Abstract
Aim: The influence of family history of diabetes, probably reflecting genetic and lifestyle factors, on the association of combined genetic and lifestyle risks with diabetes is unknown. We examined these associations.
Methods: This cross-sectional study included 9,681 participants in the Tohoku Medical Megabank Community-based Cohort Study. A lifestyle score, which was categorized into ideal, intermediate, and poor lifestyles, was given. Family history was obtained through a self-reported questionnaire. A polygenic risk score (PRS) was constructed in the target data (n=1,936) using publicly available genome-wide association study summary statistics from BioBank Japan. For test data (n=7,745), we evaluated PRS performance and examined the associations of combined family history and genetic and lifestyle risks with diabetes. Diabetes was defined as non-fasting blood glucose ≥ 200 mmHg, HbA1c ≥ 6.5%, and/or self-reported diabetes treatment.
Results: In test data, 467 (6.0%) participants had diabetes. Compared with a low genetic risk and an ideal lifestyle without a family history, the odds ratio (OR) was 3.73 (95% confidence interval [CI], 1.92–7.00) for a lower genetic risk and a poor lifestyle without a family history. Family history was significantly associated with diabetes (OR, 3.58 [95% CI, 1.73–6.98]), even in those with a low genetic risk and an ideal lifestyle. Even among participants who had an ideal lifestyle without a family history, a high genetic risk was associated with diabetes (OR, 2.49 [95% CI, 1.65–3.85]). Adding PRS to family history and conventional lifestyle risk factors improved the prediction ability for diabetes.
Conclusions: Our findings support the notion that a healthy lifestyle is important to prevent diabetes regardless of genetic risk.
Keywords: Family history, Genetic, Lifestyle, Diabetes mellitus, Epidemiology
Introduction
Diabetes is rapidly increasing in high-, low-, and middle-income countries 1 , 2) . Diabetes and its complications cause premature mortality, which is a public health burden worldwide 1 , 2) . Therefore, identifying high-risk individuals and preventing or delaying its onset are important.
Genetic and environmental factors are involved in diabetes onset 3) . Lifestyle interventions, such as diet, exercise, and weight loss, have been shown to prevent diabetes 4 - 6) . In terms of genetic factors, recent large-scale genome-wide association studies (GWASs) identified over 270 independent diabetes-related genetic loci 7 - 9) . Furthermore, a polygenic risk score (PRS) is constructed based on GWAS summary statistics and can predict the onset of type 2 diabetes in European and non-European population 10 - 14) .
Several studies have examined the joint associations of combined lifestyle adherence and genetic susceptibility with diabetes in general populations. In the UK Biobank Study, a high genetic risk was associated with a higher risk of developing diabetes independent of lifestyle and a poor lifestyle was associated with an increased risk of developing diabetes independent of the genetic risk score 15) . The Dongfeng–Tongji Cohort Study has shown that the genetic risk score is associated with a higher diabetes risk independent of lifestyle factors but a healthy lifestyle is associated with a substantially reduced diabetes risk within different genetic risk scores 16) . In two cohorts of Chinese participants, genetics and lifestyle were independently associated with a higher risk of developing diabetes 17) . However, there are no studies that examined the combination of genetic susceptibility and lifestyle adherence with diabetes among Japanese population.
Additionally, a family history of diabetes is a well-known risk factor for diabetes because it might reflect genetics and shared lifestyle. Family history is reportedly associated with a higher risk of diabetes even after adjusting for genetics and lifestyle 18 - 20) . Although a combination of genetic and lifestyle risks has identified high-risk individuals for diabetes, considering the family history may help in targeting high-risk individuals more precisely and may offer personalized prevention measures. However, to our knowledge, no studies have quantitatively examined the associations of combined genetic and lifestyle risks with diabetes in terms of the family history.
Aim
This study aimed to examine the cross-sectional associations of combined PRS, healthy lifestyle, and family history with the prevalence of diabetes in a large prospective cohort of the Tohoku Medical Megabank Community-based Cohort (TMM CommCohort) Study. Additionally, we examined whether adding a PRS to family history and conventional lifestyle risk factors could improve its ability to predict diabetes.
Methods
Study Population and Design
This study was a cross-sectional study of participants aged ≥ 20 years who live in Miyagi Prefecture, northeastern Japan, who were included in the TMM CommCohort study 21) . The details have been described elsewhere 21 , 22) . In brief, the TMM CommCohort study was conducted from May 2013 to March 2016 and recruited more than 50,000 participants according to the following: type 1 survey (n=41,097 participants), which was performed in specific municipal health check-up sites, and type 2 survey (n=13,855), which was performed in assessment centers. Participants provided information on their lifestyle and other potentially health-related aspects through a self-reported questionnaire and blood and urine samples. All participants provided written informed consent for the study (n=54,952). The type 1 survey participants were included as a control group in the GWAS, which was used to construct a PRS in this study. Therefore, to avoid overfitting, we selected the type 2 survey participants (n=13,855). Participants who withdrew from the study on July 13, 2021, failed to return a self-reported questionnaire (n=204), had no genetic information (n=3,789), had self-reported type 1 diabetes, and had missing data regarding glucose, HbA1c, height, weight, gamma-glutamyl transferase (GGT), physical activity, smoking status, and family history of diabetes (n=163) were excluded. Moreover, 18 participants with ≥ 6 standard deviations away from the means of each principal component were excluded. Finally, 9,681 participants fulfilled all inclusion criteria. These participants were randomly split into target data (n=1,936; 20%) and test data (n=7,745; 80%) ( Fig.1 ) . The target data were used to determine the P-thresholds of the best-fit PRS for HbA1c. The test data were used to examine associations of combined family history, PRS, and lifestyle with diabetes. This study was conducted in accordance with the amended Declaration of Helsinki. Local institutional review boards or independent ethics committees approved the protocol, and written informed consent was obtained from all patients. This study was approved by the Institutional Review Board of Tohoku Medical Megabank Organization (approval number: 2022-4-047; approval date: June 30, 2022).
Fig.1.
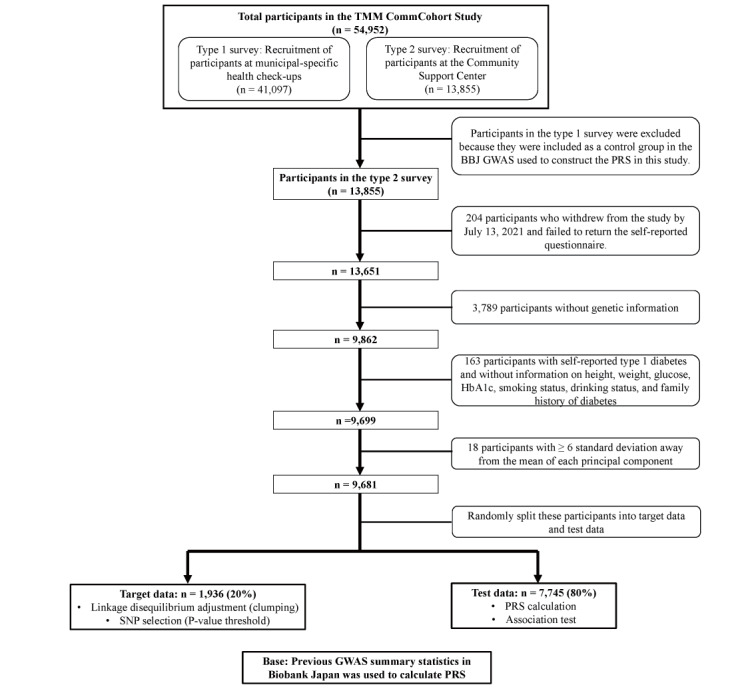
Flowchart of the study participants
Healthy Lifestyle Factors and Family History of Diabetes
In this study, we constructed a healthy lifestyle score based on the following four diabetes risk factors: never-smoking, BMI, regular physical activity, and low GGT 23 - 26) . Weight was measured in increments of 0.1 kg, and 1.0 kg was subtracted to account for the weight of the participant’s clothing using a body composition analyzer (InBody720; Biospace Co, Ltd., Seoul, Korea). BMI was calculated as weight (kg) divided by height squared (m2). Obesity was defined as BMI ≥ 25.0 kg/m2 based on the WHO criteria for Japanese individuals 27) . Never-smoking was defined as participants who had smoked <100 cigarettes during their lifetime 28) . Information regarding physical activity was collected using a self-reported questionnaire. Participants answered the hours spent for each activity (sitting, standing, walking, and strenuous work) per average day in the last years. The choices were the following: 0 min to none, 30 min to less than 1 h, 120 min to 1–3 h, 240 min to 3–5 h, 360 min to 5–7 h, 480 min to 7–9 h, 600 min to 9–11 h, and 660 min to ≥ 11 h. The average frequency (times/week) and duration (min/time) of normal walking, brisk walking, moderate-intensity exercise, and hard-intensity exercise during leisure time were obtained 29) . Frequency was classified into the following categories: less than once per month, one to three times per month, one to two times per week, three to four times per week, and almost every day. Duration was also classified into the following categories: <30 min, 30–59 min, 1–2 h, 2–3 h, 3–4 h, and ≥ 4 h. The present study defined “walking,” “normal walking,” “brisk walking,” and “moderate-intensity exercise” as moderate physical activity and “strenuous work” and “hard-intensity exercise” as vigorous physical activity 30) . The average times of moderate and vigorous physical activities during leisure time were determined by multiplying frequency and duration. Subsequently, we calculated the minutes of moderate-intensity activity per week by adding the duration of walking and average time of moderate physical activity during leisure time. Similarly, we calculated the minutes of vigorous physical activity per week by adding the duration of strenuous work and average time of vigorous physical activity during leisure time. Physical activity was defined as at least 150 min of activity with moderate intensity weekly or 75 min of vigorous activity weekly according to the WHO guideliness 31) . We collected non-fasting blood samples. The GGT levels were measured using an enzymatic method. A higher GGT was defined as GGT ≥ 50 IU/L based on health examination results in Japan 25) . GGT is commonly considered a marker of excessive alcohol consumption and visceral fat especially for hepatic steatosis 32 , 33) . However, previous studies have shown that higher GGT is associated with diabetes regardless of drinking status and BMI 25 , 26 , 34) . Therefore, we used GGT level as a surrogate marker of alcohol consumption because there is a U-shaped relation between alcohol drinking and incident diabetes 26) . Overall lifestyle was subsequently categorized into ideal (having at least three ideal lifestyle factors), poor (having at least three poor lifestyle factors), or intermediate (having two ideal lifestyle factors). We collected information on family history through a self-reported questionnaire. Participants answered whether they had a family history of diabetes through their biological father, mother, brother, and sister. If they answered that at least one of them had diabetes, they were deemed to have a family history of diabetes.
Diabetes
Non-fasting glucose levels were measured using the hexokinase method. HbA1c levels were measured using the latex agglutination turbidimetry method. Diabetes was defined as non-fasting glucose ≥ 200 mg/dL, HbA1c ≥ 6.5% (48 mmol/mol), and/or self-reported treatment for diabetes.
Genotyping, Quality Control, and PRS Derived from BioBank Japan
Study participants were genotyped using Affymetrix Axiom Japonica Array (v2) separately in 21 batches 35) . For quality control, we excluded plates with an average call rate <0.95 and removed samples with Dish QC metric <0.82 or step1 call rate <0.97 before batch genotyping. Subsequently, we removed variants with a P-value of Hardy–Weinberg equilibrium test <1.00×10−6, minor allele frequency <0.01, or missing rate >0.01 from each batch. A direct genotype dataset in PLINK BED format was obtained by merging the genotype datasets for the 21 batches using QCTOOL (v2.0.4) (https://www.well.ox.ac.uk~gavqctool). We obtained a direct genotype dataset in Oxford BGEN format from the TMM CommCohort study participants. We calculated PRS based on the summary statistics of a previous GWAS for diabetes in BioBank Japan (BBJ), which was publicly available at the National Bioscience Database Center 7) . The participants included in our studies were independent from the BBJ participants. All single nucleotide polymorphisms (SNPs) on X and Y chromosomes were removed from the base and target data to eliminate the possibility of non-autosomal sex effects. To calculate PRS, we used PLINK. As a default setting of PLINK, we performed clumping to capture the right level of causal signal using the following options: --clump-p1 1 --clump-r2 0.1 –clump-kb 250. After clumping, we calculated the PRS for each individual included in the target data (n=1936) using various variant sets according to different P-value thresholds. PRS was calculated using the default formula for PRS calculation in PLINK (https://choishingwan.github.io/PRS-Tutorial/plink/). We calculated PRS following nine different P-value thresholds: 5.0×10−8, 0.001, 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5. Among the various PRSs with different numbers of SNPs, we chose the list of variants that showed the best fit (determined by a variance explained by the PRS). The settings of the best-fit PRS for HbA1c in the target data were PRS using P<0.2 ( Supplemental Table 1 ) . Therefore, we used a PRS constructed by P<0.2 in the test data for this analysis.
Supplemental Table1. P-value threshold and related parameters on PRS construction .
| Traits | P-threshold | Number of SNP | R2 |
|---|---|---|---|
| HbA1c | 5.0 × 10-8 | 102 | 0.0083587720365206 |
| 0.001 | 1523 | 0.0069292604848436 | |
| 0.01 | 5760 | 0.0061782942260876 | |
| 0.05 | 16637 | 0.0108040782866358 | |
| 0.1 | 26742 | 0.0111353862108406 | |
| 0.2 | 42845 | 0.0111962866537727 | |
| 0.3 | 56383 | 0.0102524876262533 | |
| 0.4 | 68077 | 0.0101184426932776 | |
| 0.5 | 78302 | 0.0092796911886064 |
Bold fonts in this table indicate the best fit and settings for our analysis. PRS, polygenic risk score.
Statistical Analysis
Data were presented as means (SD) or median (interquartile range) for continuous variables and number (percentage) for categorical variables. Multivariable logistic regression analyses were performed to examine the association between lifestyle factors and diabetes prevalence. Adjusted OR and 95% CI were estimated. Participants were classified by their PRS tertile for our analysis of a potential association between PRS and diabetes. Similarly, multivariable logistic regression analyses were performed to examine the association between genetic risk scores and diabetes. Additionally, the association between family history and diabetes prevalence was examined through a multivariable logistic regression analysis. Subsequently, we combined the PRS tertile and lifestyle and made nine group categories. Multivariable logistic regression analysis was performed to test the association of genetic and lifestyle factors with diabetes prevalence. Finally, we combined family history, PRS tertile, and lifestyle and made 18 group categories. To compare the characteristics of the participants with and without a family history of diabetes, we used the Student’s t-test for continuous variables, Mann–Whitney U test for nonparametric variables, and χ2-test for categorical variables. Multivariable logistic regression analysis was performed to test the influence of family history on the associations of combined genetic and lifestyle risks with diabetes. All models were adjusted for age, sex, and the first six principal components (to adjust for population structure). To investigate the influence of family history on predictive ability, we calculated the area under the receiver operating characteristic curve (AUROC), and 95% CI was calculated before and after adding family history to the statistical model, including PRS and lifestyle risk factors. AUROCs were compared using the DeLong test.
Furthermore, we conducted several sensitivity analyses. Participants with a treatment for diabetes might change their lifestyle and habits, such as quitting smoking and drinking, and might receive interventions regarding weight loss. Therefore, to rule out diabetes treatment, we excluded participants with a treatment for diabetes (n=348). Then, we stratified the analysis according to median age (<60.0 years, ≥ 60.0 years). Finally, a similar analysis was performed only for non-obese subjects (BMI<25.0 kg/m2).
A two-sided P<0.05 was considered statistically significant. Basic statistical analysis was performed using software R version 4.1.2 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Participant Characteristics
A total of 7,745 participants included in the test data were analyzed to examine the associations of combined family history, PRS, and lifestyle risk with diabetes. The mean (SD) values for age, BMI, glucose, and HbA1c of the study participants were 57.3 (13.4) years, 22.7 (3.5) kg/m2, 88.3 (16.7) mg/dL, and 5.5% (0.5%), respectively. The median (interquartile range) value of GGT was 19.0 [14.0, 30.0]. The proportions (%) of women, non-obese, never-smoker, those who have regular physical activity, those who have low GGT (<50.0), those who have diabetes, and those who have a family history of diabetes were 5,911 (76.3%), 6,019 (77.7%), 5,167 (66.7%), 3,430 (44.3%), 6,906 (89.2%), 467 (6.0%), and 992 (12.8%), respectively ( Table 1 ) . Compared with participants without a family history of diabetes, participants with a family history of diabetes have higher BMI, glucose, HbA1c levels, and higher proportion of women. Additionally, participants with a family history of diabetes have a slightly higher PRS than those without a family history of diabetes ( Supplemental Table 2 ) .
Table 1. Characteristics of the study participants according to combined genetic and lifestyle risk and family history of diabetes.
| Family history | No (n = 6,753) | Yes (n = 992) | Overall | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic risk | Low genetic risk | Intermediate genetic risk | High genetic risk | Low genetic risk | Intermediate genetic risk | High genetic risk | |||||||||||||
| Lifestyle | Ideal | Intermediate | Poor | Ideal | Intermediate | Poor | Ideal | Intermediate | Poor | Ideal | Intermediate | Poor | Ideal | Intermediate | Poor | Ideal | Intermediate | Poor | |
| Number | 1578 | 541 | 184 | 1499 | 541 | 214 | 1419 | 570 | 207 | 193 | 65 | 21 | 206 | 90 | 31 | 240 | 107 | 39 | 7745 |
| Age, years | 57.8 (14.0) | 56.8 (13.6) | 57.9 (12.0) | 58.3 (13.5) | 56.0 (12.4) | 55.1 (12.3) | 57.9 (13.9) | 56.0 (13.6) | 55.2 (12.7) | 57.4 (12.9) | 54.1 (12.3) | 58.5 (13.4) | 56.7 (12.6) | 55.4 (13.0) | 54.0 (12.6) | 58.2 (11.8) | 56.4 (12.0) | 54.5 (12.2) | 57.3 (13.4) |
| Women, % | 1335 (84.6) | 331 (61.2) | 61 (33.2) | 1274 (85.0) | 358 (66.2) | 81 (37.9) | 1212 (85.4) | 377 (66.1) | 81 (39.1) | 174 (90.2) | 46 (70.8) | 8 (38.1) | 183 (88.8) | 66 (73.3) | 18 (58.1) | 212 (88.3) | 77 (72.0) | 17 (43.6) | 5911 (76.3) |
| BMI, kg/m2 | 21.6 (2.8) | 23.5 (3.7) | 26.1 (3.2) | 21.7 (2.6) | 23.9 (3.8) | 26.7 (3.5) | 21.8 (2.7) | 24.3 (3.9) | 26.4 (3.7) | 21.3 (2.5) | 24.0 (3.8) | 26.1 (3.9) | 21.9 (3.1) | 25.2 (4.1) | 27.5 (4.2) | 22.0 (2.8) | 24.2 (3.6) | 26.9 (3.9) | 22.7 (3.5) |
| Glucose, mg/dl | 85.5 (11.8) | 88.1 (14.4) | 91.4 (16.4) | 86.2 (12.7) | 89.6 (17.1) | 92.6 (16.9) | 87.7 (16.0) | 91.6 (23.6) | 92.5 (21.7) | 87.3 (11.0) | 89.4 (17.5) | 88.0 (11.6) | 88.9 (17.1) | 92.5 (19.5) | 97.3 (23.2) | 93.2 (28.3) | 100.1 (32.1) | 91.0 (19.0) | 88.3 (16.7) |
| HbA1c, % | 5.4 (0.3) | 5.4 (0.4) | 5.6 (0.7) | 5.5 (0.4) | 5.5 (0.5) | 5.6 (0.5) | 5.5 (0.4) | 5.6 (0.6) | 5.7 (0.7) | 5.5 (0.4) | 5.5 (0.5) | 5.8 (0.6) | 5.6 (0.5) | 5.8 (0.7) | 6.0 (0.6) | 5.8 (0.8) | 5.9 (1.1) | 5.7 (0.5) | 5.5 (0.5) |
| Prevalence of diabetes, % | 34 (2.2) | 15 (2.8) | 16 (8.7) | 64 (4.3) | 28 (5.2) | 15 (7.0) | 70 (4.9) | 54 (9.5) | 23 (11.1) | 12 (6.2) | 4 (6.2) | 5 (23.8) | 19 (9.2) | 14 (15.6) | 10 (32.3) | 53 (22.1) | 24 (22.4) | 7 (17.9) | 467 (6.0) |
| Treatment for diabetes | 22 (1.4) | 11 (2.0) | 9 (4.9) | 53 (3.5) | 20 (3.7) | 11 (5.1) | 53 (3.7) | 40 (7.0) | 9 (4.3) | 10 (5.2) | 4 (6.2) | 3 (14.3) | 17 (8.3) | 12 (13.3) | 6 (19.4) | 41 (17.1) | 16 (15.0) | 7 (17.9) | 344 (4.4) |
| METs, MET-min/week | 99.2 | 38.6 | 33.5 | 105.0 | 39.0 | 27.0 | 99.0 | 38.5 | 34.8 | 98.4 | 30.0 | 18.0 | 126.3 | 38.9 | 64.5 | 90.0 | 49.0 | 27.0 | 67.5 |
| [13.5, 248.9] | [0.0, 142.7] | [2.8, 113.0] | [18.0, 238.7] | [2.8, 115.7] | [0.0, 85.4] | [17.4, 261.3] | [0.0, 135.0] | [0.0, 87.9] | [18.0, 252.0] | [0.0, 90.0] | [2.8, 67.5] | [31.5, 247.6] | [3.2, 137.1] | [0.0, 103.5] | [18.0, 224.7] | [8.7, 129.9] | [0.0, 64.3] | [9.0, 200.0] | |
| GGT, IU/L | 17.0 | 23.0 | 56.5 | 18.0 | 24.0 | 51.0 | 18.0 | 24.0 | 56.0 | 16.0 | 22.0 | 55.0 | 19.0 | 26.0 | 54.0 | 18.0 | 23.0 | 53.0 | 19.0 |
| [13.0, 23.0] | [16.0, 37.0] | [29.8, 75.2] | [14.0, 25.0] | [16.0, 36.0] | [29.2, 79.8] | [14.0, 24.5] | [16.0, 37.0] | [36.0, 77.5] | [13.0, 22.0] | [16.0, 36.0] | [19.0, 79.0] | [14.0, 24.0] | [16.0, 37.6] | [36.5, 84.5] | [14.0, 25.0] | [16.5, 44.5] | [37.5, 91.0] | [14.0, 30.0] | |
| Smoking status, % | |||||||||||||||||||
| Never-smoker | 1355 (85.9) | 178 (32.9) | 26 (14.1) | 1273 (84.9) | 192 (35.5) | 22 (10.3) | 1204 (84.8) | 211 (37.0) | 33 (15.9) | 168 (87.0) | 22 (33.8) | 0 (0.0) | 183 (88.8) | 39 (43.3) | 4 (12.9) | 205 (85.4) | 47 (43.9) | 5 (12.8) | 5167 (66.7) |
| Ex-smoker | 149 (9.4) | 245 (45.3) | 93 (50.5) | 157 (10.5) | 248 (45.8) | 121 (56.5) | 143 (10.1) | 241 (42.3) | 115 (55.6) | 22 (11.4) | 30 (46.2) | 19 (90.5) | 19 (9.2) | 37 (41.1) | 13 (41.9) | 30 (12.5) | 40 (37.4) | 26 (66.7) | 1748 (22.6) |
| Current smoker | 74 (4.7) | 118 (21.8) | 65 (35.3) | 69 (4.6) | 101 (187) | 71 (33.2) | 72 (5.1) | 118 (20.7) | 59 (38.6) | 3 (1.6) | 13 (20.0) | 2 (9.5) | 4 (1.9) | 14 (15.6) | 14 (45.2) | 5 (2.1) | 20 (18.7) | 8 (20.5) | 830 (10.7) |
| Drinking status, % | |||||||||||||||||||
| Never-drinker | 774 (49.0) | 178 (32.9) | 47 (25.5) | 754 (50.3) | 181 (33.5) | 51 (23.8) | 700 (49.3) | 208 (36.5) | 43 (20.8) | 103 (53.4) | 20 (30.8) | 8 (38.1) | 107 (61.9) | 34 (37.8) | 9 (29.0) | 107 (44.6) | 37 (34.6) | 12 (30.8) | 3373 (43.6) |
| Ex-drinker | 33 (2.1) | 20 (3.7) | 4 (2.2) | 23 (1.5) | 18 (3.3) | 1 (0.5) | 27 (1.9) | 23 (4.0) | 8 (3.9) | 3 (1.6) | 1 (1.5) | 0 (0.0) | 2 (1.0) | 6 (6.7) | 1 (3.2) | 6 (2.5) | 3 (2.8) | 0 (0.0) | 179 (2.3) |
| Current drinker (<23g/day) | 608 (38.5) | 218 (40.3) | 56 (30.4) | 559 (37.3) | 213 (39.4) | 71 (33.2) | 527 (37.1) | 204 (35.8) | 76 (36.7) | 75 (38.9) | 28 (43.1) | 6 (28.6) | 83 (40.3) | 32 (35.6) | 10 (32.3) | 104 (43.3) | 44 (41.1) | 13 (33.3) | 2927 (37.8) |
| Current (≥ 23g/day) | 163 (10.3) | 125 (23.1) | 77 (41.8) | 163 (10.9) | 129 (23.8) | 91 (42.5) | 165 (11.6) | 135 (23.7) | 80 (38.6) | 12 (6.2) | 16 (24.6) | 7 (33.3) | 14 (6.8) | 18 (20.0) | 11 (35.5) | 23 (9.6) | 23 (21.5) | 14 (35.9) | 1266 (16.3) |
| Healthy lifestyle factors | |||||||||||||||||||
| Non-obesity | 1446 (91.6) | 341 (63..0) | 48 (26.1) | 1386 (92.5) | 319 (59.0) | 47 (22.0) | 1307 (92.1) | 329 (57.7) | 46 (22.2) | 182 (94.3) | 39 (60.0) | 5 (23.8) | 183 (88.8) | 41 (45.6) | 5 (16.1) | 218 (90.8) | 65 (60.7) | 12 (30.8) | 6019 (77.7) |
| Never-smoker | 1355 (85.9) | 178 (32.9) | 26 (14.1) | 1273 (84.9) | 192 (35.5) | 22 (10.3) | 1204 (84.8) | 211 (37.0) | 33 (15.9) | 168 (87.0) | 22 (33.8) | 0 (0.0) | 183 (88.8) | 39 (43.3) | 4 (12.9) | 205 (85.4) | 47 (43.9) | 5 (12.8) | 5167 (66.7) |
| Regular physical activity | 909 (57.6) | 108 (20.0) | 17 (9.2) | 867 (57.8) | 108 (20.0) | 22 (10.3) | 835 (58.8) | 122 (21.4) | 16 (7.7) | 106 (54.9) | 12 (18.5) | 3 (14.3) | 114 (55.3) | 25 (27.8) | 3 (9.7) | 139 (57.9) | 19 (17.8) | 5 (12.8) | 3430 (44.3) |
| GGT, <50.0 IU/L | 1540 (97.6) | 455 (84.1) | 68 (37.0) | 147 (98.3) | 463 (85.6) | 94 (43.9) | 1382 (97.4) | 478 (83.9) | 75 (36.2) | 191 (99.0) | 57 (87.7) | 9 (42.9) | 103 (98.5) | 75 (83.3) | 11 (35.5) | 236 (98.3) | 83 (77.6) | 13 (33.3) | 6906 (89.2) |
BMI, body mass index; GGT, gamma-glutamyl transferase; METs, metabolic equivalents,
Diabetes: defined as non-fasting glucose ≥ 200 mg/dL, hemoglobin A1c ≥ 6.5, and/or self-reported treatment for diabetes.
Non-obesity: defined as BMI <25.0 kg/m2 based on the Western Pacific Region of the World Health Organization criteria for Japanese individuals. Never-smoker: defined as participants who had smoked <100 cigarettes during their lifetime.
Regular physical activity: defined as meeting the American Heart Association recommendations of at least 150 min of moderate activity per week or 75 min of vigorous activity per week (or an equivalent combination).
A higher GGT was defined as GGT ≥ 50 IU/L based on health examination in Japan.
Overall lifestyle was subsequently categorized into ideal (having least three ideal lifestyle factors), poor (having at least three poor lifestyle factors), or intermediate (all other combination).
Supplemental Table 2. Characteristics of participants according to family history.
| Family history | No | Yes | P for difference |
|---|---|---|---|
| Number | 6753 | 992 | |
| Age, years | 57.4 (13.6) | 56.7 (12.4) | 0.160 |
| Women, % | 5110 (75.7) | 801 (80.7) | 0.001 |
| BMI, kg/m2 | 22.7 (3.4) | 22.9 (3.7) | 0.033 |
| Glucose, mg/dl | 87.8 (15.6) | 91.5 (22.1) | <0.001 |
| HbA1c, % | 5.5 (0.5) | 5.7 (0.7) | <0.001 |
| Prevalence of diabetes, % | 319 (4.7) | 148 (14.9) | <0.001 |
| Treatment for diabetes | 228 (3.4) | 116 (11.7) | <0.001 |
| GGT, IU/L | 19.0 [14.0, 30.0] | 20.0 [15.0, 31.0] | 0.418 |
| METs, MET-min/week | 67.5 [9.0, 200.6] | 72.0 [9.5, 197.6] | 0.828 |
| Smoking status, % | 0.033 | ||
| Never-smoker | 4494 (66.5) | 673 (67.8) | |
| Ex-smoker | 1512 (22.4) | 236 (23.8) | |
| Current smoker | 747 (11.1) | 83 (8.4) | |
| Drinking status, % | 0.140 | ||
| Never-drinker | 2936 (43.5) | 437 (44.1) | |
| Ex-drinker | 157 (2.3) | 22 (2.2) | |
| Current drinker (<23g/day) | 2532 (37.5) | 395 (39.8) | |
| Current drinker (≥ 23g/day) | 1128 (16.7) | 138 (13.9) | |
| Healthy lifestyle factors | |||
| Non-obesity | 5269 (78.0) | 750 (75.6) | 0.095 |
| Never-smoker | 4494 (66.5) | 673 (67.8) | 0.440 |
| Regular physical activity | 3004 (44.5) | 426 (42.9) | 0.380 |
| GGT, <50.0 IU/L | 6028 (89.3) | 878 (88.5) | 0.509 |
| PRS | -0.00028 (0.00005) | -0.0027 (0.00004) | <0.001 |
BMI, body mass index; GGT, gamma-glutamyl transferase; METs, metabolic equivalents; PRS, polygenic risk score
Diabetes: defined as non-fasting glucose ≥ 200 mg/dL, hemoglobin A1c ≥ 6.5, and/or self-reported treatment for diabetes.
Non-obesity: defined as BMI <25.0 kg/m2 based on the Western Pacific Region of the World Health Organization criteria for Japanese individuals. Never-smoker: defined as participants who had smoked <100 cigarettes during their lifetime.
Regular physical activity: defined as meeting the American Heart Association recommendations of at least 150 min of moderate activity per week or 75 min of vigorous activity per week (or an equivalent combination).
A higher GGT was defined as GGT ≥ 50 IU/L based on health examination in Japan.
To compare the characteristics of participants with and without family history of diabetes, we used the Student’s t-test for continuous variables, Mann-Whitney U test for nonparametric variables, and the χ2-test for categorical variables.
Association of Lifestyle with Diabetes Prevalence
A poor lifestyle was associated with increased prevalence of diabetes. The multivariate ORs were 1.00 (reference), 1.58 (1.25–1.98), and 2.37 (1.74–3.20) for ideal, intermediate, and poor lifestyles, respectively.
Associations of PRS with Diabetes Prevalence
A higher PRS indicates a higher OR for diabetes prevalence. The multivariate ORs were 1.00 for the low genetic risk, 1.96 (1.49–2.60) for the intermediate genetic risk, and 3.16 (2.45–4.12) for the high genetic risk.
Associations of a Family History of Diabetes with Diabetes Prevalence
Participants with a family history have significantly higher ORs for diabetes (OR, 4.43 [95% CI, 3.54–5.52]).
Associations of Combined Lifestyle and Genetic Risk with Diabetes Prevalence
A poor lifestyle was associated with a higher OR of diabetes within each category of genetic risk. Compared with a low genetic risk and an ideal lifestyle, participants with a low genetic risk and a poor lifestyle had a significantly higher OR (3.31; 95% CI, 1.84–5.76). Similarly, a higher genetic risk was associated with a higher OR of diabetes within each lifestyle category. Participants with a high genetic risk and an ideal lifestyle had a significantly higher OR (2.85; 95% CI, 2.03–4.08). Participants with a high genetic risk and a poor lifestyle had a significantly higher OR of diabetes (OR, 5.63 [95% CI, 3.38–9.27]) ( Supplemental Table 3 ) .
Supplemental Table 3. Associations of lifestyle and PRS with diabetes prevalence.
| DM/number of participants | % | OR, 95%CI | |||
|---|---|---|---|---|---|
| Low genetic risk | Ideal (<1 poor factors) | 46/1771 | (2.6) | Ref | |
| Intermediate (2 poor factors) | 19/606 | (3.1) | 1.53 | (0.91-2.49) | |
| Poor (>3 poor factors) | 21/205 | (10.2) | 3.31 | (1.84-5.76) | |
| Intermediate genetic risk | Ideal (<1 poor factors) | 83/1705 | (4.9) | 1.99 | (1.39-2.88) |
| Intermediate (2 poor factors) | 42/631 | (6.7) | 2.98 | (1.92-4.59) | |
| Poor (>3 poor factors) | 25/245 | (10.2) | 4.78 | (2.82-7.99) | |
| High genetic risk | Ideal (<1 poor factors) | 123/1659 | (7.4) | 2.85 | (2.03-4.08) |
| Intermediate (2 poor factors) | 78/677 | (11.5) | 4.85 | (3.31-7.16) | |
| Poor (>3 poor factors) | 30/246 | (12.2) | 5.63 | (3.38-9.27) |
Abbreviations: PRS, polygenic risk score; CI, confidence interval; DM, diabetes mellitus; OR, odds ratio.
Analysis by multivariable logistic regression model.
Adjusted for age, sex, first six principal components.
Associations of Combined Lifestyle, Genetic Risk, and Family History of Diabetes with Diabetes Prevalence
A higher genetic risk and a poor lifestyle were positively associated with diabetes prevalence independent of family history ( Table 2 ) . Family history was positively associated with diabetes prevalence even among participants with a low genetic risk and an ideal lifestyle (OR, 3.58 [95% CI, 1.76–6.98]). Even among participants without a family history, participants with a low genetic risk and a poor lifestyle had significantly higher ORs for diabetes prevalence (OR, 3.73 [95% CI, 1.92–7.00]). Furthermore, participants with a high genetic risk, poor lifestyle, and family history had significantly higher ORs for diabetes prevalence (OR, 12.00 [95% CI, 4.34–29.97]). The AUROC value (95% CI) for combined PRS and lifestyle was 0.774 (0.755–0.793) and that for combined family history, PRS, and lifestyle was 0.802 (0.783–0.821) (P for difference<0.001) ( Table 3 ) .
Table 2. Associations of combined family history and genetic and lifestyle risk with diabetes.
| DM/number of participants | % | OR, 95%CI | ||||
|---|---|---|---|---|---|---|
| Family history, no | Low genetic risk | Ideal (<1 poor factors) | 34/1578 | (2.2) | Ref | |
| Intermediate (2 poor factors) | 15/541 | (2.8) | 1.19 | (0.62-2.19) | ||
| Poor (>3 poor factors) | 16/184 | (8.7) | 3.73 | (1.92-7.00) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 64/1499 | (4.3) | 2.10 | (1.38-3.25) | |
| Intermediate (2 poor factors) | 28/541 | (5.2) | 2.81 | (1.65-4.74) | ||
| Poor (>3 poor factors) | 15/214 | (7.0) | 3.61 | (1.83-6.83) | ||
| High genetic risk | Ideal (<1 poor factors) | 70/1419 | (4.9) | 2.49 | (1.65-3.85) | |
| Intermediate (2 poor factors) | 54/570 | (9.5) | 5.21 | (3.31-8.29) | ||
| Poor (>3 poor factors) | 23/207 | (11.1) | 6.19 | (3.42-11.03) | ||
| Family history, yes | Low genetic risk | Ideal (<1 poor factors) | 12/193 | (6.2) | 3.58 | (1.73-6.98) |
| Intermediate (2 poor factors) | 4/65 | (6.2) | 4.27 | (1.22-11.49) | ||
| Poor (>3 poor factors) | 5/21 | (23.8) | 12.01 | (3.43-36.90) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 19/206 | (9.2) | 6.02 | (3.25-10.85) | |
| Intermediate (2 poor factors) | 14/90 | (15.6) | 11.25 | (5.45-22.31) | ||
| Poor (>3 poor factors) | 10/31 | (32.3) | 34.83 | (13.74-85.27) | ||
| High genetic risk | Ideal (<1 poor factors) | 53/240 | (22.1) | 16.99 | (10.59-27.60) | |
| Intermediate (2 poor factors) | 24/107 | (22.4) | 17.82 | (9.68-32.51) | ||
| Poor (>3 poor factors) | 7/39 | (17.9) | 12.00 | (4.34-29.97) |
Abbreviations: CI, confidence interval; DM, diabetes mellitus; OR, odds ratio.
Adjusted for age, sex and first six principal components.
Table 3. Area under the receiver operating characteristic curve for genetic and lifestyle risk to predict diabetes.
| Area under the receiver operating characteristic curve (95% CI) | |
|---|---|
| Model 1 a | 0.738 (0.718-0.758) |
| Model 1 + PRS | 0.764 (0.744-0.784) |
| Model 1 + lifestyle score | 0.751 (0.732-0.770) |
| Model 1 + family history | 0.776 (0.757-0.796) |
| Model 1 + PRS + lifestyle score | 0.774 (0.755-0.793) |
| Model 1 + PRS + family history | 0.793 (0.773-0.813) |
| Model 1 + lifestyle + family history | 0.788 (0.769-0.807) |
| Model 1 + PRS + lifestyle + family history | 0.802 (0.783-0.821) |
Abbreviations: CI, confidence interval; PRS, polygenic risk score.
a Model 1 was adjusted for age, sex, and the top six principal components.
Diabetes: defined as non-fasting glucose ≥ 200 mg/dL, hemoglobin A1c ≥ 6.5, and/or self-reported treatment for diabetes.
The same pattern of associations was found in the sensitivity analysis, excluding participants with a treatment for diabetes ( Supplemental Table 4 ) . Additionally, because younger participants may not yet have a clear family history, the contribution of PRS may be greater in young than in elderly participants. Although we conducted stratified analysis by age, the results were substantially unchanged compared with those using all participants ( Supplemental Tables 5 and 6 ) . This stratified analysis did not show that the increase in AUROC was greater for younger participants when PRS was added to a model that included family history and lifestyle risk ( Supplemental Table 7 ) . Finally, we restricted ourselves to non-obese participants, but the results were essentially unchanged compared with those using all participants ( Supplemental Table 8 ) .
Supplemental Table 4. Associations of family history, PRS, and lifestyle risk with diabetes prevalence among participants without treatment for diabetes.
| DM/number of participants | % | OR, 95%CI | ||||
|---|---|---|---|---|---|---|
| Family history, no | Low genetic risk | Ideal (<1 poor factors) | 11/1521 | (0.7) | Ref | |
| Intermediate (2 poor factors) | 4/517 | (0.8) | 1.03 | (0.28-3.06) | ||
| Poor (>3 poor factors) | 7/171 | (4.1) | 5.15 | (1.81-13.77) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 12/1449 | (0.8) | 1.17 | (0.51-2.71) | |
| Intermediate (2 poor factors) | 8/528 | (1.5) | 2.38 | (0.91-5.98) | ||
| Poor (>3 poor factors) | 3/202 | (1.5) | 2.25 | (0.50-7.53) | ||
| High genetic risk | Ideal (<1 poor factors) | 17/1398 | (1.2) | 1.78 | (0.84-3.93) | |
| Intermediate (2 poor factors) | 14/536 | (2.6) | 3.96 | (1.76-9.11) | ||
| Poor (>3 poor factors) | 15/203 | (7.4) | 12.64 | (5.55-29.66) | ||
| Family history, yes | Low genetic risk | Ideal (<1 poor factors) | 2/182 | (1.1) | 1.73 | (0.27-6.56) |
| Intermediate (2 poor factors) | 0/58 | (0.0) | 0.00 | (0.00-39.43) | ||
| Poor (>3 poor factors) | 2/18 | (11.1) | 15.19 | (2.11-69.20) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 2/186 | (1.1) | 1.81 | (0.28-6.87) | |
| Intermediate (2 poor factors) | 2/78 | (2.6) | 4.50 | (0.68-17.57) | ||
| Poor (>3 poor factors) | 4/24 | (16.7) | 38.77 | (9.58-134.26) | ||
| High genetic risk | Ideal (<1 poor factors) | 12/203 | (5.9) | 10.89 | (4.65-25.79) | |
| Intermediate (2 poor factors) | 8/94 | (8.5) | 17.23 | (6.37-44.87) | ||
| Poor (>3 poor factors) | 0/33 | (0.0) | 0.00 | (0.00-0.00) |
Abbreviations: PRS, polygenic risk score; CI, confidence interval; DM, diabetes mellitus; OR, odds ratio.
Analysis by multivariable logistic regression model.
Adjusted for age, sex and first 6 principal components.
Supplemental Table 5. Associations of family history, PRS, and lifestyle risk with diabetes prevalence among participants under 60 years old.
| DM/number of participants | % | OR, 95%CI | ||||
|---|---|---|---|---|---|---|
| Family history, no | Low genetic risk | Ideal (<1 poor factors) | 3/716 | (0.4) | Ref | |
| Intermediate (2 poor factors) | 3/277 | (1.1) | 2.18 | (0.40-11.97) | ||
| Poor (>3 poor factors) | 8/85 | (9.4) | 13.86 | (3.68-67.01) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 5/663 | (0.8) | 1.77 | (0.43-8.71) | |
| Intermediate (2 poor factors) | 6/290 | (2.1) | 4.44 | (1.14-21.37) | ||
| Poor (>3 poor factors) | 3/124 | (2.4) | 3.48 | (0.62-19.63) | ||
| High genetic risk | Ideal (<1 poor factors) | 5/611 | (0.8) | 2.07 | (0.50-10.18) | |
| Intermediate (2 poor factors) | 10/295 | (3.4) | 7.96 | (2.38-36.02) | ||
| Poor (>3 poor factors) | 7/118 | (5.9) | 10 | (2.63-48.26) | ||
| Family history, yes | Low genetic risk | Ideal (<1 poor factors) | 3/94 | (3.2) | 6.91 | (1.24-38.51) |
| Intermediate (2 poor factors) | 1/40 | (2.5) | 4.95 | (0.24-40.99) | ||
| Poor (>3 poor factors) | 0/10 | (0.0) | 0 | (0.00-0.00) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 2/98 | (2.0) | 4.9 | (0.63-30.24) | |
| Intermediate (2 poor factors) | 4/49 | (8.2) | 20.61 | (4.29-110.14) | ||
| Poor (>3 poor factors) | 4/18 | (22.2) | 70.71 | (12.96-425.89) | ||
| High genetic risk | Ideal (<1 poor factors) | 5/115 | (4.3) | 9.67 | (2.31-48.11) | |
| Intermediate (2 poor factors) | 7/60 | (11.7) | 27.42 | (7.25-131.94) | ||
| Poor (>3 poor factors) | 2/22 | (9.1) | 22.08 | (23.67-148.56) |
Abbreviations: PRS, polygenic risk score; CI, confidence interval; DM, diabetes mellitus; OR, odds ratio.
Analysis by multivariable logistic regression model.
Adjusted for age, sex and first 6 principal components.
Supplemental Table 6. Associations of family history, PRS, and lifestyle risk with diabetes prevalence among participants over 60 years old.
| DM/number of participants | % | OR, 95%CI | ||||
|---|---|---|---|---|---|---|
| Family history, no | Low genetic risk | Ideal (<1 poor factors) | 31/862 | (3.6) | Ref | |
| Intermediate (2 poor factors) | 12/264 | (4.5) | 1.08 | (0.52-2.11) | ||
| Poor (>3 poor factors) | 8/99 | (8.1) | 1.95 | (0.80-4.26) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 59/836 | (7.1) | 2.11 | (1.36-3.35) | |
| Intermediate (2 poor factors) | 22/251 | (8.8) | 2.5 | (1.39-4.42) | ||
| Poor (>3 poor factors) | 12/90 | (13.3) | 3.58 | (1.67-7.24) | ||
| High genetic risk | Ideal (<1 poor factors) | 65/808 | (8.0) | 2.5 | (1.62-3.93) | |
| Intermediate (2 poor factors) | 44/275 | (16.0) | 4.77 | (2.92-7.86) | ||
| Poor (>3 poor factors) | 16/89 | (18.0) | 5.15 | (2.59-9.95) | ||
| Family history, yes | Low genetic risk | Ideal (<1 poor factors) | 9/99 | (9.1) | 2.99 | (1.30-6.28) |
| Intermediate (2 poor factors) | 3/25 | (12.0) | 3.99 | (0.91-12.46) | ||
| Poor (>3 poor factors) | 5/11 | (45.5) | 15.99 | (4.24-58.17) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 17/108 | (15.7) | 6.02 | (3.12-11.30) | |
| Intermediate (2 poor factors) | 10/41 | (24.4) | 9.18 | (3.92-20.22) | ||
| Poor (>3 poor factors) | 6/13 | (46.2) | 23.93 | (7.20-77.52) | ||
| High genetic risk | Ideal (<1 poor factors) | 48/125 | (38.4) | 19.6 | (11.73-33.23) | |
| Intermediate (2 poor factors) | 17/47 | (36.2) | 15.73 | (7.65-31.95) | ||
| Poor (>3 poor factors) | 5/17 | (29.4) | 9.83 | (2.92-29.05) |
Abbreviations: PRS, polygenic risk score; CI, confidence interval; DM, diabetes mellitus; OR, odds ratio.
Analysis by multivariable logistic regression model.
Adjusted for age, sex and first 6 principal components.
Supplemental Table 7. Area under the receiver operating characteristic curve for genetic and lifestyle risk to predict diabetes according to median age.
| <60 years old | ≥ 60 years old | |
|---|---|---|
| Area under the receiver operating characteristic curve (95% CI) | ||
| Model 1 a | 0.766 (0.714-0.817) | 0.616 (0.587-0.645) |
| Model 1 + PRS | 0.775 (0.722-0.828) | 0.672 (0.644-0.700) |
| Model 1 + lifestyle score | 0.792 (0.743-0.841) | 0.635 (0.607-0.663) |
| Model 1 + family history | 0.807 (0.762-0.852) | 0.688 (0.660-0.717) |
| Model 1 + PRS + lifestyle score | 0.801 (0.752-0.851) | 0.683 (0.656-0.710) |
| Model 1 + PRS + family history | 0.810 (0.764-0.857) | 0.720 (0.693-0.748) |
| Model 1 + lifestyle + family history | 0.829 (0.783-0.875) | 0.701 (0.674-0.729) |
| Model 1 + PRS + lifestyle + family history | 0.831 (0.784-0.877) | 0.729 (0.702-0.757) |
Abbreviations: CI, confidence interval; PRS, polygenic risk score.
a Model 1 was adjusted for age, sex, and the top six principal components.
Diabetes: defined as non-fasting glucose ≥ 200 mg/dL, hemoglobin A1c ≥ 6.5, and/or self-reported treatment for diabetes.
Supplemental Table 8. Associations of family history, PRS, and lifestyle risk with diabetes prevalence among non-obesity.
| DM/number of participants | % | OR, 95%CI | ||||
|---|---|---|---|---|---|---|
| Family history, no | Low genetic risk | Ideal (<1 poor factors) | 30/1446 | (2.1) | Ref | |
| Intermediate (2 poor factors) | 3/41 | (19.5) | 0.95 | (0.39-2.05) | ||
| Poor (>3 poor factors) | 3/48 | (6.3) | 2.19 | (0.50-6.80) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 56/1386 | (4.0) | 2.12 | (1.35-3.40) | |
| Intermediate (2 poor factors) | 12/319 | (3.8) | 2.21 | (1.05-4.39) | ||
| Poor (>3 poor factors) | 3/47 | (6.4) | 2.18 | (0.48-6.98) | ||
| High genetic risk | Ideal (<1 poor factors) | 61/1307 | (4.7) | 2.53 | (1.62-4.04) | |
| Intermediate (2 poor factors) | 18/329 | (5.5) | 2.78 | (1.46-5.18) | ||
| Poor (>3 poor factors) | 5/456 | (1.1) | 4.92 | (1.53-13.31) | ||
| Family history, yes | Low genetic risk | Ideal (<1 poor factors) | 10/182 | (5.5) | 3.68 | (1.65-7.62) |
| Intermediate (2 poor factors) | 2/39 | (5.1) | 3.63 | (0.56-13.59) | ||
| Poor (>3 poor factors) | 0/5 | (0.0) | 0.00 | (0.00-0.00) | ||
| Intermediate genetic risk | Ideal (<1 poor factors) | 17/183 | (9.3) | 6.91 | (3.56-1306) | |
| Intermediate (2 poor factors) | 5/41 | (12.2) | 12.85 | (3.93-35.63) | ||
| Poor (>3 poor factors) | 0/5 | (0.0) | 0.00 | (0.00-0.00) | ||
| High genetic risk | Ideal (<1 poor factors) | 47/218 | (21.6) | 19.66 | (11.74-33.41) | |
| Intermediate (2 poor factors) | 8/65 | (12.3) | 8.59 | (3.30-20.42) | ||
| Poor (>3 poor factors) | 1/12 | (8.3) | 4.51 | (0.23-29.18) |
Abbreviations: PRS, polygenic risk score; CI, confidence interval; DM, diabetes mellitus; OR, odds ratio.
Analysis by multivariable logistic regression model.
Adjusted for age, sex and first 6 principal components.
Discussion
In this general community-based population of approximately 9,000 Japanese individuals, a high PRS was associated with a higher OR for diabetes prevalence independent of lifestyle and family history. Conversely, an ideal lifestyle was associated with a decreased OR for diabetes prevalence regardless of genetic risk and family history. Furthermore, even among participants with a low PRS and an ideal lifestyle, participants with a family history had a significantly higher OR for diabetes prevalence compared with participants without a family history. Because it is more likely that family members have not yet developed diabetes in younger individuals and that their family history is unclear, we hypothesized that the contribution of PRS to diabetes would be higher in younger that in older adults. Although we conducted a stratified analysis by age, the associations were essentially unchanged. Furthermore, the increase in AUROC was not greater for younger than older participants when PRS was added to a model that included family history and lifestyle risk.
Several previous studies have reported the associations of combined genetic and lifestyle risks with diabetes. A high genetic risk and an unfavorable lifestyle, such as obesity, physical inactivity, smoking, and unhealthy and dietary patterns, were independently associated with an increased risk of diabetes in European and US population 15 , 16 , 36 , 37) . A previous study in Asia showed that genetic factors were independently associated with the risk of diabetes. However, a healthy lifestyle was associated with a significantly lower risk of diabetes even among any genetic risk groups 17) . Asians might have lower insulin sensitivity 3 , 38) . In fact, a large-scale GWAS of a Japanese population detected several variants in genes related to pancreatic acinar cells and insulin secretion 7) . Furthermore, genetic risk allele frequencies and risk factors are different between Asian and other continental populations 3 , 39) . Despite the differences in genetic and lifestyle risks between Japanese and other populations, no studies that combined genetic and lifestyle risks to quantitatively assess the association of diabetes have been reported in a Japanese population. We observed that lifestyle and genetic factors were independently associated with the prevalence of diabetes among the Japanese population, which is consistent with the findings of previous studies.
A family history of diabetes is a well-known risk factor for the incidence of diabetes in offspring 18 - 20) . It may also likely reflect genetic and shared environmental factors among family members. However, several studies showed that family history is completely unexplained by genetic, lifestyle, and anthropometric risk factors 18 - 20) . In the European Prospective Investigation into Cancer and Nutrition study, anthropometric, genetic, and lifestyle risks, including physical activity, smoking status, and Mediterranean diet pattern, explained only 13% of the risk of diabetes associated with family history 19) . Our study found that family history was associated with higher ORs for diabetes prevalence irrespective of genetic and lifestyle risks, which is consistent with previous studies. This finding suggests that family history conveys more than heritable genetic information and lifestyle. Therefore, further studies are warranted to discover factors that may explain the association of family history with diabetes, which might provide new insights into the etiology of diabetes. In the current study, even among participants with a family history of diabetes, participants with healthier lifestyle factors had lower ORs for diabetes prevalence even within each category of genetic risk. This finding indicates that adhering to a healthy lifestyle might be important to prevent diabetes regardless of genetic risk and family history of diabetes. However, family history was positively associated with diabetes even among participants with a low genetic risk and an ideal lifestyle. Therefore, individuals with a low genetic risk and an ideal lifestyle but with a family history of diabetes must be screened frequently for prevention and early intervention. Additionally, information on lifestyle risk and family history are collected in the clinical setting, but our study showed that genetic risk is associated with the prevalence of diabetes regardless of lifestyle risk and family history. Therefore, genetic risk may need to be considered to stratify individuals at a high risk of diabetes.
Previous studies showed that adding family history, in addition to genetic and lifestyle risks, improved the predictive ability for diabetes beyond genetic and lifestyle risks 40 , 41) . In the current study, incorporating family history achieved significantly greater predictive ability than that of a model with known lifestyle risk and PRS, which is consistent with previous studies. In the clinical setting, collecting information on family history can be easily conducted. Therefore, information on the family history of diabetes may be useful for the assessment of diabetes risk and tailoring of personalized medicine using lifestyle and genetic information.
To the best of our knowledge, this is the first study that showed quantitative estimates of the associations of combined family history and genetic and lifestyle risks with diabetes in the general Japanese population. Nevertheless, our study still has several limitations. First, we could not confirm causal relationships as this was a cross-sectional study. However, genetic factors do not change because of disease or lifestyle factors. Thus, reverse causality is unlikely with respect to the genetic factor. Second, lifestyle may be influenced by diabetes treatment, which could cause reverse causality. However, our results were substantially unchanged after excluding participants with diabetes treatment. Third, family history was obtained through self-reported questionnaire. Participants with diabetes may more likely report having a family history than participants without diabetes. Therefore, prospective cohort studies are required to provide evidence regarding family history, PRS, and lifestyle. Fourth, our study population included only Japanese participants. Martin AR et al. 42) indicated that the prediction accuracy for PRS is consistently higher with GWAS summary statistics from ancestry-matches statistics because of ancestry-correlated differences in the frequency and effect of risk alleles. Additionally, genetics and lifestyle may vary by ethnicity 3 , 39) . Thus, studies on other ethnic population are required to confirm the generalizability of our findings. Finally, additional variants associated with diabetes may be identified in future GWASs and whole genome sequencing, and these variants may improve genetic risk performance. The Tohoku Medical Megabank Project is ongoing to construct whole genome sequencing data for 100,000 participants and has already completed whole genome sequencing for more than 50,000 participants. In the future, we will examine whether the use of whole genome sequencing data improves the predictive ability over the PRS in this study.
Conclusions
Our study provided quantitative estimates of the associations between lifestyle and genetic risks with diabetes in the general Japanese population. Genetic and lifestyle risks were independently associated with higher diabetes prevalence. Furthermore, family history was positively associated with diabetes prevalence even among participants with a low genetic risk and an ideal lifestyle. Adding PRS to family history and conventional lifestyle risk factors improved our ability to predict diabetes. These findings support the notion that a healthy lifestyle is important to prevent diabetes in any genetic risk groups. Additionally, genetic risk may need to be considered to stratify individuals at a high risk of diabetes. Family history must also be considered for diabetes prevention even among participants with a low genetic risk and an ideal lifestyle.
Acknowledgement
The authors thank all the volunteers who participated in this project. The authors thank the members of the Tohoku Medical Megabank Organization, including the Genome Medical Research Coordinators and the office and administrative personnel for their assistance. A complete list of members is available at https://www.megabank.tohoku.ac.jp/english/a220901/.
Funding and Assistance
This work was supported by Tohoku Medical Megabank Project from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT); the Japan Agency for Medical Research and Development [AMED; JP22tm0124005] (to A.H.); and JST SPRING, [Grand Number JPMJSP2114] (to M.T). This research used the super computer system provided by Tohoku Medical Megabank Project founded by AMED [Grant Number JP21tm0424601].
Data Available
All data used to support the findings of this study may be released upon application to the Tohoku Medical Megabank Organization (Sendai, Japan), which can be contacted through Prof. Atsushi Hozawa (email: hozawa@megabank.tohoku.ac.jp).
Conflicts of Interest
There are no conflicts of interest.
References
- 1).World Health Organization. Diabetes. Available from https: //www.who.int/news-room/fact-sheets/detail/diabetes. Accessed March 14, 2023 [Google Scholar]
- 2).Liu J, Bai R, Chai Z, Cooper ME, Zimmet PZ, Zhang L. Low- and middle-income countries demonstrate rapid growth of type 2 diabetes: an analysis based on Global Burden of Disease 1990-2019 data. Diabetologia, 2022; 65: 1339-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Hu FB. Globalization of diabetes: the role of diet, lifestyle, and genes. Diabetes Care, 2021; 34: 1249-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Tuomilehto J, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa M; Finnish Diabetes Prevention Study Group. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med, 2001; 344: 1343-1350 [DOI] [PubMed] [Google Scholar]
- 5).Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM; Diabetes Prevention Program Research Group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med, 2002; 346: 393-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Diabetes Prevention Program Research Group. Long-term effects of lifestyle intervention or metformin on diabetes development and microvascular complications over 15-year follow-up: the Diabetes Prevention Program Outcomes Study. Lancet Diabetes Endocrinol, 2015; 3: 866-875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Suzuki K, Akiyama M, Ishigaki K, Kanai M, Hosoe J, Shojima N, Hozawa A, Kadota A, Kuriki K, Naito M, Tanno K, Ishigaki Y, Hirata M, Matsuda K, Iwata N, Ikeda M, Sawada N, Yamaji T, Iwasaki M, Ikegawa S, Maeda S, Murakami Y, Wakai K, Tsugane S, Sasaki M, Yamamoto M, Okada Y, Kubo M, Kamatani Y, Horikoshi M, Yamauchi T, Kadowaki T. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat Genet, 2019; 51: 379-386 [DOI] [PubMed] [Google Scholar]
- 8).Vujkovic M, Keaton JM, Lynch JA, Miller DR, Zhou J, Tcheandjieu C, Huffman JE, Assimes TL, Lorenz K, Zhu X, Hilliard AT, Judy RL, Huang J, Lee KM, Klarin D, Pyarajan S, Danesh J, Melander O, Rasheed A, Mallick NH, Hameed S, Qureshi IH, Afzal MN, Malik U, Jalal A, Abbas S, Sheng X, Gao L, Kaestner KH, Susztak K, Sun YV, DuVall SL, Cho K, Lee JS, Gaziano JM, Phillips LS, Meigs JB, Reaven PD, Wilson PW, Edwards TL, Rader DJ, Damrauer SM, O'Donnell CJ, Tsao PS; HPAP Consortium; Regeneron Genetics Center; VA Million Veteran Program; Chang KM, Voight BF, Saleheen D. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet, 2020; 52: 680-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Spracklen CN, Horikoshi M, Kim YJ, Lin K, Bragg F, Moon S, Suzuki K, Tam CHT, Tabara Y, Kwak SH, Takeuchi F, Long J, Lim VJY, Chai JF, Chen CH, Nakatochi M, Yao J, Choi HS, Iyengar AK, Perrin HJ, Brotman SM, van de Bunt M, Gloyn AL, Below JE, Boehnke M, Bowden DW, Chambers JC, Mahajan A, McCarthy MI, Ng MCY, Petty LE, Zhang W, Morris AP, Adair LS, Akiyama M, Bian Z, Chan JCN, Chang LC, Chee ML, Chen YI, Chen YT, Chen Z, Chuang LM, Du S, Gordon-Larsen P, Gross M, Guo X, Guo Y, Han S, Howard AG, Huang W, Hung YJ, Hwang MY, Hwu CM, Ichihara S, Isono M, Jang HM, Jiang G, Jonas JB, Kamatani Y, Katsuya T, Kawaguchi T, Khor CC, Kohara K, Lee MS, Lee NR, Li L, Liu J, Luk AO, Lv J, Okada Y, Pereira MA, Sabanayagam C, Shi J, Shin DM, So WY, Takahashi A, Tomlinson B, Tsai FJ, van Dam RM, Xiang YB, Yamamoto K, Yamauchi T, Yoon K, Yu C, Yuan JM, Zhang L, Zheng W, Igase M, Cho YS, Rotter JI, Wang YX, Sheu WHH, Yokota M, Wu JY, Cheng CY, Wong TY, Shu XO, Kato N, Park KS, Tai ES, Matsuda F, Koh WP, Ma RCW, Maeda S, Millwood IY, Lee J, Kadowaki T, Walters RG, Kim BJ, Mohlke KL, Sim X. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature, 2020; 582: 240-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Inaishi J, Hirakawa Y, Horikoshi M, Akiyama M, Higashioka M, Yoshinari M, Hata J, Mukai N, Kamatani Y, Momozawa Y, Kubo M, Ninomiya T. Association Between Genetic Risk and Development of Type 2 Diabetes in a General Japanese Population: The Hisayama Study. J Clin Endocrinol Metab, 2019; 104: 3213-3222 [DOI] [PubMed] [Google Scholar]
- 11).Chikowore T, Ekoru K, Vujkovi M, Gill D, Pirie F, Young E, Sandhu MS, McCarthy M, Rotimi C, Adeyemo A, Motala A, Fatumo S. Polygenic Prediction of Type 2 Diabetes in Africa. Diabetes Care, 2022; 45: 717-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Ge T, Irvin MR, Patki A, Srinivasasainagendra V, Lin YF, Tiwari HK, Armstrong ND, Benoit B, Chen CY, Choi KW, Cimino JJ, Davis BH, Dikilitas O, Etheridge B, Feng YA, Gainer V, Huang H, Jarvik GP, Kachulis C, Kenny EE, Khan A, Kiryluk K, Kottyan L, Kullo IJ, Lange C, Lennon N, Leong A, Malolepsza E, Miles AD, Murphy S, Namjou B, Narayan R, O'Connor MJ, Pacheco JA, Perez E, Rasmussen-Torvik LJ, Rosenthal EA, Schaid D, Stamou M, Udler MS, Wei WQ, Weiss ST, Ng MCY, Smoller JW, Lebo MS, Meigs JB, Limdi NA, Karlson EW. Development and validation of a trans-ancestry polygenic risk score for type 2 diabetes in diverse populations. Genome Med, 2022; 14: 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Ashenhurst JR, Sazonova OV, Svrchek O, Detweiler S, Kita R, Babalola L, McIntyre M, Aslibekyan S, Fontanillas P, Shringarpure S; 23andMe Research Team; Pollard JD, Koelsch BL. A Polygenic Score for Type 2 Diabetes Improves Risk Stratification Beyond Current Clinical Screening Factors in an Ancestrally Diverse Sample. Front Genet, 2022; 13: 871260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Wedekind LE, Mahajan A, Hsueh WC, Chen P, Olaiya MT, Kobes S, Sinha M, Baier LJ, Knowler WC, McCarthy MI, Hanson RL. The utility of a type 2 diabetes polygenic score in addition to clinical variables for prediction of type 2 diabetes incidence in birth, youth and adult cohorts in an Indigenous study population. Diabetologia, 2023; 66: 847-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Said MA, Verweij N, van der Harst P. Associations of Combined Genetic and Lifestyle Risks With Incident Cardiovascular Disease and Diabetes in the UK Biobank Study. JAMA Cardiol, 2018; 3: 693-702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Han X, Wei Y, Hu H, Wang J, Li Z, Wang F, Long T, Yuan J, Yao P, Wei S, Wang Y, Zhang X, Guo H, Yang H, Wu T, He M. Genetic Risk, a Healthy Lifestyle, and Type 2 Diabetes: the Dongfeng-Tongji Cohort Study. J Clin Endocrinol Metab, 2020; 105: dgz325 [DOI] [PubMed] [Google Scholar]
- 17).Li H, Khor CC, Fan J, Lv J, Yu C, Guo Y, Bian Z, Yang L, Millwood IY, Walters RG, Chen Y, Yuan JM, Yang Y, Hu C, Chen J, Chen Z, Koh WP, Huang T, Li L. Genetic risk, adherence to a healthy lifestyle, and type 2 diabetes risk among 550,000 Chinese adults: results from 2 independent Asian cohorts. Am J Clin Nutr, 2020; 111: 698-707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).van 't Riet E, Dekker JM, Sun Q, Nijpels G, Hu FB, van Dam RM. Role of adiposity and lifestyle in the relationship between family history of diabetes and 20-year incidence of type 2 diabetes in U.S. women. Diabetes Care, 2010; 33: 763-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).InterAct Consortium; Scott RA, Langenberg C, Sharp SJ, Franks PW, Rolandsson O, Drogan D, van der Schouw YT, Ekelund U, Kerrison ND, Ardanaz E, Arriola L, Balkau B, Barricarte A, Barroso I, Bendinelli B, Beulens JW, Boeing H, de Lauzon-Guillain B, Deloukas P, Fagherazzi G, Gonzalez C, Griffin SJ, Groop LC, Halkjaer J, Huerta JM, Kaaks R, Khaw KT, Krogh V, Nilsson PM, Norat T, Overvad K, Panico S, Rodriguez-Suarez L, Romaguera D, Romieu I, Sacerdote C, Sánchez MJ, Spijkerman AM, Teucher B, Tjonneland A, Tumino R, van der A DL, Wark PA, McCarthy MI, Riboli E, Wareham NJ. The link between family history and risk of type 2 diabetes is not explained by anthropometric, lifestyle or genetic risk factors: the EPIC-InterAct study. Diabetologia, 2013; 56: 60-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Ding M, Ahmad S, Qi L, Hu Y, Bhupathiraju SN, Guasch-Ferré M, Jensen MK, Chavarro JE, Ridker PM, Willett WC, Chasman DI, Hu FB, Kraft P. Additive and Multiplicative Interactions Between Genetic Risk Score and Family History and Lifestyle in Relation to Risk of Type 2 Diabetes. Am J Epidemiol, 2020; 189: 445-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Hozawa A, Tanno K, Nakaya N, Nakamura T, Tsuchiya N, Hirata T, Narita A, Kogure M, Nochioka K, Sasaki R, Takanashi N, Otsuka K, Sakata K, Kuriyama S, Kikuya M, Tanabe O, Sugawara J, Suzuki K, Suzuki Y, Kodama EN, Fuse N, Kiyomoto H, Tomita H, Uruno A, Hamanaka Y, Metoki H, Ishikuro M, Obara T, Kobayashi T, Kitatani K, Takai-Igarashi T, Ogishima S, Satoh M, Ohmomo H, Tsuboi A, Egawa S, Ishii T, Ito K, Ito S, Taki Y, Minegishi N, Ishii N, Nagasaki M, Igarashi K, Koshiba S, Shimizu R, Tamiya G, Nakayama K, Motohashi H, Yasuda J, Shimizu A, Hachiya T, Shiwa Y, Tominaga T, Tanaka H, Oyama K, Tanaka R, Kawame H, Fukushima A, Ishigaki Y, Tokutomi T, Osumi N, Kobayashi T, Nagami F, Hashizume H, Arai T, Kawaguchi Y, Higuchi S, Sakaida M, Endo R, Nishizuka S, Tsuji I, Hitomi J, Nakamura M, Ogasawara K, Yaegashi N, Kinoshita K, Kure S, Sakai A, Kobayashi S, Sobue K, Sasaki M, Yamamoto M. Study Profile of the Tohoku Medical Megabank Community-Based Cohort Study. J Epidemiol, 2021; 31: 65-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Kuriyama S, Yaegashi N, Nagami F, Arai T, Kawaguchi Y, Osumi N, Sakaida M, Suzuki Y, Nakayama K, Hashizume H, Tamiya G, Kawame H, Suzuki K, Hozawa A, Nakaya N, Kikuya M, Metoki H, Tsuji I, Fuse N, Kiyomoto H, Sugawara J, Tsuboi A, Egawa S, Ito K, Chida K, Ishii T, Tomita H, Taki Y, Minegishi N, Ishii N, Yasuda J, Igarashi K, Shimizu R, Nagasaki M, Koshiba S, Kinoshita K, Ogishima S, Takai-Igarashi T, Tominaga T, Tanabe O, Ohuchi N, Shimosegawa T, Kure S, Tanaka H, Ito S, Hitomi J, Tanno K, Nakamura M, Ogasawara K, Kobayashi S, Sakata K, Satoh M, Shimizu A, Sasaki M, Endo R, Sobue K, Tohoku Medical Megabank Project Study Group T, Yamamoto M. The Tohoku Medical Megabank Project: Design and Mission. J Epidemiol, 2016; 26: 493-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Akter S, Goto A, Mizoue T. Smoking and the risk of type 2 diabetes in Japan: A systematic review and meta-analysis. J Epidemiol, 2017; 27: 553-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Smith AD, Crippa A, Woodcock J, Brage S. Physical activity and incident type 2 diabetes mellitus: a systematic review and dose-response meta-analysis of prospective cohort studies. Diabetologia, 2016; 59: 2527-2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Itabashi F, Hirata T, Kogure M, Narita A, Tsuchiya N, Nakamura T, Nakaya N, Sasaki R, Takanashi N, Sakata K, Tanno K, Sugawara J, Kuriyama S, Tsuji I, Kure S, Hozawa A. Combined Associations of Liver Enzymes and Obesity With Diabetes Mellitus Prevalence: The Tohoku Medical Megabank Community-based Cohort Study. J Epidemiol, 2022; 32: 221-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Hozawa A, Okamura T, Tanaka T, Miura K, Kikuchi Y, Kadowaki T, Yoshita K, Takebayashi T, Tamaki J, Minai J, Tada T, Chiba N, Okayama A, Ueshima H. Relation of Gamma-glutamyltransferase and alcohol drinking with incident diabetes: the HIPOP-OHP study. Relation of Gamma-glutamyltransferase and alcohol drinking with incident diabetes: the HIPOP-OHP study. J Atheroscler Thromb, 2010; 17: 195-202 [DOI] [PubMed] [Google Scholar]
- 27).WHO/IASO/IOTF. The Asia-Pacific perspective: redefining obesity and its treatment. Australia: Health Communications Australia Pty Ltd. 2000 [Google Scholar]
- 28).Hamilton CM, Strader LC, Pratt JG, Maiese D, Hendershot T, Kwok RK, Hammond JA, Huggins W, Jackman D, Pan H, Nettles DS, Beaty TH, Farrer LA, Kraft P, Marazita ML, Ordovas JM, Pato CN, Spitz MR, Wagener D, Williams M, Junkins HA, Harlan WR, Ramos EM, Haines J. The PhenX Toolkit: get the most from your measures. Am J Epidemiol, 2011; 174: 253-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Fujii H, Yamamoto S, Takeda-Imai F, Inoue M, Tsugane S, Kadowaki T, Noda M. Validity and applicability of a simple questionnaire for the estimation of total and domain-specific physical activity. Diabetology international, 2011: 2: 47-54 [Google Scholar]
- 30).Kikuchi H, Inoue S, Odagiri Y, Ihira H, Inoue M, Sawada N, Noda M, Tsugane S. Intensity-specific validity and reliability of the Japan Public Health Center-based prospective study-physical activity questionnaire. Prev Med Rep, 2020; 20: 101169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).World Health Organization. Physical activity. Available from https: //www.who.int/news-room/fact-sheets/detail/physical-activity. Accessed March 14, 2023 [Google Scholar]
- 32).Teschke R, Brand A, Strohmeyer G. Induction of hepatic microsomal gamma-glutamyltransferase activity following chronic alcohol consumption. Biochem Biophys Res Commun, 1977; 75(3): 718-724 [DOI] [PubMed] [Google Scholar]
- 33).Coku V, Shkembi X. Serum Gamma-glutamyltransferase and Obesity: is there a Link? Med Arch, 2018; 72(2): 112-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Lee DH, Silventoinen K, Jacobs DR Jr, Jousilahti P, Tuomileto J. gamma-Glutamyltransferase, obesity, and the risk of type 2 diabetes: observational cohort study among 20,158 middle-aged men and women. J Clin Endocrinol Metab, 2004; 89(11): 5410-5414 [DOI] [PubMed] [Google Scholar]
- 35).Yamada M, Motoike IN, Kojima K, Fuse N, Hozawa A, Kuriyama S, Katsuoka F, Tadaka S, Shirota M, Sakurai M, Nakamura T, Hamanaka Y, Suzuki K, Sugawara J, Ogishima S, Uruno A, Kodama EN, Fujino N, Numakura T, Ichikawa T, Mitsune A, Ohe T, Kinoshita K, Ichinose M, Sugiura H, Yamamoto M. Genetic loci for lung function in Japanese adults with adjustment for exhaled nitric oxide levels as airway inflammation indicator. Commun Biol, 2021; 4: 1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Schnurr TM, Jakupović H, Carrasquilla GD, Ängquist L, Grarup N, Sørensen TIA, Tjønneland A, Overvad K, Pedersen O, Hansen T, Kilpeläinen TO. Obesity, unfavourable lifestyle and genetic risk of type 2 diabetes: a case-cohort study. Diabetologia, 2020; 63: 1324-1332 [DOI] [PubMed] [Google Scholar]
- 37).Merino J, Guasch-Ferré M, Li J, Chung W, Hu Y, Ma B, Li Y, Kang JH, Kraft P, Liang L, Sun Q, Franks PW, Manson JE, Willet WC, Florez JC, Hu FB. Polygenic scores, diet quality, and type 2 diabetes risk: An observational study among 35,759 adults from 3 US cohorts. PLoS Med, 2022; 19: e1003972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Kodama K, Tojjar D, Yamada S, Toda K, Patel CJ, Butte AJ. Ethnic differences in the relationship between insulin sensitivity and insulin response: a systematic review and meta-analysis. Diabetes Care, 2013; 36: 1789-1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Tsugane S. Why has Japan become the world's most long-lived country: insights from a food and nutrition perspective. Eur J Clin Nutr, 2021; 75: 921-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Gim J, Kim W, Kwak SH, Choi H, Park C, Park KS, Kwon S, Park T, Won S. Improving disease prediction by incorporating family disease history in risk prediction models with large-scale genetic data. Genetics, 2017; 207: 1147-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Duschek E, Forer L, Schönherr S, Gieger C, Peters A, Kronenberg F, Grallert H, Lamina C. A polygenic and family risk score are both independently associated with risk of type 2 diabetes in a population-based study. Sci Rep, 2023; 13: 4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet, 2019; 51: 584-591 [DOI] [PMC free article] [PubMed] [Google Scholar]


