Abstract
Mammalian evolution has been influenced by viruses for millions of years, leaving signatures of adaptive evolution within genes encoding for viral interacting proteins. Synaptogyrin-2 (SYNGR2) is a transmembrane protein implicated in promoting bacterial and viral infections. A genome-wide association study of pigs experimentally infected with porcine circovirus type 2b (PCV2b) uncovered a missense mutation (SYNGR2 p.Arg63Cys) associated with viral load. In this study, CRISPR/Cas9-mediated gene editing of the porcine kidney 15 (PK15, wtSYNGR2+p.63Arg) cell line generated clones homozygous for the favorable SYNGR2 p.63Cys allele (emSYNGR2+p.63Cys). Infection of edited clones resulted in decreased PCV2 replication compared to wildtype PK15 (P<0.05), with consistent effects across genetically distinct PCV2b and PCV2d isolates. Sequence analyses of wild and domestic pigs (n>700) revealed the favorable SYNGR2 p.63Cys allele is unique to domestic pigs and more predominant in European than Asian breeds. A haplotype defined by the SYNGR2 p.63Cys allele was likely derived from an ancestral haplotype nearly fixed within European (0.977) but absent from Asian wild boar. We hypothesize that the SYNGR2 p.63Cys allele arose post-domestication in ancestral European swine. Decreased genetic diversity in homozygotes for the SYNGR2 p.63Cys allele compared to SYNGR2 p.63Arg, corroborates a rapid increase in frequency of SYGNR2 p.63Cys via positive selection. Signatures of adaptive evolution across mammalian species were also identified within SYNGR2 intraluminal loop domains, coinciding with the location of SYNGR2 p.Arg63Cys. Therefore, SYNGR2 may reflect a novel component of the host-virus evolutionary arms race across mammals with SYNGR2 p.Arg63Cys representing a species-specific example of putative adaptive evolution.
Author summary
Our research provides direct evidence of the functional role of a missense substitution in the host SYNGR2 gene (SYNGR2 p.Arg63Cys) in the replication of porcine circovirus 2, the smallest virus known to infect mammalian cells. DNA sequences and haplotype analyses of SYNGR2 across domestic and wild pig populations suggests a likely origin and subsequent positive selection of the favorable SYNGR2 p.63Cys allele post-domestication in European swine. While SYNGR2 p.Arg63Cys represents a species-specific example of putative adaptive evolution, signatures of selection detected in SYNGR2 across mammalian species suggested that this gene is a new component of the host-virus evolutionary arms race.
Introduction
The host-virus evolutionary arms race embodies an age-old battle to maintain fitness across all domains of life [1]. Recent evidence indicates that majority of genetic adaptation in host genomes occurs within viral interacting proteins (VIPs), which can interact with viruses in a strain-specific manner or across multiple members of a viral family [2]. While our knowledge of VIPs, especially in humans, has grown in recent years, the identification of adaptive genetic variants affecting host resilience remains limited. Additionally, the influence that viruses have on host evolution cannot be characterized by a single adaptive event, as the ability of viruses to rapidly counter-evolve is thought to drive repeated instances of adaptive change in VIPs [1,3]. Therefore, it’s not only important to identify adaptive host alleles, but to also understand their function and context within the host-virus evolutionary arms race. Due to the constant threat imposed by viral pathogens, there is substantial interest in elucidating the role of host genetics in disease susceptibility across diverse fields from agriculture to human health.
Porcine circovirus type 2 (PCV2) is the smallest known virus capable of infecting mammalian cells with a single-stranded circular DNA genome approximately 1.7Kb in length. The majority of PCV2 strains belong to one of three major subtypes: PCV2a, PCV2b, and PCV2d. In the early 2000’s a global genotype shift from PCV2a to PCV2b corresponded with increased incidence and severity of porcine circovirus associated diseases (PCVAD). However, evidence of increased PCV2d predominance in recent years suggests another global genotype shift may be in progress [4]. While only a fraction of pigs develop clinical disease, PCV2 is prevalent in domestic and wild populations around the world and continues to pose a constant threat to animal welfare. Previously, our group uncovered a host missense variant (p.Arg63Cys) within the host Synaptogyrin-2 (SYNGR2) gene that was associated with PCV2b viral load and immune response following in vivo experimental challenge [5]. Infection of a CRISPR-Cas9 mediated SYNGR2 knock-out porcine kidney 15 (PK15) cell line significantly reduced PCV2b replication beginning 24 hours post infection [5]. However, direct experimental evidence for the causality of the SYNGR2 p.Arg63Cys variant or consistency of its effect across PCV2 subtypes have yet to be provided.
SYNGR2 is a transmembrane protein with reported functions in vesicle biogenesis and membrane trafficking and transport [6]. Despite no known function in host immunity, SYNGR2 has been recently implicated in promoting both viral and bacterial infections. Specifically, a direct interaction was reported between SYNGR2 and the non-structural protein of a tick-borne human RNA virus, severe fever with thrombocytopenia syndrome virus (SFTSV) [7]. This interaction was found to be crucial for the formation of intracytoplasmic inclusion bodies utilized as viral factories for SFTSV replication [7]. Additionally, SYNGR2 was found necessary for internalization of the active CdtB subunit and induced toxicity following exposure to Aggregatibacter actinomycetemcomitans cytotoxin [8,9]. Together, these studies indicate that SYNGR2 may represent a non-immune VIP common across pathogens and mammalian hosts. Therefore, investigating the evolution of SYNGR2 in domestic pigs as well as other mammalian lineages could provide valuable insight and a model of host adaptation to co-evolving pathogens.
In this study, we provide direct experimental evidence for the function of the SYNGR2 p.Arg63Cys variant in the early stages of infection using in vitro models with consistent effects observed across PCV2 subtypes. Evaluation of SYNGR2 sequence data revealed a likely origin and subsequent positive selection of the SYNGR2 p.63Cys allele post-domestication in European swine. Corresponding with the location of the SYNGR2 p.Arg63Cys variant, analyses for signatures of selection across mammalian species highlighted the intraluminal loop domains as key regions of putative adaptive evolution within SYNGR2. Therefore, SYNGR2 may reflect a novel component of the host-virus evolutionary arms race across mammals with SYNGR2 p.Arg63Cys representing a species-specific example of putative adaptive evolution.
Results
The SYNGR2 p.63Cys allele confers a significant reduction in PCV2b replication
The porcine kidney 15 (PK15) cell line is of epithelial-origin and homozygous for the unfavorable SYNGR2 p.63Arg allele associated with higher viral load following in vivo experimental PCV2b challenge [5]. Edited PK15 clones homozygous for the alternate SYNGR2 p.63Cys allele were generated via CRISPR-Cas9 gene editing using a single guide RNA and DNA template homologous to the non-targeting strand (Figs A and B in S1 Text). To directly compare the effect of the SYNGR2 p.Arg63Cys variant on PCV2 infection, a SYNGR2 knock-out PK15 clone (emSYNGR2-del) [5], an edited PK15 clone homozygous for the SYNGR2 p.63Cys allele (emSYNGR2+p.63Cys), and the wildtype PK15 cell line (wtSYNGR2+p.63Arg) were simultaneously infected with a PCV2b isolate. The number of PCV2b genome copies was significantly less for the emSYNGR2-del clone compared to either cell line with predicted functional SYNGR2 proteins (P<0.05–0.001; Fig 1). However, a significant reduction in viral replication was also observed for the emSYNGR2+p.63Cys clone compared to wtSYNGR2+p.63Arg at 24 and 48 hours post infection (hpi) in the cell fraction (P<0.05–0.005) and at 72 hpi in the supernatant fraction (P<0.01; Fig 1). On average, the number of PCV2b copies after 72 hours increased by only 26-fold in emSYNGR2+p.63Cys cells and 85-fold in the supernatant compared to 77-fold and 1,177-fold in wtSYNGR2+p.63Arg cells and supernatant, respectively (P<0.05).
Fig 1. PCV2 genome copy number in wildtype and edited PK15 clones following in vitro infection.
Log10 transformed viral genome copy number per well in the cell (top) and per uL in the supernatant (bottom) fractions across timepoints post infection with PCV2b or PCV2d (MOI = 0.00025). Error bars represent one standard error from the mean for three independent infection replicates. *P<0.05, **P<0.01, ***P<0.001.
Consistent effect of SYNGR2 p.Arg63Cys variant between genetically distinct PCV2 isolates
To assess whether the effect of the SYNGR2 p.Arg63Cys variant is consistent across PCV2 strains, these same edited and wildtype cell lines were infected with another PCV2 isolate belonging to the PCV2d subtype. Nucleotide and predicted amino acid sequence comparison confirmed the PCV2d isolate is genetically distinct from the PCV2b isolate with 96.15% similarity across the genome. The first open reading frame (ORF1), encoding the viral Replicase protein, exhibited greater similarity between isolates at both the nucleotide (97.46%) and amino acid level (99.04%) than ORF2, which encodes the viral Capsid protein. Specifically, ORF2 was 93.73% and 93.99% similar between isolates at the nucleotide and amino acid levels, respectively. Alignment of the predicted Capsid amino acid sequence revealed characteristic subtype-specific substitutions for both the PCV2b and PCV2d isolates [10]. For instance, both isolates have residues at positions 59 (R/K), 206 (I), and 63 (R), which form a large surface on the head of the Capsid protein, predicted to increase host-cell binding affinity compared to PCV2a strains [10]. However, the PCV2b and PCV2d isolates differed within the Capsid tail region, with the PCV2d Capsid possessing two additional amino acids at the C-terminus, 234 (S) and 235 (E), predicted to further enhance host-cell receptor binding [10].
Following in vitro infection with the PCV2d isolate, the emSYNGR2-del cell line exhibited fewer PCV2d copies than either the wtSYNGR+p.63Arg and emSYNGR2+p.63Cys clones (P<0.05–0.001; Fig 1). A reduction in viral replication was also observed for the emSYNGR2+p.63Cys clone compared to wtSYNGR2+p.63Arg beginning at 24 hpi in the cell fraction and 48 hpi in the supernatant fraction (P<0.05; Fig 1). After 72 hpi, the amount of PCV2d copies increased 1,069-fold on average in wtSYNGR2+p.63Arg cells and 2,368-fold in the supernatant compared to only 39-fold in emSYNGR2+p.63Cys cells and 133-fold in supernatant (P<0.05).
Effect of SYNGR2 p.Arg63Cys alleles on PCV2 infection are not due to differences in gene expression
Previous work revealed no differences in SYNGR2 expression following in vitro infection of wildtype PK15 cells with PCV2b [5]. However, PCV2 has been shown to infect only a small proportion of cells in culture, which may have hindered detection of potential gene expression differences during infection. In attempt to enhance our ability to detect alterations in gene expression, the wtSYNGR2+p.63Arg and emSYNGR2+p.63Cys cell lines were inoculated with the PCV2b isolate at a higher multiplicity of infection (MOI) as well as evaluated at an additional timepoint post infection (12 hpi) to better characterize the early stages of infection. Supporting these prior results, we observed no differences in SYNGR2 expression levels between control and infected cells for either cell line (Fig C in S1 Text). However, SYNGR2 expression was lower in the emSYNGR2+p.63Cys cells compared to wtSYNGR2+p.63Arg in both control and infected cells at various timepoints post infection (P<0.05; Fig C in S1 Text).
Consistent with the previous findings, a substantial reduction in the number of PCV2b copies was observed for emSYNGR2+p.63Cys compared to the wtSYNGR2+p.63Arg cell line starting at 24 hpi in the cell fraction (P<0.05) and 48 hpi in the supernatant (P<0.01; Fig 2). After 72 hpi, the amount of PCV2b copies increased 168-fold on average in wtSYNGR2+p.63Arg cells compared to only 8-fold in emSYNGR2+p.63Cys cells (P<0.05). This fold change was even more pronounced in the supernatant fraction, with a 6,195-fold increase on average for wtSYNGR2+p.63Arg compared to only 109-fold for emSYNGR2+p.63Cys (P<0.005). Comparing the slope of the line in the cell fraction between timepoints post infection, the rate of viral replication from 12 to 24 hpi was dramatically lower in emSYNGR2+p.63Cys cells compared to wtSYNGR2+p.63Arg (P<0.05). However, this difference was not maintained at later timepoints, with similar viral replication rates between cell lines after 24 hours. The effect of the early difference in cells is reflected in the supernatant fraction at subsequent timepoints, with substantially lower replication rates for emSYNGR2+p.63Cys relative to wtSYNGR2+p.63Arg after 24 hours (P<0.005).
Fig 2. PCV2b genome copy number in wtSYNGR2+p.63Arg and emSYNGR2+p.63Cys PK15 clones following in vitro infection.
Log10 transformed viral genome copy number per well in the cell (top) and per uL in the supernatant (bottom) fractions across timepoints post infection with PCV2b (MOI = 0.00075). Error bars represent one standard error from the mean for three independent infection replicates. *P<0.05, **P<0.01, ***P<0.001, ***P<0.0001.
Sequence comparison between the emSYNGR2+p.63Cys and wtSYNGR2+p.63Arg cell lines had previously confirmed no differences in the SYNGR2 coding sequence besides the intended 63Arg (C) > 63Cys (T) allelic substitution (Fig B in S1 Text). However, additional genomic sequencing of non-coding regions revealed two intronic SNPs and one within the distal 3’UTR that differed between the two cell lines. Therefore, another edited PK15 clone homozygous for the SYNGR2 p.63Cys allele (2emSYNGR2+p.63Cys), but otherwise identical to the wtSYNGR2+p.63Arg cell line, was similarly infected with PCV2b to further validate the effect of the SYNGR2 p.Arg63Cys variant. Consistent with the first clone, a significant decrease in viral replication was observed in the 2emSYNGR2+p.63Cys cell and supernatant fractions starting at 24 hpi compared to the wtSYNGR2+p.63Arg cell line (P<0.05) along with lower overall fold-change (P<0.01) and replication rate from 12 to 24 hpi in the cell fraction (P<0.05; Fig 2). No differences in gene expression were observed between control and infected 2emSYNGR2+p.63Cys and wtSYNGR2+p.63Arg cells across timepoints (Fig C in S1 Text).
Haplotype distribution across Sus scrofa subgroups provides insight into the evolutionary origin and positive selection of the SYNGR2 p.63Cys allele in domestic swine
Previous analysis of several domestic populations revealed significant variation in the frequency of the favorable SYNGR2 p.63Cys allele between breeds [5], prompting further inquiry into both the evolutionary origin of this allele and selection within the Suidae lineage. Comparative analysis of sequences available within the European Nucleotide Archive (ENA) from wild or domestic S. scrofa (n = 731) and relative species within the Sus genus (n = 20) identified 19 SNPs within the SYNGR2 coding sequence, with SYNGR2 p.Arg63Cys being the only missense variant (Table B in S1 Text). The favorable SYNGR2 p.63Cys allele was found exclusively within domestic swine and was more prevalent in European (43.1%) than Asian (8.1%) with the highest frequencies in Duroc (68.3%) and Pietrain (41.2%) breeds (Table C and D in S1 Text). Genotypes for 15 SNPs (MAF>0.01) were used to assign haplotypes independently within three groups: S. scrofa domestic, S. scrofa wild boar, and Sus relatives. While no overlap was observed between S. scrofa (n = 10) and Sus relative haplotypes (n = 4), substantial overlap was present between domestic and wild boar with only two haplotypes, Hap1 and Hap8, exclusively found in domestic swine (Table 1).
Table 1. SYNGR2 haplotypes identified across Suidae groups.
(*187 = SYNGR2 p.Arg63Cys; DM = Domestic, WB = Wild Boar, SR = Sus Relatives).
| SYNGR2 Haplotype | SNP | Frequency | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 105 | *187 | 195 | 198 | 210 | 321 | 324 | 465 | 516 | 540 | 576 | 588 | 597 | 603 | 645 | DM (n = 676) | WB (n = 55) | SR (n = 20) | All (n = 751) | |
| Hap1 | G | T | C | C | T | T | C | C | C | C | C | G | C | C | C | 0.35 | -- | -- | 0.316 |
| Hap2 | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.309 | 0.472 | -- | 0.313 |
| Hap3 | - | C | - | T | - | C | - | - | - | T | - | - | T | T | - | 0.134 | 0.082 | -- | 0.127 |
| Hap4 | - | C | - | T | - | C | - | T | T | T | - | - | - | - | - | 0.045 | 0.069 | -- | 0.045 |
| Hap5 | - | C | - | T | - | C | - | - | - | - | - | - | - | - | - | 0.031 | 0.05 | -- | 0.03 |
| Hap6 | - | C | - | - | - | C | - | - | - | - | - | - | - | - | - | 0.017 | 0.06 | -- | 0.022 |
| Hap7 | - | C | - | - | C | C | - | - | - | - | - | - | - | - | - | 0.015 | -- | -- | 0.014 |
| Hap8 | - | C | - | T | - | C | - | - | - | T | - | - | - | - | - | 0.013 | -- | -- | 0.011 |
| Hap9 | - | C | T | T | - | C | - | - | - | - | - | - | - | - | - | 0.011 | 0.098 | -- | 0.018 |
| Hap10 | - | C | - | T | - | C | - | - | T | T | - | - | - | - | - | -- | 0.019 | -- | 0.008 |
| Hap11 | - | C | - | - | C | - | - | - | - | - | T | A | - | - | T | -- | -- | 0.5 | 0.012 |
| Hap12 | - | C | - | - | C | - | T | - | - | - | T | A | - | - | T | -- | -- | 0.125 | 0.003 |
| Hap13 | - | C | - | - | C | C | - | T | - | T | - | - | T | - | - | -- | -- | 0.125 | 0.003 |
| Hap14 | A | C | - | - | C | - | - | - | - | - | T | A | - | - | T | -- | -- | 0.1 | 0.003 |
| 0.925 | 0.85 | 0.85 | 0.925 | ||||||||||||||||
Hap1 was the most prevalent haplotype within the domestic group (35%) and the only haplotype that included the favorable SYNGR2 p.63Cys allele. The second most frequent haplotype within the domestic group (Hap2, 30.9%) and most prevalent haplotype observed in wild boar (47.2%) was identical with Hap1 except for the SYNGR2 p.63Arg allele. Both Hap1 (p.63Cys) and Hap2 (p.63Arg) were more predominant amongst European domestic (EUD) than Asian domestic (ASD) sequences, accounting for 77.6% of the haplotypes within EUD compared to only 17.7% in ASD (Fig D and Table E in S1 Text). This trend was even more dramatic across wild boar sequences, with Hap2 being nearly fixed (97.7%) in European wild boar (EUW) and completely absent from Asian wild boar (ASW; Fig D and Table E in S1 Text). The geographic separation between European predominant (Hap1 and Hap2) and Asian predominant (Hap3-Hap10) haplotypes is clearly illustrated in the haplotype network as well as the likely derivation of Hap1 from the ancestral European Hap2 background (Fig 3). Although present in the ASDO (ASD Other) group at a frequency of ~3%, Hap1 was otherwise only present in the Meishan breed with a frequency of 17.3% (Fig E in S1 Text). However, Hap1 was present in all seven EUD breeds, with the highest frequency in Duroc (67.8%) followed by Pietrain (43%; Fig E in S1 Text). EUD breeds with noted historical Asian introgression, such as Landrace and Large White/Yorkshire, exhibited greater haplotype diversity than other EUD breeds (Fig E in S1 Text). Specifically, compared to Landrace (50.8%) and Large White/Yorkshire (70.9–72.5%), Hap1 and Hap2 account for 100% of the haplotypes in Iberian, which is one of the few breeds with no written or molecular evidence of Asian introgression [11,12]. Together, these findings indicate that the favorable SYNGR2 p.63Cys allele arose post-domestication in ancestral European domestic pigs from which modern European breeds were derived and was subsequently introduced into Asian domestic swine via European introgression [13].
Fig 3. Integer-joining network of SYNGR2 haplotypes within Suidae.
Haplotype frequencies within each S. scrofa subgroup were applied to hypothetical sample sets of equal size. Node size represents haplotype frequency across S. scrofa, and different colored segments represent the proportion within each subgroup: European Domestic (EUD), European Wild Boar (EUW), Asian Domestic (ASD), Asian Wild Boar (ASW), Sus relatives (SUR). Black nodes represent inferred intermediate haplotypes not present in any samples within the dataset. Single nucleotide changes are represented as hatch marks.
To test for evidence of recent positive selection favoring the SYNGR2 p.63Cys allele in domestic swine, the level of genetic diversity within the SYNGR2 locus was compared between sequence groups homozygous for alternate SYNGR2 p.Arg63Cys alleles (Arg/Arg = 356, Cys/Cys = 173). Based on 10 SYNGR2 SNPs, the Cys/Cys group exhibited dramatically lower variation compared to the Arg/Arg group. Specifically, the observed heterozygosity within the Cys/Cys group was only 0.018 compared to 0.177 in the Arg/Arg group (Table 2). Heterogeneity was also observed across all SYNGR2 SNPs within the Arg/Arg group, with eight out of 10 SNP exhibiting minor allele frequencies (MAF) greater than 0.05 (Table 3). In contrast, none of the SNPs within the Cys/Cys group had MAF greater than 0.05, with two being completely fixed (Table 3). In fact, the SNP with the highest MAF (0.02) in the Cys/Cys group exhibited a similar level of heterogeneity as the SNP with the lowest MAF (0.018) in the Arg/Arg group (Table 3). Together, these findings emphasize a significant reduction in genetic diversity across the SYNGR2 locus in the derived Cys/Cys sequences relative to the ancestral Arg/Arg sequences, consistent with a rapid increase in frequency of the SYNGR2 p.63Cys allele via positive selection in domestic swine.
Table 2. Parameters of genetic diversity across domestic groups with alternate homozygous SYNGR2 p.Arg63Cys genotypes.
Average number of alleles (Num), effective number of alleles (Eff_num), observed heterozygosity (Ho), expected heterozygosity (Hs), and inbreeding coefficient (Gis) were estimated for each group.
| Group | Num | Eff_num | Ho | Hs | Gis |
|---|---|---|---|---|---|
| Arg/Arg | 2.1 | 1.562 | 0.177 | 0.316 | 0.439 |
| Cys/Cys | 1.8 | 1.018 | 0.018 | 0.018 | -0.013 |
Table 3. Allelic frequencies for SYNGR2 SNPs between domestic groups with alternate homozygous SYNGR2 p.Arg63Cys genotypes.
Bottom portion of table denotes the average (Avg), highest (Max), and lowest (Min) minor allele frequency (MAF) in each group.
| SNP | Allele | Arg/Arg | Cys/Cys | Overall |
|---|---|---|---|---|
| 195 | C | 0.982 | 1 | 0.988 |
| T | 0.018 | 0 | 0.012 | |
| 198 | C | 0.559 | 0.997 | 0.702 |
| T | 0.441 | 0.003 | 0.298 | |
| 210 | C | 0.031 | 0 | 0.021 |
| T | 0.969 | 1 | 0.979 | |
| 261 | C | 0.381 | 0.012 | 0.26 |
| G | 0.579 | 0.988 | 0.713 | |
| T | 0.04 | 0 | 0.027 | |
| 321 | C | 0.48 | 0.02 | 0.331 |
| T | 0.52 | 0.98 | 0.669 | |
| 465 | C | 0.921 | 0.997 | 0.946 |
| T | 0.079 | 0.003 | 0.054 | |
| 516 | C | 0.901 | 0.997 | 0.932 |
| T | 0.099 | 0.003 | 0.068 | |
| 540 | C | 0.644 | 0.98 | 0.754 |
| T | 0.356 | 0.02 | 0.246 | |
| 597 | C | 0.74 | 0.985 | 0.82 |
| T | 0.26 | 0.015 | 0.18 | |
| 603 | C | 0.744 | 0.985 | 0.823 |
| T | 0.256 | 0.015 | 0.177 | |
| MAF | Avg | 0.206 | 0.008 | 0.141 |
| Max | 0.48 | 0.02 | 0.331 | |
| Min | 0.018 | 0 | 0.012 |
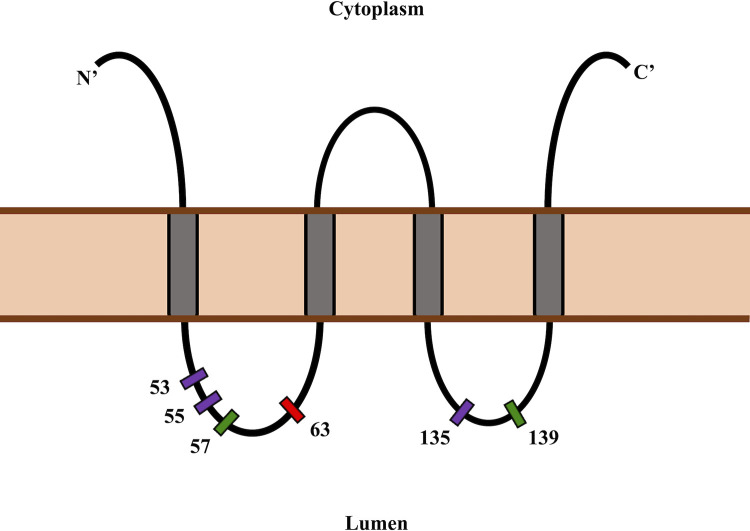
Signatures of adaptive evolution in the first intraluminal loop of the SYNGR2 protein in mammals
Host VIPs often interact with even distantly related viruses and evolve to limit viral replication or pathogenesis [2]. To test whether this host-virus coevolutionary dynamic has left signatures of positive selection across the SYNGR2 locus, we used the PAML package to fit models of molecular evolution to three taxonomic groups: mammals, primates, and even-toed ungulates [14,15]. Overall dN/dS (ω) estimates indicated that negative selection is acting to conserve the SYNGR2 protein sequence across mammalian species (ω = 0.09–0.183; Table 4). However, comparison between site-specific models supports positive selection on a subset of codons located exclusively within the two intraluminal loop domains in each taxonomic group (P<0.01; Table 4). Residue 53, which is located within the first intraluminal loop and only 10 residues upstream from the SYNGR2 p.Arg63Cys site, was identified in all three groups (Fig 4). Two other residues within the first intraluminal loop, 55 and 57, and two residues within the second intraluminal loop, 135 and 139, were also identified in the mammalian and/or even-toed ungulate groups (Fig 4). These findings highlight the intraluminal loops as sites of potential adaptive evolution to environmental cues, including host-pathogen interactions.
Table 4. Analysis of positive selection within SYNGR2 across mammalian species using site-specific models of evolution.
For each model, the dN/dS ratio was estimated for the entire gene (dN/dS) and across each site within SYNGR2 using the corresponding gene tree. A significant likelihood ratio test (LRT) statistic indicates the model allowing positive selection (M8, ω≤1 or ω>1) is a better fit than the null model (M7, ω≤1). The number of positive selection sites (PSS) is reported for each cluster.
| Mammals | Primates | Even-Toed Ungulates | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | dN/dS | PSS (ω>1) | LRT | dN/dS | PSS (ω>1) | LRT | dN/dS | PSS (ω>1) | LRT |
| M7 | 0.183 | -- | 65.79 (P<0.001) | 0.090 | -- | 13.41 (P<0.01) | 0.142 | -- | 27.14 (P<0.001) |
| M8 | 0.167 | 4 (3%) | 0.102 | 1 (0.6%) | 0.192 | 4 (3.6%) | |||
Fig 4. Position of putative positive selection sites identified across mammals within the SYNGR2 protein.
Schematic depicting the four transmembrane domains (gray boxes), two intraluminal loops, one cytoplasmic loop domain and cytoplasmic N- and C-termini. Amino acid residues identified as positive selection sites within one (green) or more (purple) taxonomic groups are represented relative to the SYNGR2 p.Arg63Cys polymorphism (red).
Discussion
Proposed mechanism of SYNGR2 function during early stages of PCV2 infection
Previously, our group uncovered a missense variant (SYNGR2 p.Arg63Cys) within the host Synaptogyrin-2 (SYNGR2) gene statistically associated with PCV2b viral load and immune response following in vivo experimental challenge [5]. In vitro infection of a predicted SYNGR2 knock-out edited PK15 clone with the same PCV2b isolate demonstrated clear involvement of SYNGR2 in viral replication [5]. However, direct experimental evidence for the causality of the SYNGR2 p.Arg63Cys variant or consistency of its effect across PCV2 subtypes had yet to be provided. In this study, we validated the function of the SYNGR2p.Arg63Cys variant in PCV2 infection using multiple in vitro models and PCV2 isolates. Specifically, CRISPR-Cas9 mediated allelic substitution for the SYNGR2 p.63Cys allele in PK15 cells resulted in significant reductions in viral replication consistent across PCV2 subtypes and inoculate concentration. These findings demonstrate a direct effect of the host SYNGR2 gene and causality of the associated SYNGR2 p.Arg63Cys substitution on PCV2 replication. Additionally, the consistent effect of SYNGR2 genotype on both PCV2b and PCV2d subtypes despite distinct infection kinetics, suggests that PCV2 evolution has yet to successfully counter the protection provided by the SYNGR2 p.63Cys allele.
While further analysis is necessary to understand the exact functional mechanism, the lag in PCV2 replication during the first 24 hours post infection in edited clones compared to wildtype cells, highlights a potential function of SYNGR2 in the early stages of PCV2 infection. This timeframe corresponds with the first intracytoplasmic phase of PCV2 morphogenesis, in which virions are internalized via endocytosis into early endosomes and subsequent translocation to the nucleus for viral replication [16,17]. SYNGR2 has been identified as a transmembrane component of cytoplasmic vesicles as well as involved in vesicle biogenesis found to be necessary for the promotion of SFTSV replication, a human RNA virus [6,7]. Additionally, SYNGR2 has been further characterized as a crucial factor for internalization of the A. actinomycetemcomitans cytotoxin active subunit (CdtB) via clathrin-mediated endocytosis [8,9]. Therefore, a function of SYNGR2 predominantly in the early stages of PCV2b infection is a logical conjecture. We propose that the detrimental effects on PCV2 replication resulting from SYNGR2 knock-out or the SYNGR2 p.63Cys allele is due to impaired endocytosis and/or slower translocation of virions to the nucleus for replication.
Origin and positive selection of the SYNGR2 p.63Cys allele within domestic swine
To investigate the evolutionary origin and history of the favorable SYNGR2 p.63Cys allele within the Suidae lineage, we evaluated SYNGR2 sequence data from 751 wild and domestic pigs available within the European Nucleotide Archive database. In this dataset, the SYNGR2 p.63Cys allele was found to be unique to domestic pigs and more prevalent in European than Asian breeds, with the highest frequencies in Duroc and Pietrain breeds. Additionally, a single SYNGR2 haplotype (Hap1) was defined by the SYNGR2 p.63Cys substitution and was likely derived form an ancestral haplotype (Hap2) nearly fixed within European wild boar and completely absent from Asian wild boar. A striking difference was also observed between European and Asian breeds, with Hap1 prevalent in all European compared to only one Asian breed (Meishan). Given the historical prominence of the Meishan breed, if the SYNGR2 p.63Cys allele had originated in this breed it would more than likely be present in many other Asian domestic breeds, which we did not observe. Additional insight is provided by the presence of only Hap1 and Hap2 in Iberian samples. Coinciding with noted continuous introgression between Iberian and European wild boar [12,18–20], Hap2 was present at the highest frequency in Iberian (88.2%) compared to all other European breeds. However, Iberian pigs are also one of the few European breeds with no known written or molecular evidence of Asian introgression [11,12]. Therefore, we propose that the favorable SYNGR2 p.63Cys allele arose post-domestication in ancestral European domestic pigs from which modern European breeds were formed (Fig 5). While many studies emphasize introgression of Asian domestic into European domestic swine during the 18th century, a few studies have also noted possible European introgression into Asian populations, particularly within Northern China [13,21,22]. Therefore, the presence of the SYNGR2 p.63Cys allele in Asian domestic swine under this proposed model can be explained by subsequent introgression of European haplotypes (Fig 5).
Fig 5. Proposed scenario of SYNGR2 p.63Cys/Hap1 origin in European domestic swine.
Solid lines reflect well-documented and experimentally supported directions of gene flow between populations throughout swine domestication. Dashed lines represent less documented directions of gene flow necessary for the proposed scenario of origin in EUD (red lines). Pie charts represent the frequency of Hap1 (orange) and Hap2 (blue) in each S. scrofa subgroup. (ASW = Asian Wild Boar, ASD = Asian Domestic, EUW = European Wild Boar, EUD = European Domestic).
Despite the seemingly recent emergence of this allele post-domestication, the extremely high frequency within Duroc and notable frequencies across European breeds suggests potential positive selection within domestic swine. As previously stated by Walker et al. (2018), the Duroc breed is noted for lean growth and has historically exhibited strong selective pressure for this trait throughout the 20th century, coinciding with the earliest detection of PCV2 strains in archival samples from the 1960’s [23]. Based on the effects that even mild PCV2 infections can have on growth, we postulated that the SYNGR2 p.63Cys allele may have been unknowingly selected for in this breed due to the relationship between PCV2 viral load and overall fitness [5]. A similar mechanism of selection, albeit of variable intensity, on other correlated production traits could also have led to the elevated frequency of this allele across European domestic breeds. To test this hypothesis, we compared the genetic diversity exhibited across 10 SYNGR2 SNP between domestic pigs with alternate homozygous SYNGR2 p.Arg63Cys genotypes. Overall, this analysis revealed a dramatic reduction in heterozygosity amongst Cys/Cys compared to Arg/Arg individuals, supporting recent positive selection favoring the SYNGR2 p.63Cys allele.
A model of host-virus coevolution across mammals
Given that SYNGR2 may represent a common mammalian VIP, we also assessed for signatures of adaptive evolution within SYNGR2 across mammalian species, which would support a model of repeated host-virus coevolution. Corresponding with reports of strong purifying selection acting on non-immune VIPs [2], we observed an overall signature of negative selection for SYNGR2 across mammals. However, we also found evidence of adaptive evolution with positively selected sites identified exclusively within the intraluminal loop domains and nearby the location of the SYNGR2 p.Arg63Cys substitution. Together, these findings indicate that SYNGR2 may reflect a component of the host-virus evolutionary arms race common across mammals with SYNGR2 p.Arg63Cys representing a species-specific example of recent putative adaptive evolution against PCV2 infection.
Materials and methods
PK15 cell culture
The porcine kidney cell line (PK15) was grown in DMEM high glucose media supplemented with 10% FBS and 1% Penicillin-Streptomycin (5,000 U/mL) at 37° C with 5% CO2. The cells were passaged every 3–4 days (1:10 split) and allowed to passage at least twice after thawing before being plated for transfection or infection protocols.
PCV2 isolates
The PCV2b strain (UNL2014001, accession KP016747.1) is the same isolate used in prior in vivo and in vitro infections [5] and was previously propagated in swine testicular (ST) cells with a final titer of 1x104 50% tissue culture infection dose (TCID50) as described by McKnite et al. (2014)[24]. The PCV2d isolate was obtained from the Kansas State University (KSU) diagnostic laboratory from an animal of unknown clinical status. The PCV2d strain was propagated in PK15 cells and harvested via three consecutive freeze-thaw cycles. To determine TCID50, the harvested virus was titrated in PK15 cells and analyzed via immunocytochemistry after 72 hours. The cells were incubated with a polyclonal anti-PCV2 primary antibody (ISU-3, 1:100) followed by a donkey anti-rabbit IgG secondary antibody conjugated with horse-radish peroxidase (AB_2534697, Invitrogen, 1:2,000). PCV2d-infected cells were stained using the peroxidase AEC substrate kit (Vector Laboratories) and visualized via light microscopy. The final PCV2d titer was estimated to be ~1x105.5 TCID50. The complete PCV2d genome was sequenced via dideoxy sequencing and compared with the complete PCV2b genome sequence to assess overall similarity. The percent identity between the two isolates for both the Replicase and Capsid viral proteins was also assessed for the corresponding nucleotide and predicted amino acid sequences.
In vitro editing of SYNGR2 in PK15 cells
The knock-out SYNGR2 clone (emSYNGR2-del) utilized in this study is the same clone (E1) previously generated by Walker et al. (2018)[5]. To generate PK15 clones homozygous for the alternate SYNGR2 p.63Cys allele, a single guide RNA was utilized targeting a protospacer adjacent motif (PAM) recognized by the Cas9 enzyme that overlapped the SYNGR2 p.Arg63Cys locus. The guide RNA was hybridized with fluorescently labeled Alt-R CRISPR-Cas9 tracrRNA ATTO 550 (IDT) and Alt-R S.p. Hifi Cas9 Nuclease V3 (IDT) following the manufacturer’s protocol to form Ribonucleoprotein (RNP) complexes. These RNP complexes were reverse transfected into PK15 cells in 12-well plates (4.0x105 cells/well) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) at a final concentration of 10nM. Cutting efficiency of this guide RNA was evaluated as previously described by Walker et al. (2018) using the T7E1 mutation detection kit (NEB). An 80 bp single stranded DNA template (IDT) homologous to the non-targeting strand and encoding the desired SYNGR2 p.63Cys allele was reverse transfected along with RNP complexes into PK15 cells. After 24 hours post transfection, the cells were collected and sorted using Fluorescence Activated Cell Sorting into 96 well plates to generate single-cell clones. Genomic DNA from each single-cell clone was extracted using QuickExtract DNA extraction solution following the manufacturer’s protocol (Lucigen). Each clone was genotyped for the SYNGR2 p.Arg63Cys variant by KASPar assay (LGC). RNA was extracted from selected clones using the RNeasy mini kit (Qiagen) and complementary DNA (cDNA) was generated using the First Strand cDNA Synthesis Kit (Cytiva). The SYNGR2 locus was sequenced via dideoxy sequencing at both the genomic and transcriptomic levels to confirm clone genotypes and assess for potential off-target effects. Sequences were aligned via clustal omega (https://www.ebi.ac.uk/Tools/msa/clustalo) and annotated using Jalview 2.11.2.7 software [25].
In vitro infection of PK15 cells
Wildtype PK15 and edited clones were cultured in 12-well plates (4 cm2) with 5.0x105 cells per well and infected when 80–100% confluent with either the PCV2b or PCV2d isolates. Infections were conducted a total of three times, each at a different time and cell passage, to obtain independent infection replicates. Cells were inoculated with PCV2d at the same multiplicity of infection (MOI) = 0.00025 used in the previous study [5] while a higher MOI = 0.00075 was used for PCV2b in attempt to better detect differences in gene expression due to infection. The inoculate was removed after a one-hour incubation period and cells were washed twice with PBS followed by supplementation with fresh media (DMEM high glucose with 2% FBS). The cells were incubated at 37° C with 5% CO2 for up to 5 days. Control cells were inoculated with plain DMEM high glucose media and maintained the same way as infected cells. Supernatant and cell samples were collected from corresponding wells at specific time points post infection and frozen at -80° C. PCV2d viral DNA was extracted from pelleted cell and supernatant (200 uL) samples using the QIAamp Blood DNA mini kit (Qiagen). PCV2b viral DNA and host cell RNA were simultaneously extracted from cell pellets using the AllPrep DNA/RNA mini kit (Qiagen). cDNA was generated from host-cell RNA samples as described above.
PCV2 genome copy number and host SYNGR2 expression profiling
Expression of SYNGR2 and PCV2 viral genome copy number were quantified across time points post infection using TaqMan Master Mix and the CFX384 Real Time PCR system (BioRad). The qPCR assays were designed using the IDT Realtime PCR Tool software (www.idt.com). A conserved viral genomic sequence within ORF2 (Capsid) was targeted to enable quantification of PCV2b and PCV2d with the same assay. PCV2 genome copy number per microliter for each viral DNA sample (50 uL) was quantified by plotting cycle crossing thresholds (CT) obtained for the technical triplicates on a standard curve generated by serial dilution of a PCR-amplified PCV2b control. The PCV2 copy number was then adjusted to reflect the total amount per well () in the cell fraction and per microliter () in the supernatant fraction. The PCV2 copy number across timepoints post infection were log10 transformed and compared between PK15 clones by Tukey’s honest significance test or t-test. Host cell cDNA samples were used to profile expression of SYNGR2 in wildtype and edited PK15 clones. The qPCR assay targeted the 3’UTR of the host SYNGR2 transcript. Simultaneous profiling of the ribosomal protein L32 (RPL32) gene was used for normalization. Mean normalized expression (MNE) values were calculated based on CT values obtained for the technical triplicates taking qPCR efficiencies into account [26]. MNE values for wildtype (wtSYNGR2+p.63Arg) and edited (emSYNGR2+p.63Cys) PK15 clones were log10 transformed and compared by t-test.
Collection, alignment, and variant discovery of Suidae SYNGR2 sequences
DNA sequences generated from individual animals with clearly defined wild or domestic purebred status were obtained from public sources as described in the Supplementary information (S1 Text). The sequencing reads for each sample were individually aligned to the coding region of the reference Sus scrofa SYNGR2 transcript sequence (XM_021066554.1, 85-759bp) using Standard Nucleotide BLAST software (NCBI). Aligned reads were visualized using the multi-sequence alignment viewer and deviations from the reference SYNGR2 sequence were recorded for each sample. Genotypes were called based on the nucleotides present across aligned reads corresponding to each variant site. To minimize inclusion of false variants due to sequencing or alignments errors, focus was placed on variants identified across multiple samples within the dataset.
Haplotype analysis
SYNGR2 haplotypes were identified using Haploview 4.2 software [27]. To account for differences in sample size, predominant haplotypes were identified separately for each of the three broad groups (Domestic, Wild Boar, and Sus Relatives). Only biallelic variants with minor allele frequencies (MAF) above 0.01 within at least one group were included in the haplotype analyses. Haplotype frequency (HAF) thresholds were independently set for each group according to sample size: Sus relatives (HAF>0.049), Wild Boar (HAF>0.018), and Domestic (HAF>0.01). The haplotypes were compared across groups to compile a set of SYNGR2 haplotypes. A haplotype network based on SYNGR2 coding sequences corresponding to each haplotype was generated using POPART software [28]. Specifically, the Integer Joining Network (IJN) method was utilized, which generates a network based on an inferred neighbor-joining tree and allows for the inclusion of nodes that reflect predicted ancestry and reduce edge length (reticulation tolerance = 0.5). To account for differences in sample size among groups, haplotype frequencies based on the actual dataset were applied to hypothetical groups of equal size (n = 100). The node size reflects the haplotype frequency across groups and the pie segments within each node represent the number of samples with that haplotype in each group.
Selection analysis within domestic swine
A total of 529 domestic samples with homozygous SYNGR2 p.Arg63Cys genotypes were divided into two groups, Arg/Arg (n = 356) and Cys/Cys (n = 173), regardless of breed or geographic origin. Genotypes for SYNGR2 SNPs with MAF>0.01 (excluding SYNGR2 p.Arg63Cys) were included in subsequent analyses. Allelic frequencies and parameters of genetic diversity were estimated using GenoDive 3.5 software [29]. Allelic frequency was calculated for each SNP both overall and within genotype groups. The genetic diversity tool was used to evaluate average number of alleles or allelic richness (num), effective number of alleles (Eff_num), observed heterozygosity (Ho), expected heterozygosity (Hs), total heterozygosity (Ht), and the inbreeding coefficient (Gis) for each genotype group.
SYNGR2 sequence and selection analyses across mammalian taxonomic groups
Reference transcript sequences encoding the common SYNGR2 isoform (224 aa) were obtained from NCBI for a total of 40 mammalian species (Table A in S1 Text). Each sequence was translated into the corresponding predicted protein sequence using an online DNA to protein translation software (http://bio.lundberg.gu.se/edu/translat.html). The sequences were categorized into three taxonomic groups: Mammals (n = 22), Primates (n = 16), and Even-Toed Ungulates (n = 10). The DNA and protein sequences within each group were aligned using Clustal Omega with codon specific alignment (PAL2NAL) as outlined in Jeffares et al. (2015)[14]. The amino acid alignments were used to estimate the phylogenetic tree or “gene” tree via the RAxML tool with parameters outlined in Jeffares et al. (2015)[14].
We used CODEML to implement a maximum likelihood approach within the PAML package [15] that fits aligned sequences to specified models of evolution by estimating the ratio of non-synonymous (dN) to synonymous (dS) substitutions (dN/dS or ω). Specifically, we analyzed each taxonomic group of aligned SYNGR2 sequences using site-specific models of evolution, in which ω is estimated for each site (i.e. codon) within the coding sequence. To test for evidence of positive selection within SYNGR2, two different models were run for the same data set representing the null hypothesis (M7), which does not allow for positively selected sites (ω≤1), and the alternate hypothesis (M8), which does allow for positively selected sites (ω≤1, ω>1). The CODEML models were run as described in Jeffares et al. (2015)[14]. The likelihood ratio test (LRT) values for the two models were then compared via chi-square distribution (df = 2) to determine which is the best fit for the data [14]. Under the M8 model, any sites within the protein sequence classified as ω>1 are specified in the output file and represent putative sites of positive selection [14]. The selection analysis was conducted twice for each group using either the gene or species tree to account for potential differences in evolutionary rates at the gene and species level. We observed a high level of concordance between the two analyses for each group so only the gene tree results are presented in this study.
Supporting information
Fig A. CRISPR-Cas9 guide RNA and template design for generation of clones homozygous for alternate SYNGR2 p.63Cys allele. The selected guide RNA (sg_AF) targeted a PAM site within exon 2 (gray box) that included the SYNGR2 p.63Arg allele (red). An 80 bp ssDNA sequence (thick black line) homologous to the non-targeting strand and encoding the alternate SYNGR2 p.63Cys allele (yellow) was used as a template for homology-directed repair. Fig B. Alignment of the SYNGR2 coding sequence for wildtype and edited PK15 cell lines. Sequences were obtained by targeted amplification of the SYNGR2 transcript and Sanger sequencing. Position of the substitution of p.63Arg (C) to p.63Cys (T) in the edited PK15 clones, emSYNGR2+p.63Cys and 2emSYNGR2+p.63Cys, is highlighted in red. The 106 bp deletion in the predicted SYNGR2 knock-out PK15 clone, emSYNGR2-del, is represented by dashes. Fig C. Expression of SYNGR2 in wildtype and edited PK15 following PCV2b infection mock-infected control cells. Expression represented as Log10 transformed mean normalized expression (MNE) across three independent replicates with error bars representing one standard error from the mean. Samples collected from control and infected cells across timepoints post PCV2b infection (MOI = 0.00075). Letters denote significant differences in gene expression between cell lines within treatment group (C = control, I = infected) or between treatment groups within cell lines (wt = wildtype, em = edited). *P<0.05, **P<0.01. Fig D. Frequency of SYNGR2 haplotypes across geographic and domestic/wild S. scrofa subgroups. The two haplotypes that differ by only the SYNGR2 p.Arg63Cys allele, Hap1 (Cys) and Hap2 (Arg), are represented as independent pie segments. Hap3-Hap10 and rare haplotypes were combined into a single category denoted as “Other”. (ASW = Asian Wild Boar, ASD = Asian Domestic, EUW = European Wild Boar, EUD = European Domestic). Fig E. Frequency of SYNGR2 haplotypes across domestic breeds. The two haplotypes that differ by only the SYNGR2 p.Arg63Cys SNP, Hap1 (Cys) and Hap2 (Arg), are represented as independent pie segments. Hap3-Hap10 and rare haplotypes were combined into a single category denoted as “Other”. (BR = Berkshire, DR = Duroc, IB = Iberian, PI = Pietrain, LR = Landrace, LW = Large White, YR = Yorkshire, EUDO = European Domestic Other, EH = Erhualian, MS = Meishan, ASDO = Asian Domestic Other). Table A. Mammalian SYNGR2 transcript sequences and taxonomic groups. Table B. SYNGR2 SNP identified across Suidae sequences. Each SYNGR2 SNP is denoted by nucleotide position within the SYNGR2 coding sequence. (*187 = SYNGR2 p.Arg63Cys; SSC12 = chromosome 12, S.scrofa11.1). Table C. Allelic frequencies for SYNGR2 SNP across Sus scrofa subgroups. (*187 = SYNGR2 p.Arg63Cys; EUD = European Domestic, EUW = European Wild, ASD = Asian Domestic, ASW = Asian Wild). Table D. Allelic frequencies for SYNGR2 SNP across domestic breeds. (*187 = SYNGR2 p.Arg63Cys; BR = Berkshire, DR = Duroc, IB = Iberian, PI = Pietrain, LR = Landrace, LW = LargeWhite, YR = Yorkshire, EUDO = European other, EH = Erhualian, MS = Meishan, ASDO = Asian other). Table E. Frequency of SYNGR2 haplotypes across S. scrofa subgroups. (EUD = European Domestic, EUW = European Wild, ASD = Asian Domestic, ASW = Asian Wild).
(DOCX)
(XLSX)
Data Availability
All the data generated by this study, including numerical data for all of our graphs and summary statistics, is available as supplementary information (S1 Table.xlsx). Public swine DNA sequences used in this study were obtained from data files available within the European Nucleotide Archive (ENA) database (https://www.ebi.ac.uk/ena/browser/home) that were generated from samples corresponding to a single individuals with defined wild or domestic status (S1 Table). The species trees were also obtained from NCBI for each of the taxonomic groups (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi).
Funding Statement
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2019-05380 from the USDA National Institute of Food and Agriculture to DCC and HV. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Meyerson NR, Sawyer SL. Two-stepping through time: mammals and viruses. Trends in microbiology. 2011;19(6):286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enard D, Cai L, Gwennap C, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. elife. 2016;5:e12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annual review of genetics. 2012;46:677–700. [DOI] [PubMed] [Google Scholar]
- 4.Xiao C-T, Halbur PG, Opriessnig T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. Journal of General Virology. 2015;96(7):1830–41. [DOI] [PubMed] [Google Scholar]
- 5.Walker LR, Engle TB, Vu H, Tosky ER, Nonneman DJ, Smith TP, et al. Synaptogyrin-2 influences replication of Porcine circovirus 2. PLoS genetics. 2018;14(10):e1007750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belfort GM, Bakirtzi K, Kandror KV. Cellugyrin induces biogenesis of synaptic-like microvesicles in PC12 cells. The Journal of biological chemistry. 2005;280(8):7262–72. Epub 2004/12/14. doi: 10.1074/jbc.M404851200 . [DOI] [PubMed] [Google Scholar]
- 7.Sun Q, Qi X, Zhang Y, Wu X, Liang M, Li C, et al. Synaptogyrin-2 Promotes Replication of a Novel Tick-borne Bunyavirus through Interacting with Viral Nonstructural Protein NSs. The Journal of biological chemistry. 2016;291(31):16138–49. Epub 2016/05/27. doi: 10.1074/jbc.M116.715599 ; PubMed Central PMCID: PMC4965563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boesze-Battaglia K, Dhingra A, Walker LM, Zekavat A, Shenker BJ. Internalization and Intoxication of Human Macrophages by the Active Subunit of the Aggregatibacter actinomycetemcomitans Cytolethal Distending Toxin Is Dependent Upon Cellugyrin (Synaptogyrin-2). Frontiers in Immunology. 2020;11:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boesze-Battaglia K, Walker LP, Dhingra A, Kandror K, Tang H-Y, Shenker BJ. Internalization of the active subunit of the Aggregatibacter actinomycetemcomitans cytolethal distending toxin is dependent upon cellugyrin (synaptogyrin 2), a host cell non-neuronal paralog of the synaptic vesicle protein, synaptogyrin 1. Frontiers in Cellular and Infection Microbiology. 2017;7:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei R, Xie J, Theuns S, Nauwynck HJ. Changes on the viral capsid surface during the evolution of porcine circovirus type 2 (PCV2) from 2009 till 2018 may lead to a better receptor binding. Virus evolution. 2019;5(2):vez026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groenen MA. A decade of pig genome sequencing: a window on pig domestication and evolution. Genetics Selection Evolution. 2016;48(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramírez O, Burgos-Paz W, Casas E, Ballester M, Bianco E, Olalde I, et al. Genome data from a sixteenth century pig illuminate modern breed relationships. Heredity. 2015;114(2):175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491(7424):393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffares DC, Tomiczek B, Sojo V, dos Reis M. A beginners guide to estimating the non-synonymous to synonymous rate ratio of all protein-coding genes in a genome. Parasite Genomics Protocols: Springer; 2015. p. 65–90. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer applications in the biosciences. 1997;13(5):555–6. [DOI] [PubMed] [Google Scholar]
- 16.Meerts P, Misinzo G, McNeilly F, Nauwynck H. Replication kinetics of different porcine circovirus 2 strains in PK-15 cells, fetal cardiomyocytes and macrophages. Archives of virology. 2005;150(3):427–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Carino C, Duffy C, Sanchez-Chardi A, McNeilly F, Allan GM, Segales J. Porcine circovirus type 2 morphogenesis in a clone derived from the l35 lymphoblastoid cell line. Journal of comparative pathology. 2011;144(2–3):91–102. Epub 2010/08/31. doi: 10.1016/j.jcpa.2010.07.001 . [DOI] [PubMed] [Google Scholar]
- 18.Dadousis C, Muñoz M, Ovilo C, Fabbri MC, Araújo JP, Bovo S, et al. Admixture and breed traceability in European indigenous pig breeds and wild boar using genome-wide SNP data. Scientific reports. 2022;12(1):7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero-Medrano JM, Megens H-J, Groenen MA, Ramis G, Bosse M, Pérez-Enciso M, et al. Conservation genomic analysis of domestic and wild pig populations from the Iberian Peninsula. BMC genetics. 2013;14:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Asch B, Pereira F, Santos L, Carneiro J, Santos N, Amorim A. Mitochondrial lineages reveal intense gene flow between Iberian wild boars and South Iberian pig breeds. Animal genetics. 2012;43(1):35–41. [DOI] [PubMed] [Google Scholar]
- 21.Kim KI, Lee JH, Li K, Zhang YP, Lee SS, Gongora J, et al. Phylogenetic relationships of Asian and European pig breeds determined by mitochondrial DNA D-loop sequence polymorphism. Animal genetics. 2002;33(1):19–25. [DOI] [PubMed] [Google Scholar]
- 22.Zhao P, Du H, Jiang L, Zheng X, Feng W, Diao C, et al. PRE-1 Revealed Previous Unknown Introgression Events in Eurasian Boars during the Middle Pleistocene. Genome biology and evolution. 2020;12(10):1751–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen B, Krueger L, Seeliger F, Bruegmann M, Segalés J, Baumgaertner W. Retrospective study on the occurrence of porcine circovirus 2 infection and associated entities in Northern Germany. Veterinary microbiology. 2009;138(1–2):27–33. [DOI] [PubMed] [Google Scholar]
- 24.McKnite A, Bundy J, Moural T, Tart J, Johnson T, Jobman E, et al. Genomic analysis of the differential response to experimental infection with porcine circovirus 2b. Animal genetics. 2014;45(2):205–14. [DOI] [PubMed] [Google Scholar]
- 25.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England). 2009;25(9):1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon MF, Rey A, Castan-Laurel I, Gres S, Sibrac D, Valet P, et al. Expression of ectolipid phosphate phosphohydrolases in 3T3F442A preadipocytes and adipocytes. Involvement in the control of lysophosphatidic acid production. The Journal of biological chemistry. 2002;277(26):23131–6. Epub 2002/04/17. doi: 10.1074/jbc.M201530200 ; PubMed Central PMCID: PMC2000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England). 2005;21(2):263–5. Epub 2004/08/07. doi: 10.1093/bioinformatics/bth457 . [DOI] [PubMed] [Google Scholar]
- 28.Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6(9):1110–6. [Google Scholar]
- 29.Meirmans PG. genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Molecular Ecology Resources. 2020;20(4):1126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]