Abstract
The Ruvb-like AAA ATPase1 (Ruvbl1; also known as Pontin) is an evolutionary conserved protein belonging to the adenosine triphosphates associated with diverse cellular activities (AAA+) superfamily of ATPases. Ruvbl1 is a component of various protein supercomplexes and is involved in a variety of cellular activities, including chromatin remodeling, DNA damage repair, and mitotic spindle assembly however, the developmental significance of this protein is unknown and needs detailed investigation. We investigated the developmental significance of Ruvbl1 in multiciliated cells of the Xenopus laevis epidermis since ruvbl1 is expressed in the multiciliated cells and pronephros during X. laevis embryogenesis. The knockdown of ruvbl1 significantly impaired cilia-driven fluid flow and basal body polarity in the X. laevis epidermis compared to control embryos, but did not affect cilia morphology. Our results suggest that Ruvbl1 plays a significant role in embryonic development by regulating ciliary beating; however, further investigation is needed to determine the mechanisms involved.
Keywords: Ruvbl1, Ciliogenesis, Fluid flow, Basal body polarity, Xenopus laevis
INTRODUCTION
Motile cilia beat in a coordinated fashion to create a fluid flow essential to development and homeostasis (Meunier & Azimzadeh, 2016). Ciliary beating is generated by a complex of axonemal dynein motors that form a bridge between adjacent double microtubules. When ATP activates the kinetic region of the dynein, it begins to travel along the adjacent double microtubules. The force generated by the motion of the dynein can induce sliding between the adjacent microtubules, but since they are connected by nexin, they cannot slide and the energy is converted into a bending motion of the microtubules (Ishikawa, 2017). Motile cilia are found in human airways, fallopian tubes, and the brain to generate mucus clearance, egg transportation, and cerebrospinal fluid circulation, respectively. Genetic mutations that impair ciliary beating are relevant to primary ciliary dyskinesia (PCD) and include mutations in axonemal dynein and factors related to dynein assembly (Fabczak & Osinka, 2019; Leigh et al., 2019).
Ruvb-Like AAA ATPase1 (Ruvbl1; also known as Pontin) is an evolutionarily conserved protein belonging to the adenosine triphosphates associated with diverse cellular activities (AAA+) superfamily of ATPases. Ruvbl1 was discovered in 1998 in large RNA polymerase II holoenzyme oligomers (Qiu et al., 1998). Ruvbl1 is associated with many cellular processes (Jha & Dutta 2009), including chromatin remodeling, transcription regulation (Jónsson et al., 2001), ribonucleoprotein complex biogenesis (Izumi et al., 2010), and mitotic assembly (Gartner et al., 2003). Ruvbl1 is highly expressed in mouse embryonic stem cells and Ruvbl1 knockout mice are lethal. It is also found in hematopoietic tissues, testes, lung, brain, and liver in adult mice and targeted knockout of Ruvbl1 in hematopoietic tissues results in bone marrow failure, suggesting that Ruvbl1 is essential for early embryogenesis as well as adults in mice (Bereshchenko et al., 2012). Various Studies have shown that Ruvbl1 and Ruvbl2 act as dynein assembly chaperones involved in cilia-associated processes (Li et al., 2017; Hartill et al., 2018). Conditional ablation of Ruvbl1 in mice results in renal disease and hydrocephalus, which suggests that Ruvbl1 is crucial for the development and function of monocilia and multicilia (Dafinger et al., 2018).
In this study, we investigated the developmental significance of Ruvbl1 in ciliogenesis using Xenopus laevis as an animal model. X. laevis embryos have several ciliated tissues, such as the gastrocoel roof plate, neural tube, and mucociliary epidermis, and are easily accessible for manipulation, observation, and analysis (Walentek & Quigley, 2017). Ruvbl1 is strongly expressed in multiciliated cells of X. laevis as investigated by whole-mount in situ hybridization and the knockdown of ruvbl1 significantly disrupts the basal body polarity, which affects cilia-driven fluid flow. Thus, we propose that Ruvbl1 affects basal body polarity during X. laevis embryogenesis and is crucial for cilia motility.
METHODS
1. Xenopus laevis growth conditions and in vitro fertilization
Mature X. laevis were obtained from the Korean Xenopus Resource Center for Research and maintained in plastic aquarium tanks circulated with dechlorinated water at 18℃ (recommended by the Institutional Review Board of Kyungpook National University, Korea). Ovulation was induced in females by injecting of 1,000 IU of human chorionic gonadotrophin (hCG) into the dorsal lymph sac during the evening before the experiment. After 16 hours, the eggs were collected in 1 X MBS (88 mM NaCl, 5 mM Hepes, 2.5 mM NaHCO3, 1 mM KCl, 1 mM MgSO4, and 0.7 mM CaCl2; pH 7.8) by squeezing the female. The eggs were fertilized after several washes with 0.1 X MBS using a sperm suspension solution from isolated testes. Following fertilization, the jelly coat was removed by swirling the embryos in 2% L-cysteine in a 0.5 X MBS solution. The embryos were washed several times with 0.5 X MBS and cultured in 0.5 X MBS at 15℃–18℃.
2. mRNA synthesis, morpholinos (MO) design, and microinjections
To visualize the basal bodies, mRNA for centrin-RFP and clamp-GFP were synthesized using the SP6 mMessage mMachine kit (Invitrogen, Carlsbad, CA, USA). For the knockdown experiment, morpholinos (MO) were synthesized to the following base composition: 5’-[CTC CTC GAT TTT CAT GGT GGA AG]-3’ (Gene Tools, Philomath, OR, USA). Finally, mRNAs or MOs were injected into one–cell staged embryos, then the embryos were incubated at 18℃– 23℃ until the development reached the desired stages.
3. Whole-mount in situ hybridization (WISH)
The embryos were fixed at the desired stages in MEMFA (4% paraformaldehyde, 0.1 M MOPS (pH 7.4), 1 mM MgSO4, 2 mM EGTA) overnight at 4℃. An antisense probe for Ruvbl1 was synthesized using the T7 mMessage mMachine kit (Invitrogen). The probe was detected using an alkaline phosphatase-labeled anti-digoxigenin antibody (1:1,000, Roche, Basel, Switzerland) and NBT/BCIP (Roche).
4. Reverse transcription – polymerase chain reaction (RT-PCR)
Total RNA was extracted from embryos using the TRIzol™ Reagent (Invitrogen). cDNA was synthesized via reverse transcription using a Primescript™ 1st strand cDNA synthesis kit (Takara, Shiga, Japan) from RNAs. PCR was performed using specific primer pairs (Table 1) and EmeraldAmp® PCR Master Mix (Takara). The PCR products were separated on a 1% agarose gel and images were captured using WiseCapture I-1000 (Daihan Scientific, Wonju, Korea). The following primers were used:
Table 1. Primer sequences for RT-PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| odc | 5’-CAG CTA GCT GTG GTG TGG-3’ | 5’-CAA CAT GGA AAC TCA CAC C-3’ |
| ruvbl1 | 5’-ATG GCG CTG AGC GAC-3’ | 5’-CGG AAG TTC CTG GAC-3’ |
5. Immunofluorescence and confocal microscopy
mRNA and MO-injected embryos were fixed at stage 32 in MEMFA. Acetylated tubulin (1:1,000, Sigma-Aldrich, St. Louis, MO, USA) was used as the primary antibody to observe the motile cilia in the embryos. The secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG (1:2,000, Invitrogen) and Alexa Fluor 568 goat anti-mouse IgG (1:2,000, Invitrogen). Imaging was performed with an Olympus FV1200 confocal microscope (Olympus, Shinjuku, Japan).
6. Velocity measurement of fluid flow and statistical analysis
The embryos were anesthetized with 0.1 X benzocaine. Then, fluorescent microbeads were added along the surface of the embryos and videos were recorded. The velocity of the fluorescent beads was measured using Tracker 6.0. Statistical analyses were performed by Prism 9 (GraphPad software). The results are means±SD of three different experiments. The student t-test was used to identify the level of significance (* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001).
RESULTS AND DISCUSSION
1. Ruvbl1 is expressed in multiciliated cells during Xenopus laevis embryogenesis
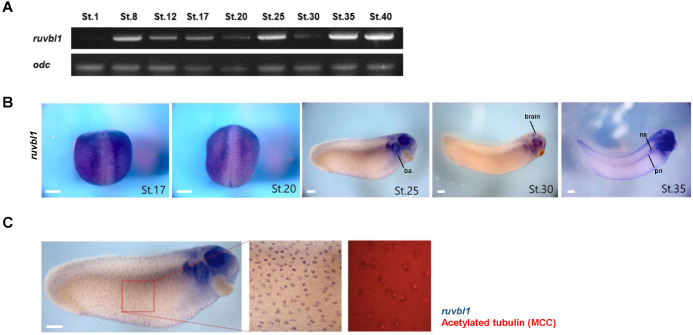
To elucidate the developmental role of Ruvbl1, we analyzed the temporal and spatial expression patterns of ruvbl1 using reverse transcription – polymerase chain reaction (RT-PCR) and whole-mount in situ hybridization (WISH) during X. laevis embryogenesis. Ruvbl1 was a maternal gene and expressed generally through all stages of embryonic development, with particularly high expression at stages 8 (blastula), 25 (early tailbud), and 35–40 (late tailbud) (Fig. 1A). Ruvbl1 was expressed as regular dots in the epidermis at stages 17, 20, 25, and 30 and was localized in the branchial arches, eyes, and brain at stages 25, 30, and 35. It was weakly expressed in the pronephros but highly expressed in the three nephrostomes at stage 35 (Fig. 1B). Based on the ruvbl1 spatial expression in the epidermis and ciliated tissues (e.g., pronephros and brain), we hypothesized that ruvbl1 is specifically expressed in multiciliated cells and performed WISH and immunofluorescence using acetylated tubulin antibodies in the same embryos. The WISH-IF data showed that ruvbl1 was localized in the multiciliated cells of the X. laevis epidermis (Fig. 1C). These results suggest that Ruvbl1 may play a crucial role in X. laevis embryogenesis and be involved in ciliogenesis.
Fig. 1. Spatio-temporal expression patterns of ruvbl1 during Xenopus laevis embryogenesis.
(A) Temporal expression patterns by RT-PCR. Ruvbl1 is expressed maternally throughout all stages. Ornithine decarboxylase (odc) is used as a loading control. (B) Spatial expression patterns via whole-mount in situ hybridization. Digoxigenin–labeled antisense probes for ruvbl1 were used. At the late neurula stage (St. 17 and 20), ruvbl1 is expressed as dots in the epidermis. At the tailbud stage, ruvbl1 is expressed in the epidermis, branchial arches, eyes, and brain. At stage 35, ruvbl1 is expressed in the pronephros and nephrostome (scale bar=250 μm). (C) Double staining with WISH of ruvbl1 (purple) and immunofluorescence for multicilaited cells using acetylated tubulin (red) on the same embryos (scale bar=250 μm). ba, branchial arch; pn, pronephros; ns, nephrostomes.
2. Ruvbl1 is required for ciliary-driven fluid flow in Xenopus laevis epidermis
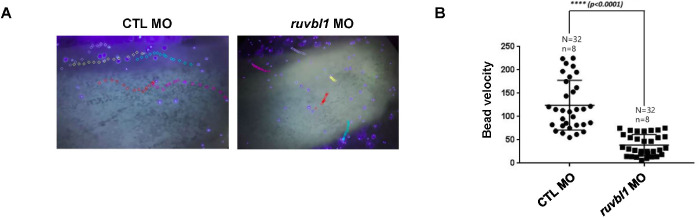
Ruvbl1 was expressed in ciliated cells during X. laevis embryogenesis. Ruvbl1 and Ruvbl2 are outer dynein arm subunits crucial for ciliary movement (Lee et al., 2020). Ciliary movement generates the directed fluid flow essential for development and physiological processes, such as brain development and kidney function (Werner & Mitchell, 2012). The fluid flow in X. laevis epidermis functions in the defense against infections and is easily visualized using fluorescent microbeads. To investigate Ruvbl1 association with ciliary-driven fluid flow, we injected ruvbl1 morpholino into X. laevis embryos at the one–cell stage to block translation and used fluorescent microbeads to observe the fluid flow in the epidermis. We found that ruvbl1 morphants had a much slower fluid flow compared to the control MO-injected embryos (Fig. 2A and B), suggesting that Ruvbl1 is required for ciliary beating in the X. laevis epidermis.
Fig. 2. Knockdown of ruvbl1 perturbed ciliary movement in Xenopus laevis epidermis.
(A) Control MO (32 ng) or ruvbl1 MO (32 ng) was injected into one–cell staged embryos. Injected embryos were collected at stage 28 and treated with fluorescent microbeads on their head side. The microbead flow from head to tail was recorded using the Tracker 6.0 program. (B) Bead velocity graph. A total of 32 beads from 8 embryos for each control MO or ruvbl1 MO-injected embryo. Ruvbl1 morphants had significantly reduced microbead flow. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
3. Knockdown of ruvbl1 disrupts basal body polarity
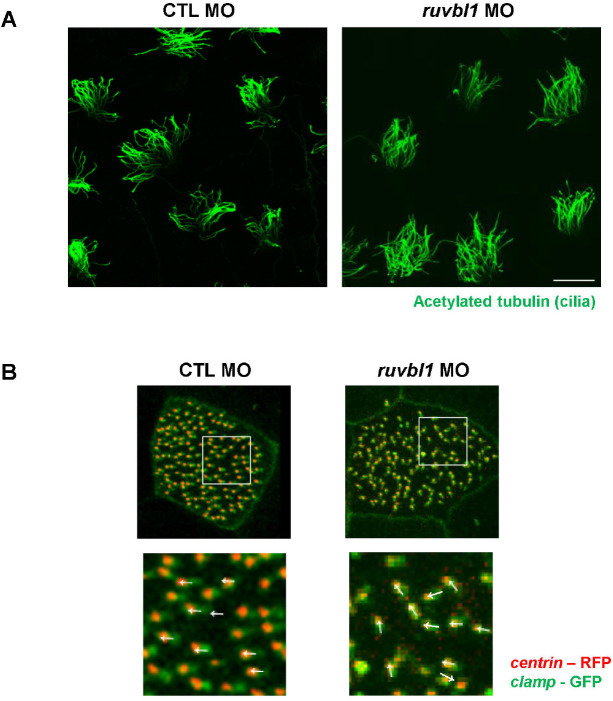
Defective fluid flow may be caused by a decreased number of cilia in multiciliated cells. To evaluate the role of Ruvbl1 in ciliogenesis during X. laevis embryogenesis, we stained Ruvbl1 morphants using acetylated tubulin and observed multicilia in the epidermis. The knockdown of ruvbl1 did not affect the length and amount of the cilia (Fig. 3A). Dafinger et al. (2018) reported that Ruvbl1 deletion in mice disrupts cilia formation and leads to renal disease and hydrocephalus. In this study, we examined the multiciliated cells only in the epidermis of ruvbl1 morphants. Thus, additional studies of cilia in the pronephros and brain are needed to reveal whether Ruvbl1 in X. laevis ciliogenesis is consistent with that of mice.
Fig. 3. Knockdown of Ruvbl1 led to defective basal body polarity in Xenopus laevis embryos.
(A) Embryos were injected with control MO or ruvbl1 MO and fixed in MEMFA at stage 32. Motile cilia were stained green with acetylated tubulin and observed via confocal microscopy. The depletion of ruvbl1 did not affect the number of cilia in multiciliated cells (scale bar=20 μm). (B) Embryos were injected with centrin-RFP and clamp-GFP with each MO to visualize basal body polarity, which is illustrated by white arrows. Ruvbl1 morphants had defective basal body polarity compared to the control MO-injected embryos.
Impaired fluid flow may also be due to disorganized basal body polarity within each multiciliated cell (Werner & Mitchell, 2012). Thus, we injected centrin - RFP and clamp - GFP with ruvbl1 MO to examine the effect of ruvbl1 knockdown on basal body polarity. In the control MO-injected embryos, the basal bodies were all polarized in a similar manner, but the ruvbl1 morphants displayed disruptive basal body polarity (Fig. 3B). These experimental results suggest that ruvbl1 knockdown does not affect cilia length or shape, but does disturb basal body polarity, which can lead to defective fluid flow in the epidermis. A recent study in zebrafish has shown that the depletion of Ruvbl1 reduced both their inner and outer dynein (Li et al., 2017). Although we did not check the axonemal dyneins in the ruvbl1 morphants, we determined that ruvbl1 knockdown disrupts basal body polarity. This suggests a novel function of Ruvbl1 in multiciliated cells. Interactome analysis of Ruvbl1 identified several ciliary proteins and the involvement of Ruvbl1 in cilia-related processes via its interactions with several ciliary proteins, such as DNAAF1 (Dafinger et al., 2018; Hartill et al., 2018). Previous studies combined with our data prompted us to hypothesize that Ruvbl1 may be involved in both axonemal dynein assembly and cilia-related processes, including basal body polarization, via interacting ciliary proteins. Further studies are needed to determine the accuracy of this hypothesis. In conclusion, this study showed that Ruvbl1 is specifically localized in multiciliated cells and plays a crucial role in muticilia motility during X. laevis embryogenesis.
Acknowledgements
This research was supported by the National Research Foundation of Korea, Ministry of Science & ICT, Korea (2021R1A2C1010408) and Korea Environment Industry and Technology Institute (KEITI) through Core Technology Development Project for Environmental Diseases Prevention and management funded by Korea Ministry of Environment (2022003310001).
ORCID
Chan Young Kim https://orcid.org/0009-0009-5147-8681
Hyun-Kyung Lee https://orcid.org/0000-0003-4989-698X
Hongchan Lee https://orcid.org/0000-0001-9009-209X
Hyun-Shik Lee https://orcid.org/0000-0002-3837-9867
Conflict of interests
The authors declare no potential conflict of interest.
Authors’ contributions
Con ceptualization: Kim CY, Lee HS.
Data curation: Kim CY, Lee HK.
Methodology: Kim CY.
Writing-original draft: Kim CY.
Wr iting-review & editing: Kim CY, Lee HK, Lee H, Lee HS.
Ethics approval
This study was approved by Kyungpook National University (2021-0017).
REFERENCES
- Bereshchenko O, Mancini E, Luciani L, Gambardella A, Riccardi C, Nerlov C. Pontin is essential for murine hematopoietic stem cell survival. Haematologica-the Hematology Journal. 2012;97:1291–1294. doi: 10.3324/haematol.2011.060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafinger C, Rinschen MM, Borgal L, Ehrenberg C, Basten SG, Franke M, Höhne M, Rauh M, Göbel H, Bloch W, Thomas Wunderlich F, Peters DJM, Tasche D, Mishra T, Habbig S, Dötsch J, Müller RU, Brüning JC, Persigehl T, Giles RH, Benzing T, Schermer B, Liebau MC. Targeted deletion of the AAA-ATPase Ruvbl1 in mice disrupts ciliary integrity and causes renal disease and hydrocephalus. Exp Mol Med. 2018;50:1–17. doi: 10.1038/s12276-018-0108-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabczak H, Osinka A. Role of the novel Hsp90 co-chaperones in dynein arms’ preassembly. Int J Mol Sci. 2019;20:6174. doi: 10.3390/ijms20246174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, Waldhäusl W, Pasternack MS, Wagner L. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskeleton. 2003;56:79–93. doi: 10.1002/cm.10136. [DOI] [PubMed] [Google Scholar]
- Hartill VL, van de Hoek G, Patel MP, Little R, Watson CM, Berry IR, Shoemark A, Abdelmottaleb D, Parkes E, Bacchelli C, Szymanska K, Knoers NV, Scambler PJ, Ueffing M, Boldt K, Yates R, Winyard PJ, Adler B, Moya E, Hattingh L, Shenoy A, Hogg C, Sheridan E, Roepman R, Norris D, Mitchison HM, Giles RH, Johnson CA. DNAAF1 links heart laterality with the AAA+ ATPase RUVBL1 and ciliary intraflagellar transport. Hum Mol Genet. 2018;27:529–545. doi: 10.1093/hmg/ddx422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harb Perspect Biol. 2017;9:a028076. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- Jha S, Dutta A. RVB1/RVB2: Running rings around molecular biology. Mol Cell. 2009;34:521–533. doi: 10.1016/j.molcel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- Lee C, Cox RM, Papoulas O, Horani A, Drew K, Devitt CC, Brody SL, Marcotte EM, Wallingford JB. Functional partitioning of a liquid-like organelle during assembly of axonemal dyneins. eLife. 2020;9:e58662. doi: 10.7554/eLife.58662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MW, Horani A, Kinghorn B, O’Connor MG, Zariwala MA, Knowles MR. Primary ciliary dyskinesia (PCD): A genetic disorder of motile cilia. Transl Sci Rare Dis. 2019;4:51–75. doi: 10.3233/TRD-190036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao L, Yuan S, Zhang J, Sun Z. Axonemal dynein assembly requires the R2TP complex component Pontin. Development. 2017;144:4684–4693. doi: 10.1242/dev.152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier A, Azimzadeh J. Multiciliated cells in animals. Cold Spring Harb Perspect Biol. 2016;8:a028233. doi: 10.1101/cshperspect.a028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- Walentek P, Quigley IK. What we can learn from a tadpole about ciliopathies and airway diseases: Using systems biology in Xenopus to study cilia and mucociliary epithelia. Genesis. 2017;55:e23001. doi: 10.1002/dvg.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ. Understanding ciliated epithelia: The power of Xenopus. Genesis. 2012;50:176–185. doi: 10.1002/dvg.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]