Abstract
Sphingomonas (formerly Pseudomonas) paucimobilis UT26 utilizes γ-hexachlorocyclohexane (γ-HCH), a halogenated organic insecticide, as a sole carbon and energy source. In a previous study, we showed that γ-HCH is degraded to 2,5-dichlorohydroquinone (2,5-DCHQ) (Y. Nagata, R. Ohtomo, K. Miyauchi, M. Fukuda, K. Yano, and M. Takagi, J. Bacteriol. 176:3117–3125, 1994). In the present study, we cloned and characterized a gene, designated linD, directly involved in the degradation of 2,5-DCHQ. The linD gene encodes a peptide of 343 amino acids and has a low level of similarity to proteins which belong to the glutathione S-transferase family. When LinD was overproduced in Escherichia coli, a 40-kDa protein was found after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Northern blot analysis revealed that expression of the linD gene was induced by 2,5-DCHQ in S. paucimobilis UT26. Thin-layer chromatography and gas chromatography-mass spectrometry analyses with the LinD-overexpressing E. coli cells revealed that LinD converts 2,5-DCHQ rapidly to chlorohydroquinone (CHQ) and also converts CHQ slowly to hydroquinone. LinD activity in crude cell extracts was increased 3.7-fold by the addition of glutathione. All three of the Tn5-induced mutants of UT26, which lack 2,5-DCHQ dehalogenase activity, had rearrangements or a deletion in the linD region. These results indicate that LinD is a glutathione-dependent reductive dehalogenase involved in the degradation of γ-HCH by S. paucimobilis UT26.
γ-Hexachlorocyclohexane (γ-HCH [also called γ-BHC and lindane]) is a halogenated organic insecticide which has been used worldwide. Because of its toxicity and long persistence in soil, most countries have prohibited its use; however, many contaminated sites remain throughout the world. Moreover, some countries are presently using γ-HCH for economic reasons, and new sites are continually being contaminated.
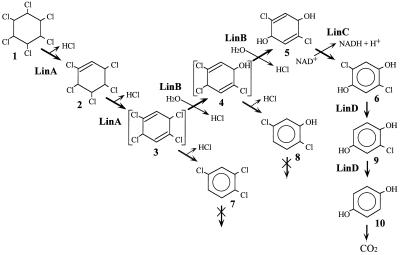
Sphingomonas (formerly Pseudomonas) paucimobilis UT26 utilizes γ-HCH as a sole source of carbon and energy (8). UT26 degrades γ-HCH through the pathway shown in Fig. 1 (14, 16, 17). γ-HCH is likely converted by two steps of dehydrochlorination via γ-pentachlorocyclohexene (γ-PCCH) to 1,3,4,6-tetrachloro-1,4-cyclohexadiene (1,4-TCDN). This is productively metabolized to 2,5-dichloro-3,5-cyclohexadiene-1,4-diol (2,5-DDOL) by two steps of hydrolytic dehalogenation. Then, 2,5-DDOL is further degraded to 2,5-dichlorohydroquinone (2,5-DCHQ), and finally 2,5-DCHQ is mineralized. Two dead-end products, 1,2,4-trichlorobenzene (1,2,4-TCB) and 2,5-dichlorophenol (2,5-DCP), have also been found in culture supernatants.
FIG. 1.
Proposed assimilation pathway of γ-HCH in S. paucimobilis UT26. Compounds: 1, γ-HCH/γ-BHC; 2, γ-PCCH; 3, 1,4-TCDN; 4, 2,4,5-DNOL; 5, 2,5-DDOL; 6, 2,5-DCHQ; 7, 1,2,4-TCB; 8, 2,5-DCP; 9, CHQ; 10, HQ.
In previous studies, we cloned and sequenced three genes involved in the early steps of γ-HCH degradation in UT26 (7, 16, 17). The linA gene encodes γ-HCH dehydrochlorinase (LinA), which converts γ-HCH to 1,2,4-TCB via γ-PCCH. LinA shows no homology to known proteins (7). The linB gene encodes 1,4-TCDN chlorohydrolase (LinB), which converts 1,4-TCDN to 2,5-DDOL via 2,4,5-trichloro-2,5-cyclohexadiene-1-ol (2,4,5-DNOL). LinB shows significant similarity to hydrolytic dehalogenase, DhlA, from Xanthobacter autotrophicus (9). The linC gene encodes 2,5-DDOL dehydrogenase, which converts 2,5-DDOL to 2,5-DCHQ (17). LinC shows homology to the members of the short-chain alcohol dehydrogenase family (19).
In this study, we describe the isolation and characterization of a gene directly involved in the degradation of 2,5-DCHQ and show that its product is a glutathione-dependent reductive dechlorinase which converts 2,5-DCHQ to hydroquinone (HQ) via chlorohydroquinone (CHQ).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Sphingomonas strains, Pseudomonas strains, and Escherichia coli were grown on Luria broth (12) or W minimal medium (8). Cultures were incubated at 30°C for Sphingomonas and Pseudomonas strains and at 37°C for E. coli strains. Antibiotics were used at final concentrations of 50 μg/ml for ampicillin and kanamycin, 25 μg/ml for nalidixic acid, and 20 μg/ml for tetracycline.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. paucimobilis strains | ||
| UT26 | Growth on γ-HCH; Nalr | 8 |
| UT102 | Nalr Kmr | 18 |
| UT103 | Nalr Kmr | 18 |
| UT116 | Nalr Kmr | 18 |
| P. putida strain | ||
| PpY101 | met Nalr | 11 |
| E. coli strains | ||
| HB101 | F−hsdS recA ara proA lacY galK rpsL xyl mtl supE | 12 |
| MV1190 | Δlac-proAB thi supE Δsrl-recA306::Tn10 F′ traD36 proAB lacIqZΔM15 | 17 |
| Plasmids | ||
| pAQN | pMB9 replicon; lacIqaqna Apr | 23 |
| pKM1 | pUC18 carrying 3-kb SacI-HindIII fragment; direction of linD is identical to that of lac promoter of pUC18 | This study |
| pKM1R | pUC18 carrying same fragment as pKM1 with opposite direction | This study |
| pKM2 | pUC18 carrying 1.2-kb EcoRI-MluI fragment of pKM170 | This study |
| pKM2R | pUC18 carrying same fragment as pKM2 with opposite direction | This study |
| pKM170 | pUC18 carrying 2.0-kb fragment of pKM1; deletion plasmid with deletion about 900 bp from 5′ end of fragment inserted in pKM1 | This study |
| pKM9372 | pUC18 carrying 8-kb EcoRI fragment of pMM9372 | This study |
| pKS13 | RK2 replicon; cos Mob+ Tcr | 11 |
| pKSM937 | pKS13 with 20 kb of UT26 DNA containing linD | This study |
| pMFY42 | RSF1010 replicon; Kmr Tcr | 4 |
| pMM9372 | pMFY42 carrying 8-kb EcoRI fragment of pKSM937 | This study |
| pMYLB1 | pAQN carrying linB in place of aqna | 16 |
| pMYLD2 | pAQN carrying linD in place of aqna | This study |
| pRK2013 | ColE1::RK2 Tra+ Kmr | 3 |
| pUC18 | pMB9 replicon of pKM9372; Apr | 26 |
aqn, aqualysin I gene of Thermus aquaticus.
Preparation of CHQ.
CHQ was prepared as follows. A solution of 2-chloro-1,4-benzoquinone (Aldrich, Milwaukee, Wis.) solution (174 mM in ethylacetate) was mixed with the same amount of ascorbic acid solution (1.1 M in 100 mM phosphate buffer [pH 7.0]) and vortexed. After centrifugation, the upper phase was diluted 10 times by ethylacetate. The resultant solution was used as a CHQ stock solution, which was diluted 1,000 times just before the assay.
Isolation of DNA.
Plasmid DNA of E. coli was isolated by the alkaline lysis method of Maniatis et al. (12) and, if needed, purified by cesium chloride-ethidium bromide density gradient centrifugation. Total DNA from Sphingomonas strains was isolated as described previously (15).
Construction of a gene library.
A gene library of S. paucimobilis in Pseudomonas putida PpY101 or E. coli HB101 was constructed by using a broad-host-range cosmid vector, pKS13 (11), as described previously (16).
Assay for 2,5-DCHQ dehalogenase activity.
A small quantity of each colony was picked and suspended in 100 μl of the assay solution (20 mM phosphate buffer [pH 7.0] containing 2,5-DCHQ at 1 μg/ml). The solution was incubated for 12 to 18 h at 30°C for P. putida and at 37°C for E. coli. Then, 100 μl of ethylacetate was added and the mixture was vortexed for 1 min. After centrifugation (15,000 × g, for 5 min), the ethylacetate layer was recovered. One microliter of this extract was analyzed by gas chromatography (GC) with an electron capture detector (ECD).
GC and GC-MS analyses.
GC analysis with an ECD was performed under the same conditions as described previously (16). GC-mass spectrometry (MS) analysis was performed with a GC-MS QP5000 spectrometer (Shimadzu, Kyoto, Japan) and a DB-1 capillary column (0.25 mm thick by 30 m long; J&W Scientific, Folsom, Calif.). The column temperature was increased from 80 to 160°C at a rate of 5°C/min and then from 160 to 260°C at a rate of 10°C/min. The carrier gas flow rate was 20 ml/min.
Nucleotide sequence determination.
Nucleotide sequences were determined by the dideoxy-chain termination method with the Applied Biosystems model 373A DNA sequencing system (Applied Biosystems, Foster City, Calif.).
Southern blot analysis.
Southern blot analysis was performed at 56°C as described previously (16). The ECL gene detection system (Amersham, Arlington Heights, Ill.) was used according to the provided protocol.
Northern blot analysis.
S. paucimobilis UT26 was grown on W medium, and 2,5-DCHQ was added during the exponential phase (optical density at 660 nm, 0.3 to 0.4). After 1 h of incubation, total RNA was isolated by the method described by Hopwood et al. (6). As a control, total RNA was isolated from cells incubated without 2,5-DCHQ. Hybridization and detection were performed by using digoxigenin-labeled DNA with the CSPD system (Boehringer, Mannheim, Germany), according to the provided protocol.
Analysis of the linD gene product.
For overexpression of the linD gene product, plasmid pMYLD2 was constructed from pAQN (23). Plasmid pAQN was digested with EcoRI and HindIII to replace the 1.8-kb aqualysin-coding fragment with the 1.2-kb EcoRI-HindIII fragment including the linD gene from pKM2 for pMYLD2. In pMYLD2, the linD gene is expressed under the control of the tac promoter. Expression is repressed tightly by the lacIq gene product, which is produced from the same plasmid when IPTG (isopropyl-β-d-thiogalactopyranoside) is not added. Overexpression of LinD was achieved as described previously (7) by using E. coli MV1190 containing pMYLD2. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was done as described previously (16).
TLC analysis.
To detect the metabolites resulting from 2,5-DCHQ by LinD, we used thin-layer chromatography (TLC). TLC plates (HPTLC Precoated Silica Gel 60) were purchased from Merck (Darmstadt, Germany). The substrate, 2,5-DCHQ (50 μg/ml [final concentration]), was added to resting cells of E. coli MV1190 (50 mg [wet weight] per ml in 20 mM phosphate buffer [pH 7.0]) overexpressing LinD or LinB as a control (16). After incubation for various time periods, 500 μl of the culture was extracted with ethylacetate. After evaporation, the reaction mixture was resuspended in a small amount of ethylacetate and the suspension was spotted onto the TLC plate. The solvent system for the separation of metabolites consisted of benzene-ethylacetate (85:15, vol/vol). After development, the TLC plate was dried and exposed to iodine vapor.
LinD activity assay using crude cell extracts.
To investigate the effect of glutathione on LinD activity, we used cell extracts prepared from E. coli MV1190 overexpressing LinD. Cells were suspended in 20 mM Tris-HCl buffer (pH 7.5) and sonicated. After centrifugation (17,000 × g) at 4°C for 10 min, the supernatant was used as the crude cell extract. Ninety microliters of 2,5-DCHQ solution (100 μM in 20 mM potassium phosphate buffer [pH 7.0] including 200 μM ascorbic acid) was mixed with 10 μl of crude extract with and without 2 mM glutathione. After incubation for 10 min at 30°C, the reaction mixture was extracted with ethylacetate and analyzed by GC-MS. One unit of LinD activity was defined as the amount of enzyme that transformed 1 μmol of 2,5-DCHQ per min. Crude cell extract of E. coli MV1190 not overexpressing LinD was used as a control. Protein concentrations of crude extracts were measured by a protein assay kit (Bio-Rad, Hercules, Calif.).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. D89733.
RESULTS
Screening of clones with 2,5-DCHQ degradation activity.
The gene library of UT26 was constructed in P. putida PpY101, and clones which showed 2,5-DCHQ degradation activity (we designated it LinD activity) were identified upon analysis of biotransformation products by GC provided with an ECD. Of 1,000 recombinants tested, six clones showed LinD activity. The positive clones completely transformed 1 ppm of 2,5-DCHQ within 12 h. The activity was not detected in E. coli HB101, which had the same positive cosmids. This suggested either that the gene responsible for the degradation of 2,5-DCHQ has a promoter which is functional in P. putida but not in E. coli HB101 or that only P. putida has some factor(s) necessary for LinD activity. None of the positive clones had LinA, LinB, or LinC activity, suggesting that the linD gene is not linked to the linA, linB, and linC genes, which are responsible for the upstream pathway of γ-HCH degradation in S. paucimobilis UT26.
Subcloning and restriction enzyme mapping of the linD gene.
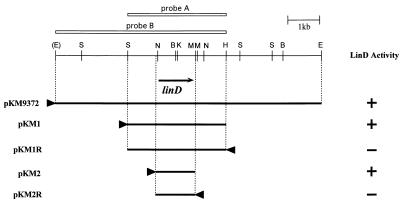
EcoRI digestion of one of the positive cosmids, pKSM937, produced three fragments (2, 7, and 8 kb) suitable for subcloning. Each of these fragments was subcloned into pMFY42, and the recombinant plasmids were introduced into P. putida PpY101. All clones which showed LinD activity had the 8-kb fragment. The resulting plasmid was called pMM9372. When the 8-kb EcoRI fragment was ligated to pUC18 and introduced into E. coli MV1190, the transformant also showed LinD activity. This may be due to the higher copy number of pUC18 than that of pKS13. A summary of the restriction enzyme mapping of this 8-kb fragment is shown in Fig. 2. Later investigation indicated that the EcoRI site, which was located at the 5′ end of pKM9732, was derived from the cloning site of the cosmid vector, pKS13. Southern blot analysis was performed with the cloned 3.0-kb SacI-HindIII fragment as a probe (probe A [Fig. 2]). Total DNA of UT26, digested with SacI, EcoRI, and BamHI, was separated on a 0.6% (wt/vol) agarose gel, blotted, and hybridized with the ECL-labeled probe. Only one signal was detected, indicating that the cloned fragment originated from UT26 and that there was no other sequence highly homologous to the linD gene in UT26 (data not shown).
FIG. 2.
Restriction map of pKM9372 and deletion analysis of pKM1. The directions of transcription by the lac promoter are indicated by arrowheads. The procedure for the measurement of LinD activity is described in Materials and Methods. The deduced location and direction of the linD gene is indicated by the arrow under the restriction map. The white bars above the map indicate the DNA fragments used as probes in Southern blot analyses. B, BamHI; E, EcoRI; H, HindIII; K, KpnI; M, MluI; N, NaeI; S, SacI.
Deletion and sequence analysis of the linD gene.
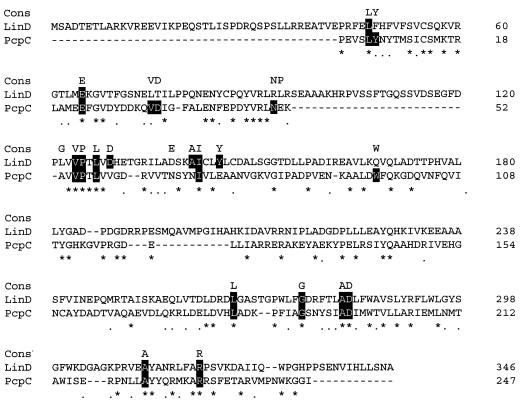
Deletion analysis of the 3-kb SacI-HindIII fragment revealed that the 1.2-kb region was necessary for LinD activity (Fig. 2). Sequence analysis of this region revealed that there was only one open reading frame of reasonable size (1,038 bp) in this region. As this open reading frame was preceded by a putative Shine-Dalgarno sequence (22), we designated it linD. The linD gene likely encodes a polypeptide of 346 amino acids, and its deduced molecular mass is 38.7 kDa. A sequence with high homology to the promoter of E. coli or which was expected to form a stem-loop structure to function as a terminator was not found around the linD gene. The G+C content of the linD gene is 60.8%, which is close to the total G+C contents of the type strain of S. paucimobilis (65%) (20), linB (64.3%), and linC (62.5%) (16, 17). Although a computer search revealed no nucleotide sequence with significant similarity to the whole linD gene, the LinD protein sequence showed a low level of similarity with some proteins which belong to the theta class of the glutathione S-transferase (GST) family, e.g., GST of E. coli (Swiss-Prot P39100) and Arabidopsis thaliana (Swiss-Prot P42769). The sequence with the most significant similarity was PcpC (Swiss-Prot Q03520) (20% identity, 31% similarity), the tetrachlorohydroquinone dechlorinase of Flavobacterium sp. strain ATCC 39723, which catalyzes the dechlorination of tetrachlorohydroquinone to 2,6-dichlorohydroquinone by two steps. The alignment of LinD with PcpC is shown in Fig. 3.
FIG. 3.
Alignment between LinD and PcpC. Identical and similar residues between LinD and PcpC are indicated by asterisks and dots, respectively. “Cons” indicates residues conserved in at least 10 of 13 bacterial GST sequences in the review by Vuilleumier (27). The amino acid residues with black backgrounds are conserved between LinD or PcpC and those in the top row (Cons). The alignment was generated by using the ClustalW sequence alignment program.
Overexpression of linD in E. coli and identification of the gene product.
To identify the linD gene product, we constructed plasmid pMYLD2, in which the linD gene was under the control of the tac promoter. E. coli MV1190 transformed with this plasmid was incubated with or without IPTG, and total proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. An overproduced protein band corresponding to about 40 kDa was observed in the IPTG-treated cells (data not shown). The molecular mass of this protein was almost equal to that deduced from the nucleotide sequence of the linD gene. Thus, it was confirmed that the product of linD is a protein with the molecular mass of about 40 kDa.
Expression of the linD gene in UT26.
We previously demonstrated that LinD activity was inducibly expressed in UT26 in the presence of 2,5-DCHQ (18). Northern blot analysis was performed with the total RNA of UT26 incubated with or without 2,5-DCHQ during the log phase. The result indicated that the linD gene was inducibly expressed by 2,5-DCHQ (data not shown).
Southern blot analysis of linD in LinD-less mutants.
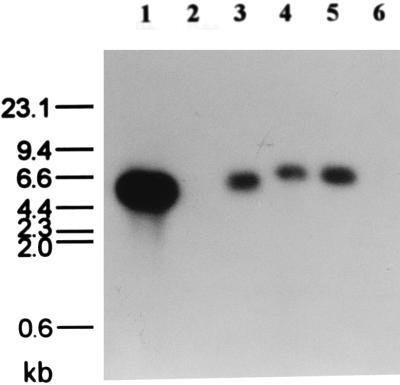
We previously isolated UT102, UT103, and UT116, Tn5-induced mutants defective in 2,5-DCHQ dehalogenase activity which showed phenotypes different from each other (18). UT102 is a mutant which constitutively expresses faint LinD activity, while UT103 and UT116 have no LinD activity at all. Southern blot analysis with the 5.0-kb EcoRI-HindIII fragment containing linD (probe B [Fig. 2]) as a probe to the total DNAs of the mutants digested with EcoRI and HindIII indicated that UT116 lacked the whole linD gene, while UT102 and UT103 had rearrangements in the linD gene or in its flanking regions (Fig. 4). This result supported the hypothesis that linD is directly involved in 2,5-DCHQ degradation activity in UT26.
FIG. 4.
Southern blot analysis of UT26 and its Tn5-induced mutants probed with the linD gene. Lanes: 1, 5-kb EcoRI-HindIII fragment containing the linD gene; 2, λ DNA digested with HindIII; 3 to 6, total DNA of UT26, UT102, UT103, and UT116, respectively, digested with EcoRI and HindIII. The conditions of the experiment are described in Materials and Methods.
Identification of metabolites of 2,5-DCHQ by the LinD protein.
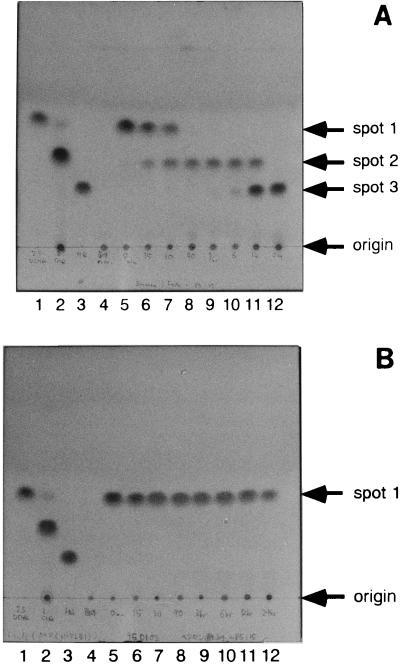
To detect the metabolites of 2,5-DCHQ catalyzed by LinD, we used TLC. 2,5-DCHQ (spot 1) was changed rapidly to spot 2, which was slowly converted to spot 3 by E. coli cells overexpressing LinD (Fig. 5A). In contrast, no change in 2,5-DCHQ was observed when E. coli cells overexpressing LinB were used as a control (Fig. 5B).
FIG. 5.
TLC analysis of the 2,5-DCHQ degradation activity of E. coli overproducing the LinD protein (A) and E. coli overproducing the LinB protein (negative control) (B). Lanes: 1, authentic 2,5-DCHQ (spot 1); 2, authentic CHQ (spot 2); 3, authentic HQ (spot 3); 4, extraction from E. coli MV1190 cells overproducing LinD or LinB; 5 to 12, extraction from E. coli MV1190 cells overproducing LinD or LinB incubated with 50 ppm of 2,5-DCHQ for 0 min, 15 min, 60 min, 90 min, 3 h, 6 h, 12 h, and 24 h, respectively. The procedure for TLC analysis was described in Materials and Methods.
Each spot was extracted by ethylacetate and analyzed by GC-MS. It was found that the retention times and mass spectra of spot 2 and spot 3 were identical to those of CHQ and HQ, respectively (data not shown). Furthermore, the mobilities of the spots corresponding to authentic compounds of 2,5-DCHQ, CHQ, and HQ were the same as spot 1, spot 2, and spot 3, respectively (Fig. 5A). To investigate whether CHQ was a substrate of LinD, CHQ was incubated with E. coli cells overexpressing LinD or LinB. The former cells slowly converted CHQ to HQ, whereas the latter cells did not (data not shown). This indicated that CHQ is also a substrate of LinD, although it seems to be a poor substrate compared with 2,5-DCHQ.
LinD activity is increased by the addition of glutathione.
Because LinD has homology to GSTs, we investigated the effect of glutathione on LinD activity. Crude cell extracts of E. coli MV1190 overexpressing LinD were prepared. When glutathione was added to a reaction mixture, LinD activity increased from 1.57 U/mg of protein to 5.85 U/mg of protein (a 3.7-fold increase). This result strongly suggested that LinD activity is glutathione dependent.
DISCUSSION
We have cloned the linD genes as a candidate for direct involvement in the degradation of 2,5-DCHQ, the intermediate of γ-HCH degradation by S. paucimobilis UT26. The fact that E. coli cells overproducing LinD converted 2,5-DCHQ to HQ and that the Tn5-induced mutants which show deficiencies in LinD activity have a deletion or rearrangements in the linD gene support the hypothesis that the linD gene is directly involved in 2,5-DCHQ degradation in UT26. Recently, we revealed that rearrangements occurred in the linD gene in UT103, which has no LinD activity (13).
Northern blot analysis revealed that linD mRNA is expressed upon induction with 2,5-DCHQ, whereas linA, linB, and linC are constitutively expressed. This suggested the existence of a regulatory system for linD gene expression.
TLC analysis revealed that LinD rapidly converts 2,5-DCHQ to CHQ and slowly converts CHQ to HQ. This indicates that LinD catalyzes the reductive dehalogenation of 2,5-DCHQ. However, the conversion of CHQ to HQ may not be essential for the degradation pathway of γ-HCH in S. paucimobilis UT26, because the conversion rate of CHQ to HQ by LinD is much lower than the rate of CHQ degradation by resting UT26 cells (13). Another degradation pathway of CHQ may exist in S. paucimobilis UT26.
D. B. Janssen classified the dehalogenation reaction of halogenated compounds into different groups according to their reaction mechanisms (10), and the reaction by LinD can be categorized into the reductive dehalogenation group. There are a few examples of enzymes from aerobic microorganisms belonging to this group, which is further categorized into two subgroups based on the presence of a reaction catalyzed by GSTs and on the presence of a reaction not catalyzed by GSTs (1, 2, 5, 21, 24, 25). To our knowledge, there is no report of the cloning of genes encoding enzymes catalyzing the latter type of reaction.
We found that LinD activity was increased by the addition of glutathione. The amino acid sequence of LinD has similarity to some known GSTs, although its molecular mass is larger than other GSTs. Bacterial GSTs were reviewed recently (27) and were placed in the theta class. Actually, LinD has similarity to GSTs of this class such as PcpC of Flavobacterium, the theta class GST of a plant (A. thaliana), and the GST of E. coli. Furthermore, LinD has 15 of 23 amino acids which are conserved in 10 of the 13 bacterial GST sequences aligned in the review article (Fig. 3). These results suggest the possibility that LinD is a novel member of the bacterial GSTs.
The purification of chlorophenol 4-monooxygenase in Burkholderia cepacia AC1100, which degrades 2,4,5-trichlorophenoxyacetic acid via 2,5-DCHQ, was reported previously (28). This enzyme converts 2,4,5-trichlorophenol to 2,5-DCHQ and then to 5-chloro-1,2,4-trihydroxybenzene in an NADH-dependent manner. Although B. cepacia AC1100 converts the same substrate as LinD, 2,5-DCHQ, its degradation pathway seems to be different from that of UT26.
From previous work and this study, we show that S. paucimobilis UT26 completely dechlorinates γ-HCH to HQ by three different types of dehalogenases, LinA, LinB, and LinD (Fig. 1). Each enzyme removes two chlorines which are located symmetrically at the γ-HCH ring.
ACKNOWLEDGMENTS
We thank K. Kimbara and M. Shimura, Railway Technical Research Institute, for technical assistance with mass spectrometry. This study was performed at the Biotechnology Research Center of The University of Tokyo.
K.M. is financially supported by research fellowships of the Japan Society for the Promotion of Science for Young Scientists. This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Apajalahti J H A, Salkinoja-Salonen M S. Complete dechlorination of tetrachlorohydroquinone by cell extracts of pentachlorophenol-induced Rhodococcus chlorophenolicus. J Bacteriol. 1987;169:5125–5130. doi: 10.1128/jb.169.11.5125-5130.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee S, Mahadevan A. Dissimilation of 2,4-D acid by Azotobacter chroococcum. Xenobiotica. 1990;20:607–618. doi: 10.3109/00498259009046876. [DOI] [PubMed] [Google Scholar]
- 3.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuda M. Ph.D. thesis. Tokyo, Japan: The University of Tokyo; 1989. [Google Scholar]
- 5.Häggblom M M, Janke D, Salkinoja-Salonen M S. Hydroxylation and dechlorination of tetrachlorohydroquinone by Rhodococcus sp. strain CP-2 cell extracts. Appl Environ Microbiol. 1989;55:516–519. doi: 10.1128/aem.55.2.516-519.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 7.Imai R, Nagata Y, Fukuda M, Takagi M, Yano K. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from γ-hexachlorocyclohexane. J Bacteriol. 1991;173:6811–6819. doi: 10.1128/jb.173.21.6811-6819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai R, Nagata Y, Senoo K, Wada H, Fukuda M, Takagi M, Yano K. Dehydrochlorination of γ-hexachlorocyclohexane (γ-BHC) by γ-BHC-assimilating Pseudomonas paucimobilis. Agric Biol Chem. 1989;53:2015–2017. [Google Scholar]
- 9.Janssen D B, Pries F, van der Ploeg J R, Kazemier B, Terpstra P, Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol. 1989;171:6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen D B, Pries F, van der Ploeg J R. Genetics and biochemistry of dehalogenating enzymes. Annu Rev Microbiol. 1994;48:163–191. doi: 10.1146/annurev.mi.48.100194.001115. [DOI] [PubMed] [Google Scholar]
- 11.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M, Yano K. Cloning of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989;171:2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 13.Miyauchi, K., Y. Nagata, and M. Takagi. Unpublished data.
- 14.Nagasawa S, Kikuchi R, Nagata Y, Takagi M, Matsuo M. Aerobic mineralization of γ-HCH by Pseudomonas paucimobilis UT26. Chemosphere. 1993;26:1719–1728. [Google Scholar]
- 15.Nagata Y, Imai R, Sakai A, Fukuda M, Yano K, Takagi M. Isolation and characterization of Tn5-induced mutants of Pseudomonas paucimobilis UT26 defective in γ-hexachlorocyclohexane dehydrochlorinase (LinA) Biosci Biotechnol Biochem. 1993;57:703–709. doi: 10.1271/bbb.57.703. [DOI] [PubMed] [Google Scholar]
- 16.Nagata Y, Nariya T, Ohtomo R, Fukuda M, Yano K, Takagi M. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J Bacteriol. 1993;175:6403–6410. doi: 10.1128/jb.175.20.6403-6410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagata Y, Ohtomo R, Miyauchi K, Fukuda M, Yano K, Takagi M. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J Bacteriol. 1994;176:3117–3125. doi: 10.1128/jb.176.11.3117-3125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata Y, Miyauchi K, Suh S, Futamura A, Takagi M. Isolation and characterization of Tn5-induced mutants of Sphingomonas paucimobilis defective in 2,5-dichlorohydroquinone degradation. Biosci Biotechnol Biochem. 1996;60:689–691. [Google Scholar]
- 19.Neidle E, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Cis-diol dehydrogenases encoded by the TOL pWW0 plasmid xylL gene and the Acinetobacter calcoaceticus chromosomal benD gene are members of the short-chain alcohol dehydrogenase superfamily. Eur J Biochem. 1992;204:113–120. doi: 10.1111/j.1432-1033.1992.tb16612.x. [DOI] [PubMed] [Google Scholar]
- 20.Palleroni N J. Genus I. Pseudomonas Migula 1894, 237AL (Nom. cons. Opin. 5, Jud. Comm. 1952, 237) In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 141–199. [Google Scholar]
- 21.Romanov V, Hausinger R P. NADPH-dependent reductive ortho dehalogenation of 2,4-dichlorobenzoic acid in Corynebacterium sepedonicum KZ-4 and coryneform bacterium strain NTB-1 via 2,4-dichlorobenzoyl coenzyme A. J Bacteriol. 1996;178:2656–2661. doi: 10.1128/jb.178.9.2656-2661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shine J, Dalgarno L. Determination of cistron specificity in bacterial ribosomes. Nature (London) 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 23.Terada I, Kwon S-T, Miyata Y, Matsuzawa H, Ohta T. Unique precursor structure of an extracellular protease, aqualysin I, with NH2- and COOH-terminal pro sequences and processing in E. coli. J Biol Chem. 1990;265:6576–6581. [PubMed] [Google Scholar]
- 24.Uotila J S, Kitunen V H, Saastamoinen T, Coote T, Häggblom M M, Salkinoja-Salonen M S. Characterization of aromatic dehalogenases of Mycobacterium fortuitum CG-2. J Bacteriol. 1992;174:5669–5675. doi: 10.1128/jb.174.17.5669-5675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 27.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xun L. Purification and characterization of chlorophenol 4-monooxygenase from Burkholderia cepacia AC1100. J Bacteriol. 1996;178:2645–2649. doi: 10.1128/jb.178.9.2645-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]