Abstract
The plant-pathogenic bacterium Pseudomonas syringae pv. glycinea PG4180.N9 synthesizes high levels of the polyketide phytotoxin coronatine (COR) at 18°C, whereas no detectable toxin is produced at 28°C. Previously, we reported that the temperature-sensitive activation of three promoters within the COR biosynthetic gene cluster might explain thermoregulation of COR biosynthesis. The present study was aimed at furthering our understanding of the transcriptional as well as the posttranslational effects of temperature on expression of cmaB, which encodes an enzyme involved in COR biosynthesis. Transcriptional fusions using a promoterless glucuronidase gene and Northern blot analyses were used to monitor promoter activities and transcript abundance for the cmaABT operon during bacterial growth at 18 and 28°C. Promoter activity and transcription rates were maximal when cells were incubated at 18°C and sampled at mid-logarithmic phase. Transcription declined moderately during the transition to stationary phase but remained higher at 18°C than at 28°C. Western blot analysis indicated that CmaB accumulated in the late stationary phase of P. syringae cultures grown at 18°C but not in cultures incubated at 28°C. Temperature shift experiments indicated that CmaB stability was more pronounced at 18°C than at 28°C. Although temperature has a strong impact on transcription of COR biosynthetic genes, we propose that thermoregulation of protein stability might also control COR synthesis.
The phytopathogenic bacterium Pseudomonas syringae pv. glycinea PG4180.N9 causes bacterial blight of soybeans, a foliar disease characterized by necrotic leaf spots surrounded by chlorotic halos. The symptoms of bacterial blight are most severe during periods of cold, humid weather (9). P. syringae pv. glycinea PG4180.N9 produces the chlorosis-inducing polyketide phytotoxin coronatine (COR) in a temperature-dependent manner (21). COR functions as an important virulence factor in P. syringae (4). Currently, its mode of action is the subject of intensive investigations (6, 10, 22, 36). COR consists of two moieties, the polyketide coronafacic acid (CFA) and a cyclized amino acid, coronamic acid (CMA) (see Fig. 2A). Both compounds function as intermediates in COR biosynthesis and are joined by an amide linkage (2, 19). Biosynthesis of COR in P. syringae is maximal at 18°C, whereas no detectable amount of COR is produced at 28 to 30°C, a temperature range otherwise optimal for growth of P. syringae (21, 25).
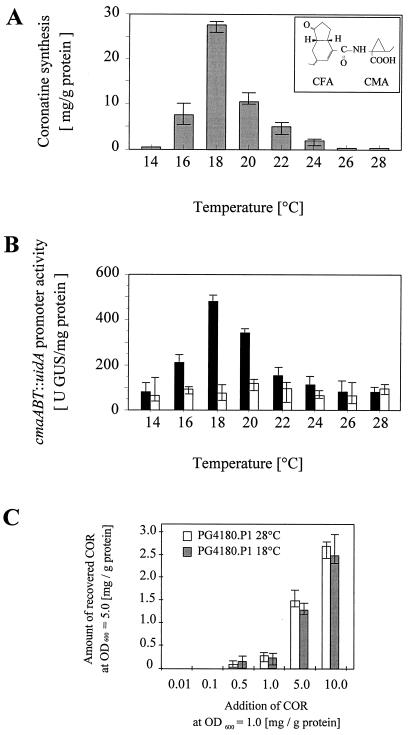
FIG. 2.
Temperature effects on COR production by PG4180.N9 (A) and on cmaABT::uidA promoter activity in PG4180.N9 (B). (A) For detection of COR, extracts from supernatants of late-stationary-phase PG4180.N9 cultures were examined by HPLC. (B) Promoter activities were determined as GUS expression rates in late-stationary-phase cultures of P. syringae pv. glycinea PG4180.N9(pRGMU1) (black bars) and PG4180.P1(pRGMU1) (white bars). Quantities represent the average of three experiments with two replicates. The structure of coronatine consisting of a polyketide component (CFA) and a cyclized amino acid derivative (CMA) is shown in the inset of panel A. (C) Determination of the detection limit of COR quantitation. Defined amounts of COR were added and later recovered from PG4180.P1 (COR−) cells grown at 18 and 28°C.
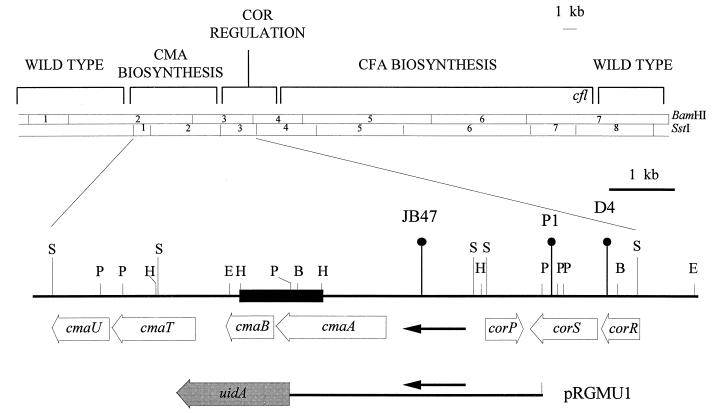
In P. syringae pv. glycinea PG4180.N9, enzymes involved in COR biosynthesis are encoded by the plasmid-borne 32-kb COR gene cluster (Fig. 1). Mutational, transcriptional, and nucleotide sequence analyses have been used to characterize the COR gene cluster (5). Based on the phenotypic characterization of transposon insertions, two separate DNA regions were associated with CMA and CFA biosynthesis. The 6.9-kb DNA region required for CMA biosynthesis was reported to contain three genes, cmaA, cmaT, and cmaU (30). Putative biosynthetic functions were suggested for the gene products of cmaA and cmaT based on sequence similarities to amino acyl-adenylating enzymes and thioesterases, respectively (30). Reevaluation of the nucleotide sequence data recently indicated errors in the published sequence of the CMA biosynthetic DNA region. When the cmaA locus was overexpressed in Escherichia coli by using the T7 promoter, two proteins with molecular masses of approximately 62 and 29 kDa were observed instead of the predicted 100-kDa gene product for cmaA (3). The N-terminal amino acid sequences of the 62- and 29-kDa proteins, designated CmaA and CmaB, were determined and will be published elsewhere (3). The translational start site of cmaA was located 399 bp downstream of the previously published start site, whereas the predicted translational stop of cmaA was located 608 bp upstream of the previously indicated stop site (Fig. 1). A second 806-bp open reading frame (ORF), designated cmaB, with a translational start site at nucleotide 3100 of the previously released sequence was identified (Fig. 1). These corrections in the nucleotide sequence were submitted to GenBank (accession no. U14657). Changes in the nucleotide sequence of cmaA did not alter its relatedness to genes encoding amino acyl-adenylating enzymes. The role of CmaB as a putative oxidative cyclase will be reported elsewhere (3).
FIG. 1.
Top, partial restriction map of the COR biosynthetic gene cluster of P. syringae pv. glycinea PG4180.N9 showing functional regions as determined by transposon mutagenesis (2). Insertions in regions marked “WILD TYPE” had no effect on COR biosynthesis. The location of the cfl gene (17, 25) is indicated. Bottom, enlargement of the CMA biosynthetic and COR regulatory region, showing the locations and orientations of biosynthetic (cmaA, cmaB, cmaT, and cmaU) and regulatory (corP, corS, and corR) genes. Vertical lines with closed circles indicate insertion mutants used in this study. The cmaABT promoter region is represented by a black horizontal arrow. The location of pRGMU1, which contains the cmaABT promoter fused to uidA, is outlined as a bar with filled arrow. The 1.2-kb HindIII fragment used as a DNA probe for Northern blot analyses is indicated by a bold black bar. Restriction enzymes used: B, BamHI; E, EcoRI; H, HindIII; P, PstI; S, SstI.
The genes required for CFA and CMA biosynthesis are located on two separate transcriptional units designated the cfl/CFA operon and the CMA operon, respectively (17, 30) (Fig. 1). Transcriptional fusions of a promoterless β-glucuronidase (GUS) gene (uidA) to promoter regions upstream of either transcriptional unit indicated that temperature affected transcription of the COR biosynthetic genes. GUS expression was optimal when P. syringae PG4180.N9 cells containing the cmaABT::uidA and cfl::uidA transcriptional fusions were incubated at 18°C; in contrast, GUS expression at 28°C was four- to sixfold lower (17, 30). However, these results do not explain why P. syringae showed a complete loss of COR synthesis at 28°C instead of the predicted four- to sixfold decrease in COR synthesis.
Previously, we demonstrated the requirement of a modified two-component regulatory system for temperature-dependent COR synthesis (32). The regulatory system is encoded within the COR gene cluster of PG4180.N9 and consists of a putative histidine protein kinase, CorS, and two proteins, designated CorR and CorP, with relatedness to transcriptional response regulators. Insertion mutations in each of the three regulatory genes abolished transcriptional activation of the COR biosynthetic genes and suggested a role for these genes in temperature-dependent transcription (32).
As described for other genes involved in virulence and pathogenicity of phytopathogenic bacteria (1, 24), regulation of COR synthesis in P. syringae PG4180.N9 is controlled by environmental factors. One aim of this study was to analyze the correlation between COR biosynthesis and steady-state rates of COR gene expression in response to slight changes in ambient temperature. To initiate investigations on the temperature-sensing and signal transduction mechanism, we tested during which stages of bacterial growth the incubation temperature had the strongest impact on COR gene expression and COR synthesis. The transcriptional fusion cmaABT::uidA was monitored in cis and in trans during all growth phases of PG4180.N9 at 18 and 28°C. Quantitative Northern and Western blot analyses, respectively, were used to examine the abundance of the cmaABT transcript and the CmaB protein. Rates of COR and CMA biosynthesis were measured in a growth-dependent manner. Our results indicate that transcriptional activation of COR biosynthetic genes at 18°C as well as decreased protein stability at 28°C contribute to the thermoregulation of COR production.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas strains were maintained on mannitol-glutamate (MG) medium (14) at 28°C. Prior to the temperature-dependent growth studies, a single colony of each PG4180.N9 derivative grown 96 h on MG agar was resuspended in 5 ml of King’s B medium (15) and incubated overnight on a rotary shaker at 280 rpm and 28°C. Two milliliters of the overnight culture was then used to inoculate 200 ml of HSC medium (21) in 1-liter Erlenmeyer flasks. Cultures were incubated under the growth conditions described for the overnight culture but with various incubation temperatures. Bacterial growth was continuously monitored by measuring the optical density at 600 nm (OD600). The protein concentration in cell lysates was determined by the Bradford assay (26). The following antibiotics were added to media (in micrograms per milliliter): kanamycin, 25; spectinomycin, 25; streptomycin, 25; and chloramphenicol, 100.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| E. coli DH5α | 26 | |
| P. syringae pv. glycinea | ||
| PG4180.N9 wild type | COR+ CMA+ CFA+ Kmr | 31 |
| PG4180.JB47 | COR− CMA− CFA+ Smr Spr; carries a mini-Tn5uidA insertion within the cmaABT operon | 27 |
| PG4180.G70 | COR− CMA+ CFA− Kmr | 2 |
| PG4180.P1 | COR− CMA− CFA− Kmr GmrcorS | 32 |
| PG4180.D4 | COR− CMA− CFA− KmrcorR | 2 |
| Plasmids | ||
| pMU2 | Tcr; contains SstI fragments 1 and 2 of the COR biosynthetic gene cluster in pLAFR3 | 31 |
| pRGMU1 | Smr Spr; contains a 2.9-kb PstI insert carrying the cmaABT promoter region derived from the COR biosynthetic gene cluster in pRG960sd | 30 |
| pRGMU3 | Smr Spr; contains a 1.4-kb SalI-EcoRV insert carrying a constitutive promoter region derived from pMU2 in pRG960sd | 30 |
| pRG960sd | Smr Spr; contains promoterless uidA with start codon and Shine-Dalgarno sequence | 33 |
GUS assays.
Plasmid pRGMU1 (30) contains a fusion of the cmaABT promoter to a promoterless GUS gene (uidA) and was used to measure transcriptional activity in P. syringae PG4180.N9 in trans. Mutant PG4180.JB47 (27) contains a promoterless uidA gene on a mini-Tn5 transposable element (37) within the cmaABT operon of P. syringae PG4180.N9. This mutant was used to measure cmaABT promoter activities in cis. Plasmid pRGMU3 (30) contains a constitutive promoter of P. syringae PG4180.N9 fused to a promoterless uidA gene and served as a control. Promoter activity was quantified by fluorometric analysis of GUS activity (38), using a Fluorolite-1000 microplate reader (Dynatech, Denkendorf, Germany) and 96-well microtiter plates.
Northern blot analysis.
Total RNA was isolated from 1.0 ml of bacterial cultures by using an RNeasy RNA isolation kit (Qiagen, Hilden, Germany). Aliquots of RNA (8 μg) were loaded on denaturing 1.5% agarose-formaldehyde gels, electrophoretically separated, and transferred to nitrocellulose filters (Schleicher & Schuell, Dassel, Germany) according to standard procedures (26). The DNA probe for Northern blot analysis was constructed by digesting plasmid pMU2 with HindIII. After electrophoretic separation, a 1.2-kb HindIII fragment was purified and radiolabeled with [α-32P]dCTP, using a random primer labeling kit (Stratagene, Heidelberg, Germany). Probes had a specific radioactivity of >4 × 108 cpm per μg of DNA. Hybridizations were carried out in hybridization solution (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10× Denhardt’s reagent, 0.5 mg of herring sperm DNA per ml, 50 mM NaH2PO4, 50% deionized formamide) for 16 h at 42°C, and posthybridization washes were performed at 50°C in 0.1× SSC–0.1% sodium dodecyl sulfate. Signals were detected and quantified with a PhosphorImager and Molecular Dynamics ImageQuant software as recommended by the manufacturer (Molecular Dynamics, Krefeld, Germany).
Western blot analysis.
Total protein extracts were isolated from cell pellets of bacterial cultures (1.5 ml) and were quantified by the Bradford assay, using standard procedures (26). Proteins were diluted, and equal amounts (1 μg/lane) were loaded on sodium dodecyl sulfate–10% polyacrylamide gels. Electrophoresis, electroblotting, and hybridizations on nitrocellulose membranes (Amersham-Buchler, Braunschweig, Germany) were conducted according to standard procedures (26). Polyclonal antisera raised against purified CmaB in mice were provided by the Oklahoma State University Hybridoma Center for Monoclonal Antibody Research. The specificity of the CmaB antiserum at a dilution of 1:500 was evaluated with recombinant CmaB overproduced in E. coli and crude protein extracts of P. syringae PG4180.N9. For signal detection, secondary anti-mouse immunoglobulin G antibodies conjugated to alkaline phosphatase (Sigma, Darmstadt, Germany) were used at a concentration of 1:4,000, and the reaction was visualized using 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium salt. Signals were quantified with an Ultroscan XL densitometer (Pharmacia-LKB, Bromma, Sweden) according to the manufacturer’s recommendations.
Detection and quantitation of COR synthesis and CMA production.
Organic acids were extracted from cell-free bacterial supernatants (1.5 ml) and analyzed for the presence of COR by a high-pressure liquid chromatography (HPLC) method described elsewhere (21). For analysis of CMA biosynthesis, cell-free bacterial supernatants of PG4180.G70 (5 ml) were used. Detection and quantitation of CMA was done as reported by Ullrich et al. (31), with most steps scaled down 1:100 to facilitate processing of many samples.
RESULTS
Steady-state COR synthesis and COR gene expression in response to temperature.
PG4180.N9 and PG4180.P1 were used in this experiment, and transcriptional activity was evaluated in the presence and absence of the cmaABT::uidA fusion contained on pRGMU1. The COR− mutant PG4180.P1 contains a gentamicin resistance cassette in corS and is impaired in the thermoresponsive transcriptional activation of COR biosynthetic genes (17, 32). All strains were grown in 200 ml of HSC medium at 14, 16, 18, 20, 22, 24, 26, or 28°C. To ensure sufficient bacterial growth and maximal COR synthesis at each incubation temperature, cells were incubated for 3 to 8 days until they reached an OD600 of approximately 4.5 to 5.0 (late stationary phase). Supernatants of PG4180.N9 and PG4180.P1 cultures were then examined for COR synthesis (Fig. 2A).
COR biosynthesis in PG4180.N9 was maximal at 18°C and decreased steadily when cells were incubated at higher or lower temperatures. No COR was detected in cultures incubated at 14, 26, and 28°C. COR synthesis in mutant PG4180.P1 was not detected throughout all of the temperatures studied (data not shown). To ensure that COR− phenotypes were due to lack of COR synthesis rather than an insufficient sensitivity of the detection system, the detection limit of the used HPLC quantitation method was determined. Defined amounts of HPLC-purified COR were added to PG4180.P1 (COR−) cultures grown to an OD600 of 1.0. Aliquots of these cultures were then incubated at 18 or 28°C until they reached an OD600 of 5.0. Cells were harvested, and amounts of COR in the supernatants were determined (Fig. 2C). Approximately 60 to 70% of the added COR was lost during the incubation and the subsequent extraction procedure. An average of 0.1 mg/g of protein was recovered from PG4180.P1 which had been treated with 0.5 mg of COR/g of protein, whereas no COR was detected when PG4180.P1 had been treated with 0.1 mg of COR/g of protein. These data indicated that the detection limit is roughly 0.1 mg of COR/g of protein. Temperature did not seem to influence the recovery efficiency. Given this detection limit, PG4180.N9 produced approximately 280 times more COR at 18°C than could minimally be detected with this technique.
Transcription of the fusion cmaABT::uidA in PG4180.N9(pRGMU1) and PG4180.P1(pRGMU1) was examined at the same temperatures (Fig. 2B). In preliminary experiments, the possibility of endogenous GUS activity was eliminated by testing PG4180.N9 and PG4180.N9(pRG960sd) for GUS activity. No activity could be measured (data not shown). GUS expression in PG4180.N9(pRGMU1) was maximal at 18°C and decreased at lower and higher incubation temperatures. Basal levels of GUS activity (above fivefold less than the amount obtained at 18°C) were observed when PG4180.N9(pRGMU1) cells were grown at 14, 26, and 28°C (Fig. 2B). GUS expression in PG4180.P1(pRGMU1) remained consistently low throughout the temperature range. Basal levels of cmaABT promoter activities in PG4180.N9 at 14 and 28°C on one hand and no COR synthesis at these temperatures on the other hand prompted investigations on temperature-dependent transcript stability and protein stability during bacterial growth (see below).
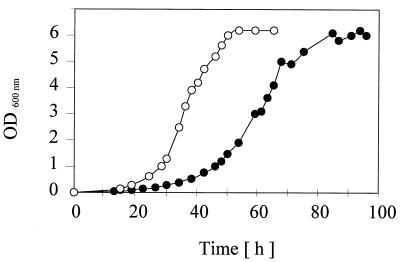
Growth of P. syringae PG4180.N9 at 18 and 28°C.
Growth curves for PG4180.N9 in HSC medium at 18 and 28°C are shown in Fig. 3. During exponential growth, changes in optical density directly correlated with changes in the total number of viable cells as determined by dilution plating (data not shown). Growth at 18°C was considerably slower than growth at 28°C, with maximum growth rates of 0.088 h−1 at 18°C and 0.139 h−1 at 28°C. The doubling times during exponential growth were about 8 h at 18°C and 5 h at 28°C. The lag phase was more pronounced at 18°C than at 28°C, indicating the possible requirement of specific acclimation to growth at the lower temperature. When incubated at 18°C, PG4180.N9 reached the stationary phase at 60 h whereas cells incubated at 28°C reached the stationary phase at 35 h. Maximal optical densities during late stationary phase were similar at the two temperatures. When these experiments were repeated with other derivatives of PG4180.N9 used in this study, temperature-dependent growth differences very similar to those reported for the wild-type strain were observed (data not shown).
FIG. 3.
Growth curves for P. syringae pv. glycinea PG4180.N9 at two different temperatures. Cells growing exponentially at 28°C were transferred to fresh HSC medium and grown at 18°C (•) or 28°C (○). Growth was monitored by measuring the OD600 in two experiments with two replicates.
Analysis of the cmaABT promoter activity during growth of PG4180.N9.
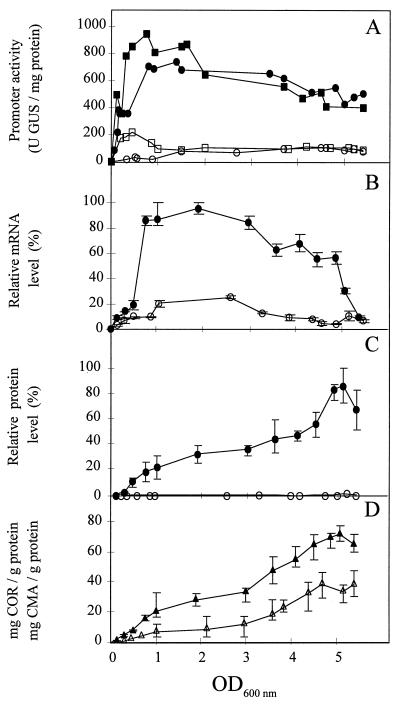
The effects of temperature on transcriptional activity of the cmaABT promoter in cis and in trans were examined by analyzing GUS activity in PG4180.N9(pRGMU1) and PG4180.JB47 incubated at 18 and 28°C. In contrast to PG4180.N9(pRGMU1), which allowed measurement of the cmaABT promoter strength in trans, PG4180.JB47 carries a promoterless uidA gene in the CMA biosynthetic region in cis (27). During the early exponential phase, GUS expression from plasmid pRGMU1 and reporter gene expression in PG4180.JB47 increased rapidly when cells were incubated at 18°C and reached maximum expression rates of 700 to 900 U of GUS/mg of protein at an OD600 of 1.0 to 1.5 (Fig. 4A). In both strains, GUS expression at 18°C remained relatively constant during the transition period from exponential growth to stationary phase (OD600 = 2.0 to 3.5). During stationary phase, GUS expression in PG4180.N9(pRGMU1) and PG4180.JB47 declined to 500 U of GUS/mg of protein (OD600 = 5.0). The results indicated that the cmaABT promoter is most active during exponential growth at 18°C. In contrast, GUS expression in PG4180.N9(pRGMU1) and PG4180.JB47 incubated at 28°C remained low (80 to 100 U of GUS/mg of protein) throughout the exponential and stationary phases (Fig. 4A). For early logarithmic growth of PG4180.JB47, an increase in cmaABT promoter activity to about 200 U of GUS/mg of protein was observed at 28°C. However, this was not true for GUS expression from plasmid pRGMU1. In general, similar results were obtained for the cmaABT promoter regardless of position (cis or trans), suggesting that copy number and positional effects were not important. Growth temperature experiments were also conducted with PG4180.N9(pRGMU3), which carries a constitutive promoter upstream of uidA (30). Expression of uidA from this promoter was not influenced by temperature and remained relatively constant during all growth phases (data not shown).
FIG. 4.
Effects of temperature on cmaABT transcriptional activities, cmaABT transcript abundance, CmaB protein synthesis, and production of COR and CMA during growth of P. syringae pv. glycinea PG4180.N9. (A) cmaABT promoter activities were measured in GUS expression units in trans in PG4180.N9(pRGMU1) at 18°C (•) and 28°C (○) and in cis in PG4180.JB47 at 18°C (▪) and 28°C (□). (B) Quantitative Northern blot analysis for cmaABT transcript abundance at 18°C (•) and 28°C (○). The relative mRNA level is related to the highest measured value, which was defined as 100%. (C) Quantitative Western blot analysis using polyclonal antibodies raised against CmaB and PG4180.N9 cultures grown at 18°C (•) and 28°C (○). The relative protein level is related to the highest measured value, which was defined as 100%. (D) Synthesis of COR (closed triangles) and CMA (open triangles) by PG4180.N9 and PG4180.G70, respectively, at 18°C. Both compounds were absent from supernatants of PG4180.N9 or PG4180.G70 grown at 28°C. Quantities represent two experiments with three replicates.
Northern blot analysis of the cmaABT operon.
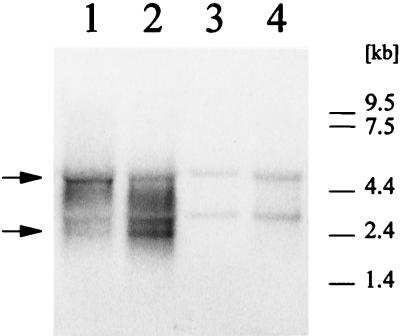
To determine the size of the cmaABT transcript, total RNA samples were prepared from P. syringae cells grown at 18°C and harvested in the late stationary phase. The DNA probe, a 1.2-kb HindIII fragment from within the cmaABT operon (Fig. 1), hybridized with two RNA species with molecular sizes of 5 and 2.5 kb (Fig. 5). A transcript size of 5 kb corresponded to the predicted polycistronic message for cmaABT (30), whereas the smaller RNA molecule might represent either a stable degradation product of the 5-kb transcript, a monocistronic messenger for the cmaA gene, or an additional mRNA of unknown origin. To test this, we examined total RNA of exponentially growing PG4180.N9 at 18 and 28°C as well as total RNA from the regulatory mutant PG4180.D4. This mutant is impaired in activation of the cmaABT promoter and carries a transposon insertion in corR, a gene involved in the transcription of cmaABT (2, 32). As shown in Fig. 5, the two bands were found in RNA samples from PG4180.N9 incubated at 18°C but not in those derived from cultures grown at 28°C. Neither signal was detected in RNA from PG4180.D4 incubated at 18 or 28°C, suggesting that these two RNA species were associated with temperature-dependent COR biosynthesis. Insertion mutants of PG4180.N9 are currently used to investigate the nature of this phenomenon in detail. When monitored throughout the entire growth of the culture, signal intensities for the cmaABT transcripts were highest during exponential growth of PG4180.N9 at 18°C (Fig. 4B). The greatest accumulation of cmaABT mRNA at 28°C reached approximately 25% of the levels observed at 18°C and occurred during the transition between exponential and stationary phases (OD600 = 2.0 to 3.5). At both temperatures, abundance of the cmaABT mRNA species proportionally decreased during stationary phase (Fig. 4B), thus confirming the results obtained with the cmaABT::uidA transcriptional fusions. The intensities of the 5- and 2.5-kb bands decreased at similar rates, suggesting a common type of regulation. These results indicated that mRNA abundance for the cmaABT operon varies in response to both ambient temperature and growth phase.
FIG. 5.
Northern blot analysis of total mRNA of P. syringae pv. glycinea PG4180.N9 grown at 18 and 28°C and PG4180.D4 grown at 18°C. Cells were grown to an OD600 of 1.0 and then subjected to total RNA extraction. RNA samples (8 μg) were electrophoretically separated, transferred to nitrocellulose filters, and hybridized with a 32P-labeled 1.2-kb HindIII fragment from the cmaABT operon. The cmaABT signals are marked with arrows. Lanes: 1, PG4180.N9, 18°C, OD600 = 0.7; 2, PG4180.N9, 18°C, OD600 = 1.0; 3, PG4180.N9, 28°C, OD600 = 1.0; 4, PG4180.D4, 18°C, OD600 = 1.0.
Western blot analysis of the CmaB protein during growth of PG4180.N9.
In preliminary experiments, the CmaB antiserum reacted in Western blots with a 35-kDa protein in cellular extracts from PG4180.N9 at 18°C but not with extracts from cells grown at 28°C (Fig. 6). To monitor CmaB production in response to temperature and growth stage, PG4180.N9 was incubated at 18 and 28°C, and total cellular proteins were extracted at various time points. Protein extracts were subjected to Western blot analyses using the CmaB antiserum (Fig. 4C). Signals for CmaB increased steadily throughout the growth period at 18°C. However, only weak signals were observed in protein extracts from cells incubated at 28°C. In contrast to expression of the cmaABT::uidA fusions and the cmaABT transcript (Fig. 4A and B), accumulation of the CmaB protein reached maximum levels in the late stationary phase, suggesting a considerable stability of this protein during stationary phase at 18°C.
FIG. 6.
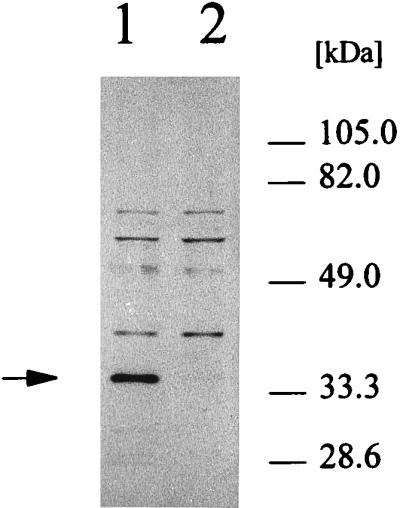
Western blot analysis of total cellular protein of P. syringae pv. glycinea grown at 18 and 28°C. Cells were grown to mid-exponential phase (OD600 = 1.0) and then subjected to total protein extraction. Protein samples of 1 μg were electrophoretically separated and transferred to nitrocellulose filters. Western blotting was performed with polyclonal antibodies raised against CmaB. The specific signal is marked with an arrow. Lanes: 1, PG4180.N9, 18°C; 2, PG4180.N9, 28°C.
COR and CMA biosynthesis in dependence of temperature and growth.
The synthesis rates for COR and CMA were analyzed in a growth phase-dependent manner to confirm the data obtained from transcriptional fusions and by Northern and Western blot analyses. COR was detected in cell-free supernatants of PG4180.N9 cultures grown at 18°C but not in those grown at 28°C (Fig. 4D). Accumulation of COR increased with bacterial growth at 18°C and correlated with CmaB abundance. Biosynthesis of CMA, one of the two precursors of COR, was monitored in supernatants of mutant PG4180.G70 throughout the bacterial growth at 18 and 28°C. PG4180.G70 is impaired in synthesis of CFA (2, 19). This mutant secretes CMA, which enabled us to quantify this compound in the bacterial supernatant. As shown in Fig. 4D, rates of CMA synthesis resembled rates of COR production at 18°C. No CMA was detected in supernatants of PG4180.G70 grown at 28°C (data not shown), supporting the idea that the biosyntheses of precursor and end product might share a common type of regulation.
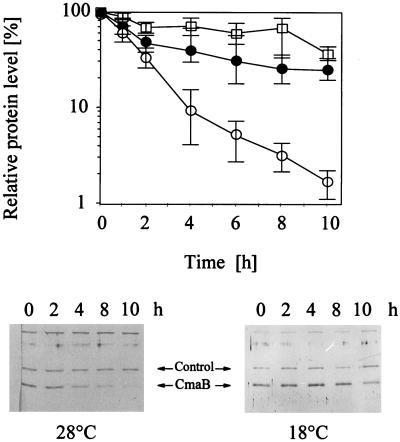
Analysis of protein stability after temperature shift.
The results of the Western blot analysis raised questions about the thermostability of proteins involved in CMA biosynthesis. To investigate this, the influence of temperature on the stability of CmaB was examined by treating aliquots of PG4180.N9 cultures (OD600 = 1.0; 18°C) with chloramphenicol and incubating them at 18 or 28°C (Fig. 7). As controls, intensities of unspecific signals which also immunoreacted with the CmaB antiserum were measured. This allowed an estimation of the general protein decay, which could be expected to occur more rapidly at 28°C than at 18°C due to increased proteolytic activities. As shown in Fig. 7, control proteins were degraded to 30 to 45% of the original values after 10 h at 28°C. Degradation to a similar extent was observed after 10 h at 18°C. Although the stability of CmaB appeared to be temperature independent for up to 2 h, the CmaB protein was more stable at 18°C than at 28°C when examined 6, 8, and 10 h after translation was interrupted. At 18°C CmaB degradation resembled the general decay of proteins, whereas at 28°C CmaB was degraded to less than 2% of the original value after 10 h. CmaB had half-lives of approximately 8 h at 18°C and 2 h at 28°C. In summary, the stability of CmaB was temperature responsive, suggesting a significant posttranslational regulation of COR biosynthetic proteins by temperature.
FIG. 7.
Effect of temperature shift from 18 to 28°C on CmaB protein stability in P. syringae pv. glycinea PG4180.N9. Cells were grown to an OD600 of 1.0 at 18°C and then treated with chloramphenicol. The culture was divided into two aliquots; one of the subcultures was incubated at 28°C (○), and the other remained at 18°C (•). Cells were harvested at the indicated time points, and total cellular protein was extracted and subjected to quantitative Western blot analysis. □, band intensities of unknown proteins which nonspecifically immunoreacted with the CmaB antiserum (control). The relative protein level is related to the highest measured value, which was defined as 100%. Quantities represent two experiments with two replicates. The photographs represent Western blots illustrating a temperature shift experiment.
DISCUSSION
Previously, coronatine synthesis by P. syringae was shown to be thermoresponsive at the transcriptional level (17, 31, 32). A modified two-component regulatory system was associated with the temperature-sensitive transcription of COR biosynthetic genes (32). In this study, we analyzed the possible influence of posttranscriptional and posttranslational factors on this system. Data reported in this study indicated that transcriptional activation is the main factor for thermosensitivity, thus confirming previous work on the regulation of COR biosynthesis (32). Furthermore, protein stability was shown to be decreased at the elevated temperature, suggesting that thermoregulation was also influenced by posttranslational factors.
We focused our attention on regulation of CmaB expression as a representative model for expression of COR biosynthetic enzymes. Transcriptional analyses of the cmaABT and cfl/CFA operons revealed that the two promoter regions had characteristics in common (17, 23, 31, 32). Recently, Rangaswamy et al. (25) demonstrated that a cfl::uidA transcriptional fusion and accumulation of Cfl, another protein involved in COR biosynthesis, were temperature dependent similarly to the system described herein.
Following synthesis, COR remains a stable component of the bacterial supernatant of P. syringae (21). Accordingly, the evaluation of late-stationary-phase supernatants of PG4180.N9 incubated at various temperatures represented the actual amount of synthesized COR. When comparing COR synthesis rates and cmaABT transcriptional activity, we identified the optimum for both at 18°C. No detectable COR and only basal levels of promoter activity were found at 14 or 28°C, confirming in part data of earlier studies (25, 30). Given the importance of COR for bacterial multiplication in planta (4, 20) and our increasing knowledge of COR as a mimic of plant signaling molecules (6, 10, 22, 36), it is tempting to speculate that P. syringae possesses a complex network for sensing optimal conditions for the energy-expensive biosynthesis of this phytotoxin. By monitoring COR biosynthetic promoter activities at various temperatures ranging from 14 to 28°C, we also attempted to rule out a possible involvement of global cold shock phenomena on our system. COR is a secondary metabolite and therefore not essential for bacterial survival at low temperatures per se. Cold shock induction of gene expression in E. coli and other organisms generally has been defined by temperature down-shifts of 13°C and more (11, 13). In the system described herein, temperature changes of less than 5°C are sufficient to change transcriptional activity, suggesting a regulation of COR biosynthesis independent of cold shock phenomena.
Interestingly, cmaABT promoter activity and cmaABT transcript abundance were highest in the early and mid-logarithmic growth phases of PG4180.N9 at 18°C and moderately decreased during the late stationary phase. This pattern of expression is different from that of genes which are activated by stress responses just before or during the transition to and the duration of the stationary phase (16). Among plant-pathogenic bacteria, a similar pattern of expression was reported for motility and virulence determinants in Ralstonia solanacearum (7). However, in R. solanacearum, expression of virulence genes was found to depend on extracellular factor-mediated cell density signals, which have not been investigated in the system described herein. In its natural habitat, a rapid response to temperature shifts resulting in the transcriptional activation of COR genes might enable P. syringae to take advantage of favorable abiotic conditions and to infect the host plant. Temperature decreases are often associated with rainy or humid weather conditions, which foster symptom development of bacterial blight of soybeans (9). As predicted for R. solanacearum (7), P. syringae may exhibit a low virulence phenotype during saprophytic growth on the leaf surface. The pathogen might shift to a highly virulent phenotype when it enters the plant tissue, where nutrients allow extensive bacterial growth.
There are several possible explanations for the basal levels of cmaABT promoter activity and transcript abundance at 28°C despite no detectable accumulation of CmaB and complete lack of COR synthesis at the same temperature. For example, a threshold level of promoter activity at 28°C might be essential for a rapid acclimation to temperature shifts in the natural setting. Alternatively, nonspecific cross talk with unrelated transcriptional activators might occur independent of the incubation temperature. Cross talk between regulatory systems in prokaryotes has been discussed intensively (8, 35). The increased instability of CmaB at 28°C may completely overcome the background of transcriptional activation, finally resulting in a very low level of CmaB and the COR− phenotype.
Why the CmaB protein was degraded more efficiently at 28°C compared to 18°C and to the control proteins remains to be elucidated. Several reviews have focused on the role of temperature in adaptive processes in prokaryotic and eukaryotic organisms (18, 28, 29).
It remains to be addressed why significant CmaB accumulation at 18°C occurred in the late stationary phase although the amount of the cmaABT transcript was relatively low at that time point. One explanation is the putative involvement of a CmaB-stabilizing factor(s) which might be present preferably in the stationary phase. Alternatively, a CmaB-destabilizing or -degrading factor(s) might be less abundant in the stationary phase. In analogy to polyketide synthesis (12) or non-ribosomal peptide synthesis (34), COR biosynthesis could be mediated by multifunctional protein complexes consisting of several enzymatic components. If CmaB is part of such a complex, it might withstand degradation more efficiently and thus accumulate in the cells toward the end of the stationary phase.
The complete loss of synthesis of COR and CMA was observed in PG4180.N9 cultures incubated at 28°C. Currently, we cannot rule out temperature-dependent enzymatic activity of CmaB or other COR biosynthetic enzymes since minor amounts of CmaB were detected at 28°C. Investigations on the enzymatic activities at 18 and 28°C will be conducted as soon as methods become available.
Our laboratories are currently investigating whether COR biosynthesis is the only virulence factor of P. syringae which is subject to such a subtle thermoregulation and whether there are global regulatory principles governing these processes.
ACKNOWLEDGMENTS
We thank M. Schergaut and J. Boch for providing mutant strain PG4180.JB47, G. W. Sundin for critical comments on the manuscript, and U. Völker for valuable instructions during the use of the PhosphorImager instrument.
C.L.B. acknowledges support from National Science Foundation grant MCB-9609681.
REFERENCES
- 1.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender C, Liyanage H, Palmer D, Ullrich M, Young S, Mitchell R. Characterization of the genes controlling biosynthesis of the polyketide phytotoxin coronatine including conjugation between coronafacic and coronamic acid. Gene. 1993;133:31–38. doi: 10.1016/0378-1119(93)90221-n. [DOI] [PubMed] [Google Scholar]
- 3.Bender, C. L., M. Ullrich, J. Patel, and R. Parry. Unpublished data.
- 4.Bender C L, Stone H E, Sims J J, Cooksey D A. Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol Mol Plant Pathol. 1987;30:273–283. [Google Scholar]
- 5.Bender C L, Palmer D A, Penaloza-Vazquez A, Rangaswamy V, Ullrich M. Biosynthesis of coronatine, a thermoregulated phytotoxin produced by the phytopathogen Pseudomonas syringae. Arch Microbiol. 1996;166:71–75. [Google Scholar]
- 6.Benedetti C E, Xie D, Turner J G. COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 1995;109:567–572. doi: 10.1104/pp.109.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clough S J, Flavier A B, Schell M A, Denny T P. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl Environ Microbiol. 1997;63:844–850. doi: 10.1128/aem.63.3.844-850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Pérez-Martín J. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol Microbiol. 1996;19:1157–1184. doi: 10.1111/j.1365-2958.1996.tb02463.x. [DOI] [PubMed] [Google Scholar]
- 9.Dunleavy J M. Bacterial, fungal, and viral diseases affecting soybean leaves. In: Wyllie T D, Scott D H, editors. Soybean diseases of the North Central Region. St. Paul, Minn: American Phytopathological Society; 1988. pp. 40–46. [Google Scholar]
- 10.Feys B J, Benedetti C E, Penfold C N, Turner J G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graumann P, Marahiel M A. Some like it cold: response of microorganisms to cold shock. Arch Microbiol. 1996;166:293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- 12.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:41–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 13.Jones P G, Inouye M. The cold-shock response—a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 14.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 15.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 16.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 17.Liyanage H, Palmer D, Ullrich M, Bender C L. Characterization and transcriptional analysis of the gene cluster for coronafacic acid, the polyketide component of the phytotoxin coronatine. Appl Environ Microbiol. 1995;61:3843–3848. doi: 10.1128/aem.61.11.3843-3848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews B W. Structural and genetic analysis of protein stability. Annu Rev Biochem. 1993;62:139–160. doi: 10.1146/annurev.bi.62.070193.001035. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell R E, Young S A, Bender C L. Coronamic acid, an intermediate in coronatine biosynthesis by Pseudomonas syringae. Phytochemistry. 1994;35:343–348. [Google Scholar]
- 20.Mittal S, Davis K R. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant-Microbe Interact. 1995;8:165–151. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 21.Palmer D A, Bender C L. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol. 1993;59:1619–1623. doi: 10.1128/aem.59.5.1619-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer D A, Bender C L. Ultrastructure of tomato leaf tissue treated with the pseudomonad phytotoxin coronatine and comparison with methyl jasmonate. Mol Plant-Microbe Interact. 1995;8:683–692. [Google Scholar]
- 23.Palmer D A, Bender C L, Sharma S B. Use of Tn5-gusA5 to investigate environmental and nutritional effects on gene expression in the coronatine biosynthetic gene cluster of Pseudomonas syringae pv. glycinea. Can J Microbiol. 1997;43:515–525. doi: 10.1139/m97-074. [DOI] [PubMed] [Google Scholar]
- 24.Rahme L G, Mindrinos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangaswamy V, Ullrich M, Jones W, Mitchell R, Parry R, Reynolds P, Bender C L. Expression and analysis of coronafacate ligase, a thermoregulated gene required for production of the phytotoxin coronatine in P. syringae. FEBS Lett. 1997;154:65–72. doi: 10.1111/j.1574-6968.1997.tb12625.x. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Schergaut, M., J. Boch, and M. S. Ullrich. Unpublished data.
- 28.Somero G N. Proteins and temperature. Annu Rev Physiol. 1993;57:43–68. doi: 10.1146/annurev.ph.57.030195.000355. [DOI] [PubMed] [Google Scholar]
- 29.Somero G N, Low P S. Temperature: a “shaping force” in protein evolution. Biochem Soc Symp. 1976;41:33–43. [PubMed] [Google Scholar]
- 30.Ullrich M, Bender C L. The biosynthetic gene cluster for coronamic acid, an ethylcyclopropyl amino acid, contains genes homologous to amino acid-activating enzymes and thioesterases. J Bacteriol. 1994;176:7574–7586. doi: 10.1128/jb.176.24.7574-7586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ullrich M, Guenzi A C, Mitchell R E, Bender C L. Cloning and expression of genes required for coronamic acid (2-ethyl-1-aminocyclopropane 1-carboxylic acid), an intermediate in the biosynthesis of the phytotoxin coronatine. Appl Environ Microbiol. 1994;60:2890–2897. doi: 10.1128/aem.60.8.2890-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ullrich M, Peñaloza-Vázquez A, Bailey A-M, Bender C L. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol. 1995;177:6160–6169. doi: 10.1128/jb.177.21.6160-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Eede G, Deblaere R, Goethals K, Montagu M V, Holster M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant-Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- 34.von Döhren H. The organization of multifunctional peptide and depsipeptide synthetases. Biochem Soc Trans. 1993;21:214–218. doi: 10.1042/bst0210214. [DOI] [PubMed] [Google Scholar]
- 35.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiler E W, Kutchan T M, Gorba T, Brodschelm W, Niesel U, Bublitz F. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 37.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology. 1995;141:1691–1505. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Y, Lu Y, Heu S, Hutcheson S W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]