Abstract
The three activities of NadR were demonstrated in purified protein and assigned to separate domains by missense mutations. The N-terminal domain represses transcription of genes for NAD synthesis and salvage. The C-terminal domain has nicotinamide ribose kinase (NmR-K; EC 2.7.1.22) activity, which is essential for assimilation of NmR, converting it internally to nicotinamide mononucleotide (NMN). The central domain has a weak adenylyltransferase (NMN-AT; EC 2.7.7.1) activity that converts NMN directly to NAD but is physiologically irrelevant. This central domain mediates regulatory effects of NAD on all NadR activities. In the absence of effectors, pure NadR protein binds operator DNA (the default state) and is released by ATP (expected to be present in vivo). NAD allows NadR to bind DNA in the presence of ATP and causes repression in vivo. A superrepressor mutation alters an ATP-binding residue in the central (NMN-AT) domain. This eliminates NMN-AT activity and places the enzyme in its default (DNA binding) state. The mutant protein shows full NmR kinase activity that is 10-fold more sensitive to NAD inhibition than the wild type. It is proposed that NAD and the superrepressor mutation exert their effects by preventing ATP from binding to the central domain.
The cofactors NAD and NADP are involved in over 300 reactions, or 17% of all classified reactions (34). Beyond its role in redox reactions, NAD acts in DNA ligation (5, 23), ADP-ribosylation of proteins (21), protein deacetylation (30), calcium signaling (33), and as a precursor of vitamin B12 (19). During oxidative stress, NADH is dangerous because it provides electrons for the formation of hydroxyl radicals by the Fenton reaction (8, 9); under the same circumstances, NADPH plays a protective role (11, 15, 16). The central, variable (and sometimes conflicting) roles of pyridine nucleotides suggest a need for tight control.
Levels of NAD and NADP in Salmonella enterica are controlled at several points. Feedback inhibition of the first biosynthetic enzyme by NAD (20) maintains the aggregate pool of NAD(P). The balance between NAD and NADP is controlled by feedback inhibition of NAD kinase by NADPH (7, 13, 35). Here, we characterize the trifunctional NadR protein, which represses the transcription of several synthetic genes when NAD levels are high and provides two assimilatory activities when NAD levels are low.
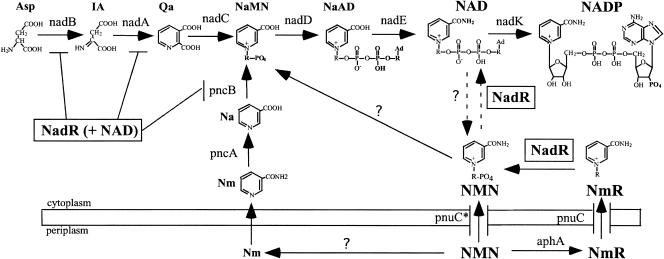
Salmonella synthesizes NAD (and NADP) either de novo or from any of several exogenous pyridine compounds (Fig. 1). The oxygen-stimulated pyridine nucleotide cycle cleaves NAD (or NADP) to NMN, which is then converted back to NAD through the last two synthetic steps (24). Salmonella assimilates exogenous NMN by two pathways (6). The more efficient route, route 1, requires only 10 μM exogenous NMN and utilizes an unknown glycohydrolase to form Nm, which enters the cell and is assimilated via PncA, PncB, and the synthetic enzymes NadD and NadE (Fig. 1). The less efficient route, route 2, requires 100 μM NMN and is apparent only when route 1 is blocked. In route 2, NMN is first converted to NmR by the periplasmic acid phosphatase AphA (6). The produced NmR enters the cell via the PnuC transporter protein.
FIG. 1.
NAD(P) synthesis and recycling in S. enterica (including contributions from this study). The gene encoding each enzyme activity is indicated if known. Question marks indicate reactions that have been assayed but not associated with a particular gene or enzyme. Intermediates include aspartic acid (Asp), iminoaspartic acid (IA), quinolinic acid (Qa), nicotinic acid ribonucleotide (NaMN), nicotinic acid adenine dinucleotide (NaAD), NAD, NADP, Na, Nm, NMN, and NmR.
The nadR gene was initially identified by mutations that eliminate repression of the biosynthetic genes nadB and nadA and the salvage genes pnuC and pncB (3, 25, 38). The repressor function of NadR is provided by an N-terminal helix-turn-helix domain, which binds a base sequence (TGTTTA, the NAD box) found in the control region of target genes (25). NadR was later found to have two enzymatic activities. Nucleoside kinase activity (NmR-K; EC 2.7.1.22) phosphorylates NmR to form NMN (17). The adenylyltransferase activity (NMN-AT; E.C. 2.7.7.1) catalyzes conversion of NMN to NAD (22, 26). In principle, both enzymatic activities of NadR contribute to assimilation of NmR, as diagrammed in Fig. 1.
Evidence is presented herein that NadR contributes to both transport and assimilation of NmR. The NmR kinase activity of NadR adds a phosphate to transported NmR, thereby trapping a charged pyridine compound and producing NMN, which is further converted to NAD by deamidation (PncC) and the last two biosynthetic enzymes (Fig. 1). The adenylyltransferase activity of NadR can convert internal NMN directly to NAD; however, evidence presented here suggests that this activity is not physiologically significant.
The three activities of NadR are demonstrated for purified protein, and each is assigned to an individual domain. All three activities are subject to feedback regulation by NAD (in conjunction with ATP). Results suggest that a high level of NAD causes NadR to lose enzymatic activity and repress several NAD synthetic genes; conversely, a low NAD level activates the assimilatory enzymatic activities and leads to derepression of biosynthetic genes.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this paper: aa, amino acids; IPTG, isopropyl-β-d-thiogalactopyranoside; Kmapp, apparent Km; LB, Luria-Bertani broth; Na, nicotinic acid; NAD, β-NAD; NADP, β-NADP; Nm, nicotinamide; NMN, β-nicotinamide mononucleotide; NMN-AT, β-nicotinamide mononucleotide adenylyltransferase; NmR, β-nicotinamide ribose; NmR-K, β-nicotinamide riboside kinase; and X-Gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside.
Bacterial strains and growth media.
Bacterial strains are listed in Table 1. Rich medium was LB (Difco) supplemented with 5 g of NaCl/liter and solidified with 1.5% agar (Baltimore Biological Laboratories). Minimal medium was E medium (32) supplemented with 0.2% glucose. The chromogenic β-galactosidase substrate X-Gal (Diagnostic Chemicals) was dissolved in N,N-dimethylformamide and diluted into aqueous solutions to a final concentration of 25 μg/ml. Ampicillin was used at 100 μg/ml, and chloramphenicol was used at 20 μg/ml. Nicotinic acid was added to minimal plates at both high (4 × 10−4 M) and low (10−6 M) concentrations. NMN was added at 10−4 M. Unless otherwise indicated, chemicals were obtained from Sigma Chemical Company (St. Louis, Mo.).
TABLE 1.
Bacterial strains
| Strain | Genotypea | Source or reference |
|---|---|---|
| Salmonella | ||
| TR10000 | Wild-type S. enterica (serovar Typhimurium LT2) | Lab collection |
| TT7466 | nadD157(Ts) zbe-1028::Tn10 | Lab collection |
| TT7467 | nadD158(Ts) zbe-1028::Tn10 | Lab collection |
| TT7468 | nadD159(Ts) zbe-1028::Tn10 | Lab collection |
| TT7469 | nadD187(Ts) zbe-1028::Tn10 | Lab collection |
| TT7470 | nadD188(Ts) zbe-1028::Tn10 | Lab collection |
| TT10740 | nadE381(Ts) nadB499::MudJ leu-4017(Del:leuA-ppsB) ara-9 gal-205 | Lab collection |
| TT11357 | nadB499::MudJ serB1463::Tn10 nadR508 (RsT−) | 39 |
| TT11358 | nadB499::MudJ serB1463::Tn10 nadR509 (RsT−) | 39 |
| TT11360 | nadB499::MudJ serB1463::Tn10 nadR511 (RsT−) | 39 |
| TT11388 | nadB499::MudJ serB1463::Tn10 nadR522 (RsT−) | 39 |
| TT12990 | nadB103 pncA278::Tn10dCm | Lab collection |
| TT12927 | nadB499::MudJ pncA278::Tn10dCm nadR511 (RsT+) | Lab collection |
| TT13007 | nadB499::MudJ pncA278::Tn10dCm | Lab collection |
| TT13150 | pncA278::Tn10dCm nadR581::MudJ (R−T−) | 39 |
| TT13498 | nadB499::MudJ pncA180::Tn10 | Lab collection |
| TT13499 | nadB499::MudJ pncA180::Tn10 nadE381(Ts) | Lab collection |
| TT14945 | nadB499::MudJ pncA278::Tn10dCm nadR299 (R+T−) | 39 |
| TT15131 | nadB103 pncA278::Tn10dCm DEL787 (serB-pnuA) | Lab collection |
| TT15459 | nadB499::MudJ pncA278::Tn10dCm nadR260 (R−T+) | 39 |
| TT15463 | nadB499::MudJ pncA278::Tn10dCm nadR276 (R−T+) | 39 |
| TT15466 | nadB499::MudJ pncA278::Tn10dCm serB1463::Tn10 nadR320 (R−T+) | 39 |
| TT15471 | nadB499::MudJ pncA278::Tn10dCm serB1463::Tn10 nadR333 (R−T+) | 39 |
| TT15872 | nadB499::MudJ pncA278::Tn10dCm nadR312 (R+T−) | 39 |
| TT15953 | nadB227::MudJ pncA278::Tn10dCm serB9 nadR277 (R−T−) | 39 |
| TT15959 | nadB227::MudJ pncA278::Tn10dCm serB9 nadR286 (R−T−) | 39 |
| TT19857 | zcd-3518::Tn10 nadB499::MudJ DEL (leu4017-ppsB) ara-9 gal-205 | Lab collection |
| TT22856 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD188(Ts) | Lab collection |
| TT22857 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD157(Ts) | Lab collection |
| TT22858 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD158(Ts) | Lab collection |
| TT22859 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD159(Ts) | Lab collection |
| TT22860 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD187(Ts) | Lab collection |
| TT22897 | nadB51 pncA15 trpA49 pnuC269 nadR609::Tn10dTc | Lab collection |
| E. coli | ||
| TT24636 | F−opmT hsdSb gal dCm trxB15::kan (DE3)pLysS (Cmr) | Novagen |
| TT24194 | BL21(DE3)pLysS/pJG201 (wild-type nadR) | This study |
| TT24195 | BL21(DE3)pLysS/pJG202 (nadR cloned from TT11360) (RsT−) | This study |
| TT24196 | BL21(DE3)pLysS/pJG203 (nadR cloned from TT14945) (R+T−) | This study |
| TT24632 | BL21(DE3)pLysS/pJG204 (nadR cloned from TT11388) (R−T+) | This study |
| TT24633 | BL21(DE3)pLysS/pJG205 (nadR cloned from TT15466) (R−T+) | This study |
| TT24634 | BL21(DE3)pLysS/pJG206 (nadR cloned from TT15872) (R+T−) | This study |
Ts, temperature sensitive.
Cloning the nadR gene.
The Salmonella nadR gene was PCR amplified and cloned into vector pet15b (Novagen) to produce a fusion protein with six N-terminal histidines and a thrombin protease cleavage site according to the method of Penfound and Foster (25). Primers were nadR14 (5′GGAATTCCATATGTCATCGTTCGACTATCTC) and nadR15 (5′CGCGGATCCGCGTTATCCCTGCTCGCCCATCA). The resulting 1,238-bp PCR product was then digested sequentially with NdeI (Fermentas) and BamHI (Fermentas) before ligation into a similarly digested and gel-purified pet15b vector (Novagen). The ligation mix was used to transform NovaBlue (Novagen) electrocompetent cells according to the manufacturer's instructions. Clones were selected on LB plates with 50 μg of ampicillin/ml, and plasmids were sequenced according to the method of Sanger et al. (27) at the University of Utah Health Sciences DNA Sequence Facility before being transformed into strain BL21(DE3)pLysS (Novagen), which carries an IPTG-inducible gene for T7 polymerase in its chromosome. The final plasmid contains an ampicillin resistance gene and expresses nadR from an ITPG-inducible T7lac promoter. Mutant NadR proteins were cloned as described above by using DNA isolated from strains TT12927 (RsT− genotype), TT11388 (RsT−), TT15872 (R+T−), TT14945 (R+T−), TT15466 (R−T+), and TT15459 (R−T+).
Procedures for gene expression and enzyme purification.
Cells were grown in 100 ml of LB medium supplemented with 100 μg of ampicillin/ml and 34 μg of chloramphenicol/ml. The culture was grown at 37°C with shaking to an A600 of 0.6, and nadR was induced by adding IPTG to a final concentration of 1 mM. Cells were harvested after 3 h of shaking at 37°C, pelleted by centrifugation, washed with 50 mM phosphate buffer containing 500 mM NaCl, and frozen at −70°C. Frozen cell pellets were resuspended in 20 mM phosphate buffer (pH 7.8) containing 500 mM NaCl and disrupted by sonication (six 15-s cycles at output 5 at 50% duty cycle in a Bronson sonifier). Debris was removed by 30 min of centrifugation at 20,000 × g (4°C), and the supernatant was used directly for assaying NmR-K activity and total protein concentration.
Histidine-tagged protein was purified by using ProBond resin (Invitrogen) according to the manufacturer's instructions for elution by an imidazole step gradient. The enzymatic activity eluted in the 500 mM imidazole fraction. Active fractions were dialyzed into buffer (50 mM phosphate buffer [pH 7.8] containing 1 mM dithiothreitol and 20% glycerol) for storage at −80°C. Purity of the final preparation was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
NmR-K assay.
The NmR kinase activity of NadR was assayed by using gamma-labeled [γ-32P]ATP (Perkin-Elmer), and products were analyzed by thin-layer chromatography. NmR was made by incubation of NMN with calf intestinal phosphatase as previously described (14). The NmR kinase reactions were run at 37°C in a 0.04-ml reaction mixture containing 1.0 mM NmR, 5 mM MgCl2, 1.0 mM ATP, and 100 mM Tris-HCl (pH 7.6). The reaction was initiated by addition of labeled ATP and stopped by heating (98°C for 90 s); 2 μl of each reaction was spotted onto the appropriate thin-layer plate for identification and quantification of products as described below. Specific activity was defined as micromoles of product produced per minute per milligram of protein.
NMN-AT assay.
NMN-AT activity was assayed by using NAD-dependent alcohol dehydrogenase from baker's yeast (Sigma) to reduce the product to NADH and monitoring the corresponding absorbance change at 339 nm as previously described (2). The reaction mix contained 3 mM NMN, 100 mM Tris-Cl (pH 8.0), 10 mM MgCl2, 1 mM ATP, 1% (vol/vol) ethyl alcohol, and 50 μg of alcohol dehydrogenase (Sigma). To increase sensitivity when determining the apparent Km of NMN-AT for ATP, the redox cycling agents phenazine methylsulfate and diphenyl tetrazolium bromide were added, and the reaction was monitored at 570 nm (35). To test feedback inhibition by NAD, NMN-AT activity was assayed by using [carbonyl-14C]NMN that was made by pyrophosphorolysis of [carbonyl-14C]NAD (Amersham Pharmacia) by nucleotide pyrophosphatase (Sigma) according to the method of Zhu et al. (36). These reactions were run at 37°C in a 0.04-ml reaction mixture containing 3.0 mM [carbonyl-14C]NMN, 5 mM MgCl2, 1.0 mM ATP, and 100 mM Tris-HCl (pH 8.0) with and without NAD. The reaction was started by addition of labeled NMN and stopped by heating (98°C for 90 s); 10 μl of each reaction mixture was spotted onto the appropriate thin-layer plate for identification and quantification of products as described below. Specific activity was defined as micromoles of product produced per minute per milligram of protein.
Thin-layer chromatography and product quantification.
Products of the NMN-AT reaction were distributed on polyetheleneimine cellulose plates using a 0.2 M potassium citrate solution (pH 5.0). Products of the NmR kinase reaction were separated by thin-layer chromatography on Cellulose F plates (Merck, Darmstadt, Germany) using a 1 M ammonium acetate (pH 5)-ethanol (30:70) solvent system as described previously (12). Reactants and products were quantified by using a Phosphoimager SI system (Molecular Dynamics). In determining Rf values, unlabeled standards were visualizing by using a UV lamp.
Determination of kinetic parameters.
The kinetic parameters for NmR-K were determined by using the assay described above and varying the concentrations of ATP and NmR. The apparent Km values of NadR for its substrates were determined by fitting a Michaelis-Menten curve to the kinetic data by using the computer program Kaleidagraph (Synergy Software).
Sequencing NadR mutations.
The NadR region was amplified by PCR from genomic DNA using primers TP1175 (5′CCCAGCGATTCCAGTACGTTGTG) and TP1210 (5′GATTGATACGGATTGATGTTGTAGG). Nested primers TP1174 (5′CCTGATAAGCGAAGCGCCATC) and TP1211 (5′CCGCGTCTTATCAGGCCTACAGTT) were used to sequence according to the method of Sanger et al. (27) at the University of Utah Health Sciences DNA Sequence Facility.
DNA binding assay.
Binding of NadR to DNA was assayed according to the mobility shift assay of Raffaelli et al. (26) using the nadB promoter region. The probe was produced by PCR amplification of bp −200 to −1 of the nadB promoter region from wild-type S. enterica (TR10000). Competitor DNA was produced by PCR amplification of the eut operon promoter region (531 bp) corresponding to bp −540 to −9. Purified NadR protein was incubated with 50 nM probe DNA, 100 mM Tris-HCl (pH 7.8), 1 mM EDTA, 0.1 mg of bovine serum albumin/ml, 3 mM MgCl2, 1 mM dithiothreitol, and 5% glycerol for 30 min at 37°C. The samples were run on a 4 to 20% gradient polyacrylamide gel (Gradipore) in Tris-borate-EDTA (pH 7.5) running buffer at 4°C and then stained with ethidium bromide.
Database search and sequence alignment.
Database searches were performed using BLAST (1) at the NCBI website as well as the ERGO database (Integrated Genomics, Inc., Chicago, Ill.). Sequences were aligned by using the Clustal W program (31).
RESULTS
NmR-K activity.
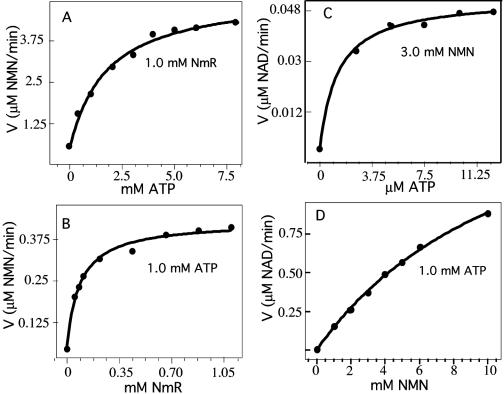
The nadR gene of S. enterica was cloned, and histidine-tagged NadR protein was purified (see Materials and Methods). Induction of the tagged nadR gene caused a 1,200-fold increase in the NmR-K activity assayed in crude extracts of Escherichia coli. The purified NadR fusion protein displayed a high NmR-K activity (1.4 μmol/min/mg), consistent with the activity reported by Kurnasov et al. (17). The apparent Km values for NmR and ATP were 0.08 ± 0.02 mM and 1.2 ± 0.2 mM, respectively (Fig. 2). The high affinity for ATP should be noted.
FIG. 2.
Michaelis-Menten plots for the determination of the apparent Km of the NadR NmR-K activity for ATP (A) and NmR (B) and the apparent Km of NadR NMN-AT activity for ATP (C) and NMN (D). Assays were performed in triplicate as described in Materials and Methods.
NadR NMN-AT activity.
An NMN-AT activity was previously demonstrated for the E. coli NadR protein, and the presence of a NadR domain belonging to the cytidinylyltransferase family was noted (26). This activity was found to be very low (0.05 μmol/min/mg at 1 mM NMN) for NadR proteins of both Salmonella (17) and E. coli (22).
The Kmapp for ATP of the Salmonella NMN-AT protein is extremely low (1.2 ± 0.4 μM) (Fig. 2), in reasonable agreement with that (1.7 μM) reported for the E. coli enzyme (26). In contrast, the Kmapp for NMN (12.8 ± 2 mM, previously determined to be 7 ± 2 mM [17]) is substantially higher for Salmonella NadR than that (0.7 mM) reported for the E. coli enzyme (26). In contrast to the low NMN-AT activity of Salmonella (0.05 μmol/min/mg), the homologous NadR enzyme of Haemophilus influenzae is highly active (0.9 μmol/min/mg) and clearly supports growth by its conversion of NMN to NAD (17). In light of this difference, the physiological relevance of Salmonella NMN-AT activity was tested.
NMN-AT activity of Salmonella NadR is neither necessary nor sufficient for the use of NMN as the sole pyrimidine source.
As described above, NMN formed by the NmR kinase can be converted to NAD by deamidation followed by the NadD and NadE reactions (Fig. 1). In principle, NMN might also be converted directly to NAD by the NMN-AT activity of NadR protein. We tested both the necessity and the sufficiency of the NadR-dependent route for growth on NMN as the sole pyrimidine source.
The NMN-AT activity is not necessary based on the observation that a nadB pncA nadR pnuC* mutant (TT22897) grows well on NMN as pyridine source. In this strain, de novo pyridine synthesis is blocked, and the pnuC* mutation allows the import of intact NMN, which can apparently be converted to NAD even without NadR protein (6). The NMN-AT activity of NadR is not sufficient to support growth, since strains lacking either NadD or NadE activity cannot grow on NMN (Table 2). These tests were done by feeding temperature-sensitive nadD or nadE mutants various pyridine sources at high and low temperatures. Addition of NMN did not allow growth of these mutants at the nonpermissive temperature, even when assimilation of NMN was enhanced by the previously described pnuC*, aphA*, or pnuD* mutations (6). NMN can support growth of nadB mutants when normal functional alleles of nadD and nadE are present or when strains with temperature-sensitive alleles are tested at permissive temperatures. Thus, the low NMN-AT activity of NadR is neither necessary nor sufficient to support growth on NMN.
TABLE 2.
Growth of nadD(Ts) and nadE(Ts) strains on NMN
| Strain | Relevant genotype | Growth ata:
|
|||||
|---|---|---|---|---|---|---|---|
| 30°C
|
37°C
|
42°C
|
|||||
| NA | NMN | NA | NMN | NA | NMN | ||
| TR10000 | Wild type | + | + | + | + | + | + |
| TT7466-7470 | nadD(Ts) | + | + | ± | ± | − | − |
| TT19857 | nadB | + | + | + | + | + | + |
| TT10740 | nadB nadE(Ts) | + | + | ± | ± | − | − |
| TT13498 | nadB pncA | + | + | + | + | + | + |
| TT13499 | nadB pncA nadE(Ts) | + | + | ± | ± | − | − |
| TT22855 | nadB pncA pnuC* | + | + | + | + | + | + |
| TT22856-22860 | nadB pncA pnuC* nadD(Ts) | + | + | ± | ± | − | − |
± signifies slow growth.
DNA binding activity of NadR.
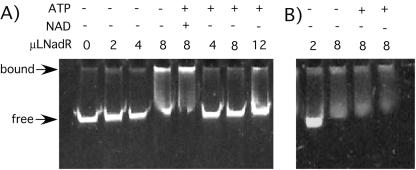
The NadR protein was previously shown to repress transcription of the nadA, nadB, and pncB promoters in response to high levels of NAD (25, 39). We confirmed the previous evidence (25) that purified NadR protein binds nadB operator sequence in the absence of ATP with or without NAD (Fig. 3A). In the presence of ATP, binding was dependent on NAD.
FIG. 3.
Gel shift assays for the characterization of the DNA binding activity of wild-type NadR protein (A) and mutant protein NadR511 (B). Assays were performed as described in Materials and Methods; the input NadR solution contained 0.6 μg/μl.
The effect of ATP on DNA binding is interesting in light of the recent discovery that NadR protein has two ATP-dependent enzymatic activities, NMN-AT and NmR-K (17, 28). Sequence analysis suggests that these activities are conferred by two distinct domains containing distinct ATP binding motifs. In addition, the Salmonella NmR-K and NMN-AT activities display different Kmapps for ATP (1.2 ± 0.2 mM and 1.2 ± 0.4 μM, respectively). While the Kmapp of NmR-K is typical of many kinases, the Kmapp of NMN-AT is surprisingly low, suggesting that the NMN-AT site is likely to be occupied under most growth conditions. Based on this, and on the location of “superrepressor” mutations described below, we propose that ATP binding to the NMN-AT domain is critical for regulation of NadR DNA binding activity.
Inhibition of NmR-K and NMN-AT by NAD.
NAD inhibits the NmR-K and NMN-AT activities of Salmonella NadR. (This inhibition must be determined in the presence of ATP, which is required for both reactions.) At an NAD concentration of 1.2 mM, the NmR-K activity is 69% inhibited (Table 3). The internal pool size of NAD is approximately 0.9 mM (18), placing inhibition of NmR-K activity in the dynamic range of in vivo NAD fluctuations. Inhibition of NmR-K explains the variation of NMN assimilation rate in response to internal levels of NAD (36); these assays depended on the conversion of NMN to NmR prior to transport and thus depended on internal kinase activity for pyridine accumulation. In contrast, the NMN-AT activity of NadR is inhibited only by concentrations of NAD well above the reported internal NAD levels, suggesting that inhibition of NmR-K activity (rather than NMN-AT activity) is responsible for the inhibitory effect of NAD on NMN assimilation.
TABLE 3.
Inhibition of NmR kinase and NMN-AT activities of NadR by NADa
| Concentration of NAD (mM) | % Inhibition of NmR kinase activity | % Inhibition of NMN-AT activity |
|---|---|---|
| 0 | 0 | 0 |
| 0.6 | 21 | 0 |
| 1.2 | 69 | 0 |
| 1.8 | 80 | 0 |
| 5.0 | 100 | 47 |
| 10 | 100 | 76 |
Assays were performed as described in Materials and Methods in triplicate with 0.14 mM NmR and 3.0 mM NMN for the NmR-K and NMN-AT assays, respectively.
Correlating NadR domains with individual activities.
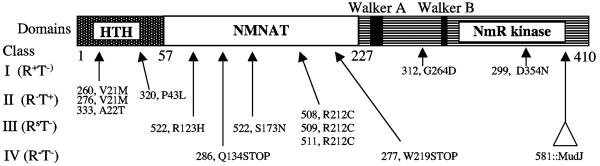
Analysis of the NadR amino acid sequence has suggested three functional domains, in agreement with the three activities reported (Fig. 4). Four types of mutants were previously classified by phenotype and mapped genetically (39). Class I mutations (at the distal end of the gene) allowed normal transcriptional control but prevented growth on exogenous NMN (called T− or transport deficient). Class II mutations (at the proximal end) did not affect growth on NMN but caused constitutive expression of nadB, which is normally repressed by high concentrations of NAD (these mutants were called R− or repression deficient). Class III mutations caused superrepression of nadB and abolished NMN utilization (RsT− or superrepressor). The superrepressor mutants appeared to trap the NadR protein in its DNA binding, enzymatically inactive form (independent of internal levels of NAD). Class IV mutations eliminated both repression and NMN assimilation activities (R−T−). Mutations of each of the above four classes (39) were sequenced (Fig. 4), and the mutant proteins were overproduced, purified, and assayed for the three NadR activities.
FIG. 4.
Mutational analysis of the NadR protein. The DNA binding domain (HTH, aa 1 to 59), the NMN-AT domain (aa 66 to 200), and the NmR-K domain (aa 232 to 345) are indicated. Mutations affecting both the repressor (R) and transport (T) functions of the NadR protein were selected by Zhu and Roth (see Table 3 in reference 39) and sequenced as described in the text. The following were the actual DNA base pair changes: alleles 508 and 509, CGC→TGC; allele 511, ATC CGC→ATT TGC; allele 522, CGC→CAC and AGC→AAC; alleles 260 and 276, GTG→ATG; allele 320, CCC→CTC; allele 333, GCT→ACT; allele 312, GAC→AAC; allele 299, GGC→GAC; allele 277, TGG→TGA; and allele 286, CAA→TAA. 581::MudJ is inserted at bp 1148.
(i) Class I mutations.
Two sequenced R+T− mutations affect conserved regions within the C-terminal NmR-K domain of NadR. The nadR312 mutation affects a glycine residue (G264D) that is predicted to be involved in substrate binding based on analysis of the structure of the H. influenzae NadR protein (28). As expected, both of the purified R+T− proteins have reduced NmR kinase activity and maintain full NMN-AT activity under the conditions tested, clearly demonstrating the importance of NmR-K activity for NMN assimilation (Table 4). In fact, mutant protein NadR312 had undetectable NmR-K activity. These proteins had normal DNA binding activity.
TABLE 4.
The phenotypes of mutant NadR strains and the NmR kinase and NMN-AT activity of the corresponding purified mutant NadR protein
| Mutant class | Relevant genotypea (strain no.) | Growth on NMNb | Expression of nadB::lac on low-nicotinatec medium | Expression of nadB::lac on high-nicotinatec medium | NmR-K activity (% wild-type activity, μmol/min/mgd) | NMN-AT activity (% wild-type activity, μmol/min/mgd) |
|---|---|---|---|---|---|---|
| Wild type (R+T+) | nadR (TT12990) | + | Blue | White | 100 (1.4 ± 0.2) | 100 (0.10 ± 0.01) |
| I (R+T−) | nadR299 (TT14945) | − | Blue | White | 13 (0.18 ± 0.1) | 100 |
| I (R+T−) | nadR312 (TT15872) | − | Blue | White | <1 (ND) | 100 |
| II (R−T+) | nadR320 (TT15466) | + | Blue | Blue | 100 | 100 |
| II (R−T+) | nadR260 (TT15459) | + | Blue | Blue | 100 | 100 |
| III (RsT−) | nadR511 (TT12927) | − | Light blue | White | 100 | <1 (ND) |
All strains contain nadB499::MudJ pncA278::Tn10dCm.
Assimilation of NMN was tested on E glucose minimal medium with 100 μM NMN.
Expression of a nadB::lac operon fusion was monitored on E glucose X-Gal minimal medium containing serine with either low nicotinate (1 μM) or high nicotinate (200 μM) levels to alter the internal levels of NAD.
NmR-K and NMN-AT activity was assayed for each of the purified mutant NadR proteins as described in Materials and Methods using 0.8 mM NmR and 3 mM NMN, respectively. Each protein was assayed in duplicate by using two independent protein preparations, with the average value being given. ND, no activity detected.
(ii) Class II mutations.
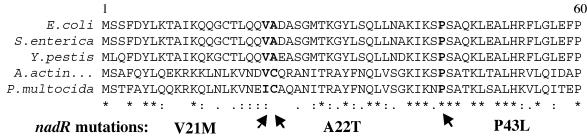
Mutants that display an R−T+ phenotype lack repression but not NMN assimilation. These mutations affect the C-terminal 60 amino acids of NadR at conserved residues in the helix-turn-helix motif (Fig. 5). The two repression-deficient NadR proteins purified showed wild-type NMN-AT and NmR-K activity (Table 4) but no DNA binding ability.
FIG. 5.
A Clustal W alignment of the DNA binding domains of NadR from different bacteria. Sequences were derived from E. coli, S. enterica (serovar Typhimurium LT2), Yersinia pestis, Actinobacillus actinomycetemcomitans, and Pasteurella multocida. Arrows indicate mutations that eliminated repressor activity in S. enterica. Asterisks and colons indicate indentical and similar residues, respectively.
(iii) Class III mutations.
Superrepressor mutations (RsT−) appear to trap the protein in the DNA binding, enzymatically inactive conformation. All of the four sequenced mutations in this class affected the central NMN-AT domain. Three identical substitutions (nadR508, nadR509, and nadR511) affected the putative NAD/ATP-binding motif of NadR (ISGA__R) (28), replacing arginine 212 with cysteine (R212C). The fourth mutant (the nadR522 mutant) has two amino acid substitutions. A histidine replaces a conserved arginine residue (R123H) in the NMN-AT active site region (as inferred from the structure of the H. influenzae NadR enzyme [28]), and an asparagine replaces serine (S173N). It will be suggested that the central NMN-AT domain is responsible for ATP-dependent regulation of DNA binding activity and that these RsT− mutations decrease the ability of this domain to bind ATP, leaving the protein in the “no-effector” or default repressing conformation. In support of this theory, we cloned, overexpressed, and purified the NadR511 (RsT−) protein and found that the protein bound DNA independently of NAD even in the presence of ATP, even when no NAD was provided (Fig. 3B). Unfortunately, protein from the nadR522 mutant was insoluble and could not be characterized.
Although the nadR511 mutant fails to grow on NMN, the purified NadR protein displayed wild-type NmR-K activity while having no detectable NMN-AT activity under the conditions tested (Table 4). This finding seemed inconsistent with the idea that the NMN-AT activity of NadR is not required for assimilation of NMN and suggested that the superrepressor mutations might have a more complex basis than simply trapping the protein in a DNA binding, enzymatically inactive conformation. Two possibilities were entertained: (i) the use of NMN might be prevented by superrepression of the PnuC transporter or (ii) the NadR511 protein might be active in vitro but not in vivo, causing failure to assimilate NmR. The first possibility is eliminated by two lines of evidence. First, pnuC is expressed from a promoter within the nadA-pnuC operon, so that even complete blockage of the main transcript by Tn10 insertions (or by superrepression) leaves pnuC expressed at a level sufficient to allow growth on NMN (37). Second, providing a constitutively expressed alternative to PnuC (PnuD*) does not allow the nadR511 mutant to grow on NMN, demonstrating that its NMN use phenotype is not due to lack of PnuC (6).
Initial evidence that the nadR511 mutant lacks NmR-K activity in vivo (but not in vitro) was the observation that some protein preparations showed low NmR-K activity that could be reactivated by dialysis or desalting, suggesting the presence of an inhibitor (presumably NAD) in some preparations (and in vivo) that reduced activity. The kinase activity of NadR511 protein is approximately 12-fold more sensitive to NAD inhibition than that of the wild type. Inhibition to 50% activity was caused by 0.08 mM NAD (compared to 1.0 mM for wild-type enzyme). We infer that the nadR511 mutant fails to grow on NMN due to superinhibition of NmR-K (by NAD). These results suggest that the single R212C mutation simultaneously eliminates ATP binding and increases the ability of NAD to inhibit NmR-K activity. It is suggested that the reduction of ATP binding causes supersensitivity to NAD, because NAD and ATP bind competitively.
The fourth class of mutants (R−T−) lacked both transport and regulatory functions and proved to be either nonsense or insertion mutations. This class is interesting because some of the nonsense mutations affect the central or C-terminal domain of the protein, yet all three activities are lost.
DISCUSSION
The trifunctional NadR protein from S. enterica was purified, and its three activities genetically correlated with particular regions of the protein. The data presented here support and add further detail to earlier suggestions that NadR is a multidomain protein (4, 39). Mutations in the N-terminal helix-turn-helix domain prevent repression of the nadB promoter when NAD levels are high (assessed in vivo) and prevent DNA binding (assessed in vitro). Mutations in the C-terminal domain reduce the NmR-K activity reported previously for NadR (17). These kinase-defective mutations have no apparent effect on the NMN-AT or DNA binding activities but abolish the ability of a nadB pncA double mutant to assimilate exogenous NMN by route 2. Thus, both in vivo and in vitro data suggest that the internal NmR kinase activity is essential for growth on exogenous NMN. Since NMN is dephosphorylated to NmR prior to uptake via PnuC (6), internal NmR-K activity is required both for driving the uptake of NmR and as the first step in its ultimate conversion to NAD(P) (Fig. 1). This model accounts for genetic evidence that some activity of NadR (other than repression) is needed for NMN uptake (29) and that this transport function of NadR is inhibited by high internal NAD levels (36).
The NadR repressor mediates several effects of NAD on de novo biosynthesis and assimilation of pyridines. The mechanistic basis of this inhibition suggests a complex historical origin. In the absence of all effectors, pure NadR protein binds operator DNA, and this binding is prevented by ATP (first described by Penfound and Foster [25]). Considered alone, ATP serves as an inducer of NAD biosynthesis, but a very low level of ATP is sufficient for this induction. NAD allows DNA binding in the presence of ATP. We propose that NAD represses transcription by preventing ATP binding. This suggests the evolutionary history of NAD control described below. While this history may be impossible to verify, it provides a way of organizing the findings reported here.
The NMN-AT activity of NadR may have once (on an evolutionary time scale) had strong kinase and transferase activities, as do NadR homologues from other bacteria, such as H. influenzae (17), but it has since been recruited to serve as a regulatory region to modulate the activities of cis DNA binding and NmR-K domains. In most bacterial genomes, the kinase and transferase activities are provided by a bifunctional protein that lacks a DNA binding domain and is likely to convert exogenous NmR to NAD in two sequential ATP-dependent reactions.
Basal levels of active NMN-AT transferase would be populated by ATP. This ATP would be displaced whenever internal concentrations of NMN allowed the catalytic event. Thus, the absence of bound ATP would correlate temporally with the presence of internal NMN and a need to repress synthesis of NAD. Such a correlation would allow development of a variable repressor function that uses the absence of ATP as a signal of pyridine influx. This mechanism became far more sophisticated when NAD (product of the reaction) rather than NMN displaced ATP and became the regulatory effector. Increased affinity for ATP might evolve as a means of increasing the concentration of NAD needed to displace ATP and signal repression. The following facts are consistent with this scenario. (i) The regulatory ATP binding site of NadR is in the NMN-AT domain. This enzyme must also have a binding site for its other substrate (NMN). We propose that NAD binds to the NMN site and displaces ATP. The NMN-AT activity has a surprisingly low apparent Km (1.2 μM) for ATP compared to the standard intracellular ATP concentration of 4.0 mM (10). This binding site would be occupied under most growth conditions, and a high NAD level would be needed to displace the ATP. (ii) The use of a single ATP binding site by both the transferase and regulatory activities is suggested by the superrepressor mutations that affect a residue within the putative ATP binding site of the NMN-AT domain. More critically, these mutations have three effects: they abolish NMN-AT activity, they lock the protein into a DNA binding conformation, and they render the kinase activity supersensitive to inhibition by NAD. (iii) This historical scenario fits with the present low NMN-AT activity of Salmonella NadR. While the NMN-AT activity is still detectable, it is not necessary or sufficient to support growth of a nadD or nadE mutant on exogenous NMN. The Salmonella NMN-AT domain may have lost much of its activity as it was recruited to serve as a regulatory region to modulate the DNA binding and NmR kinase activities. The ATP binding site was retained and tightened as an effector site, and the NMN binding site may have been coopted for occupancy by NAD (thereby decreasing its affinity for NMN).
This model contrasts with one proposed by Singh et al. based on the recently crystallized H. influenzae NadR protein (28). They observed two NAD molecules in the crystallized protein, one bound at the NMN-AT active site and the second with contacts in the NmR-K domain. Singh et al. hypothesized that this second bound NAD molecule was responsible for regulating the repressor and transport activities of NadR. Although this idea is not ruled out, the data presented here support the NMN-AT domain as the regulatory domain.
While the regulatory role of ATP may be simply historical, as outlined above, it could come into play when ATP levels drop drastically. Under normal growth conditions, the site is occupied and NAD is the predominant effector. When ATP levels become very low, it seems logical to prevent induction of NAD synthesis and recycling because both pathways require ATP. In the absence of ATP, induction of NAD synthetic enzymes would be futile.
In summary, under normal growth conditions, the NadR protein senses high internal NAD levels and represses the transcription of two enzymes involved in de novo NAD biosynthesis and one enzyme involved in scavenging and assimilation (Fig. 1). When NAD is limiting, the repression of synthesis (nadA, nadB, and pncB) is lifted and the NmR-K activity contributes to assimilation of NmR. We speculate that all pathways may shut down (regardless of the NAD level) when ATP is severely limited.
Acknowledgments
This work was supported in part by NIH grant GM23408.
In particular, we thank Sidney Velick and Baldomero Olivera for many insightful suggestions. We thank Janet Shaw, in whose lab some of the experiments were done. Laboratory members who contributed to discussions were Renee Dawson, Marian Price-Carter, Hotcherl Jeong, and Yaping Xu.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Balducci, E., M. Emanuelli, N. Raffaelli, S. Ruggieri, A. Amici, G. Magni, G. Orsomando, V. Polzonetti, and P. Natalini. 1995. Assay methods for nicotinamide mononucleotide adenylyltransferase of wide applicability. Anal. Biochem. 228:64-68. [DOI] [PubMed] [Google Scholar]
- 3.Foster, J. W., E. A. Holley-Guthrie, and F. Warren. 1987. Regulation of NAD metabolism in Salmonella typhimurium: genetic analysis and cloning of the nadR repressor locus. Mol. Gen. Genet. 208:279-287. [DOI] [PubMed] [Google Scholar]
- 4.Foster, J. W., and T. Penfound. 1993. The bifunctional NadR regulator of Salmonella typhimurium: location of regions involved with DNA binding, nucleotide transport and intramolecular communication. FEMS Microbiol. Lett. 112:179-183. [DOI] [PubMed] [Google Scholar]
- 5.Gellert, M., J. W. Little, C. K. Oshinsky, and S. B. Zimmerman. 1968. Joining of DNA strands by DNA ligase of E. coli. Cold Spring Harbor Symp. Quant. Biol. 33:21-26. [DOI] [PubMed] [Google Scholar]
- 6.Grose, J. H., U. Bergthorsson, Y. Zu, and J. R. Roth. Submitted for publication.
- 7.Grose, J. H., L. Joss, S. Velick, and J. R. Roth. Submitted for publication.
- 8.Imlay, J. A., S. M. Chin, and S. Linn. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640-642. [DOI] [PubMed] [Google Scholar]
- 9.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 10.Ingraham, J. L., O. Maaloe, and F. C. Neidhardt. 1983. Growth of the bacterial cell. Sinauer Associates, Inc., Sunderland, Mass.
- 11.Juhnke, H., B. Krems, P. Kotter, and K. D. Entian. 1996. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet. 252:456-464. [DOI] [PubMed] [Google Scholar]
- 12.Kasarov, L. B., and A. G. Moat. 1972. Convenient method for enzymic synthesis of 14 C-nicotinamide riboside. Anal. Biochem. 46:181-186. [DOI] [PubMed] [Google Scholar]
- 13.Kawai, S., S. Mori, T. Mukai, W. Hashimoto, and K. Murata. 2001. Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 268:4359-4365. [DOI] [PubMed] [Google Scholar]
- 14.Kemmer, G., T. J. Reilly, J. Schmidt-Brauns, G. W. Zlotnik, B. A. Green, M. J. Fiske, M. Herbert, A. Kraiß, S. Schlör, A. Smith, and J. Reidl. 2001. NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J. Bacteriol. 183:3974-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogoma, T., S. B. Farr, K. M. Joyce, and D. O. Natvig. 1988. Isolation of gene fusions (soi::lacZ) inducible by oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. USA 85:4799-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krapp, A. R., R. E. Rodriguez, H. O. Poli, D. H. Paladini, J. F. Palatnik, and N. Carrillo. 2002. The flavoenzyme ferredoxin (flavodoxin)-NADP(H) reductase modulates NADP(H) homeostasis during the soxRS response of Escherichia coli. J. Bacteriol. 184:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurnasov, O. V., B. M. Polanuyer, S. Ananta, R. Sloutsky, A. Tam, S. Y. Gerdes, and A. L. Osterman. 2002. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 184:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundquist, R., and B. M. Olivera. 1971. Pyridine nucleotide metabolism in Escherichia coli. I. Exponential growth. J. Biol. Chem. 246:1107-1116. [PubMed] [Google Scholar]
- 19.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 2003. Alpha-5,6-dimethylbenzimidazole adenine dinucleotide (alpha-DAD), a putative new intermediate of coenzyme B12 biosynthesis in Salmonella typhimurium. Microbiology 149:983-990. [DOI] [PubMed] [Google Scholar]
- 20.Magni, G., A. Amici, M. Emanuelli, N. Raffaelli, and S. Ruggieri. 1999. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73:135-182. [DOI] [PubMed] [Google Scholar]
- 21.Moss, J., S. Garrison, N. J. Oppenheimer, and S. H. Richardson. 1979. NAD-dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat-labile enterotoxin. J. Biol. Chem. 254:6270-6272. [PubMed] [Google Scholar]
- 22.Mushegian, A. 1999. The purloined letter: bacterial orthologs of archaeal NMN adenylyltransferase are domains within multifunctional transcription regulator NadR. J. Mol. Microbiol. Biotechnol. 1:127-128. [PubMed] [Google Scholar]
- 23.Olivera, B. M., Z. W. Hall, Y. Anraku, J. R. Chien, and I. R. Lehman. 1968. On the mechanism of the polynucleotide joining reaction. Cold Spring Harbor Symp. Quant. Biol. 33:27-34. [DOI] [PubMed] [Google Scholar]
- 24.Park, U. E., B. M. Olivera, K. T. Hughes, J. R. Roth, and D. R. Hillyard. 1989. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J. Bacteriol. 171:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penfound, T., and J. W. Foster. 1999. NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium. J. Bacteriol. 181:648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, S. K., O. V. Kurnasov, B. Chen, H. Robinson, N. V. Grishin, A. L. Osterman, and H. Zhang. 2002. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenylyltransferase and ribosylnicotinamide kinase activities. J. Biol. Chem. 277:33291-33299. [DOI] [PubMed] [Google Scholar]
- 29.Spector, M. P., J. M. Hill, E. A. Holley, and J. W. Foster. 1985. Genetic characterization of pyridine nucleotide uptake mutants of Salmonella typhimurium. J. Gen. Microbiol. 131:1313-1322. [DOI] [PubMed] [Google Scholar]
- 30.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 33.Vu, C. Q., D. L. Coyle, H. H. Tai, E. L. Jacobson, and M. K. Jacobson. 1997. Intramolecular ADP-ribose transfer reactions and calcium signalling. Potential role of 2′-phospho-cyclic ADP-ribose in oxidative stress. Adv. Exp. Med. Biol. 419:381-388. [PubMed] [Google Scholar]
- 34.You, K. S. 1985. Stereospecificity for nicotinamide nucleotides in enzymatic and chemical hydride transfer reactions. CRC Crit. Rev. Biochem. 17:313-451. [DOI] [PubMed] [Google Scholar]
- 35.Zerez, C. R., D. E. Moul, E. G. Gomez, V. M. Lopez, and A. J. Andreoli. 1987. Negative modulation of Escherichia coli NAD kinase by NADPH and NADH. J. Bacteriol. 169:184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, N., B. M. Olivera, and J. R. Roth. 1991. Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J. Bacteriol. 173:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, N., B. M. Olivera, and J. R. Roth. 1989. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J. Bacteriol. 171:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, N., B. M. Olivera, and J. R. Roth. 1988. Identification of a repressor gene involved in the regulation of NAD de novo biosynthesis in Salmonella typhimurium. J. Bacteriol. 170:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, N., and J. R. Roth. 1991. The nadI region of Salmonella typhimurium encodes a bifunctional regulatory protein. J. Bacteriol. 173:1302-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]