Abstract
We describe the characterization of a new insertion sequence, IS1515, identified in the genome of Streptococcus pneumoniae I41R, an unencapsulated mutant isolated many years ago (R. Austrian, H. P. Bernheimer, E. E. B. Smith, and G. T. Mills, J. Exp. Med. 110:585–602, 1959). A copy of this element located in the cap1EI41R gene was sequenced. The 871-bp-long IS1515 element possesses 12-bp perfect inverted repeats and generates a 3-bp target duplication upon insertion. The IS encodes a protein of 271 amino acid residues similar to the putative transposases of other insertion sequences, namely IS1381 from S. pneumoniae, ISL2 from Lactobacillus helveticus, IS702 from the cyanobacterium Calothrix sp. strain PCC 7601, and IS112 from Streptomyces albus G. IS1515 appears to be present in the genome of most type 1 pneumococci in a maximum of 13 copies, although it has also been found in the chromosome of pneumococcal isolates belonging to other serotypes. We have found that the unencapsulated phenotype of strain I41R is the result of both the presence of an IS1515 copy and a frameshift mutation in the cap1EI41R gene. Precise excision of the IS was observed in the type 1 encapsulated transformants isolated in experiments designed to repair the frameshift. These results reveal that IS1515 behaves quite differently from other previously described pneumococcal insertion sequences. Several copies of IS1515 were also able to excise and move to another locations in the chromosome of S. pneumoniae. To our knowledge, this is the first report of a functional IS in pneumococcus.
Insertion sequences (ISs) are bacterial mobile DNA elements that cause genome rearrangements, such as deletions, inversions, duplications, and replicon fusions, by their ability to transpose (24). Furthermore, a critical role of ISs in bacterial virulence is currently being recognized. Virulence genes may be located on transmissible genetic elements and form part of particular regions on the bacterial chromosomes (pathogenicity islands), which, in many cases, are flanked by ISs (14). The frequent occurrence of deletions and/or amplifications associated with pathogenicity islands may be ascribed to ISs and other transmissible genetic elements.
Streptococcus pneumoniae (pneumococcus) is an important human pathogen that has been studied intensively for many decades. Morbidity and mortality from pneumococcal infections remain high, even in regions where efficient antibacterial therapy is freely available. The role of ISs in the natural population dynamics of pneumococcus is entirely unknown, although transposable elements have been identified in the form of conjugative transposons (5, 13, 25) and ISs, namely, IS1202 (21), IS1167 (36), and IS1381 (31). Pneumococcal virulence depends upon the presence of capsular polysaccharide, since unencapsulated (rough) mutants are avirulent (12). Interestingly, recent results indicate that the genetic loci (cap) responsible for the synthesis of the pneumococcal capsules are frequently associated with ISs or IS-like sequences (1, 21, 22, 34). In particular, the cap1 locus involved in the type 1 capsule biosynthesis of S. pneumoniae is flanked by nonfunctional copies of IS1167 (22), suggesting a role of this IS element in the horizontal transfer of the capsular genetic determinants (11). Nevertheless, the transposition capacity of the ISs described in S. pneumoniae has not been demonstrated so far.
During our investigations of the cap loci of S. pneumoniae, we have characterized the mutation responsible for the unencapsulated phenotype of strain I41R, a type 1 derivative (3), and identified a segment of pneumococcal DNA that exhibited all of the hallmarks of a bacterial IS. One of the copies of this element, designated IS1515, is inserted into the pneumococcal capsular cap1E gene of the strain I41R. The sequence and characterization of IS1515, the first functional IS found in S. pneumoniae, are reported.
MATERIALS AND METHODS
Bacterial strains and plasmids.
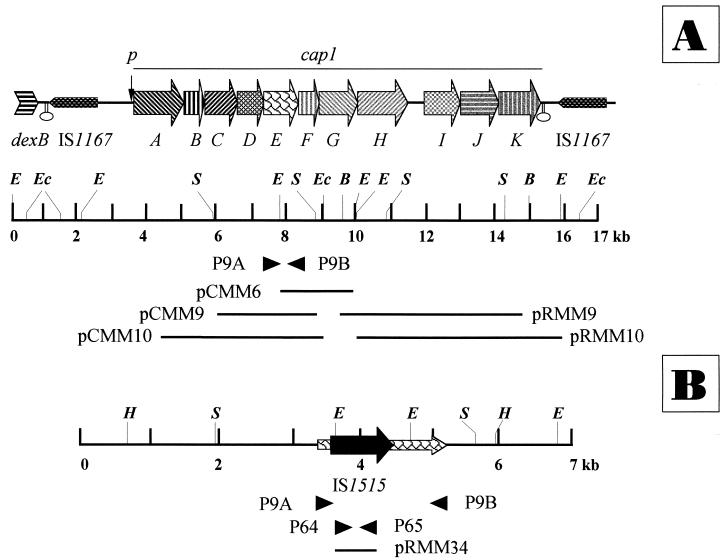
I41R (originally designated as S-I1) (3), an unencapsulated (S1−) mutant of the type 1 pneumococcal strain I41S, was kindly provided by S. Lacks (Brookhaven National Laboratory, N.Y.). The encapsulated pneumococcal strains have been described in a previous publication (22). Strains N and L are type 1 strains that were isolated in the United States in the late 1930s, whereas strain 519/43, also a type 1 strain, was isolated in Denmark in 1943; these three strains were provided by J. Henrichsen (Statens Seruminstitut, Copenhagen, Denmark). A. Fenoll (Pneumococcus Reference Laboratory, Madrid, Spain) provided all other encapsulated S. pneumoniae isolates. In particular, the type 1 pneumococci were isolated in different Spanish hospitals between 1990 and 1996. In some experiments, the pneumococcal laboratory strain M22 (28) was used as a DNA source. Escherichia coli DH5α (30) was also used. Plasmids pLSE1 (28); pGL32 (17); and pCMM6, pCMM9, pCMM10, pRMM9, and pRMM10 (22) have been described in previous publications. The insert locations, as reported in the DNA sequence of cap1 (accession no. Z83335), are as follows: pCMM6, 7864 to 9906; pCMM9, 5902 to 8732; pCMM10, 4661 to 9039; pRMM9, 9522 to 14962; and pRMM10, 10020 to 15900. The construction of pRMM34 is described below.
Growth conditions and transformation of bacteria.
S. pneumoniae was grown in liquid C medium (16) containing 0.08% yeast extract (C+Y) without shaking by procedures previously described (33) or on reconstituted tryptose blood agar base plates (Difco Laboratories) supplemented with 5% defibrinated sheep blood. E. coli cells were grown in Luria-Bertani medium (30). The preparation of pneumococcal DNA, plasmid purification, and transformation of S. pneumoniae and E. coli were carried out as described elsewhere (10). When required, capsulated transformants of S. pneumoniae were enriched by successive transfers of the transformed culture on C medium containing 0.08% bovine serum albumin and 1 μl of anti-R antiserum per ml before plating. Anti-R (antisomatic) antiserum contains group-specific agglutinins that at a convenient dilution agglutinate only rough pneumococci and was raised in rabbits as previously described (26). Lincomycin- and streptomycin-resistant S. pneumoniae transformants were selected with 0.6 and 200 μg of the antibiotic per ml, respectively. Serotyping was carried out by coagglutination (32) with a cell suspension of formalin-treated Staphylococcus aureus (Cowan strain) (Sigma Chemicals Co., St. Louis, Mo.) sensitized with type-specific pneumococcal antisera provided by the Staten Seruminstitut.
DNA techniques.
Restriction endonucleases, T4 DNA ligase, and the Klenow fragment of DNA polymerase were obtained commercially and used according to the recommendations of the suppliers. Gel electrophoresis of plasmids, restriction fragments, and PCR products was carried out in agarose gels as described previously (30). DNA was recovered from gel slices with the Gene Clean II kit (Bio 101). The NEBlot Phototope kit (Millipore) was used to construct biotin-labeled probes, and the Phototope 6K detection kit (Millipore) was used for chemiluminescent detection. Southern blotting, dot blotting, and hybridizations were carried out according to the manufacturer’s instructions. DNA sequencing was carried out with an AbiPrism377 DNA sequencer (Applied Biosystems).
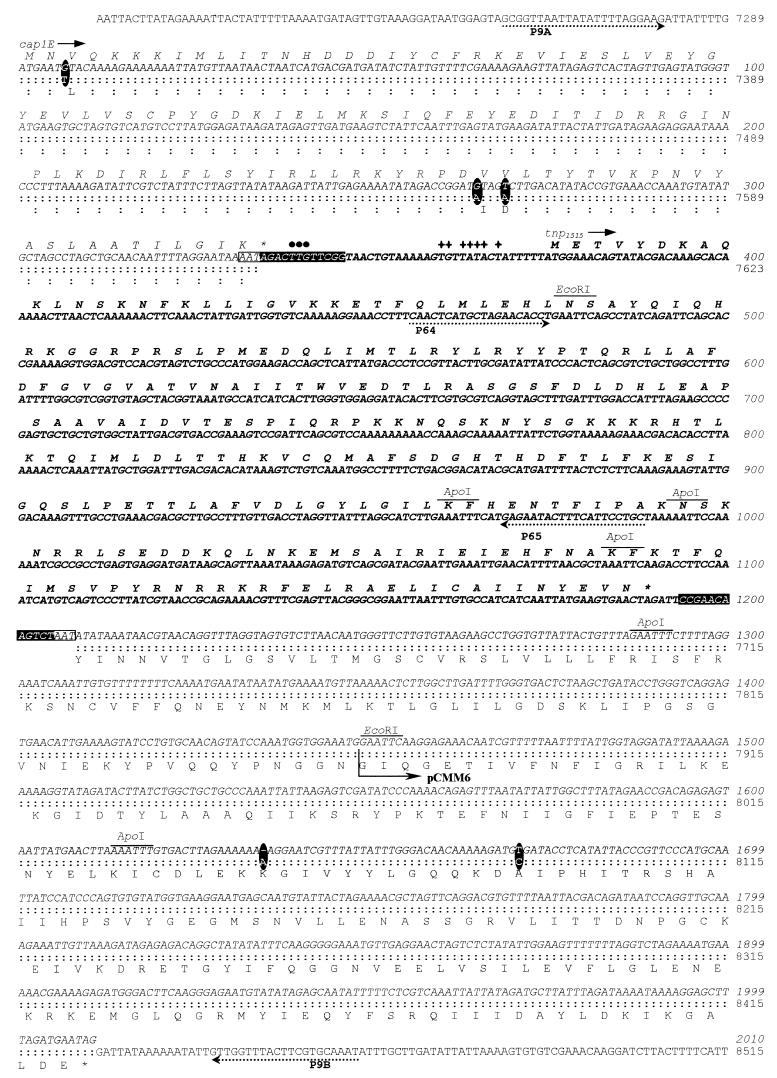
PCR amplifications were performed with 2 U of AmpliTaq DNA polymerase (Perkin-Elmer), 1 μg of chromosomal (or plasmid) DNA, 1 μM (each) synthetic oligonucleotide primer, 200 μM (each) deoxynucleoside triphosphates, and 2.5 mM MgCl2 in the buffer recommended by the manufacturer. Conditions for amplification were chosen according to the G+C content of the corresponding oligonucleotide. The following primers were used: P9A (7258), 5′-GCGGTTAATTAagcTTTAGGAAG-3′; P9B (8463/c), 5′-ATTTGCACGAAGgAtcCCAAC-3′; P64 (456), 5′-CAACTCATGCTAGAACACCT-3′; and P65 (989/c), 5′-GCAGGAATGAAAGTATTCTC-3′. The numbers in parentheses indicate the positions of the first nucleotide of the primer in the sequence shown in Fig. 2, and c means that the corresponding sequence is on the complementary strand. Lowercase letters indicate nucleotides introduced to construct the appropriate restriction sites, which are shown underlined. An internal fragment of IS1515 to be used as a probe was cloned as follows: I41R chromosomal DNA was amplified with oligonucleotides P64 and P65, the PCR product was digested with ApoI and ligated to EcoRI-digested pUC18, and the ligation mixture was used to transform E. coli DH5α. The recombinant plasmid was named pRMM34 (see below).
FIG. 2.
Nucleotide and deduced amino acid sequences of the cap1EI41R gene containing the IS1515 element. The sequence of the cap1E13868 gene (22) is shown for comparison. The sequence corresponding to the I41R strain is shown in italics, whereas that for the IS1515 element is represented in boldface italics. When nucleotides (or amino acid residues) coincide, that corresponding to strain 13868 is substituted for by a colon. The nucleotide differences between both cap1E genes are highlighted as boldface ellipses. The repeated target sequence AAT is shown inside a white box, whereas the terminal inverted repeat sequences of IS1515 are inside black boxes. Upstream of the tnp1515 gene coding for the IS1515 transposase, putative extended −10 (✚) and −35 promoter (•) regions are located. The locations and directions of oligonucleotide primers are indicated, as are EcoRI-ApoI restriction sites. One of the ends of the EcoRI insert of plasmid pCMM6 is also shown. Asterisks indicate stop codons.
Data analysis.
DNA and protein sequences were analyzed with the Genetics Computer Group software package (version 9.0) (7). Amino acid sequence homology searches were performed with the BLAST program at the National Center for Biotechnology Information server (Bethesda, Md.).
Nucleotide sequence accession number.
The nucleotide sequence for IS1515 has been deposited in the EMBL, GenBank, and DDBJ databases under accession no. Z86112.
RESULTS
Identification of IS1515.
We have recently described the molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of pneumococcus (22). cap1 is a cluster that contains 11 genes (cap1A to cap1K) arranged as a single transcriptional unit. In the course of this research, we wanted to characterize the mutation responsible for the inability of S. pneumoniae I41R to synthesize capsular polysaccharide. This strain is a rough derivative of a clinical strain isolated in the United States during the 1950s (2, 3). To determine the genetic defect responsible for the S1− phenotype, some of the plasmids constructed to sequence the type 1 locus (Fig. 1A) were tested for their ability to transform I41R to the S1+ phenotype. Encapsulated transformants were recovered only when pCMM6, pCMM9, and pCMM10 were used as donor DNAs (not shown). These findings indicated that the I41R mutation(s) mapped between positions corresponding to nucleotides 7864 and 8732 of the cap113868 locus, i.e., either in cap1E (positions 7290 to 8426) or in cap1F (positions 8419 to 8970). The genes cap1E13868 and cap1F13868 appear to code for a glycosyl transferase and a galacturonosyl acetylase, respectively (22).
FIG. 1.
Genetic organization of the cap1 cluster of S. pneumoniae 13868 containing the genes coding for type 1 capsular polysaccharide synthesis (A) and the cap1EI41R gene (B). The partial restriction maps of the corresponding regions are also shown. Thick and thin arrows represent complete or interrupted ORFs, respectively. Some of the plasmids used in this study are indicated, as are relevant restriction sites (B, BglII; E, EcoRI; Ec, Eco47III; H, HindIII; and S, ScaI). The location of the promoter of the cap1 cluster is also shown (p). Ovals represent putative transcription terminators (22). Facing solid triangles indicate the locations and directions of pairs of oligonucleotide primers.
The chromosomal region containing the cap1EI41R gene was amplified by PCR with oligonucleotides P9A and P9B (Fig. 1). Interestingly, a 2.1-kb DNA fragment was amplified instead of the 1.2-kb PCR product expected on the basis of the location of the oligonucleotide primers. Direct sequencing of the 2.1-kb PCR product confirmed the presence of an extra DNA fragment within the cap1EI41R gene (Fig. 1B). Analysis of the nucleotide sequence (Fig. 2) suggested that the foreign DNA corresponds to an IS element that was designated IS1515 (see below). Furthermore, the gene cap1EI41R showed five point mutations, i.e., four nucleotide changes and a 1-bp deletion, compared with the cap1E13868 gene previously described (22). The nucleotide changes were 7296T to G, 7554A to G, 7558A to T, and 8086C to T. The first three mutations would produce amino acid replacements in the corresponding Cap1EI41R protein, that is, 3Leu to Val, 89Ile to Val, and 90Asp to Val, respectively. In addition, a TAG stop codon (between nucleotides 334 and 336 of the cap1EI41R gene) was introduced by the presence of the IS and would lead to a truncated protein (Fig. 2). The finding that transformation of competent I41R cells with pCMM6 (Fig. 2) gave rise to S1+ transformants (see above) ruled out the possibility that those three mutations were responsible for the rough phenotype of S. pneumoniae I41R. On the other hand, even in the case of a precise excision of the IS (see below), the deletion mutation located at nucleotide 1635 of the cap1EI41R gene would also encode a defective Cap1E polypeptide as a consequence of frameshifting. Moreover, the 8086C-to-T transition together with the deletion would produce a TGA stop codon and, consequently, a prematurely truncated protein. In summary, either one of the two features, i.e., the presence of the IS1515 or the frameshift mutation, could be responsible for the rough phenotype characteristic of the I41R S. pneumoniae strain.
Structural analysis of IS1515.
The comparative analysis of the nucleotide sequences of cap1E13868 and cap1EI41R revealed an 871-bp DNA fragment in the latter, which was inserted after position 334 and flanked by a 3-bp (AAT) duplication (Fig. 2), that showed all of the features characteristic of prokaryotic ISs (9). The IS element contains 12-bp perfect inverted repeat sequences at the ends and was named IS1515. It consists of one open reading frame (ORF [provisionally designated tnp1515]) with a potential ATG start codon at positions 375 to 377 and extending to a TAG termination codon at positions 1188 to 1190. The G+C content of the tnp1515 gene (41.1%) is similar to the average G+C content of the S. pneumoniae genes (39.1%) (23). An extended −10 site (TGtTATAcT) characteristic of many S. pneumoniae promoters (29) is located 6 bp upstream of the ATG initiation codon. Eighteen base pairs upstream of the TATAcT box, a possible -35 region (TTGttc) was found. As already reported for some pneumococcal genes (29), no apparent ribosome-binding site for translation of tnp1515 was observed. The tnp1515 gene putatively encodes a 271-amino-acid peptide with a predicted pI value (9.53) characteristic of bacterial transposases (9).
Computer searches of the major databases revealed that Tnp1515 was similar to several putative transposases. Figure 3 shows a multiple alignment of the predicted amino acid sequences of the transposase of IS1515 with those of ISL2 from Lactobacillus helveticus (37), IS702 from Calothrix sp. strain PCC 7601 (19), IS1381 from S. pneumoniae (31), and IS112 from Streptomyces albus G (27). Tnp1515 was nearly 40% identical to the transposases of ISL2, IS702, and IS1381, whereas that of IS112 was the most divergent (about 25% identity). In addition, the lengths of the four transposases were very similar, ranging from 257 to 276 amino acid residues. There were no further sequence homologies to other reported IS elements, transposons, or structural genes. All of the features described above strongly suggested that the 871-bp pneumococcal DNA fragment corresponded to a new IS element.
FIG. 3.
Alignment of the deduced amino acid sequences of transposases of several IS1515-related elements. The multiple alignment was carried out with the PILEUP program. Identical amino acid residues in at least four of the proteins are shown in black boxes, and conserved substitutions in all of the transposases are shown in shaded boxes. Pairwise comparisons of the transposases were done with the BESTFIT program. The percentages of identities and similarities (in parentheses) are indicated.
Frequency and distribution of IS1515 among pneumococcal isolates.
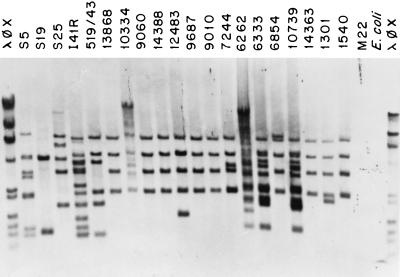
To study the distribution of IS1515 among different pneumococcal isolates, we cloned an internal PCR fragment of IS1515, to be used as a probe in hybridization experiments. The recombinant plasmid pRMM34 (Fig. 1B) harbors the 490-bp EcoRI/ApoI-ApoI fragment from IS1515 (Fig. 2). To determine the number of IS1515 copies, EcoRI-digested chromosomal DNAs from several S. pneumoniae isolates of different serogroups were hybridized with pRMM34. Since one cleavage site for EcoRI is present in the IS element but not in the probe (Fig. 2), the number of hybridization bands should reflect the copy number of IS1515. This IS (or portions of it) was found in strains of serogroups 1, 5, 19, and 25 (Fig. 4) but not in isolates of serogroups 2, 3, 4, 6, 7, 8, 9, 12, 14, 16, 17, 18, 22, 23, 31, 33, or 37 (not shown). However, since only one isolate of each group has been analyzed so far, these results cannot be taken as a proof of an association between serotype and IS carriage. The apparent copy number in the IS1515-positive strains ranged from 2 to 13. It should be mentioned that the common laboratory strain R6 (a type 2 rough derivative) and its descendants (e.g., the M22 strain) did not appear to contain the IS1515 element.
FIG. 4.
IS1515 distribution among strains of S. pneumoniae belonging to different serogroups. Southern blotting was performed with EcoRI-digested genomic DNAs from the indicated independent isolates, which were electrophoresed on agarose gels. The hybridization was carried out at 65°C with biotin-labeled pRMM34 as the probe. S5, S19, and S25 represent isolates belonging to serogroups 5, 19, and 25, respectively. M22 is a rough derivative of a type 2 strain. All other strains are type 1 isolates. λφX indicates a mixture of HindIII-digested λ DNA and the replicative form of φX174 DNA digested with HaeIII.
We have recently proposed that most (if not all) type 1 pneumococcal strains are of clonal origin (22). This suggestion came from studies showing that the molecular organizations of the cap1 gene cluster and its surrounding regions were virtually identical on otherwise unrelated type 1 S. pneumoniae isolates. Southern blot hybridization revealed that 17 of 19 DNAs prepared from S. pneumoniae type 1 clinical isolates contained several copies of the insertion sequence (Fig. 4) and that most of them shared a common pattern of bands. Only strains N and L that were isolated in the United States in the late 1930s did not hybridize with the probe. Evidence suggesting the presence of IS1515 in other gram-positive or gram-negative bacteria analyzed so far was not found (not shown).
Precise excision of IS1515.
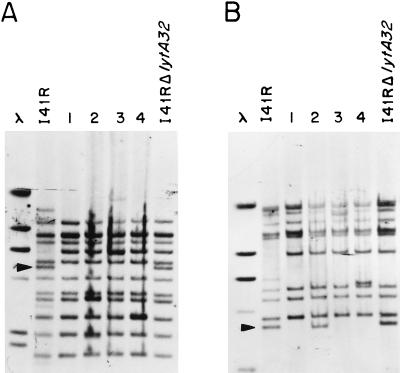
Insertion of a transposon or IS element within a gene prevents the formation of an active gene product. The activity of such a gene can be restored at a low frequency by precise excision of the IS element (9). As documented above, the cap1EI41R gene contains, in addition to the IS1515 element, a frameshift mutation located downstream of the IS copy. Consequently, to restore the S1+ phenotype in the I41R strain, both a precise excision of the IS element and the correction of the mutation are required. When competent I41R cells were transformed with pCMM6, encapsulated transformants were readily isolated (see above), indicating that this double event had been achieved. To provide insights into the detailed mechanisms underlying this finding, we constructed a lytA mutant of I41R by transformation of this strain with pGL32, a plasmid that harbors a frameshift mutation in the lytA gene coding for the S. pneumoniae major autolysin (17). The availability of such a strain (designated I41RΔlytA32 hereafter) allows long periods of incubation at 37°C by preventing the autolytic process characteristic of S. pneumoniae, thus facilitating the screening of capsule production among the transformed clones. The I41RΔlytA32 strain was transformed with pCMM6, and 10 independently isolated S1+ transformants were studied in detail. Hybridization experiments with DNA prepared from these transformants as the template and pRMM34 as the probe showed that both a 5.2-kb HindIII band (Fig. 5A) and a 3.7-kb ScaI band (Fig. 5B), corresponding to the IS1515 copy located within the cap1EI41R gene (Fig. 1), were missing in all of the transformants tested, whereas new IS1515 copies could be observed. This finding indicated that the IS1515 element is indeed a mobile genetic insertion sequence. The selected encapsulated transformants showed at least four different band patterns (Fig. 5). The cap1E gene from one of these encapsulated transformants was PCR amplified with oligonucleotides P9A and P9B, and a DNA band of 1.2 kb was obtained (not shown). This size corresponds to that expected for a cap1E gene lacking the IS1515 element (Fig. 1 and 2). Furthermore, sequencing of the PCR product confirmed the precise excision of the inserted element as well as the elimination of the frameshift mutation (not shown). However, the three cap1EI41R mutations located upstream of the insertion site were still present in the encapsulated transformant, demonstrating that those mutations were not responsible for the unencapsulated phenotype of the I41R strain.
FIG. 5.
IS1515 distribution in encapsulated transformants derived from the I41R strain. Competent cells of I41RΔlytA32 were transformed with pCMM6, and encapsulated cells were isolated as described in the text. Chromosomal DNA prepared from four independently isolated transformants (1 through 4) was digested with HindIII (A) or ScaI (B), and the fragments were separated by agarose gel electrophoresis. Southern blotting was performed at 65°C with biotin-labeled pRMM34 as the probe. The profiles of the chromosomal DNAs prepared from the parental strains I41R and I41ΔlytA32 digested with the same restriction enzymes are also shown. The DNA bands (5.2-kb HindIII and 3.7-kb ScaI fragments, respectively) corresponding to the IS1515 copy located in the cap1EI41R gene are indicated by arrowheads. λ indicates HindIII-digested λ DNA.
Interestingly, I41RΔlytA32, the strain used as the recipient in the transformation experiment described above, although containing the IS1515 copy within its cap1E gene, showed a band pattern different from that of the parental I41R strain, i.e., the IS copy located in the highest-Mr HindIII band in I41R disappeared in I41RΔlytA32, whereas this strain exhibited a new copy of the IS located in a DNA fragment with a size of about 6.5 kb (Fig. 5). This observation suggested that apart from the IS1515 element located in the cap1EI41R gene, at least an additional copy of the IS element is functional in the I41R strain, able to move along the S. pneumoniae chromosome, and capable of integrating into different sites of the bacterial genome.
Analysis of excision of IS1515 by genetic transformation.
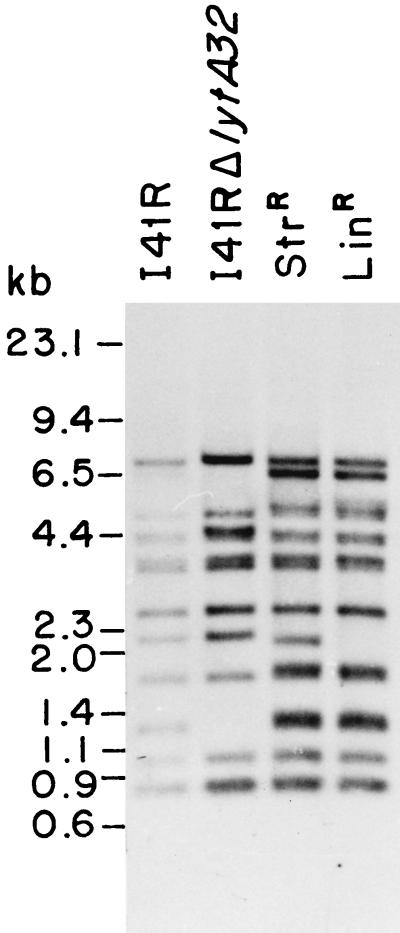
Since the I41RΔlytA32 strain was constructed by transformation of I41R, the results presented above indicated that genetic transformation could be an appropriate tool with which to analyze the excision of IS1515. Streptomycin- or lincomycin-resistant isolates were scored after transformation of competent I41R cells with either M22 chromosomal DNA or pLSE1. DNA was purified from several independently isolated transformants, and Southern blot hybridization experiments with pRMM34 as a probe showed that in every transformant, at least one copy of IS1515 had moved from its original position (Fig. 6). It should be noted that the 1-kb EcoRI band corresponding to the IS1515 copy located in cap1EI41R does not disappear in any of the unencapsulated transformants shown in Fig. 6.
FIG. 6.
IS1515 distribution in streptomycin (StrR)- or lincomycin (LinR)-resistant transformants of the I41R strain. Competent cells of I41R were transformed for the LytA− phenotype with pGL32, for streptomycin resistance with M22 chromosomal DNA, or for lincomycin resistance with pLSE1. Total DNA from one transformant of each class was digested with EcoRI, and the fragments were separated by agarose gel electrophoresis, blotted, and hybridized at 65°C with biotin-labeled pRMM34. The molecular sizes (in kilobases) of the standards (a mixture of HindIII-digested λ DNA and the replicative form of φX174 DNA digested with HaeIII) are indicated to the left.
DISCUSSION
The analysis of the nucleotide sequence of a multicopy DNA fragment identified in S. pneumoniae showed that this fragment displays the characteristic features of an IS element (24) and has been named IS1515. These features included the following. (i) The IS element is bounded by two 12-bp terminal perfect inverted repeats and flanked by two short (3-bp) direct repeats, indicating the duplication of the target sequence. (ii) There is a structure consisting of an ORF (tnp1515) encoding a putative protein with a size of 271 amino acids. This protein is rich in basic amino acid residues (pI 9.53). There is a high degree of similarity with IS1381, recently characterized in pneumococcus (31), as well as with two ISs detected in three gram-positive species, i.e., ISL2 from L. helveticus (37), IS702 from Calothrix sp. (19), and IS112 from S. albus (27). In contrast, no significant similarity was detected when IS1515 was compared to IS1202 (21) and IS1167 (36), two ISs previously described in S. pneumoniae.
We have shown here the presence of a copy of IS1515 in the cap1EI41R gene of an unencapsulated strain of pneumococcus. This gene has been identified as part of the cap1 cluster coding for the polysaccharide capsule of type 1 pneumococcus (22). Hybridization tests revealed that the genome of I41R contains at least 13 copies of IS1515, but this finding does not necessarily imply that all of these copies were identical or even functional. The IS reported here was present in multiple copies in most type 1 strains of pneumococcus (17 of 19 isolates tested) but not in the majority of pneumococci of other serotypes studied (18 of 21 serotypes analyzed). Fingerprinting analyses of type 1 pneumococci like the one carried out here with IS1515 as a marker, alone or in combination with other techniques, such as pulsed-field gel electrophoresis, may be of value in the epidemiological survey of type 1 pneumococcal infection.
One of the fundamental points in understanding the transposition is the specificity with which the ISs insert themselves into the target DNA. Specificity may result from the recognition of a specific sequence (hot spots), a structural or functional feature of the DNA, or a combination of these factors (9). The target site for the insertion of IS1515 in the cap1EI41R gene is preceded by the sequence (319) 5′-ATTTTAGGAATAAAAT-3′ (the underlined bases correspond to the direct repeat) that have a potential to form a stem-loop structure (Fig. 2). On the other hand, the region flanking the IS1515 element is particularly rich in AT base pairs (86% over 35 bp). It has been reported that the insertion hot spots for IS1 have high A+T contents (20), although this feature does not appear to be the only factor influencing the IS1 site selection (35). Very recently, a preliminary nucleotide sequence covering up to 90% of the genome of a type 4 pneumococcal strain has been released (available upon request). Only one copy of the IS1515 element, located immediately downstream of a putative ORF containing a gene encoding a PhoH homolog (contig no. 4139), appears to be present in the pneumococcal strain studied. This copy differs in two nucleotides from that reported here: a T-to-G transversion and a deletion of a TA pair at positions 418 and 423, respectively, and, thus, it codes for a truncated, putatively inactive transposase. Apart from the 3-bp target duplication (AAT) that is also conserved in this type 4 pneumococcus, no other obvious similarities in the regions flanking the IS could be found. Therefore, additional work should be carried out in order to ascertain the requirements of IS1515 for insertion into the S. pneumoniae chromosome. Precise excision of the IS presumably removes one of the direct repeats together with the inserted material. The molecular analysis of the cap1EI41R gene from DNA prepared from a type 1 encapsulated strain obtained by transformation of I41R with pCMM6 revealed that the corresponding copy of IS1515 underwent a precise excision.
Transposition frequency can be regulated indirectly through the control of transposase level or directly by modulation of the recombination reaction by alterations in transposase activity and the reactivity of the DNA substrates. The regulatory mechanisms used by transposable elements to control transpositions are numerous and act at virtually every level of gene expression (for a recent review, see reference 4). Certain proteins regulate transposase activity by interacting with transposase or by competing with transposase for DNA binding sites. This appears to be the case for the IS1 transposase, which, as reported for several IS elements, is synthesized by translational frameshifting in the −1 direction (24). The N-terminal DNA-binding domain can act as a negative regulator of transposition, presumably by excluding intact transposase from the ends of the element by competitive binding. An atypical +1 translational frameshift has recently been claimed to occur for the S. pneumoniae IS1381 transposase (31). In contrast, the IS1515 element gene encodes a complete transposase that does not require any frameshifting to be synthesized and that confers full functionality on the IS element. An attractive hypothesis is that transposition is modulated by the cellular environment, there being certain cellular conditions under which transposition will be favored and other conditions under which it will be disfavored (4). It is well known that many host mutations that increase excision are in genes implicated in DNA repair (9). The role of RecA in the transposition process, however, remains unclear, although it has been reported that transposition is a RecA-dependent process in several systems, such as IS2 (6) and IS30 (8). Transposition of IS1515 takes place during genetic transformation (Fig. 5 and 6), although we cannot conclude from these experiments whether transforming DNA or the development of competence itself (or both) is responsible for excision of the IS. It has recently been demonstrated that transcriptional activation of the S. pneumoniae recA gene occurs at competence (18). It would be interesting to test for transposition of the IS1515 element in a recA background. Unfortunately, since recA pneumococcal strains are not transformable and mutants with conditional mutation of this gene have not been isolated so far, the role of RecA in transposition in S. pneumoniae remains an open question. On the other hand, transposition of IS911, a member of the IS3 family of ISs isolated from the enterobacterium Shigella dysenteriae, exhibits a temperature-sensitive phenotype; i.e., whereas transposition was optimal at 30°C, it was greatly reduced at 42°C (15). Since the incubation of competent pneumococcal cells with transforming DNA is carried out at 30°C, we cannot discard the possibility that the transposase of the IS1515 may be also stimulated at this temperature. New experimental tools are to be designed for S. pneumoniae in order to achieve conclusive answers to the above questions.
The presence of a large number of IS elements might be expected to have a strong influence on the structure and stability of the genome, since, in addition to their transposition properties, ISs act as substrates for homologous recombination. Although other ISs have been found in pneumococci, as reported here, IS1515 represents the first example of a functional IS element in this important human pathogen.
ACKNOWLEDGMENTS
We thank E. Díaz, J. L. García, and P. García for critically reading the manuscript. The technical assistance of E. Cano, M. Carrasco, A. Díaz, G. Porras, and V. Muñoz is greatly appreciated. We also thank J. Henrichsen and A. Fenoll for kindly providing some “old” type 1 strains and the majority of clinical isolates, respectively.
This work was supported by grant PB-93-0115-C02-01 from the Programa Sectorial de Promoción General del Conocimiento. R. Muñoz was a recipient of a Contrato Temporal de Investigadores from the CSIC.
REFERENCES
- 1.Arrecubieta C, García E, López R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 2.Austrian R, Bernheimer H P. Simultaneous production of two capsular polysaccharides by pneumococcus. I. Properties of a pneumococcus manifesting binary capsulation. J Exp Med. 1959;110:571–584. doi: 10.1084/jem.110.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austrian R, Bernheimer H P, Smith E E B, Mills G T. Simultaneous production of two capsular polysaccharides by pneumococcus. II. The genetic and biochemical bases of binary capsulation. J Exp Med. 1959;110:585–602. doi: 10.1084/jem.110.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig N L. Transposition. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2339–2362. [Google Scholar]
- 5.David F, De Céspedes G, Delbos F, Horaud T. Diversity of chromosomal genetic elements and a gene identification in antibiotic-resistant strains of Streptococcus pneumoniae and Streptococcus bovis. Plasmid. 1993;29:147–153. doi: 10.1006/plas.1993.1017. [DOI] [PubMed] [Google Scholar]
- 6.Deonier R C, Mirels L. Excision of F plasmid sequences by recombination at directly repeated insertion sequence 2 elements: involvement of recA. Proc Natl Acad Sci USA. 1977;74:3965–3969. doi: 10.1073/pnas.74.9.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farkas T, Kiss J, Olasz F. The construction and characterization of an effective transpositional system based on IS30. FEBS Lett. 1996;390:53–58. doi: 10.1016/0014-5793(96)00626-6. [DOI] [PubMed] [Google Scholar]
- 9.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 10.García E, García P, López R. Cloning and sequencing of a gene involved in the synthesis of the capsular polysaccharide of Streptococcus pneumoniae type 3. Mol Gen Genet. 1993;239:188–195. doi: 10.1007/BF00281617. [DOI] [PubMed] [Google Scholar]
- 11.García E, López R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 12.Griffith F. The significance of pneumococcal types. J Hyg. 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guild W, Hazum S, Smith M D. Chromosomal location of conjugative R determinants in strain BM4200 of Streptococcus pneumoniae. In: Levy S B, Clowes R C, Koening E L, editors. Molecular biology, pathogenicity, and ecology of bacterial plasmids. New York, N.Y: Plenum; 1981. p. 610. [Google Scholar]
- 14.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 15.Haren L, Bétermier M, Polard P, Chandler M. IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol Microbiol. 1997;25:531–540. doi: 10.1046/j.1365-2958.1997.4951854.x. [DOI] [PubMed] [Google Scholar]
- 16.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in Pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 17.López R, Sánchez-Puelles J M, García E, García J L, Ronda C, García P. Isolation, characterization and physiological properties of an autolytic-defective mutant of Streptococcus pneumoniae. Mol Gen Genet. 1986;204:237–242. doi: 10.1007/BF00425504. [DOI] [PubMed] [Google Scholar]
- 18.Martin B, García P, Castanié M-P, Claverys J-P. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol Microbiol. 1995;15:367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 19.Mazel D, Bernard C, Schwarz R, Castets A M, Houmard J, Tandeau de Marsac N. Characterization of two insertion sequences, IS701 and IS702, from the cyanobacterium Calothrix species PCC 7601. Mol Microbiol. 1991;5:2165–2170. doi: 10.1111/j.1365-2958.1991.tb02146.x. [DOI] [PubMed] [Google Scholar]
- 20.Meyer J, Iida S, Arber W. Does the insertion element IS1 transpose preferentially into A+T-rich DNA segments? Mol Gen Genet. 1980;178:471–473. doi: 10.1007/BF00270502. [DOI] [PubMed] [Google Scholar]
- 21.Morona J K, Guidolin A, Morona R, Hansman D, Paton J C. Isolation, characterization, and nucleotide sequence of IS1202, an insertion sequence of Streptococcus pneumoniae. J Bacteriol. 1994;176:4437–4443. doi: 10.1128/jb.176.14.4437-4443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muñoz R, Mollerach M, López R, García E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1997;25:244–245. doi: 10.1093/nar/25.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohtsubo E, Sekine Y. Bacterial insertion sequences. Curr Top Microbiol Immunol. 1996;204:1–26. doi: 10.1007/978-3-642-79795-8_1. [DOI] [PubMed] [Google Scholar]
- 25.Provvedi R, Manganelli R, Pozzi G. Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett. 1996;135:231–236. doi: 10.1111/j.1574-6968.1996.tb07994.x. [DOI] [PubMed] [Google Scholar]
- 26.Ravin A W. Reciprocal capsular transformation of pneumococci. J Bacteriol. 1959;77:296–309. doi: 10.1128/jb.77.3.296-309.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodicio M R, Alvarez M A, Chater K F. Isolation and genetic structure of IS112, an insertion sequence responsible for the inactivation of the SalI restriction-modification system of Streptomyces albus G. Mol Gen Genet. 1991;225:142–147. doi: 10.1007/BF00282652. [DOI] [PubMed] [Google Scholar]
- 28.Ronda C, García J L, López R. Characterization of genetic transformation in Streptococcus oralis NCTC 11427: expression of the pneumococcal amidase in S. oralis using a new shuttle vector. Mol Gen Genet. 1988;215:53–57. doi: 10.1007/BF00331302. [DOI] [PubMed] [Google Scholar]
- 29.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the dpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sánchez-Beato A R, García E, López R, García J L. Identification and characterization of IS1381, a new insertion sequence in Streptococcus pneumoniae. J Bacteriol. 1997;179:2459–2463. doi: 10.1128/jb.179.7.2459-2463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smart L E, Henrichsen J. An alternative approach to typing of Streptococcus pneumoniae strains by coagglutination. Acta Pathol Microbiol Immunol Scand Sect B. 1986;94:409–413. doi: 10.1111/j.1699-0463.1986.tb03076.x. [DOI] [PubMed] [Google Scholar]
- 33.Tomasz A. Cellular metabolism in genetic transformation of pneumococci: requirement for protein synthesis during induction of competence. J Bacteriol. 1970;101:860–871. doi: 10.1128/jb.101.3.860-871.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yother J, Ambrose K D, Caimano M J. Association of a partial H-rpt element with the type 3 capsule locus of Streptococcus pneumoniae. Mol Microbiol. 1997;25:201–203. doi: 10.1046/j.1365-2958.1997.4361798.x. [DOI] [PubMed] [Google Scholar]
- 35.Zerbib D, Gamas P, Chandler M, Prentki P, Bass S, Galas D. Specificity of insertion of IS1. J Mol Biol. 1985;185:517–524. doi: 10.1016/0022-2836(85)90068-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Hui F M, Morrison D. Characterization of IS1167, a new insertion sequence in Streptococcus pneumoniae. Plasmid. 1995;33:127–138. doi: 10.1006/plas.1995.1014. [DOI] [PubMed] [Google Scholar]
- 37.Zwahlen M C, Mollet B. ISL2, a new mobile genetic element in Lactobacillus helveticus. Mol Gen Genet. 1994;245:334–338. doi: 10.1007/BF00290113. [DOI] [PubMed] [Google Scholar]