Abstract
The isolation of a Streptococcus thermophilus CNRZ368 mutant displaying a long-chain phenotype allowed us to identify the cse gene (for cellular segregation). The N terminus of Cse exhibits high similarity to Streptococcus agalactiae surface immunogenic protein (SIP), while its C terminus exhibits high similarity to S. thermophilus PcsB. In CNRZ368, deletion of the entire cse open reading frame leads to drastic lengthening of cell chains and altered colony morphology. Complementation of the Δcse mutation with a wild-type allele restored both wild-type phenotypes. The central part of Cse is a repeat-rich region with low sequence complexity. Comparison of cse from CNRZ368 and LMG18311 strains reveals high variability of this repeat-rich region. To assess the impact of this central region variability, the central region of LMG18311 cse was exchanged with that of CNRZ368 cse. This replacement did not affect chain length, showing that divergence of the central part does not modify cell segregation activity of Cse. The structure of the cse locus suggests that the chimeric organization of cse results from insertion of a duplicated sequence deriving from the pcsB 3′ end into an ancestral sip gene. Thus, the cse locus illustrates the module-shuffling mechanism of bacterial gene evolution.
Among gram-positive cocci, some species grow as single cells and others grow as grouped cells, linked together into various arrangements. In particular, cocci from several genera, such as Streptococcus, Lactococcus, Peptostreptococcus, Ruminococcus, and Coprococcus, grow as cell chains (48). The number of cells per chain varies considerably among species and strains (14). Indeed, chain length can range from two cells, as in Streptococcus pneumoniae, to around 100 cells in Streptococcus mitior (14). Few genes have been implicated in short-chain phenotypes, since their inactivation leads to an increased number of cells per chain. One of the most striking phenotypes is that of an S. pneumoniae lytA lytB double mutant, whose chain length can reach more than 100 cells, while the wild type typically grows as pairs (12). The shortening of cell chains was ascribed to the cell wall hydrolytic activity of LytA and LytB proteins in S. pneumoniae (25) and of AcmA in Lactococcus lactis (6). The increase of cell number per chain could be accompanied by obvious changes in cellular morphology. Thus, rodA, pbp2, and mreD mutants from Streptococcus thermophilus display not only long cell chains but also rod-shaped cells instead of ovoid wild-type cells (50, 52, 54). Also, an S. pneumoniae mutant depleted for expression of the essential pcsB gene forms long chains with irregularly shaped cells (33). Although the biological function of chains is unknown, it was recently demonstrated that the adhesion ability is reduced by cell chain lengthening in L. lactis (30).
In this study, we focused on a gene called cse for its role in cellular segregation, identified in S. thermophilus. This bacterium is a lactic acid bacterium used as a starter of fermentation for the conversion of milk into yogurt and many cheeses (e.g., Emmental, Gruyère, mozzarella, and cheddar). We describe the chimeric structure of cse and its central region, which is repeat rich and exhibits intraspecies sequence variability. The construction of several mutants showed that this chimeric and variable gene encodes a functional protein involved in cellular segregation and colony morphology and allowed assessment of the impact of the central part variability on cell segregation activity.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
All strains and plasmids used in this work are presented in Table 1. Depending on the experiments, S. thermophilus and its derivatives were cultivated in milk medium, TPPY (tryptose, 7 g · liter−1; Proteose peptone, 7 g · liter−1; yeast extract, 2 g · liter−1; lactose, 20 g · liter−1) (5), or M17 (51) medium without shaking. Milk medium was used for strain storage, M17 was used for mutant generation, and TPPY was employed for phenotypic analysis. Phenotypic analyses were performed at 42°C, the optimal growth temperature of S. thermophilus. Erythromycin was added at 2 μg · ml−1 when required. S. thermophilus derivative strains containing pGh9 (27) or pNST260+ plasmid derivatives were cultivated at 30°C when plasmid self-maintenance was required and at 42°C for selection of clones with the chromosome's integrated plasmid. S. thermophilus strains containing pFUN or its derivatives were cultivated at 30°C. L. lactis strains were grown at 30°C in M17 medium (51) supplemented with 0.5% glucose. Erythromycin was added at 5 μg · ml−1 when required.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant phenotype(s) or genotype(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. thermophilus | ||
| CNRZ368 | Wild-type strain | INRA-CNRZ, strain collection |
| 16D10 | EmrS. thermophilus CNRZ368 containing integrated pGh9:ISSI into cse | This work |
| CNRZ368-Δcse | S. thermophilus CNRZ368 with Δcse mutation | This work |
| CNRZ368-Δcse-T | Emr, CNRZ368-Δcse with pNST260+ integrated into the chromosome | This work |
| CNRZ368-Δcse-C | Emr, CNRZ368-Δcse with pNST260+::cse integrated into the chromosome | This work |
| LMG18311 | Wild-type strain | BCCM/LMG, strain collection |
| LMG18311-Δcse | S. thermophilus LMG18311 with Δcse mutation | This work |
| LMG18311-csevarCNRZ368 | S. thermophilus LMG18311 with the chimeric allele cseLMG18311varCNRZ368 | This work |
| L. lactis subsp. cremoris MG1363acmAΔ1 | Derivative of MG1363 wild-type strain containing 701-bp SacI-SpeI chromosomal deletion in acmA gene | 6 |
| E. coli | ||
| EC101 | supE hsd-5 thi Δ(lac-proAB) F′ (traD6 proAB+lacIqlacZΔM15) repA+, derivative of TG1 strain (47) | 23 |
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 endA1 gyrA96 thi-1 relA1 | 47 |
| BL21 (DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene I) | 47 |
| Plasmids | ||
| pGh9 | Emr, thermosensitive replication origin from pVE6002 | 27 |
| pGh9:ISS1 | pGh9 with ISS1 | 28 |
| pGh9::ΔcseCNRZ368 | pGh9 with Δcse and CNRZ368 surrounding regions | This work |
| pGh9::ΔcseLMG18311 | pGh9 with Δcse and LMG18311 surrounding regions | This work |
| pGh9::cseLMG18311varCNRZ368 | pGh9 with chimeric allele cseLMG18311varCNRZ368 | This work |
| pNST260+ | pGh9 with int and attI from ICEStI | G. Guédon, personal communication |
| pNST260+::cse | pNST260+ with CNRZ368 cse ORF, its putative promoter and terminator | This work |
| pFUN | Emr and Ampr, derivative of pIL252, with ΔSPnuc gene | 38 |
| pFUN::cse | pFUN with cse-ΔSPnuc translational fusion | This work |
| pET15b | Protein expression vector | Novagen |
| pET15b::cse | pET15b with hexa-His-Cse translational fusion | This work |
| pET15b::cse1-183 | pET15b with hexa-His-Cse1-183 translational fusion | This work |
| pET15b::cse184-461 | pET15b with hexa-His-Cse184-461 translational fusion | This work |
Recombinant plasmids derived from pGh9 were transformed into Escherichia coli EC101, a TG1 strain containing a chromosomal copy of the pWV01 repA gene (6), and selected at 37°C on Luria-Bertani (LB) (47) containing 150 μg of erythromycin ml−1. Recombinant plasmids derived from pFUN, which was kindly provided by I. Poquet from INRA (Jouy en Josas, France) (38) were transformed into E. coli DH5α and selected on LB medium containing 100 μg of ampicillin ml−1.
DNA manipulations.
Preparation of chromosomal and plasmid DNA and Southern analysis were performed according to standard protocols (47). Sequencing was performed by using dye terminator chemistry on an ABI Prism 377 genetic analyzer (PE Biosystems). Sequence data were analyzed with BLAST (1, 2), SignalP (35), Mfold (59), Dot plot (http://arbl.cvmbs.colostate.edu/molkit/dnadot/), SEG (56), and PSIPRED (29) software. GenBank, the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), and the Codon Usage Database (http://www.kazusa.or.jp/codon/) (32) were consulted. Sequence data of S. thermophilus LMG18311 strain were obtained from the UCL Life Sciences Institute website at http://www.biol.ucl.ac.be/gene/genome/. Sequencing of S. thermophilus LMG18311 was supported by the Walloon region (BIOVAL grant no. 9813866).
Nuclease assay on agar plates.
Extracellular production of nuclease activity by E. coli and S. thermophilus were detected by the metachromatic agar diffusion method, as previously described (21, 24). Briefly, following bacterial growth on solid media, plates were overlaid with toluidine blue-DNA agar and incubated at 37°C for 2 h. The presence of a pink halo around colonies indicates extracellular nuclease activity. E. coli strains were not incubated for more than 2 h, since false positives could appear after this time (38).
Recombinant DNA and mutant construction.
Oligonucleotides used in this study were purchased from MWG Biotech AG (Ebersburg, Germany) and are listed Table 2.
TABLE 2.
Oligonucleotides used in this work
| Oligonucleotide | Sequencea (5′-3′) |
|---|---|
| delcse.E5′ | CCCCCCCCAAGCTTACAAGATCAGTTTGGAAC |
| delcse.I5′ | CCCCCCGAATTCTGATAACATGTATAATTCAT |
| delcse.I3′ | CCCCCCGAATTCATTTATCCATAATGAGTACA |
| delcse.E3′ | CCCCCCCTGCAGCAGCCTGTGGTTGAAG |
| fcompl | TTGTTGGCGCGCTTTAGCCGATTTGGCTTTTGAAG |
| rcompl | TTGTTGGCGCGCCCCATTATTTTCTCAGGATGAAT |
| racse.E5′ | CCCCCCAAGCTTTGAATGTTTTGGCTAATATC |
| racse.I5′ | CCCCCCGATATCAATAGCGTCTGCAACT |
| racse.I3′ | CCCCCCGATATCAACTCCTAATACATATC |
| racse.E3′ | CCCCCTGCAGTTATGGATAAATATAATATAC |
| ravar.5′ | CATGTTACTTCAGCAACAAC |
| ravar.3′ | GCATATTCGTATGTAGCTGC |
| fnuc.cse.5′ | CCCCCCGAATTCTGATAGGGATAGGGCTT |
| fnuc.cse.15 | CCCCCCAAGCTTCATGTATAATTCATTCCTT |
| fnuc.cse.13′ | CCCCCCAAGCTTGATGAAACTTCTCACTG |
| fnuc.cse.3′ | CCCCCCGGATCCCCTGGATAAATATAATATACAG |
| sex.E5′ | CCCCCCCCATATGTTATCAAAATCTAAAAC |
| sex.I5′ | CCCCGGATCCTTAAGCTGGAGTATCTGATGC |
| sex.I3′ | CCCCCCCCATATGTACGCAGATACTGAACAA |
| sex.E3′ | CCCCGGATCCTTATGGATAAATATAATATAC |
Restriction enzyme sites are underlined.
(i) Translational fusion with Δspnuc.
To obtain the plasmid pFUN::cse, the region covering the putative promoter region and the cse open reading frame (ORF) except its stop codon, was amplified by PCR and introduced into pFUN, leading to cse-Δspnuc fusion. For pFUN::Δspcse, the regions flanking the sequence encoding the putative signal peptide (the 81 bp following the ATG putative initiation site of translation) were amplified by PCR and introduced into pFUN. This led to a Δspcse-Δspnuc fusion where the sequence encoding the putative signal peptide of Cse was replaced by a HindIII restriction site. Following selection in E. coli DH5α, both constructs were introduced into S. thermophilus by electroporation.
(ii) Deletion of cse in S. thermophilus CNRZ368 and LMG18311 strains.
In-frame cse deletion mutants were constructed as previously described (52). Briefly, the two regions flanking the locus to be deleted were independently amplified by PCR, digested by appropriate restriction enzymes, and joined by ligation together and with pGh9. After introduction of the recombinant plasmid into S. thermophilus, two crossovers, upstream and downstream of the deleted region, were selected (52). Thus, only the first and the last three codons of the cse ORF were kept, and the remainder of the ORF was replaced by an EcoRI restriction site. The cse deletion was checked by PCR and Southern hybridization (data not shown).
(iii) Δcse complementation.
Complementation of the Δcse mutation was carried out by inserting the wild-type allele in trans. For this purpose, the pNST260+ plasmid (G. Guédon, personal communication), carrying a gene encoding the ICESt1 integrase and its attI attachment site, was used (7). This plasmid can integrate into the S. thermophilus CNRZ368 chromosome at the attR site of ICESt1 (G. Guédon, personal communication). The entire cse ORF with its putative promoter and terminator was amplified by PCR and ligated into pNST260+. Following selection in E. coli EC101, the construct was introduced into S. thermophilus by electroporation. As a control, S. thermophilus was also transformed with empty pNST260+. Integration of pNST260+ and pNST260+::cse into the chromosome was selected at 42°C, the replication-restrictive temperature, in the presence of erythromycin. Plasmid integration into the chromosomal attachment site was checked by Southern hybridization (data not shown).
(iv) Allelic replacement of the var-cse region.
The approach for var-cse replacement involved the wild-type sequence containing GAT and ATC sequences localized, respectively, upstream and downstream of var-cse. These sequences allow an EcoRV restriction site formation if they are assembled. The plasmid used for replacement of the var-cse region of LMG18311 by that of CNRZ368 was constructed in a two-step cloning procedure. The var-cse flanking regions from the LMG18311 strain were amplified by PCR. After digestion by appropriate restriction enzymes, PCR fragments were ligated together, generating an EcoRV restriction site, and inserted into pGh9. This EcoRV restriction site was generated by assembling the GAT and ATC sequences that flank the wild-type var-cse sequence. The plasmid carrying the two PCR fragments was selected in E. coli EC101. Then the region localized between the GAT and ATC sequences including the var-cse region from CNRZ368 (as defined in Fig. 2D) was amplified by PCR. The PCR product was next treated at 72°C for 30 min with Pfu polymerase for generation of blunt ends. This last step removes the 3′ adenosine overhang added during the PCR. The polished fragment was inserted into the plasmid carrying the var-cse flanking regions previously digested by EcoRV to generate blunt ends. The resulting plasmid was selected in E. coli EC101. The correct insert orientation was checked by sequencing. Then the plasmid was introduced into S. thermophilus LMG18311 by electroporation. The transformed strain was checked by Southern hybridization with a specific probe for LMG18311 genomic DNA (37). Two crossover events, surrounding the var-cse region, were selected to generate the LMG18311-csevarCNRZ368 mutant.
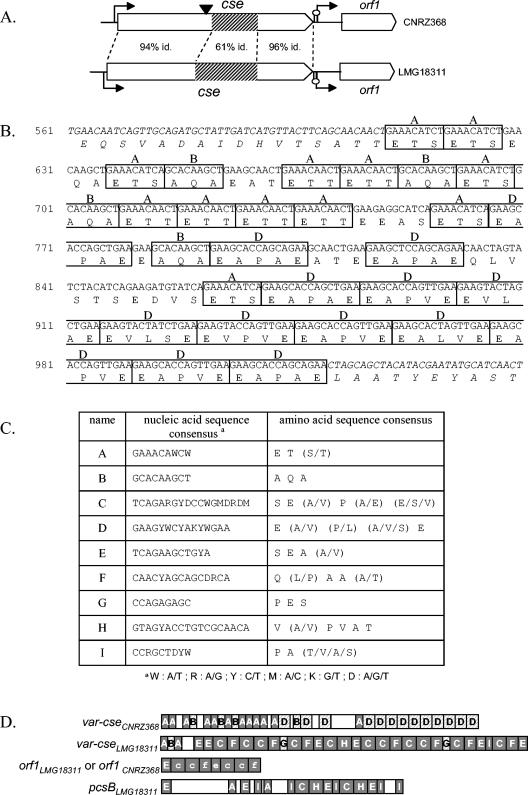
FIG. 2.
var-cse variability and repeat content of cse, pcsB, and orf1. (A) Schematic representation of the cse locus from S. thermophilus strains CNRZ368 and LMG18311. Open arrows represent ORFs and indicate their reading direction, broken arrows indicate putative promoters, and hairpin loops symbolize putative rho-independent terminators. The black arrowhead indicates the insertion site of pGh9:ISS1 within the genome of the 16D10 mutant. The percentage of sequence identity (id.) is indicated between 5′ ends, central parts, and 3′ ends of cse from CNRZ368 and LMG18311. Hatched boxes represent repeat-containing regions (var-cse). (B) Nucleic acid and amino acid sequences of var-cseCNRZ368 and the proximal region. The regions flanking var-cse are italicized. Each repeat unit is boxed, and the name of the repeat unit is indicated above the box. (C) Table of consensus repeat sequences represented in cse, pcsB, and orf1. (D) Schematic representation of repeat-containing regions. Each repeat unit is represented by a letter-containing box. Repeat units, in accordance with consensus sequences presented in panel C, are represented by capital letters, while those slightly divergent from the consensus (17% of maximum divergence) are represented by lowercase letters. Empty boxes represent regions without any repeats. Repeats in grey are common between cse from S. thermophilus strains CNRZ368 and LMG18311, orf1, and pcsB from S. thermophilus strain LMG18311. Hatched boxes represent repeats only found in cse.
(v) Plasmid constructs for protein overproduction.
The whole cse ORF and the regions encoding Cse parts from amino acid 1 to 183 and from amino acid 184 to 461 were amplified by PCR. The PCR products were independently introduced into the pET15b plasmid (Novagen), ending in an in-frame fusion downstream of the hexa-His encoding sequence. After selection in E. coli DH5α, the constructs were introduced into E. coli BL21(DE3) for protein overproduction.
Microscopy.
Colonies and cells were observed with a Nikon OPTIPHOT microscope mounted with phase contrast equipment (Ph). Colony examination was done at a magnification of ×100. Cells were observed at a magnification of ×100, with the condenser turret at position Ph4, or at a magnification of ×1,000 by phase contrast.
Protein overproduction.
E. coli BL21(DE3) strains transformed with pET15b and derivatives were grown in LB medium at 37°C supplemented with 50 μg of ampicillin ml−1. At an optical density at 600 nm (OD600) of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 100 μM during 4 h to induce expression of N-terminally hexa-His-tagged proteins. To extract proteins, cell pellets resuspended in sample electrophoresis buffer were heated for 5 min at 95°C. For cell lysis testing in nondenaturing conditions, proteins were extracted by sonication.
SDS-PAGE and renaturing SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (22) with 10% (wt/vol) polyacrylamide. Lytic activity was tested by SDS-PAGE, performed with gels containing 10% polyacrylamide (wt/vol) and 0.2% (wt/vol) S. thermophilus cells or Micrococcus lysodeikticus cells, followed by a protein renaturation step. The S. thermophilus cells used as substrates for lytic activity were prepared as previously described (39) with the following modification: after lyophilization, cells were resuspended in distilled water at a 10% final concentration and autoclaved for 20 min at 120°C. S. thermophilus proteins were extracted by glass bead disruption of cells as previously described (17). After electrophoresis, gels were gently shaken in 100 ml of water at 4°C for 1 h. Then water was replaced by 100 ml of 20 mM Tris-HCl (pH 7) containing 1% (vol/vol) Triton X-100 for overnight incubation at 42°C. Bands of lytic activity were visualized as previously described (6).
Nucleotide sequence accession numbers.
DNA sequences reported in this paper have been deposited in GenBank under accession numbers AY695844, AY730642, and AY730643.
RESULTS
cse involvement in colony morphology and cell segregation.
The 16D10 mutant, displaying a long-chain phenotype in addition to a variant colony morphology, was selected from an S. thermophilus CNRZ368 mutant collection. This collection was previously obtained by insertional mutagenesis (53) with the pGh9:ISSI plasmid (28). The locus disrupted by pGh9:ISSI in the 16D10 mutant was identified by cloning and sequencing the 2,697 bp flanking the disruption site. Sequence analysis revealed that pGh9:ISS1 disrupted an ORF of 1,386 bp, named cse for cell segregation. The CNRZ368 cse ORF encodes a putative 462-amino-acid protein of unknown function with a deduced molecular mass of approximately 48.5 kDa. The first 28 amino acids display Sec-dependent signal peptide characteristics as predicted by signalP software (35), suggesting that Cse is exported. The Cse central part, containing repeats (see below), is highly enriched in alanine, glutamate, threonine, and serine (20.4, 32.9, 15.3, and 8.3%, respectively) residues compared to the mean content of S. thermophilus proteins (6.1, 5.9, 5.7, and 4.6%, respectively), according to the Codon Usage Database (32). This central region is potentially hydrophilic (due to its high glutamate, serine, and threonine content) and acidic (due to its glutamate enrichment).
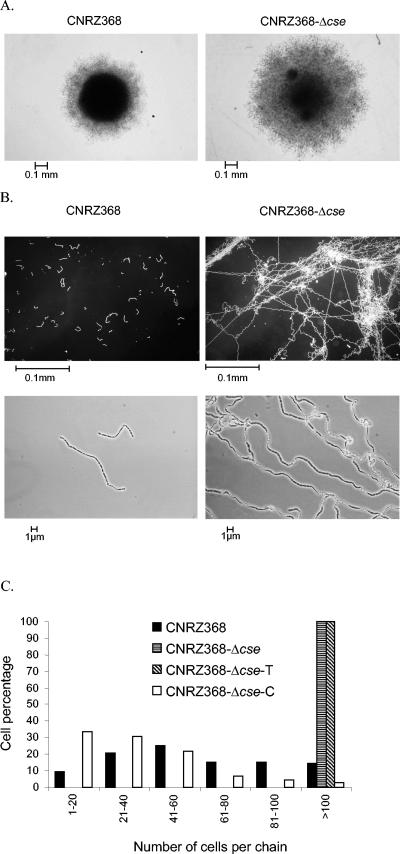
To confirm the involvement of cse in these phenotypes, a cse null mutant was constructed by in-frame deletion of the cse ORF, resulting in a CNRZ368-Δcse strain. Colonies of this mutant were flatter, larger, and less opaque than wild-type ones (Fig. 1A). Phase-contrast photonic microscopy observations revealed that CNRZ368-Δcse chains were much longer than wild-type ones (Fig. 1B). This was reinforced by counting the number of cells per chain for each strain. Only 15% of wild-type cells formed chains containing more than 100 cells in stationary phase, whereas 100% of CNRZ368-Δcse cells did (Fig. 1C). During exponential growth (OD600 = 0.2), 76.5% of the wild-type cells counted formed chains containing less than 40 cells and none of them formed chains containing more than 80 cells. On the contrary, 87.5% of the CNRZ368-Δcse cells formed chains containing more than 100 cells. These results show that the long-chain phenotype of the CNRZ368-Δcse mutant is not growth phase dependent. No major modification of the cell shape or size was observed in the CNRZ368-Δcse mutant. Moreover, mutant and wild-type strain growth curves showed no significant difference in either doubling time or in stationary phase entry OD600 value (data not shown).
FIG. 1.
Morphological consequences of cse deletion. (A) Colony morphology pictured after 20 h of growth; (B) cell chain morphology of S. thermophilus CNRZ368 strains; (C) results from counting number of cells per chain. A total of 1,000 cells were counted from three independent cultures. Cells were photographed and counted in stationary phase after 20 h of growth. After this time of growth, OD600 values reached 0.66 ± 0.074, 0.56 ± 0.038, 0.50 ± 0.029, and 0.62 ± 0.022, respectively, for strains CNRZ368, CNRZ368-Δcse, CNRZ368-Δcse-T, and CNRZ368-Δcse-C.
Complementation was performed by using the pNST260+ integrative plasmid (G. Guédon, personal communication). This plasmid can integrate into the S. thermophilus CNRZ368 chromosome in the attR site of ICESt1 (G. Guédon, personal communication), a resident integrative element (7). The Δcse mutation was complemented in trans by chromosomal integration of the cse wild-type gene carried by the pNST260+::cse plasmid, resulting in the CNRZ368-Δcse-C strain. The negative control consisted of the chromosomal insertion of empty pNST260+, resulting in the CNRZ368-Δcse-T strain. The complemented strain exhibited a wild-type phenotype with respect to colony morphology and number of cells per chain, while the negative control maintained a mutant phenotype (Fig. 1C and colony morphology data not shown). These results demonstrate that cse is involved in colony morphology and cell segregation.
Central part of cse is repeat rich and variable.
Alignment of the cse nucleotide sequence from strains CNRZ368 and LMG18311 (cseCNRZ368 and cseLMG18311) showed that the first 600 bp and the last 346 bp of these ORFs display 94 and 96% identity, respectively (Fig. 2A). In contrast, both cse allele central parts, consisting of 443 bp in cseCNRZ368 and 503 bp in cseLMG18311, show only 61% identity. Because of its variability, this region was called var-cse. Both var-cse regions show a high level of redundancy consisting of short direct repeats, with tandem or dispersed arrangement (Fig. 2B). As a consequence of its high content in short repeats, it is a low-complexity region as detected by the SEG program. Nine different repeat units (named A to I) that varied in size and sequence were distinguished (Fig. 2C). All of the repeated units have a size divisible by three and are in frame. The direct consequence is that all nucleic acid repeat sequences correspond to amino acid repeats in the cse product (Fig. 2B). The comparison of var-cse from S. thermophilus CNRZ368 and LMG18311 showed that var-cseLMG18311 is 60 bp longer than var-cseCNRZ368. Additionally, the repeat content of both var-cse alleles is qualitatively different (Fig. 2D). In conclusion, the central part of cse is almost exclusively built with repeated sequences and displays a high degree of intraspecies variability.
Secondary structures were predicted for Var-Cse by using the PSIPRED method (19). Almost the entire Var-CseCNRZ368 (93%) region and the whole Var-CseLMG18311 region are predicted to adopt coil structures, suggesting that this region is a nonglobular domain. However, the confidence value of the prediction is low (3.56 ± 1.79 for Var-CseCNRZ368 and 3.45 ± 1.83 for Var-CseLMG18311, on a 0 to 10 scale). The predicted nonglobular structure of this region could explain the tolerance for its high genetic variability.
cse is chimeric.
The nucleotide sequence of cse and its flanking regions in S. thermophilus LMG18311 was used in a BLASTN search against the genome of the same strain. Results revealed a region of 619 bp, named HRC1 for homologous region, copy 1, sharing 93% identity with another region of 618 bp, localized elsewhere in the genome and named HRC2 for homologous region, copy 2 (Fig. 3A). HRC1 includes the last 350 bp of cse, the following intergenic region, and the first 58 bp of the downstream orf1. HRC2 includes the last 350 bp of pcsB, the following intergenic region, and the first 58 bp of the downstream rppk ORF (encoding a putative ribose-phosphate pyrophosphokinase) (Fig. 3A). Thus, the C-terminal part of Cse is homologous to the C-terminal part of PcsB from S. thermophilus LMG18311 (Fig. 3A). BLAST searches within nonredundant databases showed that a PcsB orthologue is present in the Streptococcus and Lactococcus genera, whereas no protein with significant similarity to the whole Cse protein was found in these organisms. PcsB is essential in the maintenance of cell shape in Streptococcus species (9, 33, 34, 42, 43). Searches of the Conserved Domain Database show that both PcsB and Cse possess a region homologous to the CHAP domain (cysteine histidine-dependent aminohydrolase/peptidase) (Fig. 3A) which possesses a glutathionylspermidine amidase activity in E. coli (4). The CHAP domain is widely distributed in extracellular proteins, where it is supposed to be involved in peptidoglycan hydrolysis (3, 45). In contrast, the N-terminal part of Cse is homologous to the N-terminal part of the surface immunogenic protein (SIP) from Streptococcus agalactiae (Fig. 3A). Searches of the Conserved Domain Database showed that the N-terminal parts of both SIP and Cse contain regions homologous to the LysM domain (Fig. 3A), found in extracellular proteins and involved in their attachment to the cell wall (49). This domain is probably responsible for preferential localization of the SIP surface protein at the cell poles (46, 49).
FIG. 3.
cse, orf1, and their putative products are chimeric and may result from a duplication event. (A) Results from identity searches. Open arrows represent ORFs and indicate their reading directions, broken arrows indicate putative promoters, and hairpin loops symbolize putative rho-independent terminators. Hatched boxes represent repeat-containing regions, and grey color points out sequences from the cse and pcsB regions displaying high identity. Thick lines represent proteins, black boxes show putative signal peptides, and boxes with horizontal and oblique hatching symbolize putative LysM and CHAP domains, respectively. Homologous regions between proteins are linked, and positions of amino acids delimiting homologous regions are indicated. Identity (id.) and similarity (sim.) results are mentioned. (B) Duplication hypothesis. Grey and black arrows represent ORFs. Dashed lines both delimit the duplicated region and indicate the insertion site.
Interestingly, the C-terminal region of SIP is homologous to the C-terminal part of a putative protein (ORF1) (Fig. 3A) encoded by the gene immediately downstream of cse (Fig. 3A).
The CDART database contains two hypothetical proteins from Streptococcus mutans and Streptococcus intermedius with one LysM domain and one CHAP domain (GenBank accession numbers NP_720819 and BAB61101, respectively). These proteins have a LysM domain at the N terminus and a CHAP domain at the C terminus. Both proteins show significant similarity (48% identity and 63% similarity) over 92% of their length. Their CHAP domains show significant similarity with Cse (51% identity and 62% similarity for NP_720819, 60% identity and 72% similarity for BAB61101), contrary to that of the LysM-containing region, which does not exhibit significant similarity with either Cse or SIP.
pcsB and orf1 contain common repeats with cse.
Considering partial homology between pcsB and cse loci, pcsBLMG18311, orf1LMG18311, and rppkLMG18311 were scanned for repeats. The analysis revealed that the pcsBLMG18311 central region and orf1LMG18311 5′ end also contain repeats (Fig. 3A) that are common to both cse alleles (Fig. 2D). More precisely, all repeats found in pcsBLMG18311 (A, C, E, H, and I) are also represented in var-cseLMG18311, and one repeat (A) found in pcsBLMG18311 is represented in var-cseCNRZ368. The orf1 5′ end from CNRZ368 and LMG18311 strains (99.6% identity between orf1 alleles) contains C, E, and F repeated sequences, although all but one are degenerate. Additionally, the succession ECCF is found twice in orf1 and displayed once in var-cseLMG18311 (Fig. 2D).
Interestingly, var-cseLMG18311 and the orf1LMG18311 repeat-rich regions are located closed to HRC1 boundaries (Fig. 3A). Thus, HRC1 is flanked by two repeat-rich regions which contain repeated motifs common to each other and to the pcsBLMG18311 repeat-rich region.
Extracellular localization of Cse.
According to SignalP (35) analysis, Cse contains a canonical putative signal peptide, suggesting that it is exported from the cell. To test this hypothesis, cse was fused to the reporter gene Δspnuc carried by the plasmid pFUN (38). The Δspnuc gene encodes a Staphylococcus aureus nuclease lacking its signal peptide (10). The region containing the putative promoter and the entire cse ORF (except the stop codon) was cloned into the pFUN vector to generate a cse-Δspnuc fusion. The resulting pFUN::cse vector was introduced into E. coli DH5α and S. thermophilus CNRZ368. Following growth, the transformants were overlaid with TBD agar (21), allowing detection of extracellular nuclease activity by visualization of a pink halo. A pink halo was visualized around colonies of each transformed strain (data not shown), indicating that the fusion protein was extracellular. As a negative control, the same region lacking the putative signal peptide encoding sequence was cloned into pFUN to generate a Δspcse-Δspnuc fusion. Transformed by this construct, both the E. coli and S. thermophilus colony surroundings remained blue. These results indicate that Cse is exported and that its export is signal peptide dependent.
Impact of var-cse variability on Cse cell segregation activity.
The variability of var-cse raised the question of a possible variability of Cse cell segregation activity. Analysis of S. thermophilus CNRZ368 and LMG18311 cell distribution in chains revealed intraspecies variability of cell chain length (Fig. 4A). Indeed, 85% of CNRZ368 cells formed chains from 1 to 100 cells, whereas only 39% of LMG18311 cells are in this size range. Thus, a large proportion of LMG18311 cell chains were much longer than those of CNRZ368. Due to the involvement of cse in cell segregation, we hypothesized a correlation between variability of the cell chain length and var-cse variability. A Δcse mutant of LMG18311 was found to exhibit a long-chain phenotype, as the CNRZ368-Δcse strain does (Fig. 4B), confirming that the cseLMG18311 allele was functional. At the cse chromosomal locus, the var-cse region (Fig. 2D) was replaced by that of CNRZ368 in the LMG18311 genetic background. This was done by allelic replacement, taking into account that var-cse homologous flanking regions are not identical even if they show at least 94% identity (Fig. 2A). Thus, the plasmid used for allelic replacement should contain var-cse from strain CNRZ368 flanked by cse homologous regions from LMG18311 strain. No significant difference was seen between LMG18311 and LMG18311-csevarCNRZ368 strains with respect to chain length (Fig. 4A). This indicates that the cell segregation activity of Cse is not affected by the high variability of its central region.
FIG. 4.
Influence of var-cse variability on Cse cell segregation activity. (A) Results of counting numbers of cells per chain. A total of 1,000 cells were counted from three independent cultures. (B) Cell morphology of S. thermophilus strain LMG18311. Cells were photographed or counted in stationary phase after 20 h of growth. After this period of growth, OD600 values reached 0.66 ± 0.074, 1.1 ± 0.014, 1.1 ± 0.032, and 0.87 ± 0.049, respectively, for strains CNRZ368, LMG18311, LMG18311-csevarCNRZ368, and LMG18311-Δcse.
Cell wall hydrolase activity.
Since it was previously reported that cell segregation results from cell wall hydrolytic activity detectable by zymography (6, 8, 18, 36, 41, 58), Cse was tested for this activity. Thus, wild-type and CNRZ368-Δcse strain protein extracts were tested by zymography. Under our conditions, no difference was detected between the two lytic activity profiles, with either autoclaved S. thermophilus or M. lysodeikticus cells as a substrate (data not shown). Therefore, we tried to overproduce the Cse protein in E. coli with an N-terminal hexa-His tag. However, this approach was unsuccessful for the full-length Cse protein, as in the case of the full-length AcmB protein from L. lactis previously reported (16). The authors were able to circumvent this issue by overproducing AcmB fragments. Using the same alternative way, we were able to overproduce two Cse fragments in E. coli, one comprising the region from amino acid 1 to 183 and the other from amino acid 184 to 461. Thus, the N-terminal region comprised the LysM putative domain, and the C-terminal region comprised both the Var-Cse region and the CHAP domain. Renaturing SDS-PAGE, performed with crude protein extract, either on autoclaved S. thermophilus or M. lysodeikticus cells, did not allow detection of any lytic activity attributable to either the N-terminal or the C-terminal fragment. To exclude that this negative result was not due to the incorrect refolding of the Cse fragments, a cell lysis test was performed under nondenaturing conditions. For this purpose, proteins of E. coli Cse fragment overproducer strains were extracted by sonication. The control crude protein extract was obtained from an E. coli strain carrying the pET15b plasmid without an insert. After checking that the overproduced proteins were recovered in the soluble fraction, 200 μg of crude protein extract were added to a TPPY suspension of autoclaved S. thermophilus or M. lysodeikticus cells and incubated at 42°C. OD600 measurements over time did not reveal any significant differences between crude protein extracts containing the overproduced Cse fragments and the control.
Finally, we tried an indirect way, by checking whether cse would be able to suppress a long-chain phenotype resulting from a murein hydrolase defect. For this purpose, L. lactis ΔacmA was transformed with pNST260+::cse, and the number of cells per chain was counted. No difference in chain length was noticed between ΔacmA and ΔacmA transformed with pNST260+::cse (data not shown).
DISCUSSION
Three genes of S. thermophilus (rodA, pbp2b, and mreD) have been previously shown to be involved in cell chain length (50, 52, 54). These genes are also involved in cell shape, since mutants in these genes display rod-shaped cells instead of ovoid cells. In this study, we identified a fourth gene involved in the shortening of cell chains, called cse for its role in cellular segregation. Deletion of cse and complementation experiments have shown that the cse chimeric gene is involved in cell segregation. The Cse extracellular protein contains a putative LysM domain and a putative CHAP domain. LysM domains of AcmA, the major peptidoglycan hydrolase of L. lactis, are directly involved in cell wall attachment (49), and CHAP domains are widely found in surface proteins where they are supposed to be involved in peptidoglycan hydrolysis (3, 45). Therefore, Cse may be attached to the cell wall and may exhibit peptidoglycan hydrolase activity.
To test whether Cse has a peptidoglycan hydrolase activity, protein extracts of S. thermophilus wild type, S. thermophilus Δcse, and E. coli Cse fragment overproducer strains were tested by zymography with embedded S. thermophilus or M. lysodeikticus cells as a substrate. In addition, cell lysis activity was tested in liquid medium, with native crude protein extracts of E. coli Cse fragment overproducer strains. In both experiments, no Cse lytic activity was detected. Finally, cse was not able to complement the long-chain phenotype resulting from a peptidoglycan hydrolase defect of an L. lactis ΔacmA mutant. The absence of a detectable lytic activity was previously reported for PcsB from S. agalactiae (42, 43), another protein with a CHAP domain. These results suggest that either Cse has no peptidoglycan hydrolase activity or Cse activity could not be detected by these assays.
The results reported here show that cse is a chimeric gene and suggested that its 5′ and 3′ ends originated from sip-like and pcsB genes, respectively. Indeed, the region comprising the 3′ end of cse, the following intergenic region, and the beginning of orf1 (region named HRC1) shows 93% identity with the region comprising the pcsB 3′ end, the following intergenic region, and the beginning of rppk (region named HRC2) (Fig. 3A). Moreover, HRC1 is flanked by two regions encoding amino acid sequences homologous to parts of the SIP protein from S. agalactiae (Fig. 3A). Furthermore, SIP-encoding genes had been found in the genomes of all streptococci except that of S. thermophilus and S. mutans, and a PcsB-encoding gene had been found in the genomes of all streptococci, including S. thermophilus. Taken together, all of these data suggest that HRC2 had been subjected to a duplication event. The resulting duplicated sequence would have been inserted into a sip ancestral gene, thus creating two chimeric genes, cse and orf1 (Fig. 3B).
Interestingly, S. mutans and S. intermedius possess one LysM-CHAP protein. The S. mutans genome, whose complete sequence is available, does not encode a protein with significant similarity to SIP of other streptococci and encodes a protein, named GbpB, homologous to whole PcsB (9). Thus, the genomic context of S. mutans would be similar to that of S. thermophilus, suggesting that Cse and the S. mutans LysM-CHAP protein have a common ancestor that would have been generated by module shuffling. However, the S. mutans LysM-CHAP protein does not show significant similarity to SIP and shows significant similarity with Cse only from its putative CHAP domain. Moreover, the C terminus of the LysM-CHAP protein harbors only 61% identity to the C terminus of GbpB, whereas the C terminus of Cse shows 93% identity with the C terminus of S. thermophilus PcsB (Fig. 3A). If the LysM-CHAP protein of S. mutans and Cse had been generated by the same DNA duplication event, it would be expected that the percentage of divergence between these LysM-CHAP proteins and their PcsB/GbpB genomic counterparts would be the same. These data support an alternative hypothesis where the LysM-CHAP protein from S. mutans and Cse have different origins.
The consequence of the suspected DNA rearrangement that originated cse is the association of a putative LysM-encoding sequence to a putative CHAP-encoding sequence, giving rise to a new protein generation (Cse) by domain shuffling. Domain shuffling has been proposed as a mechanism of gene evolution in bacteria (13, 15, 25, 44) and implies considering genes to be associations of modules. Each gene module would encode a protein domain which has been defined as “part of a protein that can fold up independently of neighboring sequences” (11). These modules could undergo rearrangements, ending in the creation of new module associations and, consequently, new protein domain associations. As an example, extracellular proteins from S. pneumoniae that bind the choline component of the cell wall appear to evolve by domain shuffling. These multidomain proteins consist of a choline-binding domain, triggering protein association to the cell wall, and a catalytic domain that differs from one protein to another (25). Another example is the peptidoglycan hydrolase domain of Bacillus anthracis AmiA that is also found in other proteins where it is fused to different attachment signals (31).
The cse central region and the central region of its product are variable and repeat rich. The existence of variable tandemly repeated sequences is a common characteristic of extracellular proteins of gram-positive bacteria (20). This variability may facilitate adaptation to environmental changes (20), for instance, variability in the number of repeats in the alpha C protein allows group B streptococci to escape host immunity (26). In other cases, not restricted to extracellular proteins, genetic variability of repeat-rich regions directly modifies intrinsic biochemical activity of the protein, such as in restriction and modification enzymes encoded by EcoR124 and EcoR124/3 genes (40). These type I restriction enzymes recognize sites GAA(N6)RTCG and GAA(N7)RTCG, respectively, differing only in the length of a spacer. This difference in their specificity is due to two or three copies of a 12-bp sequence localized in their respective gene central parts (40). Therefore, we could not exclude that variations in the Cse repeat-rich region have consequences on cell segregation activity. However, replacement of var-cseLMG18311 by var-cseCNRZ368 at the LMG18311 cse chromosomal locus did not induce significant changes in cell segregation activity on the criterion of cell number per chain. Thus, it is likely that the high variability of the cse central region was not counter-selected because it does not affect the cell segregation activity of Cse.
The question of the role of the Cse central part in cell segregation remains open. This region could be a linker joining the N-terminal part to the C-terminal part of Cse. Such a function is assumed, for instance, by Q-linkers that join functionally distinct domains in nitrogen regulatory proteins (55). Variations in length and sequence of NifA and NtrC Q-linkers have no consequence on the activity of these proteins (55). Similarly, the replacement of var-cseLMG18311 by var-cseCNRZ368 ending in a 60-bp shortening of var-cse, since var-cseCNRZ368 is 60 bp shorter than var-cseLMG18311, did not have consequences on Cse cell segregation activity. A linker role of the Cse central part could explain why its high variability, and especially its length variability, does not affect cellular segregation. This possible function of the Cse central part would also be consistent with its predicted nonglobular structure.
Two repeat-rich regions sharing common repeat motifs flank HRC1. This suggests that the insertion event of the duplicated region occurred within an ancestral repeat-rich region localized in the central part of the sip-like gene ancestor. Thus, SIP-encoding genes of other streptococci could also contain a repeat-rich region. Therefore, repeat content and sequence complexity level were analyzed in the S. agalactiae SIP protein and orthologous counterparts in S. pneumoniae, Streptococcus pyogenes, Streptococcus gordonii, and Streptococcus suis. No repeat was found in any of these sequences. However, analysis of the amino acid composition showed that these regions have a low sequence complexity. Moreover, as observed for cse, their alignment revealed strong interspecies divergence of the central region (data not shown). Thus, although SIP proteins do not contain repeats, their central part showed low sequence complexity and interspecies variability. These data are in agreement with the existence of a low-complexity region in the central part of the sip-like gene ancestor of cse. We hypothesize that this low-complexity region could have been the site of an HRC1 insertion event responsible for cse creation. Interestingly, other multidomain proteins exhibit junctions with low-complexity sequences between domains (44, 55). For instance, the multiphosphoryl transfer protein of Rhodobacter capsulatus, a permease from a phosphotransferase system, is composed of three domains connected by two similar linker regions of 17 residues. These two linkers are rich in glycine, alanine, and proline residues, lowering the complexity of these regions (57). It has been proposed that the evolution of PTS permease occurred by interdomain shuffling and that this shuffling was allowed by genetic recombination between linker-encoding sequences (57). This example, together with cse, raises the question of a possible evolutionary advantage, at the genetic level, of DNA regions with a low-complexity sequence. Recombination events occurring inside domain-encoding sequences will probably inactivate, in many cases, the domain functionality and will therefore give rise to inefficient domain associations. We speculate that low-complexity regions can be more tolerant targets for genetic recombination events, responsible for domain shuffling, because of their low requirement in sequence and size.
Acknowledgments
We thank Isabelle Poquet and Emmanuelle Maguin for providing the pFUN and pGh9:ISS1 plasmids, respectively, and Valérie Legué for photographic contributions. We also thank Jan Kok for providing L. lactis MG1363acmAΔ1 and Gérard Guédon for allowing us to use pNST260+ prior to publication. We are grateful to Paul Hoskisson for help in preparing the manuscript.
S.L. and A. F. were supported by grants from the Ministère de l'Education Nationale de l'Enseignement Supérieur et de la Recherche. F.B. was supported by a grant from the Institut National de la Recherche Agronomique. P.H. is Research Associate at FNRS.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 4.Bollinger, J. M., Jr., D. S. Kwon, G. W. Huisman, R. Kolter, and C. T. Walsh. 1995. Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J. Biol. Chem. 270:14031-14041. [DOI] [PubMed] [Google Scholar]
- 5.Bracquart, P. 1981. An agar medium for the differential enumeration of Streptococcus thermophilus and Lactobacillus bulgaricus in yogurt. J. Appl. Bacteriol. 51:303-305. [Google Scholar]
- 6.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrus, V., Y. Roussel, B. Decaris, and G. Guedon. 2000. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 66:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, S. A., T. Hain, U. Technow, A. Darji, P. Pashalidis, S. W. Joseph, and T. Chakraborty. 2003. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J. Bacteriol. 185:6801-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chia, J. S., L. Y. Chang, C. T. Shun, Y. Y. Chang, Y. G. Tsay, and J. Y. Chen. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect. Immun. 69:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuatrecasas, P., S. Fuchs, and C. B. Anfinsen. 1967. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J. Biol. Chem. 242:1541-1547. [PubMed] [Google Scholar]
- 11.Doolittle, R. F. 1995. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64:287-314. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1277. [DOI] [PubMed] [Google Scholar]
- 13.Gubler, M., D. Braguglia, J. Meyer, A. Piekarowicz, and T. A. Bickle. 1992. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 11:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardie, J. M. 1986. Genus Streptococcus Rosenbach 1884, 22al, p. 1043-1047. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. William & Wilkins, Baltimore, Md. [Google Scholar]
- 15.Houghton, J. E., G. A. O'Donovan, and J. R. Wild. 1989. Reconstruction of an enzyme by domain substitution effectively switches substrate specificity. Nature 338:172-174. [DOI] [PubMed] [Google Scholar]
- 16.Huard, C., G. Miranda, F. Wessner, A. Bolotin, J. Hansen, S. J. Foster, and M. P. Chapot-Chartier. 2003. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149:695-705. [DOI] [PubMed] [Google Scholar]
- 17.Husson-Kao, C., J. Mengaud, L. Benbadis, and M. P. Chapot-Chartier. 2000. Mur1, a Streptococcus thermophilus peptidoglycan hydrolase devoid of a specific cell wall binding domain. FEMS Microbiol. Lett. 187:69-76. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 20.Kehoe, M. A. 1994. Cell-wall-associated proteins in Gram-positive bacteria, p. 217-261. In J.-M. G. A. R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 21.Lachica, R. V., C. Genigeorgis, and P. D. Hoeprich. 1971. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl. Microbiol. 21:585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Leenhouts, K. 1995. Integration strategies and vectors. Dev. Biol. Stand. 85:523-530. [PubMed] [Google Scholar]
- 24.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1994. Direct screening of recombinants in gram-positive bacteria using the secreted staphylococcal nuclease as a reporter. J. Bacteriol. 176:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez, R., M. P. Gonzalez, E. Garcia, J. L. Garcia, and P. Garcia. 2000. Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol. 151:437-443. [DOI] [PubMed] [Google Scholar]
- 26.Madoff, L. C., J. L. Michel, E. W. Gong, D. E. Kling, and D. L. Kasper. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. USA 93:4131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 30.Mercier, C., C. Durrieu, R. Briandet, E. Domakova, J. Tremblay, G. Buist, and S. Kulakauskas. 2002. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol. Microbiol. 46:235-243. [DOI] [PubMed] [Google Scholar]
- 31.Mesnage, S., and A. Fouet. 2002. Plasmid-encoded autolysin in Bacillus anthracis: modular structure and catalytic properties. J. Bacteriol. 184:331-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, W. L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 34.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Ohnishi, R., S. Ishikawa, and J. Sekiguchi. 1999. Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis. J. Bacteriol. 181:3178-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluvinet, A., F. Charron-Bourgoin, C. Morel, and B. Decaris. 2004. Polymorphism of eps loci in Streptococcus thermophilus: sequence replacement by putative horizontal transfer in S. thermophilus IP6757. Int. Dairy J. 14:627-634. [Google Scholar]
- 38.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potvin, C., D. Leclerc, G. Tremblay, A. Asselin, and G. Bellemare. 1988. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol. Gen. Genet. 214:241-248. [DOI] [PubMed] [Google Scholar]
- 40.Price, C., J. Lingner, T. A. Bickle, K. Firman, and S. W. Glover. 1989. Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes. J. Mol. Biol. 205:115-125. [DOI] [PubMed] [Google Scholar]
- 41.Qin, X., K. V. Singh, Y. Xu, G. M. Weinstock, and B. E. Murray. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 42:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinscheid, D. J., K. Ehlert, G. S. Chhatwal, and B. J. Eikmanns. 2003. Functional analysis of a PcsB-deficient mutant of group B streptococcus. FEMS Microbiol. Lett. 221:73-79. [DOI] [PubMed] [Google Scholar]
- 43.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reizer, J., and M. H. Saier, Jr. 1997. Modular multidomain phosphoryl transfer proteins of bacteria. Curr. Opin. Struct. Biol. 7:407-415. [DOI] [PubMed] [Google Scholar]
- 45.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 46.Rioux, S., D. Martin, H. W. Ackermann, J. Dumont, J. Hamel, and B. R. Brodeur. 2001. Localization of surface immunogenic protein on group B streptococcus. Infect. Immun. 69:5162-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schleifer, K. H. 1986. Gram-positive cocci, p. 999-1002, In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. William & Wilkins, Baltimore, Md. [Google Scholar]
- 49.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278:23874-23881. [DOI] [PubMed] [Google Scholar]
- 50.Stingele, F., and B. Mollet. 1996. Disruption of the gene encoding penicillin-binding protein 2b (pbp2b) causes altered cell morphology and cease in exopolysaccharide production in Streptococcus thermophilus Sfi6. Mol. Microbiol. 22:357-366. [DOI] [PubMed] [Google Scholar]
- 51.Terzaghi, B., and W. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thibessard, A., F. Borges, A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2004. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl. Environ. Microbiol. 70:2220-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thibessard, A., A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2002. Transposition of pGh9:ISS1 is random and efficient in Streptococcus thermophilus CNRZ368. Can. J. Microbiol. 48:473-478. [DOI] [PubMed] [Google Scholar]
- 54.Thibessard, A., A. Fernandez, B. Gintz, N. Leblond-Bourget, and B. Decaris. 2002. Effects of rodA and pbp2b disruption on cell morphology and oxidative stress response of Streptococcus thermophilus CNRZ368. J. Bacteriol. 184:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wootton, J. C., and M. H. Drummond. 1989. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 2:535-543. [DOI] [PubMed] [Google Scholar]
- 56.Wootton, J. C., and S. Federhen. 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266:554-571. [DOI] [PubMed] [Google Scholar]
- 57.Wu, L. F., J. M. Tomich, and M. H. Saier, Jr. 1990. Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J. Mol. Biol. 213:687-703. [DOI] [PubMed] [Google Scholar]
- 58.Wuenscher, M. D., S. Kohler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]