Abstract
PURPOSE
Homologous recombination deficiency (HRD) is highly prevalent in triple-negative breast cancer (TNBC) and associated with response to PARP inhibition (PARPi). Here, we studied the prevalence of HRD in non-TNBC to assess the potential for PARPi in a wider group of patients with breast cancer.
METHODS
HRD status was established using targeted gene panel sequencing (360 genes) and BRCA1 methylation analysis of pretreatment biopsies from 201 patients with primary breast cancer in the phase II PETREMAC trial (ClinicalTrials.gov identifier: NCT02624973). HRD was defined as mutations in BRCA1, BRCA2, BRIP1, BARD1, or PALB2 and/or promoter methylation of BRCA1 (strict definition; HRD-S). In secondary analyses, a wider definition (HRD-W) was used, examining mutations in 20 additional genes. Furthermore, tumor BRCAness (multiplex ligation-dependent probe amplification), PAM50 subtyping, RAD51 nuclear foci to test functional HRD, tumor-infiltrating lymphocyte (TIL), and PD-L1 analyses were performed.
RESULTS
HRD-S was present in 5% of non-TNBC cases (n = 9 of 169), contrasting 47% of the TNBC tumors (n = 15 of 32). HRD-W was observed in 23% of non-TNBC (n = 39 of 169) and 59% of TNBC cases (n = 19 of 32). Of 58 non-TNBC and 30 TNBC biopsies examined for RAD51 foci, 4 of 4 (100%) non-TNBC and 13 of 14 (93%) TNBC cases classified as HRD-S had RAD51 low scores. In contrast, 4 of 17 (24%) non-TNBC and 15 of 19 (79%) TNBC biopsies classified as HRD-W exhibited RAD51 low scores. Of nine non-TNBC tumors with HRD-S status, only one had a basal-like PAM50 signature. There was a high concordance between HRD-S and either BRCAness, high TIL density, or high PD-L1 expression (each P < .001).
CONCLUSION
The prevalence of HRD in non-TNBC suggests that therapy targeting HRD should be evaluated in a wider breast cancer patient population. Strict HRD criteria should be implemented to increase diagnostic precision with respect to functional HRD.
Some non–triple-negative breast cancers are HRD and could be considered for PARP inhibition therapy.
INTRODUCTION
Following the pioneer studies defining a role for PARP inhibition (PARPi) in BRCA1/2-mutated breast1 and ovarian cancers,2 several studies have confirmed the benefit of such targeted therapy in early and metastatic breast cancers.3-7 Although studies in metastatic disease have reported limited efficacy to patients carrying germline BRCA1/2 mutations,8 recently we reported an objective response rate above 50% for unselected triple-negative breast cancers (TNBCs) receiving neoadjuvant olaparib monotherapy.9 Moreover, responders could be identified by markers predicting homologous recombination deficiency (HRD). These encouraging results point to the potential benefit of PARPi also for patients with HRD-positive non-TNBCs.
CONTEXT
Key Objective
To evaluate the prevalence of homologous recombination repair deficiency (homologous recombination deficiency [HRD]) in primary non–triple-negative breast cancers.
Knowledge Generated
We find that a substantial fraction of primary non–triple-negative breast cancers reveal HRD. Applying a strict definition, restricted to mutations in either of five genes and/or BRCA1 methylation, 5% were found to be HRD while in a wider analysis, including 20 additional genes, the corresponding percentage was 23%. The strict definition had superior concordance with functional HRD measured by lack of RAD51 foci formation.
Relevance
Therapy targeting HRD should be considered for a wider range of breast cancers than triple-negatives.
Although genomic aberrations associated with HRD have been identified in non-TNBCs,3-5,7,10-12 the prevalence of HRD in large stage II-III breast cancers evaluated for neoadjuvant therapy remains to be elucidated. In addition, the definition of HRD is not a univocal term, and studies in other types of cancer have put into question which genomic aberrations are associated with HRD status, as well as the need for biallelic inactivation (ie, loss of heterozygosity [LOH]) to cause functional HRD.13-17 The aim of this study was to explore mutations across a panel of homologous recombination repair (HRR) genes, including BRCA1 epimutations9 and LOH, in concert with other assays defining HRD (BRCAness multiplex ligation-dependent probe amplification (MLPA); MLPA and RAD51 nuclear foci staining) to identify potential markers for PARP inhibitor sensitivity in breast cancer.
METHODS
Study Design and Patients
In the phase II PETREMAC trial, patients with stage II/III breast cancer were stratified to eight different neoadjuvant treatment regimens on the basis of ER, progesterone receptor, and human epidermal growth factor receptor 2 (HER2) expression as well as TP53 mutation status (for details; see the Data Supplement, Supplementary methods, study Protocol, and Data Supplement, Fig S1). The trial was conducted at seven trial sites in Norway, and pretreatment biopsies were collected for centralized molecular analyses. The findings among patients with TNBCs have been reported previously.9 The trial was originally powered for assessment of predictive markers within the treatment arms. No formal power estimate for the present assessment of HRD across arms was performed, and this study should be considered retrospective and exploratory.
As primary analysis, HRD was defined applying a strict definition (HRD-S; see below). In secondary analyses, a wider definition (HRD-W) was used (see below). In addition, tumor BRCAness by MLPA, RAD51 nuclear foci to test functional HRD, PAM50 subtyping, and tumor-infiltrating lymphocyte (TIL) and PD-L1 analyses were performed retrospectively.
HRR Mutations, BRCA1 Methylation, and BRCAness Analyses
Targeted DNA sequencing was performed on pretreatment tumor biopsies using a 360-gene panel,18 as a preplanned analysis per protocol.9 BRCA1 promoter methylation analysis was performed applying an amplicon-based next-generation sequencing assay as described previously19-21 (see the Data Supplement, Supplementary Methods). BRCA1 promoter methylation was combined with HRR mutations to evaluate the prevalence of HRD by either wide (HRD-W) or strict definitions (HRD-S).
HRD-W included BRCA1 methylation or mutations in either of the following HRR-related genes: ABL1, ATM, ATR, ATRX, BARD1, BLM, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, EMSY, ERCC4, FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG, MEN1, MRE11, NBN, PALB2, PTEN, and SETD2, where mutations were called by predefined criteria.9
HRD-S included BRCA1 methylation or mutations within a restricted subset of HRR genes: BARD1, BRCA1, BRCA2, BRIP1, and PALB2, as recommended by a recent European expert consensus.13 However, in contrast to the consensus gene list, RAD51C and RAD51D were not included as they were not part of the 360-gene panel used in our trial.9 Furthermore, to call HRD-S status, the HRR mutations had to be deleterious, and somatic HRR mutations or BRCA1 methylation had to be accompanied by LOH (defined as lack of the opposite allele) to ascertain biallelic gene inactivation. Allele-specific copy number analysis was performed on matched tumor-normal pair sequencing (binary alignment/map, BAM files) from the 360-gene panel using FACETS (Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing).22 In cases where there were too few single-nucleotide polymorphisms in the genomic region to estimate allele-specific copy numbers, LOH status was defined as not available.
Additionally, a preplanned analysis of tumor samples for BRCAness by MLPA was performed using the SALSA MLPA kit P376 BRCAness probemix (MRC Holland) following the manufacturer's instructions where BRCAness status was assigned using a nearest shrunken classifier as previously described.9,23,24
RAD51 Staining to Assess Functional HRD Status
RAD51, BRCA1, and γH2AX immunostaining, as well as geminin and 4′,6-diamidino-2-phenylindole (DAPI) counterstaining, were post hoc assessments performed as described elsewhere9,25 (see thee Data Supplement, Supplementary Methods). Here, pretreatment biopsies from all patients with non-TNBCs included at the trial's coordinating cancer center (Bergen) were analyzed blinded to the treatment arm.
PAM50 and HRR Gene Expression Analyses
Bulk RNA sequencing was performed on RNA extracted from snap-frozen, whole-tumor biopsies. In brief, 300-500 ng total RNA was used for library preparation, and global RNA sequencing was performed on a NovaSeq 6000 (Illumina, San Diego, CA) using 2 × 100 cycles, providing a minimum of 70 million reads per sample. For details on RNA extraction, library preparation, sequencing, and bioinformatic processing see the Data Supplement, Supplementary Methods.
A post hoc gene expression analysis was performed on pretreatment biopsies, on the basis of the RNA sequencing data, to assign the tumors to PAM50 breast cancer subtypes and to examine the expression of HRR-S genes (for details, see the Data Supplement, Supplementary Methods).
PD-L1 and Tumor-Infiltrating Lymphocyte Analyses
PD-L1 immunostaining and quantification of tumor-infiltrating lymphocytes (TILs) in hematoxylin and eosin–stained sections were conducted as post hoc assessments of all pretreatment biopsies (all trial centers), as described in the Data Supplement (Supplementary Methods). PD-L1 staining and TIL quantification results for TNBCs in the trial were presented previously,9 and these results are included here for comparison with the non-TNBC cases. For PD-L1 analysis, the combined positive score method was used, as outlined in the Data Supplement (Supplementary Methods).
Ethics and Approvals
The PETREMAC trial was approved by the Regional Ethical Committee of the Western health region in Norway (2015/1493) and The Norwegian Drug Agency (2015/8463). The trial was registered at ClinicalTrials.gov identifier: NCT02624973 and with EudraCT (2015-002816-34). The study was conducted in accordance with the protocol, good clinical practice guidelines, provisions of the Declaration of Helsinki, and local regulations. All patients signed an informed consent form before inclusion.
Statistics
Statistical tests used for each data set are given in the figures and tables. All P values reported are two-tailed. No P value was corrected for multiple testing. All statistical analyses were performed using R, version 4.1.3 or the SPSS 15.0/PASW 17.0 software package (SPSS Inc, Armonk, NY).
RESULTS
Baseline patient and tumor characteristics are given in Table 1. The detailed results of all genomic and immunostaining analyses are given in the Data Supplement (supplemental data).
TABLE 1.
Baseline Patient and Tumor Characteristics in the PETREMAC Trial

Applying a wide HRD definition (mutations in HRR-related genes and/or BRCA1 methylation; see Methods), we found a 23% prevalence of HRD-W in the non-TNBC subset (n = 39 of 169). Among these 39 non-TNBC tumors, 35 had HRR mutations while five had BRCA1 promoter methylation (Table 2), including one tumor with concordant HRR mutation and BRCA1 methylation. The prevalence of HRD-W was reported previously as 59% (n = 19 of 32) in the TNBC subgroup9 and is summarized for both non-TNBC and TNBC in Figure 1 and Table 2.
TABLE 2.
HRD Status Across the PETREMAC Treatment Arms
FIG 1.
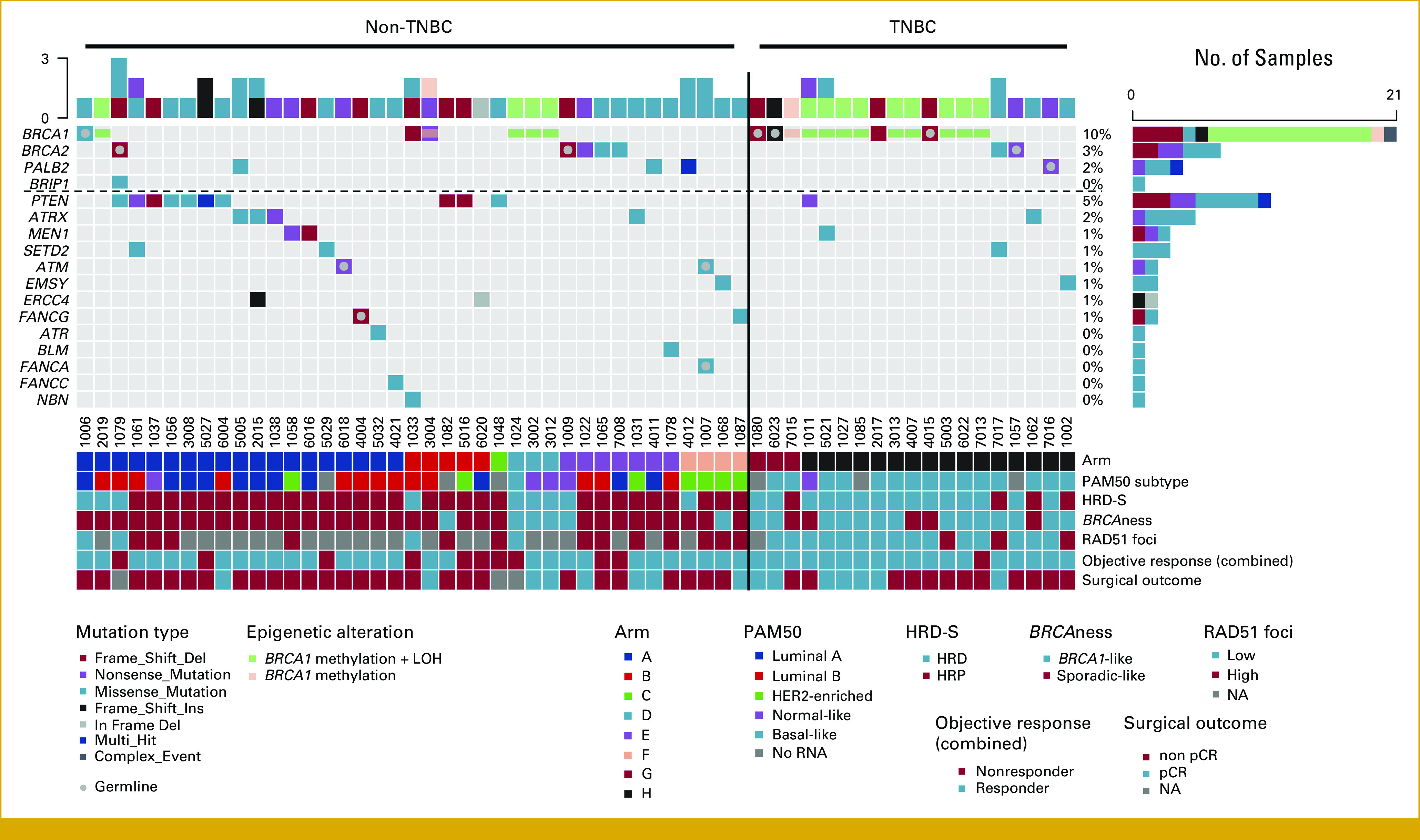
Oncoplot of HRR mutations, HRD status, response, and surgical outcome in the PETREMAC trial. All breast cancers with HRD-W are displayed. The HRD-S subset includes tumors with HRR mutations by strict definition (listed above the dotted line) and/or BRCA1 methylation and includes gene silencing on the opposite allele. BRCAness was assessed by MLPA analysis. Objective response (combined) and patients with a partial or complete response to treatment based on combined radiological and clinical evaluation. HRD, homologous recombination deficiency; HRD-S, HRD-strict definition; HRD-W, HRD scored by wide definition; HRP, homologous recombination proficiency; HRR, homologous recombination repair; LOH, loss-of-heterozygosity; MLPA, multiplex ligation-dependent probe amplification; NA, not analyzed; pCR, pathological complete response; TNBC, triple-negative breast cancer; wt, wild type.
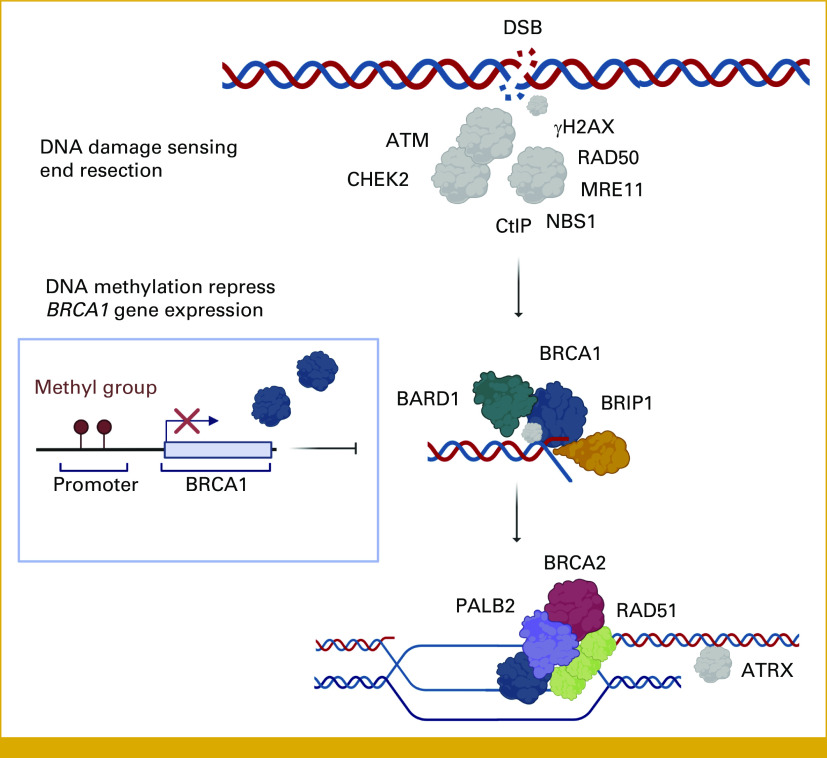
Applying the consensus guidelines,13 we implemented a stricter definition (HRD-S) including mutations only in BARD1, BRCA1, BRCA2, BRIP1, PALB2, and/or BRCA1 methylation, with concomitant LOH on the opposite allele. Among the non-TNBC tumors, 5% (n = 9 of 169) were classified as HRD-S (Fig 1 and Table 2). Three of these HRD-S–positive tumors were associated with a germline mutation (one BRCA1 and two BRCA2 mutations), one had a somatic BRCA1 and one a somatic PALB2 mutation, whereas four revealed BRCA1 promoter methylation. In contrast, the prevalence of HRD-S among TNBCs was 47% (n = 15 of 32; Table 2). HRR genetic or epigenetic mutations inactivating key HRR proteins involved in DNA double-strand break repair are visualized in Figure 2.
FIG 2.
Signaling molecules involved in homologous recombination to repair DNA DSBs. HRR of DNA DSB. HRD results from germline, somatic, or epigenetic gene alterations inactivating either of the key proteins: BRCA1, BRCA2, PALB2, RAD51C, RAD51D, BARD1, and BRIP1. The HRR proteins localize to the DSB site, acting as a helicase and opening the chromatin to allow pairing of BRCA2 and RAD51 with the undamaged DNA strand from the alternate chromosome. The undamaged copy serves as a template for DNA polymerase and PCNA to repair the damaged strand. DSB, double-strand break; HRD, homologous recombination deficiency; HRR, homologous recombination repair. Figure created with BioRender.com.
BRCAness evaluation using MLPA-based copy number variation analysis,24 identified 10 of 169 (6%) non-TNBC as BRCA1-like (Fig 1 and Data Supplement, Table S1). BRCA1-like status was observed for four of nine HRD-S/non-TNBC, contrasting six of 160 homologous recombination proficient (HRP)/non-TNBC tumors (P = .0007). Applying the same analysis to TNBC, 12 of 15 HRD-S/TNBC, contrasting six of 17 HRP/TNBC tumors were defined as BRCA1-like (P = .02).
In total, 70 non-TNBCs from the PETREMAC trial underwent RAD51 staining, but 12 tumors were nonevaluable because of either a low number of geminin-positive cells or low γH2AX staining. Of 58 non-TNBC biopsies evaluable for RAD51 status, 17 were classified as HRD-W, and only four of these (24%) had RAD51 low scores (Fig 1).9 Of nine non-TNBC tumors classified as HRD-S, four were analyzed by RAD51 staining, and all had a low RAD51 score (Fig 1). For these four cases, one was luminal A, two were luminal B, and one was a basal-like subtype by PAM50 analysis (Fig 1), and all had either BRCA1 or BRCA2 inactivation (germline or somatic). Including all nine non-TNBC tumors with HRD-S status, one was luminal A, three were luminal B, three were normal-like, one was HER2-enriched, and one had a basal-like PAM50 signature (Fig 1). Previously, we reported RAD51 low scores to predict HRD status among TNBC tumors in the PETREMAC trial, revealing that 15 of 19 tumors fulfilling HRD-W criteria exhibited RAD51 low status.9 When implementing the more accurate Illumina sequencing method to detect BRCA1 methylation in the current analysis, as compared with the qPCR method used previously,9 one tumor changed status from BRCA1 methylated to nonmethylated. In this one tumor, which was RAD51 high, BRCA1 methylation was detected, but it was below the predefined cutoff. Thus, HRD-W was present in 15 of 18 tumors with RAD51 low status, using Illumina sequencing both for the 360-gene panel and BRCA1 methylation analyses. Using the HRD-S criteria, 13 of 14 TNBC tumors classified as HRD-S had RAD51 low scores (Fig 1). The concordance between HRD-S and functional HRD status by RAD51 low immunostaining, for TNBC and non-TNBC tumors combined, was 95.5%, with sensitivity, specificity, and positive predictive values of 0.94, 0.96, and 0.85, respectively.
For non-TNBC and TNBC cases combined, BRCA1 mRNA expression was significantly lower in the BRCA1 methylated as compared with nonmethylated tumors (P < .001, Data Supplement, Fig S2) while BRCA1 mutations did not influence BRCA1 mRNA expression. When split by subtype, BRCA1 mRNA expression was also significantly lower in BRCA1 methylated as compared with nonmethylated tumors, both for non-TNBC and TNBC tumors (P = .02 and P = .0002; Data Supplement, Fig S2).
The content of TILs and PD-L1 expression in non-TNBC tumors is given in the Data Supplement (Fig S3) to compare with our previous findings in TNBC.9 TIL content and PD-L1 expression were both significantly lower in non-TNBC, as compared with TNBC tumors. For the whole trial combined, basal-like tumors had significantly higher TIL content and PD-L1 expression than other intrinsic subtypes (Data Supplement, Fig S4). Furthermore, there were high concordances between HRD-S status and high TIL or PD-L1 levels (Data Supplement, Table S2).
DISCUSSION
This report adds important new data within two areas; the prevalence of HRD in treatment-naïve, primary non-TNBC and the correlation between genomic and functional HRD status, with the use of recently defined, strict HRD criteria.13 First, among 201 patients commencing neoadjuvant treatment in the PETREMAC trial, the prevalence of HRD by strict criteria was 5% among 169 non-TNBC tumors, contrasting a prevalence of 47% among 32 TNBC tumors.9 Second, using the consensus list of core HRR germline and somatic mutations,13 as well as BRCA1 methylation to define HRD, we established a high concordance between HRD by strict criteria and functional HRD status by RAD51 low immunostaining.
Identifying other parameters that may characterize HRD-positive non-TNBCs is an important and remaining challenge. Here, HRD-positive non-TNBC tumors revealed gene expression signatures of all subtypes including luminal A/B as well as the HER2-enriched class, with only one tumor classified as basal-like. Although the basal-like signature aligns with BRCA1 defects and is prevalent in TNBC,27 about 31%-39% of breast cancers in BRCA1 germline mutation carriers are non-TNBCs,28,29 and tumors harboring other gene defects, such as BRCA2, present gene expression profiles with a closer resemblance to spontaneous breast cancers.30
The predictive impact of HRD and optimal testing strategies to identify potential PARPi responders, beyond gBRCA mutations, are being evaluated across different cancer entities.15,25,31,32 A limitation of our test panel is that RAD51C/D mutations were not included, which is generally recommended.13 However, we included BRCA1 promoter methylation status, shown to predict response to olaparib in patients with TNBC participating in the PETREMAC trial.9 Among our TNBCs treated with olaparib, using wider criteria for HRD testing,9 we identified 16 of 18 PARPi responders within the TNBC cohort to be HRD positive, whereas the current stricter HRD-S criteria in pretreatment biopsies only identified 12 subsequent PARPi responders. Although our current work demonstrates that strict HRD criteria are more precise in identifying breast cancers with functional HRD, the explanation for PARPi responsiveness among TNBC cases outside of the HRD-S subset remains to be elucidated and could be due to targeting of non-HRD–related mechanisms. Notably, PARPi can cause PARP1 trapping on DNA, which is shown to be toxic for both HRD and HRP cells.33 Furthermore, PARPi can modulate immune cells to become more cytotoxic,34 and the potential impact on the immune system has been further underlined by the findings that even in BRCA-associated cancers, the effect of PARPi is enhanced by STING agonism.35
Although the 5% prevalence of HRD status in our non-TNBC cohort is less than in previous reports of 18%-21% prevalence of HRR mutations, these reports have had wider test criteria.10-12 Assessing HRD status in non-TNBC by other biomarkers/algorithms such as HRDetect has identified 7%-9% as HRD36 while using SigMA to classify the mutational Signature 3 (Sig3) associated with HRD identified 7% of ER+ breast cancers as Sig3+ (ie, HRD).37
On the basis of our current data, we believe that the HRD-S criteria, in concert with RAD51 foci staining, are the most accurate and suitable predictive biomarkers to select patients for studies evaluating PARPi in non-TNBCs. Alternative criteria, that is, BRCAness by MLPA or basal-like subtypes by PAM50 analyses, are less accurate and are less useful for such screening.
Finally, we established that HRD-positive, stage II-III breast cancers have higher TILs and PD-L1 percentages than non-HRD tumors, confirming previous data in breast and ovarian cancers.38-40 However, the number of combined HRD-S and non-TNBC cases was too small for further analysis of the immune status in this subset. Immune activation is a frequent observation in HRD-positive tumors,38,39,41 which suggests that such breast cancers could be good candidates for anti-PD–1/PD-L1–based therapy. In the neoadjuvant setting, combined chemotherapy and immune checkpoint inhibition is highly effective for early TNBC,42,43 and immune activation, measured as an increased content of TILs, is associated with improved survival outcome.44-46 Yet, the predictive impact of HRD status toward response to immunotherapy remains to be elucidated.
In conclusion, while the fraction of non-TNBCs revealing HRD is low, the fact that non-TNBCs account for 85%-88% of all breast cancers indicates that a significant number of patients with non-TNBC may benefit from PARPi. Moreover, using our strict HRD criteria implemented herein, we could accurately identify breast cancers with functional HRD, likely to respond to HRD targeted therapy.
ACKNOWLEDGMENT
We gratefully acknowledge the contribution made by the patients included in the PETREMAC trial. Furthermore, the trial could not have been conducted without the skillful expertise of colleagues in the Departments of Oncology, Pathology, Radiology, and Surgery at the participating hospitals and the technical assistance from Nhat K. Duong, Silje Bjørneklett, and Christine Eriksen (Bergen).
Christina Engebrethsen
Stock and Other Ownership Interests: ArcticZymes Technologies, AstraZeneca, Demant, Devyser Diagnostics AB, Illumina, Next Biometrics Group, Photocure, Teva Pharmaceutical Industries ADR, Xbrane Biopharma AB
Bjørnar Gilje
Consulting or Advisory Role: Astellas Pharma, AstraZeneca, Roche
Egil S. Blix
Stock and Other Ownership Interests: Novo Nordisk
Consulting or Advisory Role: AstraZeneca, Daiichi Sankyo Nordics, Lilly, Gilead Sciences, MSD, Novartis, Pfizer, Pierre Fabre, Roche
Research Funding: Roche (Inst), Novartis (Inst), Lilly (Inst), Incyte (Inst), Merck (Inst), AstraZeneca (Inst)
Hildegunn S. Aase
Stock and Other Ownership Interests: Novo Nordisk
Alba Llop-Guevara
Patents, Royalties, Other Intellectual Property: Patent pending (WO2019122411A1)
Violeta Serra
Honoraria: AstraZeneca Spain
Consulting or Advisory Role: GlaxoSmithKline
Research Funding: AstraZeneca, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Patent based on a method to identify PARP inhibitor sensitive tumors
Per E. Lønning
Stock and Other Ownership Interests: Cytovation Ltd
Honoraria: Dagens Medisin
Consulting or Advisory Role: Laboratorios Farmaceuticos Rovi
Speakers' Bureau: Akademikonferens
Stian Knappskog
Honoraria: AstraZeneca, Pierre Fabre, Pfizer, Novartis
Research Funding: AstraZeneca (Inst), Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Patent EP2389450 A1, Patent WO 2012/010661
Hans P. Eikesdal
Honoraria: Amgen, Astra Zeneca, Bristol-Myers Squibb, Dagens Medisin, HAI interactive AS, Novartis, Pfizer, Pierre Fabre, Roche
Consulting or Advisory Role: Aptitude Health, Daiichi Sankyo, Eli Lilly, Gilead, medac, MSD, Novartis, Pfizer, Pierre Fabre, Roche
Research Funding: Astra Zeneca, Illumina, Novartis, Pfizer
Expert Testimony: Pfizer
Travel, Accommodations, Expenses: Astra Zeneca, Pierre Fabre
No other potential conflicts of interest were reported.
DISCLAIMER
The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. All authors had access to the data and vouch for its accuracy and completeness. All authors were involved in the decision to submit the manuscript for publication.
PRIOR PRESENTATION
Presented as a poster at the San Antonio Breast Cancer Symposium, San Antonio, TX, December 9, 2022.
SUPPORT
Supported by unrestricted grants from The K.G. Jebsen Foundation [SKGJ-MED-020 to H.P.E., S.K., P.E.L.], Helse Vest [912008 to P.E.L.], The Norwegian Research Council [273354 to P.E.L.] and The Norwegian Cancer Society [190281-2017 to S.K., 190275-2017 to P.E.L.]. Research funding was provided by Grieg Foundation (to H.P.E. and T.A.), Helse Vest [912252; Clinical researcher fellowship to H.P.E.], Generalitat de Catalunya [PERIS, SLT017/20/000081 to A.H.R.], “la Caixa” Foundation and European Institute of Innovation and Technology/Horizon 2020 [LCF/TR/CC19/52470003 to A.L.G.], Asociación Española Contra el Cáncer [AECC, INVES20095LLOP to A.L.G.], Generalitat de Catalunya (PERIS Fellowship SLT002/16/00477 to A.L.G.) ERA PerMed [2019-215 to V.S.] and Miguel Servet fellowship (CPII19/00033 to V.S.). Additional funding and study medication was provided by Illumina [grant number 9529854], Pfizer [WI206347] and AstraZeneca [ESR-14-10077] to H.P.E., S.K., P.E.L.
DATA SHARING STATEMENT
Haukeland University Hospital and the University of Bergen support the dissemination of research data that has been generated and increased cooperation between investigators. Trial data are collected, stored, and disseminated according to institutional guidelines and in accordance with national laws and regulations to ensure the quality, integrity, and use of clinical data. Study Protocol is available online. Signed informed consent forms are stored at each participating hospital and are available for monitoring by regulatory authorities. After publication and on formal request, raw data, including deidentified individual participant data and a data dictionary defining each field in the data set, will be shared according to institutional procedures. Requests are via a standard pro forma describing the nature of the proposed research and extent of data requirements. Data recipients are required to enter a formal data sharing agreement which describes the conditions for release and requirements for data transfer, storage, archiving, publication, and intellectual property. Requests are reviewed by the PETREMAC study team in terms of scientific merit and ethical considerations, including patient consent. Data sharing is permitted if proposed projects have a sound scientific or patient benefit rationale, as agreed by the study team and with approval from the PETREMAC coinvestigators as required.
AUTHOR CONTRIBUTIONS
Conception and design: Synnøve Yndestad, Per E. Lønning, Stian Knappskog, Hans P. Eikesdal
Financial support: Per E. Lønning, Stian Knappskog, Hans P. Eikesdal
Administrative support: Per E. Lønning, Stian Knappskog, Hans P. Eikesdal
Provision of study materials or patients: Olav K. Vintermyr, Gjertrud T. Iversen, Bjørnar Gilje, Stian Knappskog, Hans P. Eikesdal
Collection and assembly of data: Synnøve Yndestad, Christina Engebrethsen, Andrea Herencia-Ropero, Oleksii Nikolaienko, Olav K. Vintermyr, Reidun K. Lillestøl, Laura Minsaas, Beryl Leirvaag, Gjertrud T. Iversen, Bjørnar Gilje, Egil S. Blix, Helge Espelid, Hildegunn S. Aase, Turid Aas, Einar G. Gudlaugsson, Emiel A. M. Janssen, Stian Knappskog, Hans P. Eikesdal
Data analysis and interpretation: Synnøve Yndestad, Christina Engebrethsen, Oleksii Nikolaienko, Reidun K. Lillestøl, Laura Minsaas, Beryl Leirvaag, Gjertrud T. Iversen, Alba Llop-Guevara, Violeta Serra, Per E. Lønning, Stian Knappskog, Hans P. Eikesdal
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Christina Engebrethsen
Stock and Other Ownership Interests: ArcticZymes Technologies, AstraZeneca, Demant, Devyser Diagnostics AB, Illumina, Next Biometrics Group, Photocure, Teva Pharmaceutical Industries ADR, Xbrane Biopharma AB
Bjørnar Gilje
Consulting or Advisory Role: Astellas Pharma, AstraZeneca, Roche
Egil S. Blix
Stock and Other Ownership Interests: Novo Nordisk
Consulting or Advisory Role: AstraZeneca, Daiichi Sankyo Nordics, Lilly, Gilead Sciences, MSD, Novartis, Pfizer, Pierre Fabre, Roche
Research Funding: Roche (Inst), Novartis (Inst), Lilly (Inst), Incyte (Inst), Merck (Inst), AstraZeneca (Inst)
Hildegunn S. Aase
Stock and Other Ownership Interests: Novo Nordisk
Alba Llop-Guevara
Patents, Royalties, Other Intellectual Property: Patent pending (WO2019122411A1)
Violeta Serra
Honoraria: AstraZeneca Spain
Consulting or Advisory Role: GlaxoSmithKline
Research Funding: AstraZeneca, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Patent based on a method to identify PARP inhibitor sensitive tumors
Per E. Lønning
Stock and Other Ownership Interests: Cytovation Ltd
Honoraria: Dagens Medisin
Consulting or Advisory Role: Laboratorios Farmaceuticos Rovi
Speakers' Bureau: Akademikonferens
Stian Knappskog
Honoraria: AstraZeneca, Pierre Fabre, Pfizer, Novartis
Research Funding: AstraZeneca (Inst), Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Patent EP2389450 A1, Patent WO 2012/010661
Hans P. Eikesdal
Honoraria: Amgen, Astra Zeneca, Bristol-Myers Squibb, Dagens Medisin, HAI interactive AS, Novartis, Pfizer, Pierre Fabre, Roche
Consulting or Advisory Role: Aptitude Health, Daiichi Sankyo, Eli Lilly, Gilead, medac, MSD, Novartis, Pfizer, Pierre Fabre, Roche
Research Funding: Astra Zeneca, Illumina, Novartis, Pfizer
Expert Testimony: Pfizer
Travel, Accommodations, Expenses: Astra Zeneca, Pierre Fabre
No other potential conflicts of interest were reported.
REFERENCES
- 1.Tutt A, Robson M, Garber JE, et al. : Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 376:235-244, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Audeh MW, Carmichael J, Penson RT, et al. : Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 376:245-251, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Litton JK, Scoggins ME, Hess KR, et al. : Neoadjuvant talazoparib for patients with operable breast cancer with a germline BRCA pathogenic variant. J Clin Oncol 38:388-394, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spring LM, Han H, Liu MC, et al. : Neoadjuvant study of niraparib in patients with HER2-negative, BRCA-mutated, resectable breast cancer. Nat Cancer 3:1138, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tung NM, Robson ME, Ventz S, et al. : TBCRC 048: Phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol 38:4274-4282, 2020 [DOI] [PubMed] [Google Scholar]
- 6.Robson M, Im SA, Senkus E, et al. : Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 377:523-533, 2017 [DOI] [PubMed] [Google Scholar]
- 7.Gruber JJ, Afghahi A, Timms K, et al. : A phase II study of talazoparib monotherapy in patients with wild-type BRCA1 and BRCA2 with a mutation in other homologous recombination genes. Nat Cancer 3:1181-1191, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelmon KA, Tischkowitz M, Mackay H, et al. : Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: A phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 12:852-861, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Eikesdal HP, Yndestad S, Elzawahry A, et al. : Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann Oncol 32:240-249, 2021 [DOI] [PubMed] [Google Scholar]
- 10.Ballot E, Galland L, Mananet H, et al. : Molecular intrinsic subtypes, genomic, and immune landscapes of BRCA-proficient but HRD-high ER-positive/HER2-negative early breast cancers. Breast Cancer Res 24:80, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heeke AL, Xiu J, Elliott A, et al. : Actionable co-alterations in breast tumors with pathogenic mutations in the homologous recombination DNA damage repair pathway. Breast Cancer Res Treat 184:265-275, 2020 [DOI] [PubMed] [Google Scholar]
- 12.Luen SJ, Viale G, Nik-Zainal S, et al. : Genomic characterisation of hormone receptor-positive breast cancer arising in very young women. Ann Oncol 34:397-409, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vergote I, Gonzalez-Martin A, Ray-Coquard I, et al. : European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann Oncol 33:276-287, 2022 [DOI] [PubMed] [Google Scholar]
- 14.de Bono J, Mateo J, Fizazi K, et al. : Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 382:2091-2102, 2020 [DOI] [PubMed] [Google Scholar]
- 15.de Bono JS, Mehra N, Scagliotti GV, et al. : Talazoparib monotherapy in metastatic castration-resistant prostate cancer with DNA repair alterations (TALAPRO-1): An open-label, phase 2 trial. Lancet Oncol 22:1250-1264, 2021 [DOI] [PubMed] [Google Scholar]
- 16.Sokol ES, Pavlick D, Khiabanian H, et al. : Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol 10.1200/PO.19.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura K, Aimono E, Tanishima S, et al. : Olaparib monotherapy for BRIP1-mutated high-grade serous endometrial cancer. JCO Precis Oncol 10.1200/PO.19.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yates LR, Gerstung M, Knappskog S, et al. : Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21:751-759, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonning PE, Nikolaienko O, Pan K, et al. : Constitutional BRCA1 methylation and risk of incident triple-negative breast cancer and high-grade serous ovarian cancer. JAMA Oncol 8:1579-1587, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolaienko O, Lønning P, Knappskog S: epialleleR: an R/Bioconductor package for sensitive allele-specific methylation analysis in NGS data. GigaScience 12:giad087. 10.1093/gigascience/giad087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolaienko O, Eikesdal HP, Gilje B, et al. : Prenatal BRCA1 epimutations contribute significantly to triple-negative breast cancer development. medrxiv. https://www.medrxiv.org/content/10.1101/2023.05.14.23289949v1, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen R, Seshan VE: FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44:e131, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R, Hastie T, Narasimhan B, et al. : Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 99:6567-6572, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lips EH, Laddach N, Savola SP, et al. : Quantitative copy number analysis by Multiplex Ligation-dependent Probe Amplification (MLPA) of BRCA1-associated breast cancer regions identifies BRCAness. Breast Cancer Res 13:R107, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llop-Guevara A, Loibl S, Villacampa G, et al. : Association of RAD51 with homologous recombination deficiency (HRD) and clinical outcomes in untreated triple-negative breast cancer (TNBC): Analysis of the GeparSixto randomized clinical trial. Ann Oncol 32:1590-1596, 2021 [DOI] [PubMed] [Google Scholar]
- 26. Reference deleted.
- 27.Sorlie T, Tibshirani R, Parker J, et al. : Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100:8418-8423, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavaddat N, Barrowdale D, Andrulis IL, et al. : Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: Results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol Biomarkers Prev 21:134-147, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Copson ER, Maishman TC, Tapper WJ, et al. : Germline BRCA mutation and outcome in young-onset breast cancer (POSH): A prospective cohort study. Lancet Oncol 19:169-180, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen MJ, Kruse TA, Tan Q, et al. : Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One 8:e64268, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller RE, Leary A, Scott CL, et al. : ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann Oncol 31:1606-1622, 2020 [DOI] [PubMed] [Google Scholar]
- 32.Chopra N, Tovey H, Pearson A, et al. : Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat Commun 11:2662, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopkins TA, Ainsworth WB, Ellis PA, et al. : PARP1 trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol Cancer Res 17:409-419, 2019 [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Wang D, Sonzogni O, et al. : PARP-inhibition reprograms macrophages toward an anti-tumor phenotype. Cell Rep 41:111462, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantelidou C, Jadhav H, Kothari A, et al. : STING agonism enhances anti-tumor immune responses and therapeutic efficacy of PARP inhibition in BRCA-associated breast cancer. NPJ Breast Cancer 8:102, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies H, Glodzik D, Morganella S, et al. : HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 23:517-525, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batalini F, Gulhan DC, Mao V, et al. : Mutational signature 3 detected from clinical panel sequencing is associated with responses to olaparib in breast and ovarian cancers. Clin Cancer Res 28:4714-4723, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strickland KC, Howitt BE, Shukla SA, et al. : Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 7:13587-13598, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morse CB, Toukatly MN, Kilgore MR, et al. : Tumor infiltrating lymphocytes and homologous recombination deficiency are independently associated with improved survival in ovarian carcinoma. Gynecol Oncol 153:217-222, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolan E, Savas P, Policheni AN, et al. : Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med 9:eaal4922, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruand M, Barras D, Mina M, et al. : Cell-autonomous inflammation of BRCA1-deficient ovarian cancers drives both tumor-intrinsic immunoreactivity and immune resistance via STING. Cell Rep 36:109412, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid P, Cortes J, Pusztai L, et al. : Pembrolizumab for early triple-negative breast cancer. N Engl J Med 382:810-821, 2020 [DOI] [PubMed] [Google Scholar]
- 43.Mittendorf EA, Zhang H, Barrios CH, et al. : Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): A randomised, double-blind, phase 3 trial. Lancet 396:1090-1100, 2020 [DOI] [PubMed] [Google Scholar]
- 44.Denkert C, von Minckwitz G, Darb-Esfahani S, et al. : Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 19:40-50, 2018 [DOI] [PubMed] [Google Scholar]
- 45.Loi S, Drubay D, Adams S, et al. : Tumor-infiltrating lymphocytes and prognosis: A pooled individual patient analysis of early-stage triple-negative breast cancers. J Clin Oncol 37:559-569, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacob SL, Huppert LA, Rugo HS: Role of immunotherapy in breast cancer. JCO Oncol Pract 19:167-179, 2023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Haukeland University Hospital and the University of Bergen support the dissemination of research data that has been generated and increased cooperation between investigators. Trial data are collected, stored, and disseminated according to institutional guidelines and in accordance with national laws and regulations to ensure the quality, integrity, and use of clinical data. Study Protocol is available online. Signed informed consent forms are stored at each participating hospital and are available for monitoring by regulatory authorities. After publication and on formal request, raw data, including deidentified individual participant data and a data dictionary defining each field in the data set, will be shared according to institutional procedures. Requests are via a standard pro forma describing the nature of the proposed research and extent of data requirements. Data recipients are required to enter a formal data sharing agreement which describes the conditions for release and requirements for data transfer, storage, archiving, publication, and intellectual property. Requests are reviewed by the PETREMAC study team in terms of scientific merit and ethical considerations, including patient consent. Data sharing is permitted if proposed projects have a sound scientific or patient benefit rationale, as agreed by the study team and with approval from the PETREMAC coinvestigators as required.