Abstract
The tetQ-rteA-rteB operon of the Bacteroides conjugative transposon CTnDOT is responsible for tetracycline control of the excision and transfer of CTnDOT. Previous studies revealed that tetracycline control of this operon occurred at the translational level and involved a hairpin structure located within the 130-base leader sequence that lies between the promoter of tetQ and the start codon of the gene. This hairpin structure is formed by two sequences, designated Hp1 and Hp8. Hp8 contains the ribosome binding site for tetQ. Examination of the leader region sequence revealed three sequences that might encode a leader peptide. One was only 3 amino acids long. The other two were 16 amino acids long. By introducing stop codons into the peptide coding regions, we have now shown that the 3-amino-acid peptide is the one that is essential for tetracycline control. Between Hp1 and Hp8 lies an 85-bp region that contains other possible RNA hairpin structures. Deletion analysis of this intervening DNA segment has now identified a sequence, designated Hp2, which is essential for tetracycline regulation. This sequence could form a short hairpin structure with Hp1. Mutations that made the Hp1-Hp2 structure more stable caused nearly constitutively high expression of the operon. Thus, stalling of ribosomes on the 3-amino-acid leader peptide could favor formation of the Hp1-Hp2 structure and thus preclude formation of the Hp1-Hp8 structure, releasing the ribosome binding site of tetQ. Finally, comparison of the CTnDOT tetQ leader regions with upstream regions of five tetQ genes found in other elements reveals that the sequences are virtually identical, suggesting that translational attenuation is responsible for control of tetracycline resistance in these other cases as well.
CTnDOT is a Bacteroides conjugative transposon (CTn) that carries two antibiotic resistance genes, tetQ and ermF. CTns related to CTnDOT have been found in many Bacteroides strains (24). The transfer of CTnDOT is stimulated 100- to 1,000-fold by exposure of donors to tetracycline (23, 26). Both excision and transfer of CTnDOT are under tetracycline control. Tetracycline stimulation of excision and transfer functions is mediated by a three-gene operon, which consists of the tetracycline resistance gene, tetQ, and two regulatory genes, rteA and rteB (20, 25). RteA and RteB appear to be members of a two-component regulatory system that controls the expression of a third regulatory gene, rteC, which in turn controls expression of excision and transfer genes (4, 30).
Genes conferring resistance to tetracycline have been found to be regulated in a variety of ways. For example, expression of tetM appears to be controlled by a transcriptional attenuation mechanism (27). Expression of the tetA gene is controlled by a repressor protein that is encoded by a divergently transcribed gene, tetR (2, 13). Recently, we reported that the CTnDOT tetQ gene, unlike these other tet genes, appears to be controlled at the translational level (29). There is an mRNA leader region that lies between the tetQ promoter and the start codon of tetQ. This region contains a hairpin structure, whose stem is composed of two sequences, Hp1 and Hp8 (Fig. 1). Previously, we used site-directed mutagenesis to show that this hairpin structure is essential for tetracycline-dependent regulation of the tetQ-rteA-rteB operon (29). Mutations that made the hairpin structure less stable caused constitutive or nearly constitutive expression of the operon, whereas mutations that restored the hairpin structure but changed its sequence restored regulated expression.
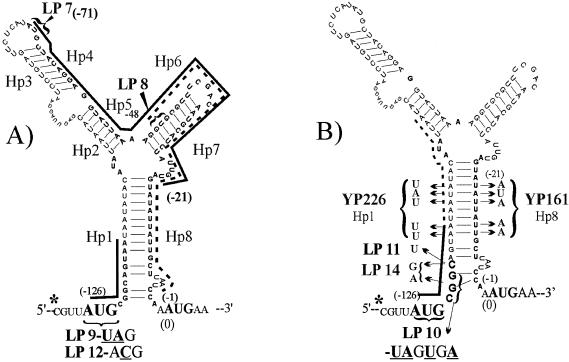
FIG. 1.
Diagram of the RNA secondary structure of Pq and the locations of mutations that affect the translation of three possible leader peptides within the tetQ leader sequence and/or disrupt the secondary structure. The RNA secondary structure was predicted by the MFOLD version 3.1 program of Zucker (http://www.bioinfo.rpi.edu/applications/mfold/rna.forml.cgi). The asterisk indicates the transcription initiation site at −130 bp upstream of the tetQ start codon that was identified by Bayley et al. (1). In panel A, three possible leader peptides (LPs) are indicated by solid or dashed lines. The first putative peptide is only 3 amino acids long and starts at bp −126 relative to the AUG of tetQ. The triangles and LP designations indicate the positions of stop codons that were inserted in the sequence at positions −71 (LP7) and −48 (LP8) relative to the tetQ start codon (Table 1). The effect of each mutation in the tetQ operon promoter on the tetracycline regulation of the uidA (GUS) reporter gene fused within the tetQ is shown in Table 1. In panel B, the site-directed mutations in Hp1 and Hp8 sequences are indicated. YP161 contains the 5-nucleotide changes in Hp8 cloned in the GUS fusion, and YP226 contains the same YP161 Hp8 sequences with complementary changes in the Hp1 sequence to restore the hairpin formation, as indicated by the brackets. The mutations in Hp1 on YP226 also increased the coding sequence of the tripeptide (indicated by the solid line) from 3 to 8 amino acids (indicated by the dotted line extension). There are three additional site-directed mutations involving the tripeptide sequence, which are in addition to the two shown in panel A. LP10 changes the sequences of the first two codons to stop codons, and this mutation also interferes with the pairing of the Hp1-Hp8 adjacent to the AUG (position 0) of tetQ. LP11 shortens the mini-peptide to only 2 amino acids by mutating the third codon, CAG, to UAG. LP14 changes the sequences of the 2nd and 3rd amino acids with minor effects on the pairing of Hp1 and Hp8. The effects of the mutations on the GUS activity, plus and minus tetracycline, compared to that of the wild-type promoter are shown in Table 1.
In translational attenuation, there is usually a leader peptide encoded within the leader region. This peptide is constantly translated, but if ribosomes stall during translation of the peptide, alternate structures can form in the leader mRNA. Since the sequence which should contain the ribosome binding site for tetQ is within Hp8, formation of the Hp1-Hp8 structure could tie up this site, preventing ribosomes from translating tetQ. Our hypothesis is that exposure of cells to tetracycline causes ribosomes to stall on a leader peptide, preventing the Hp1-Hp8 hairpin structure from forming. In this report, we identify the leader peptide. We had noted previously that between Hp1 and Hp8 there were other sequences that could form hairpin structures (Fig. 1), but it was not clear whether these sequences had any role in tetracycline regulation. We show here that only one of these intervening sequences, which we designate as Hp2 in Fig. 1, is critical for regulation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in the text and Table 1 and/or shown in a relevant figure. The recA muntant Escherichia coli strains DH5αMCR (Gibco BRL), HB101 (3, 21), and DH10B (21) were grown in Luria broth (LB) or on LB agar plates. Bacteroides strains were grown in chopped meat medium (Remel) for subculture or in TYG (Trypticase-yeast extract-glucose) medium (11). Bacteroides thetaiotaomicron 4001ΩQABC (Rifr), a derivative of strain BT4001 which has one copy of the tetQ-rteA-rteB operon, along with a copy of another CTn gene, rteC, integrated into its chromosome, was used as the background for assessing regulation of the operon (30). A translational fusion between tetQ and the reporter gene, uidA (a gene encoding a β-glucuronidase [GUS]) (8, 29), was introduced into this strain and used to assess production of GUS. Previously, we had shown that none of the gene products encoded by tetQ, rteA, rteB, or rteC had any effect on tetracycline-dependent regulation (29), but the presence of tetQ in the strain was convenient because it allowed exposure of the strain to tetracycline without inhibiting its growth. The antibiotic concentrations used were as follows: ampicillin, 100 μg/ml; erythromycin, 10 μg/ml; gentamicin, 200 μg/ml; rifampin, 10 μg/ml; trimethoprim, 10 μg/ml; and tetracycline, 3 μg/ml to select for transconjugants and 1 μg/ml for induction of CTn activity. In the case of donors or recipients that required thymidine, thymidine was provided at 100 μg/ml.
TABLE 1.
Site-directed mutations and deletions in the tetQ promoter region and their effect on gene activity measured by translational fusions with uidA (Pq::GUS fusions)a
| Constructb | Description of mutation(s)d | GUS activity (U/mg)c
|
Fold increase | |
|---|---|---|---|---|
| −TET | +TET | |||
| Wild-type Pq::GUS | PCR product from bp −279 upstream of ATG start of tetQ to 33 bp within tetQ gene cloned into pMJF2 (8, 29) | 17 | 669 | 39 |
| Site-directed mutagenesis | ||||
| LP7 | Mutations of first two codons in putative leader peptide (LP) bp −71 to bp −21; AUG CUA changed to uaG ugA (2 stop codons) as shown in Fig. 1A | 19 | 614 | 32 |
| LP8 | Mutations of first two codons in the putative LP from bp −48 to −1: GUG CGU changed to uaG uGa (2 stop codons) positions indicated in Fig. 1A | 12 | 243 | 20 |
| LP9 | AUG changed to uaG in 3-aa peptide sequence shown in Fig. 1A | 11 | 10 | 1 |
| LP10 | LP 9 with additional stop codon in 2nd of 3 codons shown in Fig. 1B; disrupts bottom of stem between Hp1 and Hp8 just upstream of tetQ AUG start at position 0 | 128 | 137 | 1 |
| LP11 | Point mutation 3rd of 3 codons, CAG to stop codon uAG, which reduces size of the 3-aa peptide to 2 aa and disrupts pairing of Hp1-Hp8 shown in Fig. 1B | 16 | 67 | 4 |
| LP12 | AUG of the 3-aa peptide changed to AcG shown in Fig. 1A; Prevents start of peptide and is different mutation from LP 9 | 1 | 1 | 1 |
| LP14 | The 2nd and 3rd codons of the 3-aa peptide changed from CGG CAG (Arg Gln) to aGG gAG (Arg Glu) shown in Fig. 1B; mutations change the 3rd aa and cause two disruptions in pairing between Hp1 and Hp8 | 10 | 13 | 1 |
| YP161 (Hp8muts) | 5 nucleotide substitutions in sequence of Hp8 that disrupts pairing with Hp1 shown in Fig. 1B | 1003 | 1303 | 1 |
| YP226 (Hp1comp) | Complementary mutations in Hp1 sequence to restore pairing between two regions | 100 | 796 | 8 |
| YP239 | Contains 4 nucleotide changes in Hp 1 that disrupt possible pairing with Hp 2 as shown in Fig. 3; changes do not affect the length of the 3-aa peptide (UAA stop changed to UgA stop) and still allow slightly altered pairing with Hp-8 with ΔG = −33.4 kcal/mol (not shown) (See Fig. 3 for location of mutations) | 50 | 420 | 8 |
| YP241 | Contains sequence of Hp1 contained in YP 239 with complementary mutations in Hp2 that reestablished Hp1::Hp2 pairing with GCs instead of AUs. (Fig. 3) | 880 | 781 | 1 |
| Deletions | ||||
| SM Del-1 | Deletion of region containing Hp6 and Hp7 as shown in Fig. 2A | 22 | 547 | 25 |
| SM Del-2 | Deletion of region containing Hp4 and Hp5 as shown in Fig. 2B | 18 | 526 | 29 |
| SM Del-3 | Deletion of all sequences between Hp1 and Hp8 (Hp2-Hp7) as shown in Fig. 2A | 12 | 11 | 1 |
| Del Hp2 | Deletion of Hp2 sequences, which eliminates pairing to Hp5 as shown in Fig. 2B | 10 | 32 | 3 |
| Del Hp2-3 | Deletion of Hp2 and Hp3 sequences as shown in Fig. 2B | 30 | 90 | 3 |
| Del Hp3-4 | Deletion of Hp3 and Hp4 sequences as shown in Fig. 2B | 19 | 548 | 29 |
| Del Hp2-5 | Deletion of Hp2-Hp5 sequences as shown in Fig. 2A | 9 | 9 | 1 |
Variation between values in the independent replicate assay was about 10%.
All of the mutations were made to wild-type Pq sequence cloned in the pGEM-T vector, and the resultant region was then cloned into pMJF2, which put the uidA in frame with the tetQ coding region forming a translational fusion.
The GUS activity is units per milligram of protein, where 1 U is the increase of 0.01 U at A415 per min at 37°C. TET, tetracycline.
Changed residues in codons are lowercase.
Site-directed mutagenesis.
Since all of the tetracycline derivatives we have tested are inducers of the tetQ operon, it was not possible to use a noninducing form of tetracycline to select for constitutive mutants. Accordingly, we turned to site-directed mutagenesis to generate mutations in the tetQ leader region. Site-directed mutagenesis was done by the Stratagene QuikChange method to construct deletions or point mutations in the region upstream of the tetQ start codon. The desired mutations were incorporated into the primers. pYP84TA, which has the wild-type Pq leader region cloned into the PCR cloning vector pGEM-T (Promega), was used as template. The PCR products (which were the entire vector plus the target sequence) were treated with DpnI for 3 h to destroy the parental nonmutated vector sequences. Five to 10 μl of the reaction mixture was used to transform E. coli DH10B with the surviving plasmids. Each Pq mutation on the pGEM-T vector was confirmed by sequencing. The mutated Pq region was isolated from an agarose gel on an SphI-HincII fragment and cloned into the SphI-SmaI site of pMJF2 (8) to generate a translational fusion between the Pq region and the uidA (GUS) gene.
RNA secondary structure prediction.
Predictions of RNA secondary structure and calculation of free energies were determined with the MFOLD version 3.1 program of Zucker (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/forml.cgi).
Triparental matings.
pMJF2::Pq clones containing mutated versions of the Pq region fused to uidA were moved by triparental matings into Bacteroides strains to determine the effect of mutated Pq region on production of GUS. In each case, the two donor strains were E. coli DH10B, which contained the pMJF2::Pq derivative, and E. coli HB101, which contained the IncPα plasmid RP1. Matings were done on nitrocellulose filters as described previously (22). RP1 cannot replicate in Bacteroides spp., but it can mobilize pMJF2 derivatives from E. coli donors to Bacteroides recipients. The Bacteroides recipient was BT4001ΩQABC, a strain that contains a copy of the tetQ-rteA-rteB operon and a copy of the rteC gene that have been integrated into the chromosome (30). The transconjugants were selected on Gen200Em10 plates.
GUS assays.
The uidA (GUS) reporter gene on pMJF2 encodes an E. coli GUS. GUS assays were done by the procedure of Feldhaus et al. (8). In this study, all of the GUS activities are expressed as units per milligram of protein and are the average of the specific activities from at least three different experiments. One unit was defined as 0.01 A415 unit per min at 37°C. Protein concentrations were determined by the method of Lowry et al. (17). GUS assays were done in triplicate in at least three separate experiments. The variation in values of GUS activity from experiment to experiment was ±10%.
RESULTS
Involvement of a small leader peptide in tetracycline regulation of the tetQ operon.
In the case of genes regulated by attenuation, a leader peptide contributes by providing a place for ribosomes to stall, thus affecting mRNA structures that form as RNA polymerase proceeds to transcribe the leader region of the message (6, 7, 9, 16, 18). Initially, two possible leader peptides were identified in the tetQ leader region (29). Both were 16 amino acids in length (Fig. 1A). The first two codons in each of these putative peptide coding sequences were replaced with stop codons (LP7 and LP8; Fig. 1A and Table 1). In both cases, the GUS activity in cell extracts was the same as that associated with the wild-type fusion (Table 1). Thus, neither of these two putative peptides plays a role in tetracycline regulation of tetQ operon expression. Further examination of the leader region sequence revealed a third possible leader peptide whose start codon was at position −126. This small peptide had been ignored initially because it was only 3 amino acids in length. Replacement of the start codon of this peptide with a stop codon (mutant LP9; Fig. 1A) reduced GUS activity to near background (1 to 10 U/mg) and made it independent of tetracycline stimulation (Table 1).
Since the 3-amino-acid leader peptide sequence was at the 5′ end of the hairpin formed by Hp1 and Hp8, it was possible that the mutation that created the stop codon had destabilized the Hp1-Hp8 hairpin by changing the mRNA sequence. Accordingly, we also constructed mutation LP12 (Fig. 1A), in which the start codon was replaced with ACG to abolish the translation of the peptide without changing the hairpin structure. Even though the GUS values for LP12 are slightly lower than those for LP9, they are close to background and exhibit no tetracycline regulation. Both results show that translation of the peptide or at least the binding of the ribosome to start translation is necessary for regulation of tetQ expression.
As a further test of the involvement of the 3-amino-acid leader peptide, mutations were made to change either its amino acid sequence or its size. In mutant LP14 (Fig. 1B), the codons specifying the second and the third amino acids were changed, changing Arg Gln to Arg Glu, but without changing the hairpin structure formed by Hp1 and Hp8. GUS activity in crude extracts from this strain was very low and was not affected by exposure to tetracycline (Table 1), suggesting that the amino acid sequence of the leader peptide is important for tetracycline induction. In LP11, a stop codon replaced the codon for the third amino acid of the leader peptide. This mutant, which could now produce a peptide only 2 amino acids long, had a 10-fold-lower level of GUS expression under inducing conditions compared to the wild-type strain but still exhibited a response to tetracycline (Table 1).
In a our previous study (29), the effects of mutations that changed the sequence of the hairpin formed by Hp1 and Hp8 but maintained the hairpin structure were tested. In all but one case, these compensatory mutations restored the full regulated phenotype of the wild-type strain. An exception was the strain that contained YP226 (Fig. 1B). YP226 had a reestablished hairpin structure but only partially recovered the wild-type phenotype. The uninduced level of this mutant was about sixfold higher than that of the wild-type strain. The mutation in YP226 extended the leader peptide to 8 amino acids in length, further supporting the hypothesis that the length of the leader peptide is important for the tetracycline effect (Table 1).
Results of our previous study had suggested that the putative ribosome binding site was sequestered when the portion of Hp8 that contains the region immediately upstream of the start codon of tetQ formed the hairpin structure with Hp1. In this connection, it is worth noting the GUS phenotype of cells containing LP10, in which two stop codons replaced the first 2 amino acids of the leader peptide by changing AUGCGG to UAGUGA (Fig. 1B and Table 1). The two stop codons partially disrupted the bottom part of the hairpin formed by Hp1 and Hp8, so that the corresponding nucleotides on Hp8 would be exposed to the ribosome. In this case, the GUS activity in the absence of tetracycline stimulation (128 U/mg) was much higher than that for the wild-type sequence (17 U/mg). This finding is consistent with our hypothesis that the ribosome binding site is within the end of the Hp8 region but that only part of it was exposed in the LP10 mutant.
Effects of deletions in the sequence between Hp1 and Hp8.
The sequence between Hp1 and Hp8 could have formed other stem-loop structures (Fig. 2). To assess the involvement of these sequences, deletions were made in this region (Fig. 2A and B). A large deletion in mutant SM Del-3 eliminated all the sequences between the end of Hp1 and the beginning of Hp8, except for 3 bp (SM Del-3; Fig. 2A). This deletion caused GUS to be produced constitutively at a very low level (Table 1). This result supports the hypothesis that some sequences between Hp1 and Hp8 are involved in tetracycline-dependent regulation.
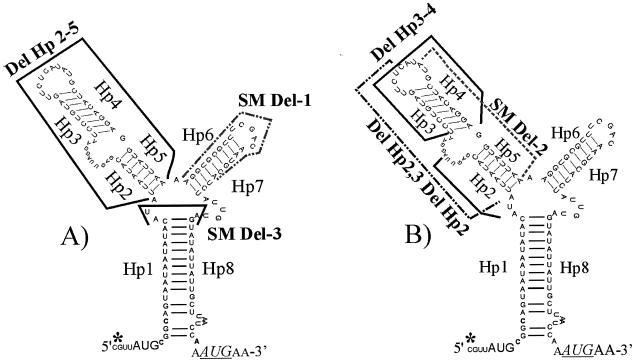
FIG. 2.
Effects of deletions in the Hp2-Hp7 region. The hypothetical secondary structure of the tetQ leader region is shown. The transcriptional start site determined by Bayley et al. (1) is indicated by an asterisk. The sequences of hairpin structures (Hps) that were deleted are indicated by dotted or dark lines. In panel A, the solid line above and between Hp1 and Hp8 indicates the sequences remaining in the SM Del-3 construct that leaves only the 3 nucleotides, AUA, between the hairpin formed by Hp1 and Hp8. Del Hp2-5 deletes all the sequences in the left loop, and SM Del-1 removes the sequences of Hp6 and Hp7. The effects of the deletions on the resultant GUS activities of the Pq::GUS fusions, with and without tetracycline induction, are shown in Table 1. In panel B, deletions of various sequences in the left loop are shown. The deletions are indicated by various solid and dashed lines, and the effects of these deletions on the GUS activity of the Pq::GUS fusions are shown in Table 1.
To test the hypothesis that sequences other than Hp1 and Hp8 are involved in regulation, additional deletion mutants were constructed and tested for their effect on GUS production. Deletion of the hairpin formed by Hp6 and Hp7 (SM Del-1; Fig. 2A) had the same GUS production pattern as the wild type (Table 1). Deletion of Hp2 through Hp5 (Del Hp2-5; Fig. 2A) was associated with low GUS expression even under tetracycline-induced conditions (Table 1), behavior similar to that seen in the case of the mutant SM Del-3. Deletion of Hp4 and Hp5 (SM Del-2; Fig. 2B) and of Hp3 and Hp4 (Del Hp3-4; Fig. 2B) resulted in mutants that exhibited tetracycline-regulated GUS activity, similar to that of the wild type (Table 1). Only the mutants that lacked the Hp2 region, Del Hp2-3, Del Hp2, and Del Hp2-5, exhibited the pattern of GUS expression associated with SM Del-3, the deletion of the entire region from Hp2 through Hp7 (Fig. 2B and Table 1). Thus, Hp2 is essential for tetracycline regulation.
Role of an Hp1-Hp2 hairpin structure in tetracycline regulation.
When the tetQ leader sequence in the region from Hp1 through Hp3 was examined for alternative RNA secondary structures, a possible hairpin structure involving Hp1 and Hp2 was identified (Fig. 3). This hairpin, however, was predicted to be less stable (ΔG = −4.81 kcal/mol) than that formed by Hp1 and Hp8 (ΔG = −33.24 kcal/mol). To test the hypothesis that a hairpin structure involving Hp1 and Hp2 might form, mutant YP241 was constructed, in which the hairpin formed by Hp1 and Hp2 was made more stable by changing four ATs to GCs without changing other hairpins or the sequence and length of the 3-amino-acid leader peptide. As a control, we first assessed the effects of the changes in Hp1 on formation of the Hp1-Hp8 hairpin structure. We started with a mutant (YP239) that had only the 4-nucleotide changes in Hp1. This mutant exhibited only a slightly higher basal level of GUS expression compared to the wild-type stain (50 versus 17 U/mg; Table 1), thus confirming that these changes in Hp1 did not have a major effect on the ability of Hp1 to pair with Hp8. When four corresponding nucleotide changes were then made in Hp2 to produce mutant YP241, there was a high level of GUS activity in the absence of tetracycline (880 U/mg; Fig. 3 and Table 1). This was the result expected if formation of the Hp1-Hp2 hairpin occurred and prevented the formation of the Hp1-Hp8 hairpin.
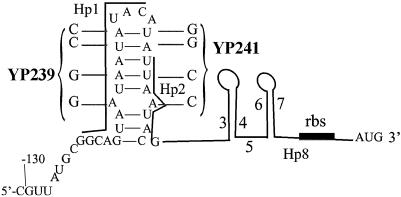
FIG. 3.
Putative hairpin structure formed by Hp1 and Hp2. If the formation of Hp1 and Hp8 as a hairpin is prevented, the MFOLD version 3.1 program predicts the formation of a hairpin between Hp1 and Hp2. The calculated energy of this putative structure is ΔG = −4.81 kcal/mol. Site-directed mutations to test the Hp1-Hp2 pairing are shown. YP239 contains the mutations in Hp1. In addition to the base substitutions in Hp1 in YP239, complementary base substitutions were made in Hp2 to construct YP241. The mutations in Hp2 reestablish the Hp1-Hp2 pairing with GC base pairs instead of AU base pairs. The effects of the mutated Hp1 and Hp2 regions on the regulation of the GUS activity in the Pq::GUS fusions are shown in Table 1.
Translational attenuation may be a general regulatory mechanism for tetQ genes.
Previous studies have shown that virtually identical alleles of the tetQ gene (>95% DNA sequence identity) are found in many different species, especially Bacteroides and Prevotella species (19). More recently, the tetQ gene has been found by DNA-DNA hybridization in gram-positive bacteria (14). To determine whether these other tetQ genes might also be regulated similarly to the tetQ on CTnDOT, the DNA sequence of the CTnDOT tetQ promoter and leader regions was aligned with promoter and leader regions of five other tetQ genes (Fig. 4).
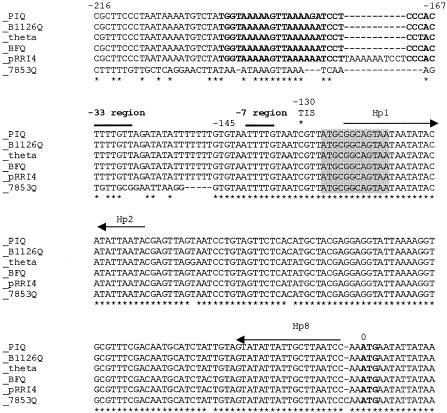
FIG. 4.
DNA sequence alignment of the tetQ leader and promoter sequences from different alleles of tetQ. The ClustalW sequence alignment of tetQ leader regions found in Bacteroides and Prevotella spp. is shown. In all cases, the ATG start codon of tetQ (boldface) is numbered 0. The negative numbers above the sequence indicate the relative upstream positions of the nucleotides to the ATG of tetQ of CTnDOT (theta). The asterisks below the aligned sequences indicate that all residues at that position were identical. The consensus transcription initiation sites (1) are indicated by the asterisk at −130. The horizontal arrows labeled Hp1, Hp2, and Hp8 indicate the positions of the two stems of the hairpin that are important for tetracycline control in the case of the TnDOT tetQ operon. The 3-amino-acid leader peptide region (shaded) is identical in all sequences. PIQ is a tetQ leader sequence from Prevotella intermedia (accession no. U73497), B1126Q is a tetQ leader sequence from Bacteroides fragilis (accession no. Z21523), theta is the CTnDOT tetQ leader sequence (accession no. X58717), BFQ is a tetQ leader from B. fragilis (accession no. Y08615), pRRI4 is a tetQ leader sequence from a plasmid that was isolated from P. ruminicola (accession no. L33696), and 7853Q is the tetQ leader sequence from the Bacteroides CTn, CTn7853 (19).
All six sequences were virtually identical from the start codon of tetQ to position −145. In four of the six cases, the identity extended even further, through position −216. One exception was the pRRI4 sequence, which contained a 10-bp insertion upstream of the −33 sequence region. Even more differences were seen in the entire region upstream of the −7 consensus sequence in the case of the tetQ gene from CTn7853 (19). To determine the effects of the differences in the CTn7853 tetQ upstream region, the same region of the tetQ promoter-leader sequence region CTn7853 that had been cloned from the largest fusion to CTnDOT tetQ was fused in frame to uidA in frame with the same amino acid. This fusion (pYP65) exhibited a tetracycline-dependent 27-fold increase in GUS activity (from 14 to 380 U/mg), although the fully induced activity was only about half of the fully induced activity associated with the fusion to the upstream region from tetQ of CTnDOT. The similarities in expression patterns of the CTn7853 tetQ gene and the CTnDOT tetQ gene are consistent with the hypothesis that most or all of the tetracycline regulation is being mediated by a translational attenuation mechanism similar to that of the CTnDOT tetQ gene and that differences in the −33 region of the promoter have at most a minor effect on gene expression.
DISCUSSION
Previously, we had shown that a hairpin structure in the tetQ mRNA that could be formed by Hp1 and Hp8 was essential for tetracycline-regulated expression of tetQ and the genes in the operon (29). We also found that tetQ is transcribed at high levels even in the absence of tetracycline and that translational fusions of tetQ to uidA gave a much higher GUS activity than transcriptional fusions to the same site, 1 bp removed, in tetQ. Based on these findings, we suggested that production of the proteins encoded by the tetQ operon might be controlled by a translational attenuation mechanism. The results reported here provide important new support for this hypothesis. In particular, the identification of a small leader peptide, whose translation is essential for tetracycline induction, and the presence of an alternative hairpin structure that could preclude formation of the Hp1-Hp8 hairpin provide evidence that the mechanism of tetracycline regulation is translational attenuation.
This model assumes that prevention of Hp1-Hp8 hairpin formation allows a ribosome binding site to be available for the translation of TetQ. There is, however, no obvious ribosome binding site sequence found in this region, as compared to AGAAAGGAG, the complementary sequence to the 3′ end of the 16S rRNA of Bacteroides fragilis or B. thetaiotaomicron. Possible ribosome binding sequences with various identities to this sequence have been found upstream of ATG start sites for the mobilizable Bacteroides transposon Tn4555 (28). In the case of tetQ, there is a possible ribosome binding sequence upstream of the GTG that is in frame with the ATG of tetQ, but there was no detectible GUS activity with a protein fusion to this site (29), indicating that the GTG was not part of the translated tetQ.
A model based on our results is shown in Fig. 5. Normally, the ribosome translates the small leader peptide and then leaves the mRNA. Since the region upstream of the start codon of the leader peptide is only 4 nucleotides long, the affinity of ribosomes for this region is probably low. Thus, in the absence of tetracycline, a small amount of translation of the leader peptide would not preclude the formation of the Hp1-Hp8 hairpin structure, which is predicted to be much more stable than the alternative Hp1-Hp2 hairpin that could also form. Tetracycline entering the cell, however, would cause at least some of the ribosomes to stall on the leader peptide, giving the Hp1-Hp2 structure a chance to form. Our finding that the length of the leader peptide affected the basal level of protein production, with a longer version being associated with a higher basal level and a shorter length being associated with a lower fold increase in protein production, is consistent with this linkage between the amount of leader sequence translated by the ribosome and its ability to destabilize the Hp1-Hp8 hairpin.
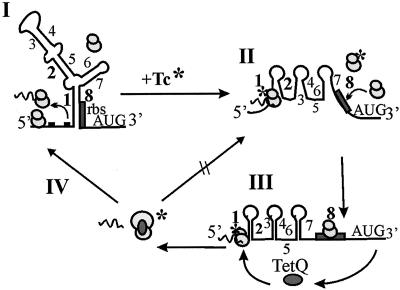
FIG. 5.
Proposed model for tetracycline regulation of the tetQ-rteA-rteB operon. In the absence of tetracycline (step I), ribosomes bind to the mRNA and translate the leader tripeptide (dashed line). The region containing the putative ribosome binding site for tetQ (rbs; shaded box) is tied up in the Hp1-Hp8 hairpin and is thus not available for ribosome binding. In the presence of tetracycline (indicated by an asterisk), those ribosomes that have tetracycline bound to them stall on the leader peptide mRNA (step II), allowing alternate mRNA structures to form (e.g., Hp1-Hp2), thus making the ribosome binding site for tetQ available. The tetQ gene can then be translated, presumably by those ribosomes that do not have bound tetracycline. Once TetQ is made, it alters the conformation of the ribosome and actually causes the release of bound tetracycline (5) and the conformation changes reduce the number of ribosomes that can bind tetracycline (step III). The effect of the altered ribosomes on the translation of Pq, indicated by the arrow (step IV), is the return of the situation in step I. Because the ribosomes are no longer sensitive to tetracycline, the mRNA structures which embed the ribosome binding site can be reformed. Thus, the alternate structures shown in steps II and III are once again prevented from forming. The percentage of ribosomes protected by interaction with TetQ will then determine the level of promoter modulation observed. This fact would explain why the concentration of TetQ produced from a multicopy vector would have a stronger negative regulatory effect than the amount of TetQ produced from a single copy of tetQ in the chromosome.
A puzzling finding from our previous study is explained by the ribosome stalling part of the model. We had found that the presence of the single copy of tetQ actually decreased expression of a fusion of the tetQ promoter with the uidA gene compared to expression seen with subinhibitory levels of tetracycline and no intact tetQ gene present (29). Expression was decreased even further if multiple copies of tetQ were present. Of course, the level of resistance, as indicated by the tetracycline MIC, increased with increasing levels of TetQ, but production of TetQ was substantially decreased. TetQ is one of the ribosomal protection type resistance proteins. The ribosomal protection tetracycline resistance proteins function by interacting with the ribosome in a GTP-dependent way which causes conformational changes in the ribosome which prevent tetracycline from binding and causes the release of ribosome-bound tetracycline (5). In our model, protected ribosomes would no longer stall on the leader peptide or would stall less often. Increasing amounts of TetQ would allow increasing numbers of ribosomes to be protected, thus decreasing induced expression of the operon. This mechanism for limiting production of TetQ and other proteins encoded as the level of TetQ increases may prevent unnecessary production of the tetQ-rteA-rteB operon proteins. RteA and RteB are presumably needed only in the first period after exposure to tetracycline to trigger excision of CTnDOT (4) and to regulate its transfer (30). We have observed previously that excision of CTnDOT occurs in only a small fraction of potential Bacteroides donor cells (4). This may be the effect of having RteA and RteB produced at submaximal levels due to the presence of TetQ.
Translational attenuation involving Hp1-Hp8 and Hp1-Hp2 structures in the mRNA appears to be entirely responsible for the tetracycline regulation of TetQ and the other operon-encoded proteins, but sequences in the promoter region also affect expression. In fact, this mode of tetracycline regulation seems to be typical of all known examples of tetQ genes in Bacteroides and Prevotella species because the leader regions of the mRNA are virtually identical in all of the leader regions of these genes. In particular, Hp1 and Hp8 are absolutely conserved. Translational attenuation has been suggested as the mechanism of regulation for other antibiotic resistance genes, such as the chloramphenicol-inducible cat and cmlA genes and the erythromycin-inducible ermC gene in Bacillus subtilis (10, 15, 16, 18). Hoshino et al. (12) noted that a leader region of a tetracycline resistance gene from a thermophilic Bacillus strain contained some possible stem-loop structures and predicted from this observation that this gene might be regulated by translational attenuation, although this hypothesis was not tested by mutational analysis of the leader region. Thus, translational attenuation appears to be a fairly common mechanism of regulating a variety of antibiotic resistance genes.
Acknowledgments
This work was supported by grant no. AI22383 from the National Institutes of Health.
REFERENCES
- 1.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 2.Berens, C., and W. Hillen. 2003. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 270:3109-3121. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, H. B., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification system of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for the excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 5.Connell, S. R., C. A. Trieber, G. P. Dinos, E. Einfeldt, D. E. Taylor, and K. H. Nierhaus. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubnau, D. 1985. Induction of ermC requires translation of the leader peptide. EMBO J. 4:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubnau, D. 1984. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. Crit. Rev. Biochem. 16:103-132. [DOI] [PubMed] [Google Scholar]
- 8.Feldhaus, M. J., V. Hwa, Q. Cheng, and A. A. Salyers. 1991. Use of an Escherichia coli β-glucuronidase gene as a reporter gene for investigation of Bacteroides promoters. J. Bacteriol. 173:4540-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrod, R., and P. S. Lovett. 1997. Leader peptides of inducible chloramphenicol resistance genes from Gram-positive and Gram-negative bacteria bind to yeast and Archaea large subunit rRNA. Nucleic Acids Res. 25:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrod, R., and P. S. Lovett. 1995. Peptide inhibitors of peptidyltransferase alter the conformation of domains IV and V of large subunit rRNA: a model for nascent peptide control of translation. Proc. Natl. Acad. Sci. USA 92:8650-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holdeman, L. V., and W. E. C. Moore. 1975. Anaerobe laboratory manual, 4th ed. Virginia Polytechnical Institute and State University, Blacksburg, Va.
- 12.Hoshino, T., T. Ikeda, N. Tomizuka, and K. Furukawa. 1985. Nucleotide sequence of the tetracycline resistance gene of pTHT15, a thermophilic Bacillus plasmid: comparison with staphylococcal TcR controls. Gene 37:131-138. [DOI] [PubMed] [Google Scholar]
- 13.Kisker, C., W. Hinrichs, K. Tovar, W. Hillen, and W. Saenger. 1995. The complex formed between Tet repressor and tetracycline-Mg2 ihsbop + reveals mechanism of antibiotic resistance. J. Mol. Biol. 247:260-280. [DOI] [PubMed] [Google Scholar]
- 14.Leng, Z., D. E. Riley, R. E. Berger, J. N. Krieger, and M. C. Roberts. 1997. Distribution and mobility of the tetracycline resistance determinant tetQ. J. Antimicrob. Chemother. 40:551-559. [DOI] [PubMed] [Google Scholar]
- 15.Lovett, P. S. 1996. Translation attenuation of chloramphenicol resistance in bacteria—a review. Gene 179:157-162. [DOI] [PubMed] [Google Scholar]
- 16.Lovett, P. S., and E. J. Rogers. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60:366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosebrough, A. S. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 143:265-275. [PubMed] [Google Scholar]
- 18.Mayford, M., and B. Weisblum. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J. 8:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolich, M. P., N. B. Shoemaker, G. R. Wang, and A. A. Salyers. 1994. Characterization of a new type of Bacteroides conjugative transposon, Tcr Emr 7853. J. Bacteriol. 176:6606-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salyers, A. A., N. B. Shoemaker, and A. M. Stevens. 1995. Tetracycline regulation of conjugal transfer genes, p. 393-400. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 21.Sambrook, J., and D. B. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Shoemaker, N. B., and A. A. Salyers. 1987. Facilitated transfer of IncPβ R751 derivatives from the chromosome of Bacteroides uniformis to Escherichia coli recipients by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 169:3160-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoemaker, N. B., and A. A. Salyers. 1988. Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J. Bacteriol. 170:1651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens, A. M., N. B. Shoemaker, L.-Y. Li, and A. A. Salyers. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens, A. M., N. B. Shoemaker, and A. A. Salyers. 1990. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J. Bacteriol. 172:4271-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, Y. A., P. He, and D. B. Clewell. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid 42:1-12. [DOI] [PubMed] [Google Scholar]
- 29.Wang, Y., N. B. Shoemaker, and A. A. Salyers. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 186:2548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]