlund A., et al. Establishing microbial composition measurement standards with reference frames. Nat. Commun. 2019;10(1):2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Knights D., Kuczynski J., Charlson E.S., Zaneveld J., Mozer M.C., Collman R.G., et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods. 2011;8(9):761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shenhav L., Thompson M., Joseph T.A., Briscoe L., Furman O., Bogumil D., et al. FEAST: fast expectation-maximization for microbial source tracking. Nat. Methods. 2019;16(7):627–632. doi: 10.1038/s41592-019-0431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zakrzewski M., Proietti C., Ellis J.J., Hasan S., Brion M.-J., Berger B., et al. Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics. 2017;33(5):782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang C., Hu J., Blaser M.J., Li H. Estimating and testing the microbial causal mediation effect with high-dimensional and compositional microbiome data. Bioinformatics. 2020;36(2):347–355. doi: 10.1093/bioinformatics/btz565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Emiola A., Oh J. High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat. Commun. 2018;9(1):4956. doi: 10.1038/s41467-018-07240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.de Nies L., Lopes S., Busi S.B., Galata V., Heintz-Buschart A., Laczny C.C., et al. PathoFact: a pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data. Microbiome. 2021;9(1):49. doi: 10.1186/s40168-020-00993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Su X., Jing G., McDonald D., Wang H., Wang Z., Gonzalez A., et al. Identifying and predicting novelty in microbiome studies. mBio. 2018;9(6):e02099. doi: 10.1128/mBio.02099-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gonzalez A., Navas-Molina J.A., Kosciolek T., McDonald D., Vázquez-Baeza Y., Ackermann G., et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods. 2018;15(10):796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sun Y., Li Y., Yuan Q., Fu X. Identifying composition novelty in microbiome studies: improvement for prediction accuracy. mBio. 2019;10(4):e00892. doi: 10.1128/mBio.00892-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., et al. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu B., Zheng D., Jin Q., Chen L., Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ching J., Mills G., Bechtel B., See L., Feddema J., Wang X., et al. WUDAPT: an urban weather, climate, and environmental modeling infrastructure for the anthropocene. Bull. Am. Meteorol. Soc. 2018;99(9):1907–1924. [Google Scholar]
- 177.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Neveu V., Nicolas G., Salek R.M., Wishart D.S., Scalbert A. Exposome-Explorer 2.0: an update incorporating candidate dietary biomarkers and dietary associations with cancer risk. Nucleic Acids Res. 2020;48(D1):D908–D912. doi: 10.1093/nar/gkz1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rinschen M.M., Ivanisevic J., Giera M., Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019;20(6):353–367. doi: 10.1038/s41580-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guijas C., Montenegro-Burke J.R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., et al. METLIN: a Technology platform for identifying Knowns and unknowns. Anal. Chem. 2018;90(5):3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.MacKinnon D.P., Fairchild A.J., Fritz M.S. Mediation analysis. Annu. Rev. Psychol. 2007;58:593–614. doi: 10.1146/annurev.psych.58.110405.085542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mackay D., Haw S., Ayres J.G., Fischbacher C., Pell J.P. Smoke-free legislation and hospitalizations for childhood asthma. N. Engl. J. Med. 2010;363(12):1139–1145. doi: 10.1056/NEJMoa1002861. [DOI] [PubMed] [Google Scholar]
- 184.Frazer K., Callinan J.E., McHugh J., van Baarsel S., Clarke A., Doherty K., et al. Legislative smoking bans for reducing harms from secondhand smoke exposure, smoking prevalence and tobacco consumption. Cochrane Database Syst. Rev. 2016;2(2) doi: 10.1002/14651858.CD005992.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bo L., Li J., Tao T., Bai Y., Ye X., Hotchkiss R.S., et al. Probiotics for preventing ventilator-associated pneumonia. Cochrane Database Syst. Rev. 2014;(10) doi: 10.1002/14651858.CD009066.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Güvenç I.A., Muluk N.B., Mutlu F.Ş., Eşki E., Altıntoprak N., Oktemer T., et al. Do probiotics have a role in the treatment of allergic rhinitis? A comprehensive systematic review and metaanalysis. American J Rhinology Allergy. 2016;30(5):e157–e175. doi: 10.2500/ajra.2016.30.4354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.