Abstract
Brucellosis is a worldwide disease of humans and livestock that is caused by a number of very closely related classical Brucella species in the alpha-2 subdivision of the Proteobacteria. We report the complete genome sequence of Brucella abortus field isolate 9-941 and compare it to those of Brucella suis 1330 and Brucella melitensis 16 M. The genomes of these Brucella species are strikingly similar, with nearly identical genetic content and gene organization. However, a number of insertion-deletion events and several polymorphic regions encoding putative outer membrane proteins were identified among the genomes. Several fragments previously identified as unique to either B. suis or B. melitensis were present in the B. abortus genome. Even though several fragments were shared between only B. abortus and B. suis, B. abortus shared more fragments and had fewer nucleotide polymorphisms with B. melitensis than B. suis. The complete genomic sequence of B. abortus provides an important resource for further investigations into determinants of the pathogenicity and virulence phenotypes of these bacteria.
Brucellosis is a bacterial disease of animals that can be transmitted to humans. The primary impact of brucellosis stems from losses due to reproductive failure in food animals and the loss of human productivity. Since brucellosis threatens the food supply and causes undulant fever, a long, debilitating disease in humans, Brucella species are recognized as potential agricultural, civilian, and military bioterrorism agents. Brucellosis in food animals is controlled by vaccination. Human brucellosis is treatable with antibiotics, though the course of antibiotic treatment must be prolonged due to the intracellular nature of Brucella.
Analysis of 16S rRNA sequences places Brucella spp. as members of the alpha-2 Proteobacteria (31). The genus Brucella has six recognized species, all of which exhibit distinct host preferences (25, 26). The high degree of similarity among the brucellae (1, 3, 13, 33) lends support to the proposal that the classical species of Brucella are actually strains of Brucella melitensis (40). However, this view conflicts with the hypothesized evolutionary isolation of these classical species due to their intracellular existence and host preference (29). Common host-pathogen associations among the classical Brucella species are as follows: B. abortus, cattle; B. suis, swine; B. melitensis, goats; B. ovis, sheep; B. canis, dogs; B. neotomae, desert wood rats. Although these host-pathogen associations represent the norm in nature, cross-species infections do occur. Recently, brucellae have also been isolated from marine mammals (11, 34). Brucellae may be more widespread than previously recognized.
Brucellae can be rapidly identified by PCR assays, such as those based on the insertion sequence IS711 (6, 7). Though the sequences of brucellae are very similar, biovars of some of the classical species were differentiated by DNA sequence determination of several outer membrane proteins (OMPs) (8, 12, 41). Strains of B. abortus biovar 1 were distinguishable by analysis of a multilocus variable nucleotide tandem repeat (4).
Pulsed-field gel electrophoresis (PFGE) maps of the classical Brucella spp. genomes are composed of two circular chromosomes of approximately 2.1 and 1.2 Mbp (22, 27, 28), with the exception of B. suis biovar 3, which has a single chromosome of 3.1 Mbp. PFGE studies revealed other differences, including a 640-kb inversion in the small chromosome of B. abortus 544 and a deletion in the small chromosome of B. ovis. The two chromosomes of brucellae differ in important ways (33). The origin of replication of the large chromosome (Chr I) is typical of bacterial chromosomes, while that of the small chromosome (Chr II) is plasmid like. Further, most of the essential genes are located on Chr I. The G+C content of the two chromosomes is nearly identical, consistent with the assertion that the assimilation and stabilization of a plasmid was an ancient event (33) in brucellae.
The genome sequences of B. melitensis and B. suis have been determined (10, 33). Comparative analyses revealed both that the two genomes are extremely similar and that they have many similarities to both bacterial plant and animal pathogens and symbionts (33, 38). The sequence identity for most open reading frames (ORFs) was 99% or higher. Nevertheless, unique fragments were reported to exist between these two genomes (33). Prior to sequencing the B. abortus genome, a large number of short sequences were available in GenBank. Many of these sequences were derived from analyses of plasmids estimated to cover 20% of the genome from a random shotgun library of B. abortus S2308 (36). In this study, we present the completed B. abortus genome sequence and compare it to the genomes of B. melitensis and B. suis. Taken together, the genome sequences of these classical Brucella species provide a firm foundation for further research into the genetic bases for host preference, pathogenesis, virulence, and biotype differences.
MATERIALS AND METHODS
Strain information.
B. abortus strain 9-941 was obtained from the National Animal Disease Center culture collection. It was originally isolated from a serologically detected, infected cattle herd in northwestern Wyoming. The isolate was identified as B. abortus biovar 1 by the National Veterinary Services Laboratory based on morphology, bacteriologic characteristics, and phage typing. The isolate is nonmotile, nonhemolytic, A-antigen dominant, catalase positive, oxidase positive, urease positive at 3.5 h, nitrate reduction positive, citrate utilization negative, H2S production positive after 2 days of incubation at 37°C, sensitive to thionin dye (1:25,000), and resistant to basic fuchsin, thionin blue (1:500,000), penicillin, and erythritol.
Purification of genomic DNA.
Total genomic DNA was purified from B. abortus strain 9-941 by a modification of a previously described method (17). Bacteria were harvested from agar plates in saline and killed by the addition of two volumes of methanol. Approximately 1010 bacteria were pelleted, washed in TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0), and treated with 0.5% Zwittergent 3-14 in TE buffered with citrate at pH 4.0 for 1 h at 50°C. The treated bacteria were washed in TE, lysed in a solution containing 4% sarcosine, 0.5% sodium dodecyl sulfate, 125 mg of proteinase K/ml, 10 mM EDTA, and 20 mM Tris (pH 8.0) for 20 min at 65°C, and the lysate was treated with RNase A. The DNA was precipitated in ethanol, removed by spooling, and resuspended in DNAzol (catalog no. 10503-027; Life Technologies, Grand Island, N.Y.). The DNA was precipitated with ethanol a second time, dissolved in 8 mM sodium hydroxide, and adjusted to pH 7.4 with 10 mM HEPES for storage at 4°C.
Library construction.
A random 2-kb insert library of B. abortus 9-941 was constructed by shearing whole genomic DNA using a nebulizer and compressed nitrogen according to protocols developed by Bruce Roe's laboratory and posted at The University of Oklahoma's Advanced Center for Genome Technology website (http://www.genome.ou.edu). The sheared DNA fragments were separated by gel electrophoresis, and fragments of 2 to 3 kb were excised from the gel and purified. The ends of the purified fragments were polished by the addition of nucleotides and Klenow fragment (New England BioLabs, Beverly, Mass.) and ligated to a SmaI-restricted calf intestinal alkaline phosphatase-treated pUC18 vector for cloning by electroporation into Escherichia coli. The library, which consisted of >90% recombinant clones, was used to construct a culture collection for sequence determination.
Sequence determination.
Plasmid DNA was extracted using the QIAprep 96 Turbo kit (QIAGEN, Santa Clarita, Calif.), quantitated using PicoGreen (dsDNA quantitation kit; Molecular Probes, Eugene, Oreg.), and labeled (DyeDeoxy Terminator cycle sequencing kit; ABI automated DNA sequencing chemistry guide, ABI, Foster City, Calif.) for sequencing in the presence of dimethyl sulfoxide. The sequence was determined (ABI Prism 3700 DNA analyzer) and assembled using Phred/Phrap/Consed software obtained from the University of Washington Genome Center (http://www.genome.washington.edu/UWGC/) and the MacVector 7.0 DNA analysis package (Accelrys Inc., San Diego, Calif.). Contigs were linked and gaps were filled by predicting linkages based on putative colinearity of sequences with B. suis. Linkages were confirmed by amplifying and sequencing genomic DNA from B. abortus 9-941. The genome sequence was derived from 37,718 plasmids, and coverage was 10-fold. Mean Phrap quality score was 86. The confirmed mean read length was 819 bp.
Annotation.
Artemis (releases 4 and 5; The Sanger Centre [www.sanger.ac.uk/software/ACT/]) (35) was used to identify putative genes by determining which of the ORFs with 50 or more amino acids (aa) encoded homologs in GenBank searches using BLASTP (2). B. abortus ORF annotations were modeled after those of the homologous ORFs in GenBank, especially those of the B. suis genome. B. abortus ORFs that were truncated due to premature stops or had frameshifts compared to homologs from B. suis or other entries in GenBank were designated pseudogenes. Pseudogenes of B. suis were obtained from the GenBank accession numbers for the two chromosomes of B. suis 1330. For B. melitensis, pseudogenes were identified by visual inspection of BLASTP searches of GenBank with Artemis and by cross-comparisons of the three genomes' DNA sequences using Act version 2 (The Sanger Centre).
Identification of differential ORFs.
ORFs that differed in length among the Brucella genomes due to frameshifts or premature stops were labeled as differential ORFs. ORFs were not categorized as differential if their lengths differed solely due to selection of alternative start codons during the annotation processes.
Transposon search.
ORFs from The Institute for Genomic Research transposon role category database of the comprehensive microbial resource website (http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl) for B. suis were used to identify shared transposon-related sequences among B. melitensis and B. abortus by use of MacVector 7.0.
Unique genes.
Putative genes were designated as unique if no homolog was identified by aligning ORFs from the three Brucella genomes with each other (BLASTP version 2.2.6, -e 0.01, -F F) and to GenBank. Products of unique genes and ORFs were denoted as hypothetical proteins.
SNPs.
Single nucleotide polymorphisms (SNPs) were identified using pairwise chromosome alignment data generated with the MUMmer 3.0 run-mummer3 script (24). No minimum separation distance restrictions were placed on neighboring SNPs. SNP totals were used as a measure of genetic distance for the neighbor-joining tree [Ps = (ΣSNP count/1,000)] construction with MEGA2 (23). Mesorhizobium loti (GenBank accession numbers NC_002678, pA:NC_002679, and pB:NC_002682) was used as an outgroup for rooting the tree.
Nucleotide sequence accession numbers.
The GenBank accession numbers for B. abortus 9-941 Chr I and Chr II are AE017223 and AE017224, respectively. The genome accession numbers used for B. suis 1330 were AE014291 and AE014292, and for B. melitensis M16 they were NC_003317 and NC_003318.
Raw data are accessible at http://patric.vbi.vt.edu/. Note that a later annotation of the B. melitensis genome sequence is available at the website http://serine.urbm.fundp.ac.be/∼seqbruce/GENOMES/.
RESULTS AND DISCUSSION
General features of the genome.
The whole genome sequence of a B. abortus biovar 1 field isolate was determined by the shotgun method. The genome is 3.3 Mb and is composed of two circular chromosomes of 2,124,242 (Chr I) and 1,162,780 bp (Chr II). The chromosome sequences of B. abortus 9-941 were assigned the same strand orientation and origin as those of B. suis. The G+C contents of Chr I and Chr II are 57.2 and 57.3%, respectively. This is identical to that found for the two chromosomes of B. suis (33) and is in agreement with that of B. melitensis (10). It is consistent with that determined in early hybridization studies (20, 21). The B. abortus genome contains 3,296 ORFs annotated as genes, 2,158 on Chr I and 1,138 on Chr II. This is similar to the annotated ORF counts for B. suis (3,388) and B. melitensis (3,197). Differences in the number of ORFs found among the three Brucella genomes derived primarily from differences in annotation of short ORFs and from large insertions-deletions (indels). Similarity among the Brucella orthologs was high, with an average amino acid sequence identity of greater than 99%. The B. abortus sequence confirmed PFGE maps (27) with regard to genome size, chromosome number, and presence of an inversion described in Chr II relative to other genomes.
Many of the annotation differences among the genomes are related to small ORFs. The number and annotation of short hypothetical ORFs of 100 aa or less are similar between B. suis and B. abortus. The annotated genome of B. melitensis has fewer short ORFs than B. abortus and B. suis. A total of 551 ORFs of less than 100 aa are annotated in B. abortus, while in B. melitensis there are only 304. The disparity in the number of small ORFs is larger when those less than 50 aa are considered. While 161 of the 551 short ORFs in B. abortus are less than 50 aa, only 11 of the 304 short ORFs in B. melitensis are. Functional assays such as microarrays and proteomic studies will be necessary to identify genes and their products.
Occurrence of fragments in B. abortus not shared between B. suis and B. melitensis.
Sequences larger than 100 bp that were previously reported to be unique in either B. suis or B. melitensis (see Table 1 in reference 33) were aligned to the B. abortus genome to determine their presence or absence (Tables 1 and 2). Many of these sequences had been shown to be related to mobile genetic elements, while others had not. Table 1 lists loci of fragments related to phages, transposable elements, or plasmids. The loci of the remaining fragments are in Table 2. Many of the fragments that were unique between B. suis and B. melitensis were found in B. abortus. The genome of B. abortus shared more fragments with B. suis and B. melitensis than B. suis and B. melitensis did with each other. B. abortus shared more fragments with B. melitensis than B. suis. This agrees with other analyses that showed B. abortus and B. melitensis being more closely related than B. abortus and B. suis (22, 27, 28).
TABLE 1.
Occurrence in B. abortus of fragments unique to either B. suis or B. melitensis that have homology to phage, transposable elements, or plasmids
| Size (bp) | Locus (intact or partial)a | Chr | Presence in:
|
||
|---|---|---|---|---|---|
| B. suis | B. melitensis | B. abortus | |||
| 16,150 | BR0512-BR0536, Tn and IS region | I | + | + | + |
| 3952 | BR1852-BR1854; Tn2020 related | I | + | − | + |
| 3538 | BR0588-BR0593; phage related | I | + | − | + |
| 842 | BR1672-BR1673, IS711 | I | + | − | − |
| 162 | BR0262Δ, putative phage fragment | I | + | − | − |
| 162 | BR1080Δ, putative phage fragment | I | + | − | − |
| 842 | BMEI1163-BME1164, IS711 | I | − | + | − |
| 20,883 | BMEI1674-BMEI1702/BMEI1703 | I | − | + | + |
| 238 | BMEI0904Δ, putative phage fragment | I | − | + | + |
| 238 | BMEI1659, putative phage fragment | I | − | + | + |
| 276 | BMEI0899, phage-related DNA binding protein | I | − | + | + |
| 18,290 | BRA0362-BRA0379, includes tra genes | II | + | − | − |
| 25,064 | BRA0419-BRA0439/BRA0418, BRA0439 | II | + | + | − |
| 48,245 | BRA1073-BRA1116/BRA1072& | II | + | + | + |
| 189 | BRA1080, dipeptide ABC transporter-permease& | + | − | − | |
| 231 | BRA1096, transcriptional regulator& | + | − | + | |
| 6851 | BRA0551-BRA0559, Tn and IS region | II | + | + | ±b |
| 842 | BruAb2_0692-BruAb2_0691, IS711 | II | − | − | + |
BR and BRA numbers designate loci in B. suis, and BMEI or BMEII numbers designate loci in B. melitensis. Δ, intergenic region; &, within Tn1953 (acc. no. AF454951).
±, partial presence (see deletion section).
TABLE 2.
Occurrence in B. abortus of regions unique between B. suis or B. melitensis without homology to phage, transposable elements, or plasmids
| Region | Chr | Size (bp) | Presence ina:
|
Putative function | ||
|---|---|---|---|---|---|---|
| S | M | A | ||||
| BR1895 | I | 111 | + | − | + | Cell division protein, FtsK |
| BR1622 | I | 232 | + | − | + | OMP31-like outer membrane protein |
| BR1182-1183 | I | 149 | + | − | + | Putative intergenic region |
| BR1059-1060 | I | 780 | + | − | − | HlyD family protein, multidrug resistance transporter |
| BR0951-0955 | I | 2,653 | + | − | − | Amino acid ABC transporter-binding protein, hypothetical, ABC transporter-permease, putative GST |
| BR0404 | I | 113 | + | − | + | Glycyl-tRNA synthetase, beta subunit |
| BR0389-0391 | I | 425 | + | − | + | Hypothetical |
| BR0355 | I | 135 | + | − | + | Hydroxypyruvate reductase |
| BR0221 | I | 174 | + | − | + | Transcriptional regulator |
| BMEI0217-0218 | I | 107 | − | + | + | Putative intergenic region |
| BMEI0801 | I | 764 | − | + | + | Propionyl-coenzymeA carboxylase beta chain |
| BMEI1554-1555 | I | 105 | − | + | + | Transporter, MFS superfamily |
| BMEI1742 | I | 194 | − | + | + | ABC transporter ATP-binding protein |
| BMEI1873 | I | 339 | − | + | + | Cell surface protein |
| BMEI1659 | I | 240 | − | + | + | Hypothetical |
| BRA0173 | II | 528 | + | − | + | Polymorphic, OMP |
| BRA0541-0542 | II | 969 | + | − | + | Hypothetical, NAD-dependent epimerase family |
| BRA0630-0636 | II | 7,738 | + | − | + | Oxidoreductase, amino acid ABC transporter-binding protein, transcriptional regulator, B-ketoadipyl coenzyme A thiolase, conserved hypothetical |
| BRA0749-0750 | II | 581 | + | − | + | ABC transporter-permease |
| BRA0907 | II | 699 | + | − | + | Conserved hypothetical |
| BMEII0466 | II | 108 | − | + | − | Tetratricopeptide repeat family |
S, B. suis; M, B. melitensis; A, B. abortus.
Large fragments shared by B. suis and B. melitensis but missing in B. abortus.
Two fragments shared by B. suis and B. melitensis were not found in B. abortus. A 2,774-bp fragment encoding a probable surface protein and two partial ORFs with homology to the insertion sequences IS711 and ISBm1 is missing from B. abortus. The probable surface protein is annotated in B. suis as a cell wall surface protein (BRA0553) and in B. melitensis as a hemagglutinin (BMEII0717). Though they are highly similar, they differ slightly in length. The second fragment is a 25-kb sequence that had previously been identified as missing in B. abortus 544, a biovar 1 strain and the type species (42). This sequence was shown to encode a number of ORFs that may be involved in polysaccharide synthesis and was predicted to potentially affect phenotypes of brucellae, such as host preference (42).
Regions containing sequences specific to B. abortus.
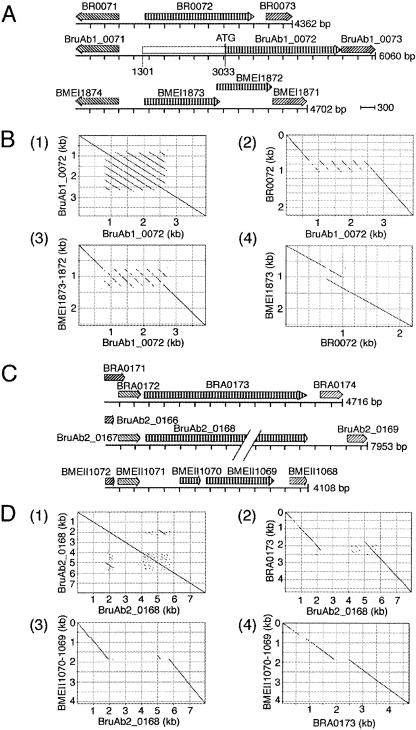
The loci BruAb1_0072 and BruAb2_0168 (Fig. 1A and C) have sequences specific to B. abortus (Fig. 1B and D) relative to B. suis and B. melitensis and contain sequences that are repeated. There are eight copies of a 250-bp sequence in the 2-kb region and two copies of a 500-bp sequence in the 4-kb region. These sequences are not homologous.
FIG. 1.
Comparison of region containing B. abortus-specific BruAb1_0072 and BruAb2_0168 to homologs in B. suis and B. melitensis. (A) Representation of ORFs from B. suis (top line), B. abortus (middle line), and B. melitensis (bottom line) in the region with homology to BruAb1_0072. Arrows show direction and extent of the ORF, and hatching shows the ORF annotation. Like patterns of hatching indicate homologs. (B) Pustell (MacVector 7.2) comparisons of BruAb1_0072 and contiguous upstream ORF to same (1); BruAb1_0072 and contiguous upstream to ORF BR0072 (2); BruAb1_0072 and contiguous upstream ORF to BMEI1873 and BMEI1872 (3); and BR0072 to BMEI1873 and BMEI1872 (4). The window size for Pustell comparisons was 20 and minimum score is 90, with a hash value of 6 and jump of 1. (C) Representation of ORFs from B. suis (top line), B. abortus (middle line), and B. melitensis (bottom line) in the region containing homology to the locus BruAb2_0168. Arrows show directions of ORFs, and hatching shows annotation of ORFs. Like patterns of hatching indicate homologs. (D) Pustell comparisons of regions represented in panel C: BruAb2_0168 to same (1); BruAb2_0168 to BR0173 (2); BruAb2_0168 to BMEII1070 and BMEII1069 (3); and BR0173 to BMEII1070 and BMEII1069 (4). The window size for Pustell comparisons was 20 and minimum score is 90, with a hash value of 6 and jump of 1.
The 2-kb region is in Chr I and encodes a similar-size putative OMP in B. abortus and B. suis, BruAb1_0072 (756 aa) and BR0072 (740 aa), respectively. Though there is an ORF of over 4,000 bp in B. abortus containing a number of possible start codons, the start codon selected for BruAb1_0072 is near a putative ribosome binding site that is more than 1,700 bp from the start of the 4,000-bp ORF (Fig. 1A). The selected start codon produces a product that is similar in size to BR0072 (Fig. 1A). In B. melitensis, there are two ORFs, BMEI1873 (366 aa) and BMEI1872 (506 aa); however, due to a frameshift in B. melitensis relative to B. suis, they appear to be pseudogenes (Fig. 1A). In the 2-kb region in B. abortus, there are eight highly similar copies of a 250-bp sequence that occur as direct tandem repeats (Fig. 1B, graph 1). The region containing the eight repeats is only found in B. abortus. In B. suis, there is a single copy of a sequence similar to the 250-bp repeat in B. abortus (Fig. 1B, graph 2). In B. melitensis, there are three direct copies of a sequence similar to the repeated 250-bp sequence in B. abortus (Fig. 1B, graph 3). The repeated sequence in B. melitensis is more similar to that in B. suis than to that in B. abortus (Fig. 1B, graphs 2 to 4).
The 4-kb region is on Chr II and encodes an autotransporter in B. abortus and B. suis, BruAb2_0168 (1,983 aa) and BRA0173 (1,113 aa), respectively (Fig. 1C). There were several possible start codons in the 6,062-bp ORF in B. abortus with homology to BRA0173. The start codon selected generating BruAb2_0168 was near the beginning of the large ORF and near a putative ribosome binding site. Experimental studies will be necessary to establish if this is an authentic start codon. In B. melitensis, a frameshift relative to the other genomes results in two relatively short ORFs, BMEII1069 (488 aa) and BMEII1070 (114 aa) (Fig. 1C). As neither of these ORFs encodes the domains of autotransporters (19), they appear to be pseudogenes. The B. abortus sequence has two direct 500-bp repeats separated by approximately 2,750 bp of sequence; the 2,750-bp sequence was B. abortus specific (Fig. 1D, graphs 1 to 3). There is a single copy of the B. abortus 500-bp repeat in B. suis (Fig. 1D, graph 2), while in B. melitensis there is only a partial copy (180 bp) (Fig. 1D, graph 3). The amino termini of BruAb2_0168 and BMEII1070 are more similar to each other than either is to the amino termini of BRA1073 (Fig. 1D, graphs 2 to 4).
The two regions encoded OMPs that were previously suggested as potentially affecting the pathogenicity or host preference of the brucellae (30).
Inversions.
Sequences surrounding and at the site of the large inversion previously described in Chr II of B. abortus 544 relative to the other Brucella (27) were analyzed in B. abortus. The inversion disrupted B. abortus homologs of ORFs BRA1003 and BRA0235 of B. suis, resulting in four pseudogenes (BruAb2_0230, BruAb2_0231, BruAb2_0943, and BruAb2_0944). The ORFs BRA1003 and BRA0235 encode a putative GAF/GGDEF prokaryotic signaling domain protein and a hypothetical protein, respectively. The sites disrupted in the large inversion relative to B. melitensis are between ORFs BMEII0292 and BMEII0293 and within BMEII1009, a homolog of BRA1003. In B. abortus, a short distance downstream of the large inversion there is an indel of 838 bp. This affected a locus with homology to B. suis ORFs BRA1004 and BRA1005 and resulted in the pseudogene BruAb2_0945 in B. abortus. The finding of a single copy of the sequence 5′-CCA-GCA-CCG-CCT-GC-3′ (bp 949172 to bp 949185) in B. abortus and two copies in both B. suis and B. melitensis separated by 810 bp is consistent with the indel in B. abortus arising from either homologous recombination or slipped-strand mispairing during replication. The inversion site and the 838-bp indel were described recently in B. abortus 2308 (37). The large inversion in the small chromosome is not found in all biovars of B. abortus (27). Though it was found in biovars 1, 2, 3, and 4 by PFGE, we detected it by PCR (15) only in biovars found in the United States, biovars 1, 2, and 4 (1).
Two small inversions were found in B. abortus. An inversion of 2,185 bp that was unique to B. abortus is near a 780-bp indel unique to B. suis (Table 2). This inversion occurs in a homolog of the B. suis proline dipeptidase BR1062, creating pseudogenes BruAb1_1065 and BruAb1_1067. A second smaller inversion of 2,150 bp was found in both B. abortus and B. melitensis, disrupting B. suis homologs BRA0485 and BRA0487 in these genomes. These ORFs encode a putative protein and a glycosyl transferase family 25 protein, respectively. The glycosyl transferase family 25 protein could possibly affect lipopolysaccharide structure.
Polymorphic regions encoding outer membrane proteins.
Two regions in the Brucella genomes encoding homologs of OMPs predicted to be virulence associated in Brucella (30, 33) were found to have greater sequence variation than that calculated for the genomes as a whole. One of the regions encoded a putative bacterial immunoglobulin-like protein with a group 1 domain (PFAM protein family PF02369) common to bacterial surface proteins invasins and adhesins. The sequence variation affected the sizes of the homologs among the three genomes and shifted the ORF in the carboxy end in B. abortus. The OMP in B. suis, BR2009, is 500 aa, while the OMPs in B. melitensis and B. abortus, BMEI0063 and BruAb1_1984, respectively, are less than 400 aa. The sequence variation resulted in the truncation of the amino terminus of BMEI0063 relative to BR2009 and the carboxy terminus of BruAb1_1984 relative to BR2009. BR2009 and BMEI0063 have proline-rich regions in their carboxy ends. In the proline-rich stretches, 21 of 25 aa are proline in B. suis and 20 of 25 aa are proline in B. melitensis. Due to a frameshift and sequence differences in the carboxy end of BruAb1_1984 from B. abortus, there is a leucine- and histidine-rich region rather than a proline one. Only a few proline- and leucine-rich regions were found by BLASTP, and these are in disparate proteins. These adhesins may affect pathogenicity and host preference (30, 33).
The second variable region encodes an autotransporter in B. suis, BR2013, and putative pseudogenes in B. melitensis and B. abortus. This region had two in-frame stop codons in B. abortus relative to B. suis and is annotated as a pseudogene (BruAb1_1988). In B. melitensis, the homolog of BR2013 (BMEI0058) appeared to be a pseudogene also, due to an in-frame stop codon. Among the three genomes, only the B. suis locus encoded all the functional domains of an autotransporter, which may represent a unique virulence factor of B. suis.
Variable sizes of ORFs.
All ORFs that varied in size among the three genomes were compiled (Tables 3 and 4). If the differences in sizes of ORFs resulted solely from selection of an alternative start codon in the annotation process, the ORFs were not labeled as being variable. The genome of B. suis was used as the reference for determining which ORFs were variable, because the annotation of its ORFs was more similar with annotation of protein homologs identified by BLASTP searches than the annotation of B. melitensis. There were almost as many variable ORFs on the large chromosomes as there were on the smaller chromosomes, even though the large chromosome has approximately twice the number of ORFs as the small chromosome. As the larger chromosome has many of the genes that encode core metabolic functions of the bacterium (33), mutations in the large chromosome may be selected against or lethal. Furthermore, the small chromosome, which appears to be a stabilized megaplasmid, may have more genes that were acquired horizontally. These genes may not be essential and, thus, might not be under positive selective pressure.
TABLE 3.
Variable ORFs among B. suis, B. abortus, and B. melitensis: Chr I (BR)
| Locus in B. suis | ORF ina:
|
Locus in B. suis | ORF ina:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. suis | B. melitensis | B. abortus | Categoryb | B. suis | B. melitensis | B. abortus | Categoryb | |||
| 1. 0014 | − | − | + | 13 | ||||||
| 2. 0017 | + | − | − | 7 | ||||||
| 3. 0021 | − | + | + | 5 | ||||||
| 4. 0030 | − | + | + | 3 | ||||||
| 5. 0043 | − | − | + | 7 | ||||||
| 6. 0072 | − | + | − | 4 | ||||||
| 7. 0083 | − | − | + | 9 | ||||||
| 8. 0089 | − | − | + | 4 | ||||||
| 9. 0130 | + | − | − | 7 | ||||||
| 10. 0146 | + | − | − | 7 | ||||||
| 11. 0172 | P | P | P | 11 | ||||||
| 12. 0174 | − | + | + | 15 | ||||||
| 13. 0197 | + | − | − | 11 | ||||||
| 14. 0201 | + | − | − | 11 | ||||||
| 15. 0202 | − | + | + | 7 | ||||||
| 16. 0205 | + | − | − | 11 | ||||||
| 17. 0206 | + | + | + | 18 | ||||||
| 18. 0231 | − | − | + | 7 | ||||||
| 19. 0258 | − | − | + | 15 | ||||||
| 20. 0278 | − | − | + | 10 | ||||||
| 21. 0279 | − | − | + | 5 | ||||||
| 22. 0281 | − | − | + | 6 | ||||||
| 23. 0339 | − | − | + | 4 | ||||||
| 24. 0367 | − | + | + | 7 | ||||||
| 25. 0396-0397 | − | + | + | 9-13 | ||||||
| 26. 0423 | + | − | − | 17 | ||||||
| 27. 0469 | − | − | = | 2 | ||||||
| 28. 0471 | − | − | + | 7 | ||||||
| 29. 0498 | − | − | + | 4 | ||||||
| 30. 0512 | + | ‡ | + | 18 | ||||||
| 31. 0514 | + | ‡ | + | 18 | ||||||
| 32. 0516 | + | ‡ | + | 14 | ||||||
| 33. 0525 | + | ‡ | + | 18 | ||||||
| 34. 0532 | + | ‡ | + | 18 | ||||||
| 35. 0535 | + | ‡ | + | 18 | ||||||
| 36. 0536 | + | ‡ | + | 18 | ||||||
| 37. 0550 | − | ‡ | + | 11 | ||||||
| 38. 0560 | − | ‡ | + | 11 | ||||||
| 39. 0561 | − | ‡ | + | 13 | ||||||
| 40. 0578 | − | = | = | 15 | ||||||
| 41. 0661 | − | − | = | 12 | ||||||
| 42. 0730 | − | + | + | 14 | ||||||
| 43. 0741 | − | − | + | 13 | ||||||
| 44. 0743 | − | − | + | 13 | ||||||
| 45. 0744 | − | = | + | 13 | ||||||
| 46. 0858 | − | − | = | 7 | ||||||
| 47. 0862 | + | − | − | 13 | ||||||
| 48. 0870 | − | − | + | 15 | ||||||
| 49. 0917 | + | − | − | 9 | ||||||
| 50. 0950 | − | − | + | 15 | ||||||
| 51. 0951 | − | + | + | 4 | ||||||
| 52. 0956 | − | + | + | 2 | ||||||
| 53. 0960 | − | − | + | 15 | ||||||
| 54. 0971 | − | − | + | 3 | ||||||
| 55. 0976 | − | + | + | 18 | ||||||
| 56. 0980 | − | + | + | 17 | ||||||
| 57. 1027 | + | ‡ | + | 7 | ||||||
| 58. 1046 | − | + | + | 15 | ||||||
| 59. 1051 | − | + | + | 1 | ||||||
| 60. 1059 | − | + | + | 4 | ||||||
| 61. 1060 | − | + | + | 13 | ||||||
| 62. 1061 | − | − | + | 7 | ||||||
| 63. 1062 | − | + | + | 9 | ||||||
| 64. 1081 | − | = | + | 17 | ||||||
| 65. 1083 | − | + | + | 15 | ||||||
| 66. 1084 | − | = | + | 15 | ||||||
| 67. 1086 | − | + | + | 11 | ||||||
| 68. 1122 | − | + | + | 15 | ||||||
| 69. 1171 | − | = | − | 17 | ||||||
| 70. 1175 | − | − | + | 7 | ||||||
| 71. 1188 | + | − | − | 18 | ||||||
| 72. 1255 | + | − | − | 15 | ||||||
| 73. 1289 | − | = | = | 2 | ||||||
| 74. 1291 | − | + | + | 15 | ||||||
| 75. 1292 | − | − | + | 15 | ||||||
| 76. 1313 | + | − | − | 8 | ||||||
| 77. 1320 | + | − | + | 15 | ||||||
| 78. 1322 | + | − | − | 3 | ||||||
| 79. 1340 | − | − | + | 13 | ||||||
| 80. 1350 | − | + | + | 13 | ||||||
| 81. 1359 | − | − | + | 5 | ||||||
| 82. 1401 | − | + | + | 4 | ||||||
| 83. 1448 | + | ‡ | − | 3 | ||||||
| 84. 1449 | + | − | − | 13 | ||||||
| 85. 1455 | − | + | + | 5 | ||||||
| 86. 1463 | ‡ | − | − | 13 | ||||||
| 87. 1512 | − | = | − | 6 | ||||||
| 88. 1544 | + | − | − | 13 | ||||||
| 89. 1572 | − | ‡ | + | 11 | ||||||
| 90. 1586 | + | + | + | 13 | ||||||
| 91. 1610 | − | − | + | 13 | ||||||
| 92. 1613 | − | + | + | 11 | ||||||
| 93. 1627 | + | ‡ | + | 15 | ||||||
| 94. 1636 | + | + | − | 18 | ||||||
| 95. 1670 | + | + | + | 13 | ||||||
| 96. 1718 | + | − | − | 11 | ||||||
| 97. 1740 | − | − | + | 13 | ||||||
| 98. 1770 | + | − | − | 11 | ||||||
| 99. 1788 | + | − | − | 13 | ||||||
| 100. 1789 | − | − | + | 13 | ||||||
| 101. 1793 | + | − | − | 7 | ||||||
| 102. 1804 | − | + | − | 6 | ||||||
| 103. 1829 | − | − | + | 5 | ||||||
| 104. 1873 | − | − | + | 3 | ||||||
| 105. 1894 | + | − | − | 11 | ||||||
| 106. 1908 | − | ‡ | + | 13 | ||||||
| 107. 1948 | − | − | = | 3 | ||||||
| 108. 1954 | + | − | + | 13 | ||||||
| 109. 1962 | − | ‡ | + | 15 | ||||||
| 110. 1967 | + | − | + | 1 | ||||||
| 111. 2009 | − | + | + | 4 | ||||||
| 112. 2013 | − | − | − | 3 | ||||||
| 113. 2046 | − | + | − | 8 | ||||||
| 114. 2047 | − | ‡ | + | 7 | ||||||
| 115. 2115 | − | − | + | 13 | ||||||
| 116. 2132 | − | + | + | 15 | ||||||
| Conserved hypothetical proteins | ||||||||||
| 117. 0047 | − | − | + | |||||||
| 118. 0128 | − | − | + | |||||||
| 119. 0131 | − | − | + | |||||||
| 120. 0470 | − | = | + | |||||||
| 121. 0473 | − | − | + | |||||||
| 122. 0573 | P | P | P | |||||||
| 123. 0592-0593 | ‡ | X | − | |||||||
| 124. 0599 | − | − | + | |||||||
| 125. 0601 | + | + | + | |||||||
| 126. 0695 | − | − | = | |||||||
| 127. 0713-0714 | − | − | +, F | |||||||
| 128. 0734 | − | − | + | |||||||
| 129. 0736 | − | = | + | |||||||
| 130. 0828 | − | + | + | |||||||
| 131. 0869 | − | − | + | |||||||
| 132. 0940 | − | − | + | |||||||
| 133. 0962 | − | − | + | |||||||
| 134. 1163 | − | − | + | |||||||
| 135. 1269 | − | = | + | |||||||
| 136. 1309 | − | = | = | |||||||
| 137. 1327 | − | + | + | |||||||
| 138. 1376 | + | − | − | |||||||
| 139. 1392 | − | = | − | |||||||
| 140. 1407 | − | − | + | |||||||
| 141. 1487 | − | + | + | |||||||
| 142. 1511 | ‡ | − | − | |||||||
| 143. 1620 | − | − | + | |||||||
| 144. 1669 | − | − | + | |||||||
| 145. 1732 | − | = | = | |||||||
| 146. 1882 | − | − | + | |||||||
| 147. 1909 | + | ‡ | + | |||||||
| 148. 2114 | − | + | + | |||||||
| 149. 2170 | − | + | + | |||||||
| Hypothetical proteins | ||||||||||
| 150. 0192 | − | + | + | |||||||
| 151. 0207 | − | + | + | |||||||
| 152. 0261 | − | + | + | |||||||
| 153. 0262 | − | + | + | |||||||
| 154. 0300 | − | + | + | |||||||
| 155. 0327 | − | − | + | |||||||
| 156. 0380 | − | + | + | |||||||
| 157. 0447 | − | = | − | |||||||
| 158. 0485 | S | S | S | |||||||
| 159. 0502 | S | S | S | |||||||
| 160. 0503 | P | P | P | |||||||
| 161. 0567 | − | − | = | |||||||
| 162. 0731 | − | + | + | |||||||
| 163. 0794 | − | − | + | |||||||
| 164. 0829 | S | S | S | |||||||
| 165. 0901 | S | S | S | |||||||
| 166. 0918 | S | S | S | |||||||
| 167. 0943 | − | + | + | |||||||
| 168. 0967 | − | + | + | |||||||
| 169. 0970 | − | + | + | |||||||
| 170. 0986 | − | − | + | |||||||
| 171. 0979 | − | − | + | |||||||
| 172. 1025 | S | S | S | |||||||
| 173. 1038 | − | ‡ | + | |||||||
| 174. 1080 | S | N | N | |||||||
| 175. 1101 | − | + | + | |||||||
| 176. 1113 | − | + | + | |||||||
| 177. 1180 | − | + | + | |||||||
| 178. 1290 | − | + | − | |||||||
| 179. 1353 | − | + | + | |||||||
| 180. 1382 | S | S | S | |||||||
| 181. 1459 | − | + | + | |||||||
| 182. 1496 | S | S | S | |||||||
| 183. 1552 | + | ‡ | + | |||||||
| 184. 1578 | S | S | S | |||||||
| 185. 1665 | − | − | + | |||||||
| 186. 1686 | − | + | + | |||||||
| 187. 1924 | S | S | S | |||||||
| 188. 1937 | − | − | + | |||||||
| 189. 1950 | − | + | + | |||||||
| 190. 1968 | − | − | + | |||||||
| 191. 2096 | − | − | + | |||||||
| 192. BMEI 1688 | X | − | = | |||||||
| 193. BMEI 1691 | X | − | + | |||||||
| 194. BMEI 1695 | X | − | = | |||||||
| 195. BMEI 1696 | X | − | + | |||||||
+ denotes ORF annotated as a pseudogene in B. suis or B. abortus genomic annotation or identified here as a pseudogene in B. melitensis; − denotes ORF not recognized as a pseudogene; ‡ denotes pseudogene as per annotation in this report; = denotes that annotation varies from GenBank but no pseudogene is recognized per comparison, different start codon used; P denotes polymorphic ORFs, some limited to amino terminus; N denotes polymorphic sequence or no ORF identified; X denotes locus not present; S denotes short ORFs (no comparisons made usually due to highly variable annotation among genomes); F denotes fused ORFs.
Functional categories: 1, amino acid biosynthesis; 2, biosynthesis of cofactors, prosthetic groups, and carriers; 3, cell envelope; 4, cellular processes; 5, central intermediary metabolism; 6, DNA metabolism; 7, energy metabolism; 8, fatty acid and phospholipid metabolism; 9, protein synthesis and fate; 10, purines, pyrimidines, nucleosides, and nucleotides; 11, regulatory functions and signal transduction; 12, transcription; 13, transport and binding proteins; 14, other categories; 15, unknown function; 16, hypothetical proteins; 17, no database match; 18, disrupted reading frame.
TABLE 4.
Variable ORFs among B. suis, B. abortus, and B. melitensis: Chr II (BRA)
| Locus in B. suis | ORF ina:
|
Locus in B. suis | ORF ina:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. suis | B. melitensis | B. abortus | Categoryb | B. suis | B. melitensis | B. abortus | Categoryb | |||
| 1. 0010 | − | + | − | 3 | ||||||
| 2. 0012 | − | − | + | 2 | ||||||
| 3. 0084 | + | − | − | 7 | ||||||
| 4. 0104 | − | = | + | 3 | ||||||
| 5. 0116 | − | − | + | 13 | ||||||
| 6. 0117 | + | − | + | 13 | ||||||
| 7. 0122 | + | ‡ | + | 4 | ||||||
| 8. 0125 | − | − | + | 4 | ||||||
| 9. 0149 | + | − | + | 7 | ||||||
| 10. 0156 | = | = | + | 4 | ||||||
| 11. 0173 | − | + | + | 3 | ||||||
| 12. 0190 | + | − | − | 13 | ||||||
| 13. 0206 | + | − | − | 15 | ||||||
| 14. 0208 | + | ‡ | + | 9 | ||||||
| 15. 0222 | − | − | + | 8 | ||||||
| 16. 0223 | − | + | + | 8 | ||||||
| 17. 0224 | − | = | + | 15 | ||||||
| 18. 0225 | + | − | − | 15 | ||||||
| 19. 0229 | + | − | − | 11 | ||||||
| 20. 0233 | − | − | + | 15 | ||||||
| 21. 0235 | − | − | + | 15 | ||||||
| 22. 0266 | − | − | + | 13 | ||||||
| 23. 0275 | − | + | − | 5 | ||||||
| 24. 0282 | = | − | − | 7 | ||||||
| 25. 0292 | − | − | + | 2 | ||||||
| 26. 0300 | − | − | + | 13 | ||||||
| 27. 0305 | − | + | + | 13 | ||||||
| 28. 0306 | − | + | + | 13 | ||||||
| 29. 0311 | − | + | + | 13 | ||||||
| 30. 0334 | − | + | + | 4 | ||||||
| 31. 0338 | + | ‡ | + | 7 | ||||||
| 32. 0339 | + | − | − | 13 | ||||||
| 33. 0340 | − | + | − | 7 | ||||||
| 34. 0346 | − | + | − | 11 | ||||||
| 35. 0353 | −, P | P | P | 7 | ||||||
| 36. 0393 | − | + | + | 13 | ||||||
| 37. 0394 | + | + | − | 13 | ||||||
| 38. 0407 | − | − | = | 13 | ||||||
| 39. 0408 | P | P | P, + | 13 | ||||||
| 40. 0412 | + | = | − | 3 | ||||||
| 41. 0418 | − | + | + | 3 | ||||||
| 42. 0438 | − | + | X | 15 | ||||||
| 43. 0442 | − | − | + | 13 | ||||||
| 44. 0443 | − | + | + | 7 | ||||||
| 45. 0444 | − | + | + | 13 | ||||||
| 46. 0449 | − | − | + | 8 | ||||||
| 47. 0450 | − | − | + | 7 | ||||||
| 48. 0464 | + | − | + | 13 | ||||||
| 49. 0470 | + | − | − | 11 | ||||||
| 50. 0471 | − | + | − | 13 | ||||||
| 51. 0487 | − | + | + | 3 | ||||||
| 52. 0516 | − | + | + | 13 | ||||||
| 53. 0527 | − | = | = | 7 | ||||||
| 54. 0528 | P | P | P | 15 | ||||||
| 55. 0535 | − | − | = | 13 | ||||||
| 56. 0544 | − | + | − | 3 | ||||||
| 57. 0547 | − | + | − | 15 | ||||||
| 58. 0550 | P | P | P | 11 | ||||||
| 59. 0551 | + | + | + | 14 | ||||||
| 60. 0554 | + | + | + | 18 | ||||||
| 61. 0559 | + | + | + | 18 | ||||||
| 62. 0569 | − | + | − | 13 | ||||||
| 63. 0571 | − | + | + | 7 | ||||||
| 64.0618 | − | − | + | 15 | ||||||
| 65. 0628 | − | = | + | 11 | ||||||
| 66. 0634 | − | X | + | 11 | ||||||
| 67. 0636 | − | X | + | 7 | ||||||
| 68. 0638 | − | + | − | 7 | ||||||
| 69. 0641 | − | − | + | 7 | ||||||
| 70. 0650 | − | + | + | 13 | ||||||
| 71. 0657 | − | + | − | 13 | ||||||
| 72. 0659 | P | − | − | 13 | ||||||
| 73. 0661 | − | + | − | 13 | ||||||
| 74. 0667-0668 | + | + | − | 1-17 | ||||||
| 75. 0687 | − | − | + | 13 | ||||||
| 76. 0690 | + | − | − | 13 | ||||||
| 77. 0691 | − | − | + | 13 | ||||||
| 78. 0696 | + | − | − | 7 | ||||||
| 79. 0702 | − | + | + | 15 | ||||||
| 80. 0713 | + | − | − | 7 | ||||||
| 81. 0716 | + | − | − | 15 | ||||||
| 82. 0722 | + | − | − | 7 | ||||||
| 83. 0729 | − | + | − | 15 | ||||||
| 84. 0740 | − | − | + | 13 | ||||||
| 85. 0760 | − | − | P | 3 | ||||||
| 86. 0776 | + | P | − | 13 | ||||||
| 87. 0780 | + | − | − | 7 | ||||||
| 88. 0783 | + | ‡ | + | 13 | ||||||
| 89. 0798 | + | + | + | 11 | ||||||
| 90. 0804 | − | − | + | 13 | ||||||
| 91. 0808 | + | + | − | 13 | ||||||
| 92. 0811 | + | − | − | 7 | ||||||
| 93. 0812 | − | + | + | 7 | ||||||
| 94. 0815 | + | − | + | 4 | ||||||
| 95. 0816 | − | − | + | 13 | ||||||
| 96. 0854 | + | − | − | 15 | ||||||
| 97. 0855 | − | − | + | 2 | ||||||
| 98. 0881 | + | = | − | 7 | ||||||
| 99. 0937 | − | ‡ | + | 13 | ||||||
| 100. 0944 | + | − | − | 11 | ||||||
| 101. 0979 | − | − | + | 15 | ||||||
| 102. 0982 | − | + | + | 15 | ||||||
| 103. 0985 | + | + | + | 11 | ||||||
| 104. 0996 | − | + | + | 13 | ||||||
| 105. 1007 | + | + | + | 9 | ||||||
| 106. 1033 | + | − | − | 9 | ||||||
| 107. 1083 | + | − | − | 7 | ||||||
| 108. 1085 | + | − | − | 7 | ||||||
| 109. 1089 | − | + | − | 4 | ||||||
| 110. 1090 | − | − | + | 13 | ||||||
| 111. 1093 | − | + | − | 13 | ||||||
| 112. 1095 | + | − | − | 13 | ||||||
| 113. 1100 | + | − | − | 13 | ||||||
| 114. 1104 | + | − | − | 13 | ||||||
| 115. 1107 | + | − | − | 13 | ||||||
| 116. 1114 | + | ‡ | + | 7 | ||||||
| 117. 1132 | − | + | + | 4 | ||||||
| 118. 1143 | + | = | − | 4 | ||||||
| 119. 1146 | − | + | − | 4 | ||||||
| 120. 1148 | − | + | − | 3 | ||||||
| 121. 1160 | + | − | − | 7 | ||||||
| 122. 1165 | − | − | + | 15 | ||||||
| 123. 1171 | − | + | − | 7 | ||||||
| 124. 1181 | − | − | + | 13 | ||||||
| 125. 1194 | − | − | + | 13 | ||||||
| 126. 1195 | + | − | − | 13 | ||||||
| 127. 1197 | − | + | − | 13 | ||||||
| 128. 1198 | − | − | + | 13 | ||||||
| 129. 0120 | − | + | + | |||||||
| 130. 0157 | − | + | + | |||||||
| 131. 0184 | − | = | + | |||||||
| 132. 0252 | = | − | − | |||||||
| 133. 0349 | = | − | − | |||||||
| 134. 0440 | − | + | + | |||||||
| 135. 0549 | − | − | + | |||||||
| 136. 0663 | + | − | − | |||||||
| 137. 0731 | − | P | P | |||||||
| 138. 0742 | − | − | + | |||||||
| 139. 0766 | − | = | − | |||||||
| 140. 0845 | + | − | − | |||||||
| 141. 0904 | − | = | + | |||||||
| 142. 0967 | − | + | + | |||||||
| 143. 0978 | + | − | − | |||||||
| 144. 1003 | − | − | + | |||||||
| 145. 1004-5 | − | − | + | |||||||
| 146. 1019 | − | − | + | |||||||
| 147. 1029 | − | − | + | |||||||
| 148. 1061 | − | − | + | |||||||
| 149. 1109 | − | + | − | |||||||
| 150. 1111 | − | − | + | |||||||
| 151. 0035 | − | − | + | |||||||
| 152. 0038 | = | = | = | |||||||
| 153. 0085 | + | − | − | |||||||
| 154. 0094 | − | + | + | |||||||
| 155. 0095 | − | + | + | |||||||
| 156. 0171 | S | S | S | |||||||
| 157. 0273 | = | − | − | |||||||
| 158. 0310 | = | ‡ | − | |||||||
| 159. 0461 | S | S | S | |||||||
| 160. 0477 | − | + | + | |||||||
| 161. 0485 | − | + | + | |||||||
| 162. 0541 | − | − | + | |||||||
| 163. 0611 | − | A | A | |||||||
| 164. 0741 | − | − | + | |||||||
| 165. 0772 | − | = | + | |||||||
| 166. 0796 | − | P | P | |||||||
| 167. 0829 | − | − | + | |||||||
| 168. 0836 | − | + | + | |||||||
| 169. 0850 | − | + | + | |||||||
| 170. 1112 | − | − | + | |||||||
| 171. 1126 | − | + | − | |||||||
+ denotes ORF annotated as a pseudogene in B. suis or B. abortus genomic annotation or identified here as a pseudogene in B. melitensis; − denotes ORF not recognized as a pseudogene; ‡ denotes pseudogene as per annotation this report; = denotes that annotation varies from GenBank but no pseudogene is recognized per comparison, different start codon used; P denotes polymorphic ORFs, some limited to amino terminus; N denotes polymorphic sequence or no ORF identified; X denotes locus not present; S denotes short ORFs (no comparisons made usually due to highly variable annotation among genomes); F denotes fused ORFs.
Functional categories: 1, amino acid biosynthesis; 2, biosynthesis of cofactors, prosthetic groups, and carriers; 3, cell envelope; 4, cellular processes; 5, central intermediary metabolism; 6, DNA metabolism; 7, energy metabolism; 8, fatty acid and phospholipid metabolism; 9, protein synthesis and fate; 10, purines, pyrimidines, nucleosides and nucleotides; 11, regulatory functions and signal transduction; 12, transcription; 13, transport and binding proteins; 14, other categories; 15, unknown function; 16, hypothetical proteins; 17, no database match; 18, disrupted reading frame.
Few of the variable ORFs appear on a list assembled by Letesson and colleagues (9) of 184 genes that were identified in large-scale random screens as affecting the pathogenicity and virulence of at least one Brucella classical species. Two genes that were only variable in B. melitensis are on the list, homologs of B. suis ORF BRA1146 (fliF; M-S ring) and BR1401 (bicA; macrolide efflux). Seven B. abortus genes were on the list: homologs of B. suis BRA0156 (flgI; P-ring), BR0161 (glnL; nitrogen regulatory IIA), BRA0804 (nikA; Ni2+ uptake), BRA0443 (glpK; glycerol kinase), BRA0599 (pyrB; pyrimidine synthesis), BR1084 (caiB domain; CAIB/BAIF family), and BR0181 (cysI; sulfite reductase). Three variable ORFs in both B. abortus and B. melitensis were on this list, homologs of B. suis BR1401 (bicA; macrolide efflux), BRA1132 (flhA; flagellum-related putative export protein), and BRA0311 (hypothetical protein).
Unique ORFs.
Several Chr I ORFs of B. abortus had no homologs in GenBank. These ORFs are referred to here as unique ORFs and are associated with regions containing homologs of phage or insertion sequences. Unique BruAb1_1085 is near a homolog of a site-specific integrase, phage family protein. Unique BruAb1_1088 flanks a resolvase family protein. Unique BruAb1_1833 is within Tn2020 (18). Though B. suis has a homolog of this ORF, it was not annotated. Most of the unique ORFs occur in a 20-kb phage-associated fragment shared only by B. melitensis (BMEI1674-BMEI1703) and B. abortus (BruAb1_0274-BruAb1_0242). BruAb1_0272 and BMEI1774 were annotated at homologous loci but on opposing strands. BruAb1_0272 was annotated on the opposing strand because it appeared to occur in an operon. BruAb1_0246 and BruAb1_0263 in B. abortus have no protein homologs in B. melitensis. Though a number of ORFs had homology to phages, the function of the encoded peptides was rarely known. Thus, the contribution of the phage- and plasmid-related regions to brucella metabolism and infectious cycle are unknown.
Metabolic capabilities.
Two genomic fragments of 7,738 and 2,653 bp identified as B. suis specific relative to B. melitensis on Chr II by Paulsen and colleagues (33) correlated with the ability of B. suis but not B. melitensis to oxidize ornithine, citrulline, arginine, and lysine and were aligned with the genome of B. abortus. Although the 2,653-bp fragment is present in B. abortus, the 7,738-bp fragment is not. Like B. melitensis, B. abortus also does not oxidize these compounds (1). This supports that the 7,738-bp fragment plays a vital role in these reactions.
B. abortus Chr I has two urease clusters, as described for B. suis (33). B. abortus ORFs BruAb1_0267-BruAb1_0273 and BruAb1_1356-BruAb1_1363 are homologs of B. suis urease cluster 1 ORFs BR0267-BR0273 and urease cluster 2 ORFs BR1356-BR1362, respectively. While there were no pseudogenes in cluster 1 of B. abortus, ureE and ureA are pseudogenes in cluster 1 in B. suis and B. melitensis. While there are no pseudogenes in cluster 2 of B. suis and B. melitensis, ureE is a pseudogene and ureD has a 6-bp insert in the urease cluster 2 in B. abortus. The sequence differences among the clusters correlate with differences in the rate of urea hydrolysis among the three bacteria. Urea is hydrolyzed in 1 to 2 h by B. abortus, compared to 0 to 30 min by B. suis (1). The rate of hydrolysis of urease in B. melitensis is variable, suggesting that at least another locus influences hydrolysis of urea. B. suis infects the urinary tract, while B. abortus and B. melitensis do not. The ability to quickly hydrolyze urea by B. suis may aid in its infection of, and excretion from, the urinary tract and subsequent spread in swine herds.
Bru-RS elements.
The number and orientation relative to ORFs of the small, palindromic Bru-RS1 and Bru-RS2 elements were determined. Twenty-two whole or partial Bru-RS1 elements (14) were identified on Ch I and 18 were identified on Chr II. Short ORFs comprised largely of Bru-RS1 or Bru-RS2 elements were not annotated in B. abortus as ORFs but are in B. suis. The Bru-RS elements were not clustered, and their orientation relative to ORFs appeared to be random. A copy of the Bru-RS1 element in B. abortus was identified in one of the homologs of proline racemase (BruAb1_0363) and in a probable transcription regulator (BruAb1_1398). There were nine whole or partial Bru-RS2 elements on Chr I and five on Chr II. None of these elements occurs within genes of known function. The elements, which are just over 100 bp, could affect gene expression.
Transposable elements.
There were only minor differences in the distribution and presence of transposase-related ORFs among the genomes, and those differences were associated with IS711, also known as IS6501 (16, 32). It was known from previous studies (6, 16, 32) that sequences of IS711 elements are not identical and, though the genomes have copies at the same loci, they have at least one copy at a unique locus also. The unique insertion locus of IS711 in B. suis is on Chr I, whereas the unique insertion loci of IS711 in B. melitensis and B. abortus are on Chr II (Tables 1 and 2). As described above, one of the common insertion site copies of IS711 is truncated along with ORF C of an ISBm1 copy in B. abortus relative to B. suis (BRA0551-BRA0559) and B. melitensis.
An analysis of the Brucella sequences suggests that the only mobile transposable element in Brucella is IS711. The B. abortus 9-941 genome has the same number of copies of IS711 as that found in B. abortus 544 (7). There is an additional copy in B. abortus 2308 (7), a biovar 1 strain that is commonly used as a vaccine challenge strain. The rough vaccine strain B. abortus RB51 has one more copy than its parental strain, S2308 (7, 39). Other brucellae have many more copies of IS711 than the sequenced genomes. In Southern blot analyses, B. ovis was estimated to have at least 30 copies (17), and Southern blot analyses have shown that the marine isolates have even more copies than that (5). This insertion sequence has not been identified in other bacterial genera and is most closely related to IS427 from the phylogenetically related bacterium Agrobacterium tumefaciens (32).
Sequence of field isolate versus vaccine challenge strain.
The sequence of B. abortus 9-941 was analyzed for loci detected by suppressive subtractive hybridization (SSH) in B. melitensis 16 M but not B. abortus S2308 (37). Homologs of B. melitensis 16 M loci BMEI0888, BMEI0943, BMEI0971, BMEI1055, BMEI1331, and BMEI919 missing in B. abortus S2308 were also missing in B. abortus 9-941. Several homologs not identified in B. abortus S2308 in initial SSH studies were identified in the sequence of B. abortus 9-941, namely BMEI0943, BMEI0971, BMEI1055, BMEII25, BMEI1331, BMEI1380, and BME1919.
Evolutionary and genomic analyses.
SNPs were identified for the shared sequences of the genomes of B. abortus, B. suis, and B. melitensis (Table 5) by using the Mummer whole genome comparison tool. This analysis identified 7,208 SNP mutations between B. abortus and B. suis genomic sequences, 6,342 SNP mutations between B. abortus and B. melitensis, and 7,844 SNP mutations between B. suis and B. melitensis. The mean for the genomes was 1 SNP for approximately 463 nucleotides.
TABLE 5.
SNPs among the three brucella genomes
| Chromosome and species | Nucleotide | No. of SNPs at:
|
||||||
|---|---|---|---|---|---|---|---|---|
| A | C | G | T | |||||
| Chr I | ||||||||
| B. abortus |
B. suis
|
|||||||
| A | 270 | 915 | 76 | |||||
| C | 132 | 142 | 841 | |||||
| G | 798 | 156 | 100 | |||||
| T | 69 | 805 | 251 | |||||
| B. abortus |
B. melitensis
|
|||||||
| A | 252 | 649 | 72 | |||||
| C | 153 | 138 | 769 | |||||
| G | 782 | 162 | 148 | |||||
| T | 73 | 637 | 252 | |||||
| B. suis |
B. melitensis
|
|||||||
| A | 146 | 820 | 73 | |||||
| C | 188 | 171 | 957 | |||||
| G | 1,085 | 183 | 196 | |||||
| T | 92 | 871 | 151 | |||||
| Chr II | ||||||||
| B. abortus |
B. suis
|
|||||||
| A | 109 | 500 | 35 | |||||
| C | 56 | 70 | 531 | |||||
| G | 512 | 67 | 78 | |||||
| T | 52 | 527 | 116 | |||||
| B. abortus |
B. melitensis
|
|||||||
| A | 117 | 355 | 33 | |||||
| C | 81 | 78 | 408 | |||||
| G | 440 | 78 | 103 | |||||
| T | 44 | 387 | 121 | |||||
| B. suis |
B. melitensis
|
|||||||
| A | 85 | 536 | 45 | |||||
| C | 100 | 88 | 617 | |||||
| G | 568 | 100 | 92 | |||||
| T | 56 | 549 | 75 | |||||
Phylogenetic tree.
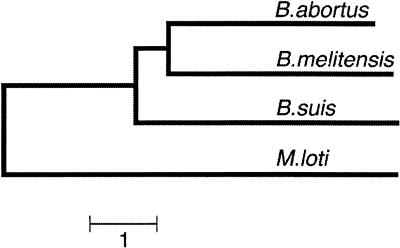
A rooted neighbor-joining tree showing the evolutionary relationships between the sequenced brucellae genomes was constructed using SNP data with MEGA2. Mesorhizobium loti data were included for use as an outgroup (Fig. 2). By this method, B. abortus was most closely related to B. melitensis, and B. suis was more closely related to B. abortus than to B. melitensis. These results, which were obtained from whole genomic DNA, are consistent with those found from PFGE studies of whole genomic DNA (27). When specific loci are used, the clustering of brucellae is dependent on the loci. For example, clustering of the three brucellae was dependent on whether results from 10 enzymes or 16 enzymes were used to construct a dendrogram using results from multilocus enzyme electrophoresis studies (13). In these studies, B. melitensis and B. abortus clustered when a dendrogram was constructed from 10 loci, but not one with 16 loci.
FIG. 2.
Evolutionary relationships among the sequenced brucellae, as shown in a neighbor-joining dendrogram generated from an SNP-based genetic distance data tree and rooted using the related rhizobial M. loti genome as the outgroup. The scale bar represents 1,000 SNPs.
In summary, the genomes of B. suis, B. melitensis, and B. abortus are very similar in sequence, organization, and structure. Few fragments are unique among the genomes. B. melitensis and B. abortus share more sequences than either does with B. suis. A comparison of the three genome sequences of Brucella gives us a foundation to further our understanding of the Brucella genus and provides the groundwork to investigate the contribution of various pathways to the relative pathogenicity and virulence of these bacteria. The genome sequences allow construction of general brucella microarrays to observe the dance between microbe and host in understanding the course of brucellosis infection.
Acknowledgments
We thank Nancy Koster, Elizabeth Schmerr, Lindsey Engelby, Kai Tanaka, Richard Thielen, Stan Strum, Donnie Brooks, Aileen Duit, Dani Umbaugh, Daryl Pringle, Scott Farris, Chad Rienke, Rick Hornsby, Xiaowu Gai, and TIGR for technical assistance. We thank Darla Ewalt for biotyping B. abortus 9-941.
Mention of trade names or commercial products herein is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Alton, G. G., L. M. Jones, and D. E. Pietz. 1975. Laboratory techniques in brucellosis. World Health Organization, Geneva, Switzerland. [PubMed]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bricker, B. J. 2002. PCR as a diagnostic tool for brucellosis. Vet. Microbiol. 90:435-446. [DOI] [PubMed] [Google Scholar]
- 4.Bricker, B. J., D. R. Ewalt, and S. M. Halling. 2003. Brucella “HOOF-Prints”: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker, B. J., D. R. Ewalt, A. P. MacMillan, G. Foster, and S. Brew. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J. Clin. Microbiol. 38:1258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bricker, B. J., and S. M. Halling. 1995. Enhancement of the Brucella AMOS PCR assay for differentiation of Brucella abortus vaccine strains S19 and RB51. J. Clin. Microbiol. 33:1640-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., J. M. Verger, M. Grayon, J. Y. Paquet, B. Garin-Bastuji, G. Foster, and J. Godfroid. 2001. Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3:729-738. [DOI] [PubMed] [Google Scholar]
- 9.Delrue, R.-M., P. Lestrate, A. Tibor, J.-J. Letesson, and X. De Bolle. 2004. Brucella pathogenesis, genes identified from random large-scale screens. FEMS Microbiol. Lett. 231:1-12. [DOI] [PubMed] [Google Scholar]
- 10.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewalt, D. R., J. G. Payeur, B. M. Martin, D. R. Cummins, and G. J. Miller. 1994. Characteristics of a Brucella species from a bottlenose dolphin (Tursiops truncates). J. Vet. Diagn. Investig. 6:448-452. [DOI] [PubMed] [Google Scholar]
- 12.Ficht, T. A., S. W. Bearden, B. A. Sowa, and H. Marquis. 1990. Genetic variation at the omp2 porin locus of the brucellae: species-specific markers. Mol. Microbiol. 4:1135-1142. [DOI] [PubMed] [Google Scholar]
- 13.Gandara, B., A. L. Merino, M. A. Rogel, and E. Martinez-Romero. 2001. Limited genetic diversity of Brucella spp. J. Clin. Microbiol. 39:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halling, S. M., and B. J. Bricker. 1994. Characterization and occurrence of two repeated palindromic DNA elements of Brucella spp.: Bru-RS1 and Bru-RS2. Mol. Microbiol. 14:681-689. [DOI] [PubMed] [Google Scholar]
- 15.Halling, S. M., N. Gibas, and S. M. Boyle. 2004. Comparative genomics B. melitensis, B. suis, and B. abortus, p. 85-102. In I. Lopez-Goni and I. Moriyon (ed.), Molecular and cellular biology of Brucella. Horizon Scientific Press, Norwich, United Kingdom.
- 16.Halling, S. M., F. M. Tatum, and B. J. Bricker. 1993. Sequence and characterization of an insertion sequence, IS711, from Brucella ovis. Gene 133:123-127. [DOI] [PubMed] [Google Scholar]
- 17.Halling, S. M., and E. S. Zehr. 1990. Polymorphism in Brucella spp. due to highly repeated DNA. J. Bacteriol. 172:6637-6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halling, S. M., and R. L. Zuerner. 2002. Evidence for lateral transfer to Brucella: characterization of a locus with a Tn-like element (Tn2020). Biochim. Biophys. Acta 1574:109-116. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 20.Hoyer, B. H., and N. B. McCullough. 1968. Polynucleotide homologies of Brucella deoxyribonucleic acids. J. Bacteriol. 95:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoyer, B. H., and N. B. McCullough. 1968. Homologies of deoxyribonucleic acids from Brucella ovis, canine abortion organisms, and other Brucella species. J. Bacteriol. 96:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jumas-Bilak, E., S. Michaux-Charachon, G. Bourg, M. Ramuz, and A. Allardet-Servent. 1998. Unconventional genomic organization in the alpha-subgroup of the Proteobacteria. J. Bacteriol. 180:2749-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 24.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, M. E. 1964. The epizootiology of brucellosis and its relationship to the identification of Brucella organisms. Am. J. Vet. Res. 25:553-557. [PubMed] [Google Scholar]
- 26.Meyer, M. E. 1966. Host-parasite relationships in brucellosis. I. Reservoirs of infection and interhost transmissibility of the parasite. Proc. 70th Annu. Meet. U.S. Livestock Sanitary Assoc., p. 129-136.
- 27.Michaux, S., J. Paillisson, M. J. Carles-Nurit, G. Bourg, A. Allardet-Servent, and M. Ramuz. 1993. Presence of two independent chromosomes in the Brucella melitensis 16M genome. J. Bacteriol. 175:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaux-Charachon, S., G. Bourg, E. Jumas-Bilak, P. Guigue-Talet, A. Allardet-Servent, D. O'Callaghan, and M. Ramuz. 1997. Genome structure and phylogeny in the genus Brucella. J. Bacteriol. 179:3244-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno, E., and I. Moriyón. 2001. Genus Brucella. In M. Dworkin (ed.), The prokaryotes: an evolving microbiological resource for the microbiological community. Springer, New York, N.Y. [online.]
- 30.Moreno, E., and I. Moriyón. 2002. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. USA 99:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno, E., E. Stackebrandt, M. Dorsch, J. Wolters, M. Busch, and H. Mayer. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouahrani, S., S. Michaux, J. Sri Widada, G. Bourg, R. Tournebize, M. Ramuz, and J. P. Liautard. 1993. Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp.: relationship between genomic structure and the number of IS6501 copies. J. Gen. Microbiol. 139:3265-3273. [DOI] [PubMed] [Google Scholar]
- 33.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross, H. M., G. Foster, R. J. Reid, K. L. Jahans, and A. P. MacMillian. 1994. Brucella species infection in sea-mammals. Vet. Rec. 134:359. [DOI] [PubMed] [Google Scholar]
- 35.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, D. O., R. O. Zandomeni, S. Cravero, R. E. Verdun, E. Pierrou, P. Faccio, G. Diaz, S. Lanzavecchia, F. Aguero, A. C. Frasch, S. G. Andersson, O. L. Rossetti, O. Grau, and R. A. Ugalde. 2001. Gene discovery through genomic sequencing of Brucella abortus. Infect. Immun. 69:865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsoktouridis, G., C. A. Merz, S. P. Manning, R. Giovagnoli-Kurtz, L. E. Williams, C. V. Mujer, S. Hagius, P. Elzer, R. J. Redkar, G. Patra, and V. G. DelVecchio. 2003. Molecular characterization of Brucella abortus chromosome II recombination. J. Bacteriol. 185:6130-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsolis, R. M. 2002. Comparative genome analysis of the alpha-proteobacteria: relationships between plant and animal pathogens and host specificity. Proc. Natl. Acad. Sci. USA 99:12503-12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vemulapalli, R., J. R. McQuiston, G. G. Schurig, N. Sriranganathan, S. M. Halling, and S. M. Boyle. 1999. Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin. Diagn. Lab. Immunol. 6:760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verger, J. M., F. Grimont, P. A. D. Grimont, and M. Grayon. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35:292-295. [Google Scholar]
- 41.Vizcaino, N., A. Cloeckaert, J. Verger, M. Grayon, and L. Fernandez-Lago. 2000. DNA polymorphism in the genus Brucella. Microbes Infect. 2:1089-1100. [DOI] [PubMed] [Google Scholar]
- 42.Vizcaino, N., A. Cloeckaert, M. S. Zygmunt, and L. Fernandez-Lago. 2001. Characterization of a Brucella species 25-kilobase DNA fragment deleted from Brucella abortus reveals a large gene cluster related to the synthesis of a polysaccharide. Infect. Immun. 69:6738-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]