Abstract
The min locus encodes a negative regulatory system that limits formation of the cytokinetic Z ring to midcell by preventing its formation near the poles. Of the three Min proteins, MinC is the inhibitor and prevents Z-ring formation by interacting directly with FtsZ. MinD activates MinC by recruiting it to the membrane and conferring a higher affinity on the MinCD complex for a septal component. MinE regulates the cellular location of MinCD by inducing MinD, and thereby MinC, to oscillate between the poles of the cell, resulting in a time-averaged concentration of MinCD on the membrane that is lowest at midcell. MinC can also be activated by the prophage-encoded protein DicB, which targets MinC to the septum without recruiting it first to the membrane. Previous studies have shown that the C-terminal domain of MinC is responsible for the interaction with MinD, DicB, and the septal component. In the present study, we isolated mutations in the C-terminal domain of MinC that affected its interaction with MinD, DicB, and the septal component. Among the mutations isolated, R133A and S134A are specifically deficient in the interaction with MinD, E156A is primarily affected in the interaction with DicB, and R172A is primarily deficient in the interaction with the septum. These mutations differentiate the interactions of MinC with its partners and further support the model of MinCD- and MinC-DicB-mediated cell division inhibition.
In Escherichia coli, the min system cooperates with the nucleoid to direct the placement of the division septum to midcell (25). The three gene products of the min system, MinC, MinD, and MinE, act in concert to prevent Z-ring formation at the cell poles (5). MinC is the division inhibitor since overexpression of MinC alone, but not overexpression of MinD or MinE, causes filamentation (5). In vitro studies revealed that MinC interacts directly with FtsZ and antagonizes FtsZ assembly (15). Unlike another cell division inhibitor, SulA, which is induced as part of the SOS response following DNA damage, MinC inhibits FtsZ polymerization without blocking its GTPase activity (15). In vivo, the cell division inhibition caused by MinC is enhanced by MinD, which is a peripheral membrane ATPase (4). MinD recruits MinC to the membrane (11, 32) and confers a higher affinity for some septal component (18), which is yet to be identified. The combined expression of MinCD makes a potent division inhibitor, leading to severe filamentation and cell death if MinE, the topological regulator, is not present. However, in the presence of MinE, this cell division inhibitor is spatially regulated such that Z-ring formation is allowed only at midcell (5).
The spatial regulation of MinCD by MinE is achieved through a coupled oscillation of the Min proteins (33). MinD recruits MinC to the membrane in one half of the cell in the shape of a test tube. MinE forms a ring at the rim of the test tube near midcell and stimulates MinD ATPase activity (12). This activity of MinE progressively removes the MinCD complex from the membrane as it moves toward the pole. Meanwhile, a new MinCD test tube forms in the other half of the cell. As the MinE ring approaches the old pole, a new MinE ring forms at the rim of the new test tube, and the cycle continues (8, 10-12, 31, 32, 36). Through this oscillation, MinC and MinD mostly occupy the poles, leaving the midcell free of MinC, so that the Z ring can form there (20, 27).
Sequence analysis of MinC from various bacteria suggested that it has two separate domains, and this was confirmed by genetic and biochemical studies (14). Overexpression of the N-terminal domain (residues 1 to 115) fused to MalE caused filamentation in the absence of MinD as effectively as the full-length fusion. It was also just as efficient at inhibiting Z-ring assembly defining the N-terminal domain of MinC as the anti-FtsZ domain (14). The yeast two-hybrid assay revealed a strong interaction between MinC and MinD, as well as MinC self-interaction. In both cases, the C-terminal domain was as active as the full-length MinC, indicating that the C-terminal domain (residues 116 to 231) is sufficient for interaction with MinD and self-association (14). Consistent with this, both MalE-MinC and MalE-MinC116-231 eluted as dimers in gel filtration chromatography (14, 38), and both were recruited to membrane vesicles by MinD (16, 22). The two-domain concept was fully supported by the crystal structure of MinC from Thermotoga maritima (3). MinC crystallized as a dimer with each monomer folded into two separate domains connected by a flexible linker. The C-terminal domain is very compact, consisting of a series of β-strands that form a triangular β-helix with three sides designated A, B, and C. The A surface forms a hydrophobic dimer interface. The two N-terminal domains in the dimer are not in close contact with each other and are in different orientations in different dimers. This variable orientation is accomplished by a flexible linker that connects the N- and C-terminal domains and that varies in length in different MinC proteins (14).
Various types of evidence indicate that MinD activates MinC to inhibit division through at least two steps. First, MinD recruits MinC to the membrane (11, 32), and second, the MinCD complex is targeted to a septal component (18). The result of these two steps is an increase in the concentration of MinC in the vicinity of the Z ring which antagonizes its assembly. These steps can be bypassed by overexpressing MinC 25- to 50-fold (6, 15). The fact that a MinD E126A mutant can target MinC to the septum but cannot fully activate MinC indicates that there is a step beyond septum targeting (40).
The cell division-inhibitory activity of MinC can also be enhanced by DicB (7, 22), a protein encoded by prophage Kim (2). Under normal conditions, expression of DicB is repressed (1). However, induction of DicB in the presence of MinC severely blocks cell division, and this inhibition is not regulated by MinE (7). By using an N-terminally truncated MinC mutant that cannot disrupt Z rings, Johnson et al. (18) showed that like MinD, DicB targets this MinC mutant to the septum. However, the targeting of MinC by DicB is not preceded by recruitment of the complex to the membrane (18). This difference from MinD-promoted targeting reflects the fact that DicB-stimulated inhibition is not spatially regulated. Whereas MinD targeting to the septum is normally masked by the oscillation which places the MinCD complex at the poles, DicB directs MinC directly to the Z ring. Interestingly, DicB targeting requires ZipA, whereas the targeting of MinC by MinD does not require ZipA (19). The septal target of the MinCD complex is unknown but does not appear to be ZipA or FtsA.
Previously, several mutations were isolated in minC that prevent it from being a division inhibitor (8, 21). One of the mutant proteins, encoded by minC19 (G10D) which alters an amino acid near the N terminus of MinC, fails to interact with FtsZ and prevent Z-ring assembly, although it still interacts with MinD and with itself (15). Mutants with mutations in the C-terminal domain appeared to have lost the ability to interact with MinD and/or DicB; however, most of these mutations resulted in an unstable MinC (35). Thus, there is little information about critical residues in the C-terminal domain.
MinD and DicB both activate MinC by binding to the C-terminal domain and targeting it to a septal component (18). In this study we set out to identify residues in MinC responsible for interactions with these two different activators. In addition, if MinC is the component in the MinCD and MinC-DicB complexes that contacts the septum, it should be possible to identify residues in MinC that interact with the septal component. In this study, we utilized the location of conserved residues within the crystal structure of MinC from T. maritima as a guide to introduce point mutations. These mutations were then analyzed to determine their effects on the interaction with MinD and/or DicB and to determine if they affected interaction with the septum.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids utilized in this study are listed in Table 1. To construct plasmid pHJZ102, in which the C-terminal part of MinC was fused to green fluorescent protein (GFP), minC116-231 was amplified from pJPB210 by using primers 5′-TAGCATGTCGACTGGAGCCTGCGGTGTGGGAGCT-3′ and 5′-TAGCATAAGCTTTCAATTTAACGGTTGAACGGTC-3′. The amplified fragment was digested with SalI and HindIII (underlined sequences in the primers) and cloned into pJC106. To construct pZH118 containing a GFP-DicB fusion, a fragment containing dicB was amplified from chromosomal DNA prepared from strain W3110. The primers used were 5′-TAGCATTCTAGAGAAACTATGAAAACGTTATTACCAAACGTTA-3′ and 5′-TAGCAATAAGCTTAACTGTCAGAACAAGCACAAATGCTG-3′. The amplified fragment was digested with XbaI and HindIII (underlined sequences) and cloned into the vector pJC106. Plasmid pZH119 containing an inducible dicB gene was constructed in the following way. dicB was amplified from chromosomal DNA with primers 5′-TAGCATGGATCCGGAGAGAAACTATGAAAACGTTATTACCAAACGTTA-3′ and 5′-AACTGTCAGAACAAGCACAAATGCTG-3′. The amplified fragment was digested with BamHI (underlined sequence in the first primer) and EcoRI, which cut downstream of the second primer, and cloned into the same sites in pAM239. To construct pCS104CD, which contained minCD under control of the PBAD promoter (41), the SacI-HindIII fragment from pSEB104 was replaced by a minCD fragment amplified from pJPB210 by using the following primers containing these sites (underlined): 5′-ATAGAGCTCGCTAATTGAGTAAGGC-3′ and 5′-TACTAAGCTTAACTTATCCTCCGAAC-3′. Plasmid pHJZ117 contains minC under the control of the Ptac promoter. The minC fragment was amplified from pZH110 by using primers 5′-GCTAGAATTCGCTAATTGAGTAAGGCCAGGATGTC-3′ and 5′-TAGCATAAGCTTTCAATTTAACGGTTGAACGGTC-3′. The amplified fragment was digested with EcoRI and HindIII (underlined sequences in the primers) and cloned into pJF118EH (12). The following derivatives containing mutations were constructed similarly: pHJZ117-1 (R172A) and pHJZ117-2 (R133A). All PCR products were sequenced to verify that no mutations had been introduced.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| JS964 | MC1061 malP::lacIq Δmin::kan | 28 |
| W3110 | Prototroph | Lab collection |
| PS88 | W3110 Δmin::kan | P1(JS964) × W3110, select for Kanr |
| Plasmids | ||
| pJC106 | pBAD18::GFP (mut2) | 10 |
| pAM239 | XmnI plus FnuD2 fragment of pUC19 containing the lac promoter in pGB2 | 8 |
| pJPB210 | minCDE on pGB2 | 28 |
| pZH106 | PBAD::gfp-minD in pJC106 | 10 |
| pZH108 | Plac::gfp-minCDE | 10 |
| pZH118 | PBAD::gfp-dicB on a pBAD18 derivative | Z. Hu and J. Lutkenhaus, unpublished data |
| pZH119 | Plac::dicB in pAM239 | Z. Hu and J. Lutkenhaus, unpublished data |
| pZH110 | minC (pJPB210 ΔminDE) | 10 |
| pJC22 | minC in pGBT9 (BD-minC) | 16 |
| pJC41 | minD in pGAD424 (AD-minD) | 16 |
| pJC51 | dicB in pGAD424 (AD-dicB) | Z. Hu and J. Lutkenhaus, unpublished data |
| pZH112 | PBAD::malE-minC116-231 | 13 |
| pSEB12 | minCDE in mini-F plasmid | 28 |
| pHJZ109 | Plac::gfp-minC116-231minD | 39 |
| pHJZ102 | PBAD::gfp-minC116-231 | This study |
| pSEB104 | PBAD::ftsZ-gfp in a pGB2 derivative | 29 |
| pCS104CD | PBAD::minCD in a pGB2 derivative | 41 |
| pJF118EH | Expression vector with Ptac | 11 |
| pHJZ117 | Ptac::minC in pJF118EH | This study |
Site-directed mutagenesis.
Mutations R133A, S134A, D144A, E210A, E156A, R170A, R172A, D180A, E193A, and D205A were introduced into minC on pZH110 by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). R133A, E156A, R170A, and R172A were also introduced into minC on the single-copy vector pSEB12 by the same method. In addition, minC mutations were introduced into different vectors to generate derivatives of pJC22 (BD-minC), pZH112 (malE-minC116-231), pZH108 (gfp-minCDE), pHJZ102 (gfp-minC116-231), pHJZ109 (gfp-minC116-231 minD), and pCS104CD (minCD) by using the same primers that were used for cloning wild-type minC.
Yeast two-hybrid assay.
To examine interactions between various MinC mutants and MinD or DicB, the corresponding plasmids described above were transformed in various combinations into the reporter strain SFY526 as described in the CLONTECH manual (BD Bioscience Clontech, Palo Alto, Calif.). Double transformants were selected on media without tryptophan and leucine. Transformants were examined for β-galactosidase activity qualitatively by a colony lift assay and quantitatively by a liquid β-galactosidase assay, as described in the CLONTECH manual.
Immunoblot analysis.
JS964 (Δmin), JS219 (28), and JS964 (Δmin) containing pZH110 or derivatives with various minC alleles were grown in Luria-Bertani (LB) medium to the exponential phase. JS964 (Δmin) containing pCS104CD (PBAD::minCD) was grown to an optical density at 600 nm (OD600) of 0.05 and induced with 0.2% arabinose for 2 h. Cells were collected by centrifugation, resuspended in sodium dodecyl sulfate (SDS) sample buffer, boiled for 5 min, and subjected to SDS-12.5% polyacrylamide gel electrophoresis (PAGE). Proteins were electrophoretically transferred to a nitrocellulose membrane, and MinC was detected by using a rabbit antiserum raised against His-MinC as the primary antibody and goat alkaline phosphatase-conjugated anti-rabbit immunoglobulin (Bio-Rad) as the secondary antibody.
Fluorescence microscopy.
To determine if MinC mutants could still be targeted to the septum by MinD, plasmid pHJZ109 (gfp-minC116-231 minD) derivatives containing R133A, E156A, R170A, or R172A were transformed into JS964 (Δmin) by selection for Spcr. Cultures were grown at 37°C until the OD600 was 0.02. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a concentration of 100 μM, and samples were examined 1 h later by fluorescence microscopy. To examine the targeting of MinC mutants by DicB, plasmid pHJZ102 and the various MinC derivatives were cotransformed into JS964 with pZH119 (Plac dicB). Cotransformants were obtained by selection for both Spcr and Ampr. Arabinose (0.0002%) and IPTG (1 mM) were added to induce the expression of gfp-minC116-231 and DicB, respectively. Samples were taken 2 h later and fixed with 0.2% glutaraldehyde. Cells were photographed with a Nikon fluorescence microscope equipped with a MagnaFire charge-coupled device camera (Optronics). To assess the oscillation of MinC and the various mutants, plasmid pZH108 (Plac gfp-minCDE) or derivatives containing R133A or R172A were transformed into JS964. Single colonies were selected on plates containing spectinomycin and 0.2% glucose. Colonies were inoculated into liquid cultures and grown at 37°C until the OD600 was 0.02. IPTG (100 μM) was added, and 0.7 μl of each culture was removed for examination 1.5 h after induction. Cells were examined by fluorescence microscopy, and pictures were taken at 8- to 12-s intervals. Images were imported into Adobe PhotoShop for assembly.
Phenotypic analysis of minC mutants.
To test the ability of MinD or DicB to activate the various MinC mutants to block cell division, pZH106 (gfp-minD) or pZH118 (gfp-dicB) was cotransformed with pZH110 (minC) and mutant derivatives into JS964 and selected on LB medium plates containing ampicillin, spectinomycin, and 0.2% glucose at 37°C. Cell morphology was examined by using a Nikon phase-contrast microscope. To test the effects of the R133A, E156A, R170A, and R172A mutations, they were introduced into the single-copy plasmid pSEB12, which contained the complete min operon. The resultant plasmids, as well as the wild-type plasmid, were transformed into JS964 and selected on LB medium plates containing chloramphenicol. The morphology of cells was examined as described above. To further examine the ability of these MinC mutants to be activated by MinD to block cell division, plasmid pCS104CD (PBAD::minCD) and derivatives of this plasmid with mutations were transformed into JS964. Single colonies on plates containing spectinomycin and 0.2% glucose were resuspended in 300 μl of LB medium and serially diluted 10-fold. Samples (4 μl) were spotted on plates with spectinomycin and different concentrations of arabinose as indicated below and incubated at 37°C overnight.
To analyze the ability of MinC mutants to inhibit cell division in the absence of MinD, plasmid pHJZ117 (Ptac::minC) and derivatives of this plasmid with minC mutations were transformed into PS88 (Δmin). Single colonies from plates containing ampicillin and 0.2% glucose were inoculated into liquid cultures in the presence of 0.2% glucose and incubated overnight. Each overnight culture was diluted 1,000-fold into LB medium and grown at 37°C until the OD600 was 0.05. Then 1 mM IPTG was added, and samples were fixed with 1% formaldehyde 2 h after induction. Cell morphology was recorded with a Nikon phase-contrast photomicroscope, and images were imported into Adobe Photoshop for assembly.
Protein purification and membrane binding assay.
MalE-MinC116-231 was purified as described previously (15, 40). Mutants R133A and R172A were purified by using the same procedure. Sedimentation assays were carried out as described previously (40). Briefly, MalE-MinC116-231 and various mutants (4 μM), MinD (4 μM), multilaminar large vesicles (400 μg/ml), and nucleotide (1 mM ADP or ATP) were mixed at room temperature in 50 μl of ATPase buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2). The reaction mixtures were incubated at room temperature for 10 min and centrifuged at 10,000 × g at room temperature in a tabletop centrifuge for 2 min. The supernatants were carefully removed, and the pellets were resuspended in 50 μl of SDS sample buffer. Aliquots (20 μl) of the samples were electrophoresed on SDS-12.5% PAGE gels and stained with Coomassie brilliant blue.
RESULTS
Site-directed mutagenesis of the C-terminal domain of MinC.
Site-directed mutagenesis of the C-terminal domain of MinC was carried out to determine if important residues involved in the interaction with MinD, DicB, and septal components could be identified. Previously isolated missense mutations in the C-terminal region of MinC resulted in instability due to increased sensitivity to the Lon protease and provided little information about the interaction of MinC with its partners (35). Since sequence analysis of MinCs from various bacteria revealed that the C-terminal domain of MinC is conserved, we assumed that the structures are likely to be similar (Fig. 1). Point mutations were designed to alter highly conserved amino acids; however, the A interface was mostly avoided since it is involved in dimerization. Charged residues at the turns between β-strands or other charged residues that are expected to be solvent exposed were chosen and changed to alanine. The mutations included R133A, S134A, D144A, E156A, R170A, R172A, D180A, E193A, D205A, and E210A. The mutations were introduced on a low-copy-number plasmid (pZH110) in which the minC gene is under the control of its own promoter.
FIG. 1.
Alignment of MinC proteins from E. coli (Ec), Salmonella enterica serovar Typhimurium (St), Yersinia pestis (Yp), Vibrio cholerae (Vc), Neisseria gonorrhoeae (Ng), Thermotoga maritima (Tm), and Bacillus subtilis (Bs). Residues that are identical in four or more sequences are indicated by a black background, and conserved residues are indicated by a gray background. The arrowheads indicate residues that were altered by mutation in this study. The RSGQ motif is underlined.
As a first test to determine the effects of these minC mutations on MinC's function, we examined their abilities to be activated by either MinD or DicB to inhibit division. To do this, the plasmids carrying the minC mutations (derivatives of pZH110) were cotransformed into a min deletion strain (JS964) with either pZH106 (gfp-minD) or pZH118 (gfp-dicB). Cotransformants were selected on plates with spectinomycin, ampicillin, and glucose. Glucose was added to repress the expression of GFP-MinD or GFP-DicB, so that the cellular concentration of the fusion protein was low, nearer the physiological level. In this test a lack of cotransformants indicated that the mutation did not affect the ability of MinC to be activated to inhibit division and prevent colony formation. The presence of cotransformants indicated that the MinC mutant was attenuated. Also, any transformants obtained could be screened for cell morphology to determine to what extent the minC mutation was attenuated. A minicell phenotype indicated that the minC mutation prevented activation, whereas a filamentous phenotype indicated that activation occurred to some extent. The results are summarized in Table 2. No cotransformants were obtained with plasmids expressing GFP-MinD (pZH106) and wild-type MinC (pZH110). Also, no cotransformants were obtained with plasmids expressing GFP-MinD and plasmids carrying minC mutations D144A, E210A, E193A, and D205A, suggesting that they were fully responsive to MinD activation. However, plasmids carrying R133A and D180A yielded as many cotransformants as a control plasmid lacking GFP-MinD. These cotransformants exhibited a minicell phenotype with no sign of filamentation, implying that the mutations could not be activated by MinD. Also, plasmids carrying S134A, R172A, and E156A yielded cotransformants, but there was some degree of filamentation, indicating that these mutations attenuated MinC to some extent. A plasmid carrying R170A yielded transformants, but the filamentation was very pronounced, indicating that this mutation had less effect on MinC.
TABLE 2.
Effects of minC mutations on MinD and DicB interactions
| MinC mutant | Phenotypic assaya
|
Yeast two-hybrid assayb
|
||
|---|---|---|---|---|
| MinD activation (pZH106/pZH110) | DicB activation (pZH118/pZH110) | MinC-MinD | MinC-DicB | |
| R133A | − | +++ | − (0) | +++ (49) |
| S134A | + | +++ | +/− | +++ |
| D144A | +++ | +++ | +++ | +++ |
| E210A | +++ | +++ | +++ | +++ |
| E156A | + | − | + (1) | − (0) |
| R170A | ++ | +++ | +++ (69) | + (1) |
| R172A | + | + | ++ (35) | + (6) |
| D180A | − | − | − | − |
| E193A | +++ | +++ | ND | ND |
| D205A | +++ | +++ | ND | ND |
| Wild type | +++ | +++ | +++ (63) | +++ (48) |
+++, lethal; ++, severe filamentation; +, slight filamentation; −, no effect.
+++, dark blue developed in 30 min; ++, dark blue developed in 1 to 2 h;+, blue developed in 3 to 4 h; +/−, pale blue developed overnight; −, no color change. The numbers in parentheses are Miller units determined from the liquid assay. ND, not determined.
In a parallel experiment, cotransformation of JS964 with a plasmid carrying wild-type MinC (pZH110) and a plasmid expressing GFP-DicB (pZH118) did not yield cotransformants. Most of the plasmids carrying minC mutations did not yield cotransformants, indicating that the mutations did not affect DicB activation. However, plasmids carrying E156A, R172A, and D180A yielded cotransformants. The cotransformants with E156A and D180A had a minicell phenotype with no sign of filamentation, while R172A yielded cotransformants with a slight degree of filamentation. Thus, E156A and D180A could not be activated by DicB, and R172A was only weakly activated by DicB.
To examine the possibility that the attenuation of the MinC mutants could have been due to lower levels of the mutant proteins, cell lysates of JS964 (Δmin) containing plasmids with various minC alleles were analyzed by immunoblotting by using antibody against MinC (Fig. 2). D180A could not be detected on the Western blot, indicating that this mutation resulted in a very unstable protein. The marked instability of D180A would account for its failure to respond to either MinD or DicB in the assays described above. The other mutants examined were expressed at a level comparable to the level of wild-type MinC. Having ruled out instability, we concluded that R133A and S134A could be activated by DicB but not by MinD. E156A appeared to be defective in activation by DicB, although it showed some deficiency in response to MinD. R170A responded well to both DicB and MinD, whereas R172A displayed deficiency in activation by both DicB and MinD. On the other hand, D144A, E210A, E193A, and D205A behaved like wild-type MinC in these assays, indicating that these residues are not critical for MinD- or DicB-mediated inhibition of cell division.
FIG. 2.
Stability of MinC mutant proteins. The effects of minC mutations on the stability of MinC were determined by immunoblot analysis. Samples from exponentially growing cultures of JS964 (Δmin) containing pZH110 (minC) or derivatives of this plasmid with various minC mutations were analyzed. The first two lanes contained controls (JS964 [Δmin] and JS219 [min+]). The third through eighth lanes contained JS964 (Δmin) with pZH110 or derivatives of pZH110 with minC mutations. Of the mutations examined, only D180A produced an unstable protein. The ninth lane contained JS964 (Δmin) with pCS104CD (CD). The latter sample was taken 2 h after addition of 0.2% arabinose to induce expression of minCD.
Assessment of MinC mutants in the context of the min operon.
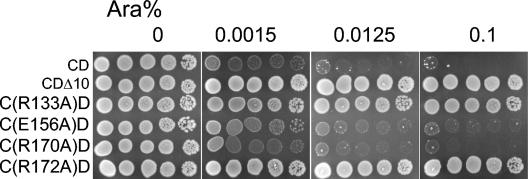
To various extents, the minC mutations R133A, E156A, R170A, and R172A displayed deficiencies in their ability to respond to MinD and inhibit division in the assay described above in which the two min genes were carried on separate plasmids (Table 2). To examine this under more physiological conditions, we introduced these four mutations into pCS104CD. This plasmid has the minCD genes in tandem under control of the PBAD promoter on a low-copy-number vector (41). The basal expression of these genes in the absence of arabinose was sufficiently low that the cell morphology of a Δmin strain was not affected (data not shown). In the presence of arabinose (>0.0125%) cell division was inhibited and colony formation was prevented (Fig. 3). Immunoblot analysis indicated that the level of MinCD with 0.2% arabinose was similar to the wild-type level (Fig. 2, compare lanes 2 and 9). When minD was replaced with minDΔ10, the growth of JS964 was not inhibited by arabinose (Fig. 3). This minD allele does not encode the last 10 amino acids of MinD that are essential for membrane binding and efficient activation of minC (13). Introduction of either the R133A or R172A mutation into pCS104CD resulted in a loss of arabinose sensitivity, indicating that these two minC mutations were defective in responding to minD and blocking division at physiological levels. On the other hand, introduction of E156A or R170A had little effect as arabinose-dependent killing was still observed. Titration of the arabinose revealed that these mutations had little effect of the ability of MinC to be activated by MinD (Fig. 3). We are not sure why E156A showed significant attenuation in the trans test done initially (Table 2), whereas it displayed little attenuation in the titration test. However, the latter test was performed under more physiological conditions and should indicate the relevant activity of E156A.
FIG. 3.
Effects of MinC mutations on cell viability in the presence of MinD. pCS104CD (PBAD::minCD) (CD) and derivatives of this plasmid containing minC mutations were transformed into JS964 (Δmin). Colonies appearing on plates with spectinomycin and 0.2% of glucose were resuspended in 300 μl of LB medium and serially diluted (10-fold steps). A 4-μl sample of each dilution was spotted on plates with spectinomycin and different concentrations of arabinose (as indicated at the top) and incubated at 37°C overnight.
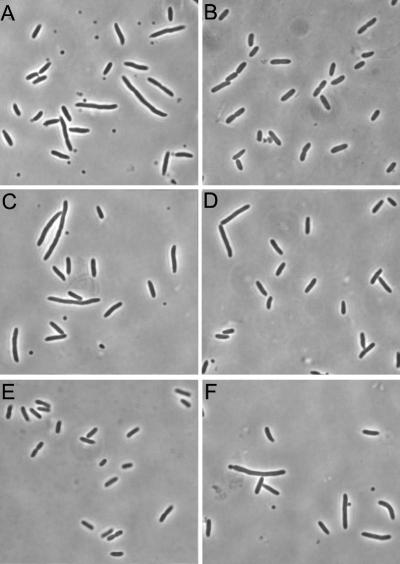
To further examine the various minC mutations and their effect on the regulation of division, each mutation was introduced into the single-copy plasmid pSEB12 (minCDE). This plasmid restored the wild-type phenotype when it was present in the min deletion strain JS964 (Fig. 4B). The amount of the Min proteins expressed from this single-copy plasmid is similar to the amount in a wild-type strain (data not shown), which allowed good assessment of the effects of min mutations on the ability of Min to properly regulate division. pSEB12 derivatives carrying the mutations R133A, E156A, R170A, and R172A were introduced into JS964. JS964 containing plasmids with the R133A and R172A mutations (Fig. 4C and F, respectively) exhibited a minicell phenotype indistinguishable from that of the strain lacking a plasmid (Fig. 4A). This test confirmed that these two mutations prevent MinC from participating in the spatial regulation of the placement of the division septum when the Min proteins are present at physiological levels. Cells with E156A had mostly a wild-type phenotype, and there were only occasional minicells and elongated cells (Fig. 4D), suggesting that E156A had only a slight defect. Cells with R170A had a wild-type phenotype (Fig. 4E).
FIG. 4.
Complementation of JS964 by a single-copy plasmid. pSEB12(minCDE) and derivatives of this plasmid containing minC mutations were transformed into JS964. Single colonies were inoculated into liquid cultures and grown at 37°C until the mid-log phase. Cell morphology was examined by phase-contrast microscopy, and pictures were taken with a MagnaFire charge-coupled device camera. (A) JS964; (B) JS964/pSEB12; (C) JS964/pSEB12 minC-R133A; (D) JS964/pSEB12 minC-E156A; (E) JS964/pSEB12 minC-R170A; (F) JS964/pSEB12 minC-R172A.
Interaction of MinD and DicB with MinC mutants in the yeast two-hybrid assay.
The simplest explanation for the failure of various MinC mutants to respond to either MinD or DicB is that they did not interact with these proteins. To test this possibility, the interaction between MinC and MinD or DicB was assessed by using the yeast two-hybrid system. In this system both MinD and DicB interact strongly with MinC (17, 18). The effects of the minC mutations are summarized in Table 2. Immunoblot analysis revealed that the levels of the mutant proteins in the yeast two-hybrid system were comparable to the levels produced by the wild-type construct (data not shown).
Mutants D144A and E210A were tested, and as expected, they behaved like wild-type MinC and interacted strongly with both MinD and DicB. R133A showed no interaction with MinD but interacted as strongly as wild-type MinC with DicB. S134A behaved similarly, interacting strongly with DicB and only weakly with MinD. These results correlate very well with the cell division inhibition test results described above (Table 2), as these two mutants inhibited division in the presence of DicB but not in the presence of MinD. Therefore, R133A and S134A were primarily defective in interaction with MinD.
E156A displayed no interaction with DicB, and the interaction with MinD was significantly attenuated. The first result is consistent with the in vivo test results, as E156A did not respond to DicB. Even though the interaction with MinD was attenuated, E156A behaved similar to wild-type MinC in the bacterial test systems, in which it was expressed at nearly physiological levels. It showed only a slight defect in MinD activation (Fig. 3) and was able to largely complement the Min phenotype when it was present on a single-copy plasmid (Fig. 4D). R170A interacted strongly with MinD but only weakly with DicB. Nonetheless, R170A could be activated by both of these inhibitors to block division.
R172A interacted with MinD and DicB in the yeast two-hybrid assay, although to a lesser extent than the wild type, especially for DicB. However, the interaction with DicB was stronger than that of R170A, which could still be activated by DicB. Thus, R172A is of particular interest since it interacted with MinD and DicB. One possibility is that although R172A can interact with both MinD and DicB, it cannot be targeted to the septum. Another possibility is that it can still be targeted but cannot be fully activated after targeting.
R172A, but not R133A, is recruited to vesicles by MinD.
To further explore the defects of the various minC mutants, we examined their abilities to interact with MinD in vitro. MalE-MinC116-231 is recruited to phospholipid vesicles by MinD in the presence of ATP (16). To test the effects of the minC mutations, the C-terminal domains of R133A and R172A were fused in frame at the C terminus of the maltose binding protein (MalE) and were purified by amylose affinity chromatography. Gel filtration chromatography of the purified proteins revealed that the mutants eluted at the same position as the wild-type fusion, indicating that these proteins were dimers, as expected, since the mutations were away from the dimer interface (data not shown).
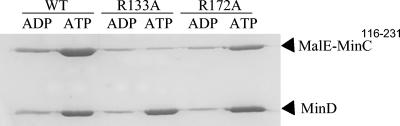
The small amount of MinD and MalE-MinC116-231 in the pellet in the presence of ADP represented the background in this assay (Fig. 5). In the presence of ATP, more than 50% of MinD was in the pellet, and it efficiently recruited MalE-MinC116-231. The R172A mutant was also recruited to the vesicles by MinD, although to a lesser extent than the wild type. This is consistent with the yeast two-hybrid results. In contrast, R133A was not recruited to the vesicles by MinD. The inability of MinD to recruit R133A is also consistent with the lack of interaction between R133A and MinD determined by the yeast two-hybrid test.
FIG. 5.
Sedimentation assay for MinD recruitment of MalE-MinC116-231 to mutilaminar large vesicles. MalE-MinC116-231 and various mutants (4 μM), MinD (4 μM), mutilaminar large vesicles (400 μg/ml), and nucleotide (1 mM ADP or ATP) were mixed at room temperature for 10 min in 50 μl of ATPase buffer. The samples were centrifuged at 10,000 × g at room temperature for 2 min, and the pellets were resuspended in 50 μl of SDS sample buffer. Aliquots (20 μl) of the samples were analyzed by SDS-12.5% PAGE and stained with Coomassie brilliant blue. WT, wild type.
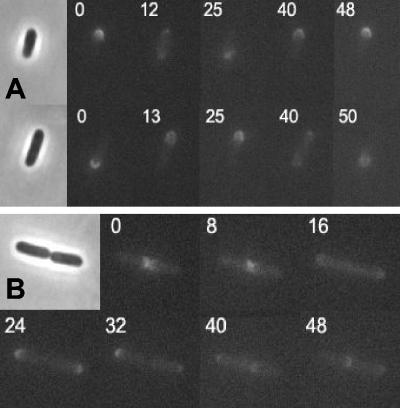
Oscillation of MinC R172A in the presence of MinD and MinE.
The tests described above indicated that R172A interacted with MinD but that the interaction was somewhat less than that of wild-type MinC. To test if the slight reduction in the interaction R172A and MinD affected the behavior of R172A, we examined its ability to oscillate in the presence of MinD and MinE. In a previous study, time-lapse experiments were carried out to monitor the localization of GFP-MinC in the presence of MinD and MinE in a min deletion background (11, 32). GFP-MinC oscillates with the same pattern that is observed for the dynamic localization of GFP-MinD in the presence of MinE (31). MinC is a cytoplasmic protein and is recruited to oscillate by its interaction with MinD and has no known role in regulating the oscillation. The R172A mutation was introduced into plasmid pZH108 (gfp-minCDE), which was used in the previous study (11). R133A was also introduced into this plasmid as a control. The plasmids were transformed into JS964 (Δmin), and exponential cultures were induced with 100 μM IPTG. Between 1 and 2 h after induction, examination of live cells by fluorescence microscopy revealed that GFP-MinC oscillated between two cell poles, with a complete cycle taking approximately 50 s, just as described previously (Fig. 6A). At this time the cells had a wild-type size distribution, indicating that at this point the level of GFP-MinC, MinD, and MinE was sufficient to restore the wild-type phenotype. GFP-R172A also oscillated, and the periodicity was the same as that of the wild type (Fig. 6B). Thus, R172A was able to interact with MinD and MinE and oscillate; however, it was unable to cooperate with MinD to spatially regulate cell division. In the control with R133A, the fluorescence was present throughout the cytoplasm, as expected, since this mutant was unable to interact with MinD. Also, the cells exhibited a minicell phenotype (data not shown).
FIG. 6.
Oscillation of R172A in the presence of MinD and MinE. Plasmid pZH108 (gfp-minCDE) and a derivative of this plasmid carrying R172A were transformed into JS964. Cells were inoculated into liquid cultures and grown at 37°C until the OD600 was 0.02. IPTG (100 μM) was added, and samples were taken 1.5 h after induction. Cells were examined by fluorescence microscopy, and pictures were taken 8 to 15 s apart (times are indicated in the images). (A) JS964/pZH108. Two cells are shown. (B) JS964/pZH108 minC-R172A.
Septal ring targeting of MinC mutants by MinD.
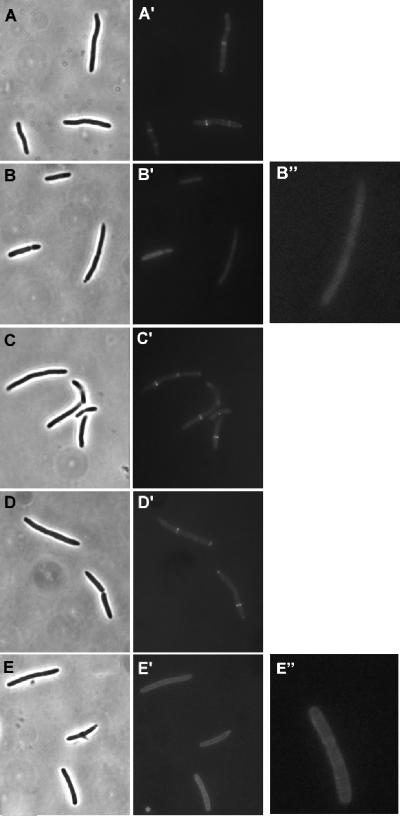
The MinC R172A mutant interacted with MinD in the yeast two-hybrid assay, was recruited to phospholipid vesicles in vitro, albeit somewhat less than the wild-type protein, and oscillated like the wild-type protein in the presence of MinD and MinE. However, R172A was unable to cooperate with MinD to direct the correct placement of the division septum, as indicated by the single-copy complementation assay (Fig. 4F). It has been shown previously that a MinC mutant missing the N terminus can be targeted to the division septum by MinD or DicB (18). Also, it has been proposed that this targeting is an important step in activation of MinC by MinD following recruitment of MinC to the membrane (19). To see if this was the step at which R172A was affected, the C-terminal domain of R172A was fused to GFP, and its localization was compared to that of the wild-type C-terminal fusion present in pHJZ109 under lac promoter control. Constructs containing the R133A, E156A, and R170A mutations were also made for comparison.
Following induction, GFP-MinC116-231 localized to the septum (Fig. 7A), which appeared as bright bands or sometimes as two bright spots transverse to the long axis of the cell. Cells expressing E156A and R170A also displayed sharp bands (Fig. 7C and D), indicating that these mutants were targeted to the septum. R133A (Fig. 7B), as expected due to its inability to interact with MinD, was present throughout the cytoplasm with no sign of localization. On the other hand, R172A yielded a halo-like appearance, indicating that it was recruited to the cytoplasmic membrane (Fig. 7E). However, no transverse bands were seen, indicating a lack of affinity for the septum. Z rings were present, as visualized by indirect immunofluorescence staining with antibody against FtsZ (data not shown). Thus, R172A was recruited to the membrane by MinD, as expected, but it could not be targeted to the septum. This result, combined with the failure of R172A to restore the wild-type division pattern, indicated that R172A was deficient in targeting and that the targeting step is important for the spatial regulation of division.
FIG. 7.
Septal ring targeting of MinC mutants by MinD. Plasmid pHJZ109 (gfp-minC116-231 minD) and derivatives of this plasmid containing R133A, E156A, R170A, and R172A were transformed into JS964. Cultures were grown at 37°C until the OD600 was 0.02, and then IPTG (100 μM) was added. One hour later, cells were fixed with 0.2% glutaraldehyde and examined by fluorescence microscopy. (A) JS964/pHJZ109; (B) JS964/pHJZ109 minC-R133A; (C) JS964/pHJZ109 minC-E156A; (D) JS964/pHJZ109 minC-R170A; (E) JS964/pHJZ109 minC-R172A.
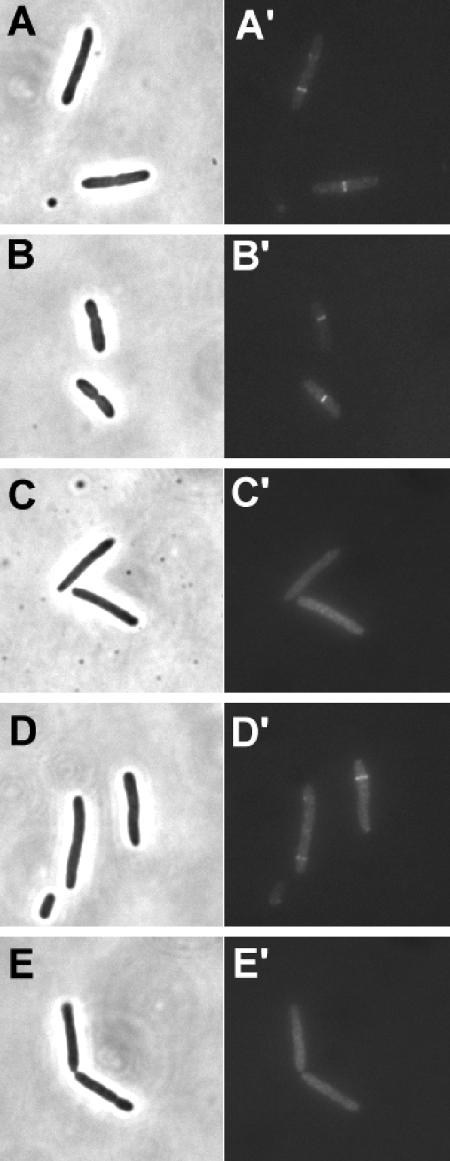
Targeting of MinC mutants to the septum by DicB.
DicB also activates MinC by targeting it to the septum. We therefore examined the MinC mutants to see whether they could be targeted by DicB. JS964 was cotransformed with pHJZ102 (PBAD::gfp-minC116-231) and pZH118 (Plac::dicB). Arabinose and IPTG were added to induce the expression of GFP-MinC116-231 and DicB, respectively. In the cells obtained, GFP-MinC116-231 was present in transverse bands, indicating that there was targeting to the septum (Fig. 8A). R133A and R170A behaved the same as the wild type (Fig. 8B and D, respectively), which is consistent with their ability to be activated by DicB (Table 2). In contrast, E156A and R172A were not targeted and appeared only in the cytoplasm (Fig. 8C and E, respectively). Since E156A could be targeted by MinD, it was specifically impaired in targeting by DicB. This lack of targeting reflected the lack of interaction between E156A and DicB in the yeast two-hybrid system. R172A, on the other hand, could not be targeted by either MinD or DicB, although it interacted with both, so it is possible that it was deficient in interaction with a common target within the septal ring.
FIG. 8.
Targeting of MinC mutants by DicB. Plasmid pHJZ102 (gfp-minC116-231) or derivatives of this plasmid carrying minC mutations were cotransformed into JS964 with pZH119 (DicB) by selecting for both Spcr and Ampr. Arabinose (0.0002%) and IPTG (100 μM) were added to induce the expression of GFP-MinC116-231 and DicB, respectively. IPTG (1 mM) was added 2 h later. Samples were taken after 30 min and fixed with 0.2% glutaraldehyde. Cells were examined by fluorescence microscopy. (A) JS964/pZH119/pHJZ102; (B) JS964/pZH119/pHJZ102 minC-R133A; (C) JS964/pZH119/pHJZ102 minC-E156A; (D) JS964/pZH119/pHJZ102 minC-R170A; (E) JS964/pZH119/pHJZ102 minC-R172A.
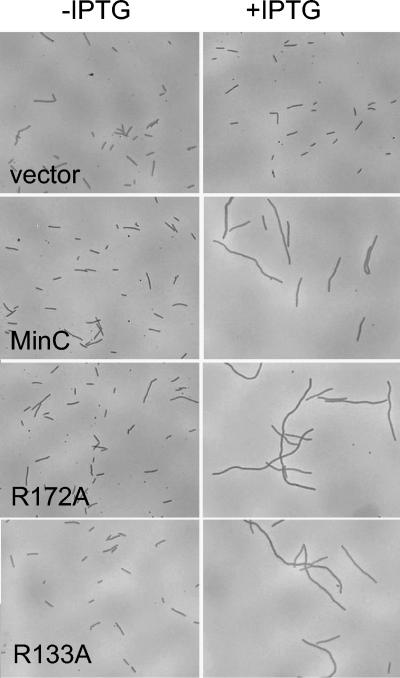
Overexpression of R172A inhibits division.
Although MinC requires an activator to efficiently inhibit division, it can inhibit division when it is overexpressed (6). This untargeted inhibition is due to the activity of the N-terminal domain and does not require the C-terminal domain (14). Since the mutations examined in this study were all in the C-terminal domain, they should not have affected the ability of MinC to inhibit division when they were overexpressed. This was of interest for R172A, which could not be activated by MinD or DicB. To confirm this expectation, minC, R172A and R133A were cloned into an expression vector (pJF118EH) downstream of the inducible tac promoter. PS88 (Δmin) containing the plasmids and a vector control were examined microscopically after growth in liquid cultures with and without IPTG. As shown in Fig. 9, overexpression of R172A and R133A in the absence of minD inhibited division to the same extent as wild-type MinC. This result confirmed that these mutations in the C-terminal domain of MinC did not affect the untargeted aspect of MinC's inhibitory activity.
FIG. 9.
Overexpression of R172A inhibits division. The ability of overexpressed R172A to inhibit division was assessed by inducing a culture of PS88 (Δmin) containing pHJZ117-1 (R172A) with IPTG for 2 h. PS88 (Δmin) containing pHJZ117 (minC), PS88 (Δmin) containing pHJZ117-2 (R133A), and PS88 (Δmin) containing the vector pJF118EH were also analyzed for comparison. For the panels on the left the medium did not contain IPTG, and for the panels on the right the medium contained 1 mM IPTG.
DISCUSSION
Central to our understanding of the regulation of cell division by the min system is an understanding of the interaction between MinC and its two activators, MinD and DicB, which prevents the formation of Z rings. Mutations isolated here differentiated interactions between MinC and its various partners and provided further evidence that the activation of MinC by MinD and DicB occurs through several steps. This study also indicated that DicB and MinD have distinct, but possibly overlapping, binding sites in the C-terminal domain of MinC but that MinCD and DicB-MinC complexes may have a common target in the septum. This study also confirmed that the targeting of MinC to the septum by MinD is necessary for efficient regulation of cell division by the min system.
MinD-MinC interaction.
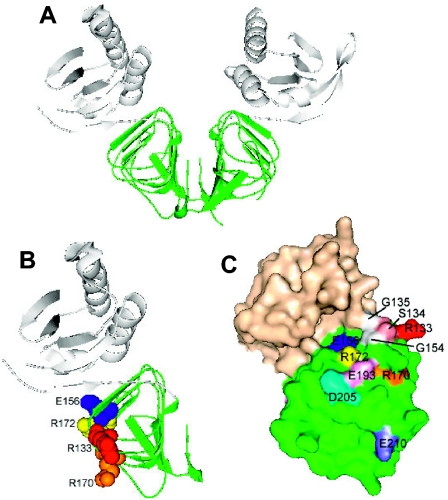
Our results indicate that residues at the junction of the B and C surfaces, especially the conserved RSGQ136 motif (Fig. 1 and 10), play a critical role in the MinC-MinD interaction. Two of the mutants analyzed in this study had alanine substitutions in this motif, R133A and S134A, and did not interact with MinD. R133A was studied in detail. It did not interact with MinD in vivo or in vitro. It also did not interact with MinD in the yeast two-hybrid system, was not recruited by MinD to the membrane in vivo, and was not recruited by MinD to phospholipid vesicles in vitro. However, this mutant interacted strongly with DicB and could be activated by DicB to inhibit division. The S134A mutant behaved similarly, although it was slightly less attenuated than R133A. The equivalent residues (R109 and S110) in T. maritima MinC lie near the top of the β-barrel structure near the junction of the B and C faces. We assume that in the E. coli protein these highly conserved residues are in a similar position. Since these residues are required for MinD interaction but not for DicB-mediated division inhibition, it is likely that they interact with MinD directly.
FIG. 10.
Model of MinC, indicating the locations of residues investigated in this study. The MinC protein structure is the structure for T. maritima MinC (PDB accession no. 1HF2). The residues corresponding to the residues of E. coli MinC examined in this study are indicated. The N terminus of MinC is white, and the C terminus is green. (A) MinC dimer; (B) MinC monomer; (C) Surface view of MinC monomer, rotated with respect to the view in panel B.
Recently, Ramirez-Arcos et al. (30) examined the effects of replacing highly conserved glycine residues with negatively charged amino acids. They reported that replacing G135, which is also within the conserved RSGQ motif, prevented interaction with MinD. Also, replacing G154, which is adjacent to the RSGQ motif in the structure (Fig. 10), had a similar effect. Together, these results indicate strongly that MinD interacts with this part of MinC. Consistent with this, mutations that were in other parts of the C-terminal domain, such as D144A, E193A, D205A, and E210A, did not significantly affect the MinD-MinC interaction. Also, the D180A mutation was near the dimer interface and resulted in a very unstable protein.
Interestingly, in the MinC dimer structure, the RSGQ motifs are on opposite ends of the dimer (Fig. 10). One possible model for the interaction of MinD with MinC is that a putative MinD dimer, which was proposed based upon comparison to the Fe-protein NifH dimer (23), interacts with a MinC dimer in a manner analogous to the Fe-protein's interaction with the MoFe-protein in the nitrogenase complex (34). In this complex the head-to-head dimer of the Fe-protein interacts with the head-to-head dimer of the MoFe-protein. However, it is not possible for one MinD dimer to contact the RSGQ motifs located on opposite ends of the MinC dimer. Thus, the Fe-protein complex is not a model for the MinD-MinC interaction.
Ramirez-Arcos et al. (30) proposed that MinC interacts with hydrophobic residues that are located in helix 7 of MinD. Recent support for this proposal comes from the independent mutagenesis studies of Ma et al. (24). Replacement of several of the hydrophobic residues in helix 7 led to a loss of interaction with MinC in the yeast two-hybrid system. Also, we previously isolated mutations in the MinD switch I (G42A and D44R) and switch II (I125E) regions that were specifically deficient in MinC interactions. The mutants could still self-interact, bind to the membrane, and be stimulated by MinE (40). The altered residue in the switch II region is adjacent to the hydrophobic residues identified in the study of Ma et al. (24), and the introduction of the negatively charged amino acid could interfere with protein-protein interactions. The switch I residues, however, are farther away in the monomer. It was suggested that they are in the signal transduction pathway that links the γ-phosphate binding domain to a region on the surface that directly interacts with MinC. In the proposed dimer model these residues come into close contact with helix 7 (41), which has been suggested to be the site for interaction with MinC (30).
DicB-MinC interaction.
DicB is not expressed under normal conditions, but when it is induced along with the C-terminal part of MinC, the complex is targeted to septal rings (18). Of the mutants which we isolated, only E156A was completely defective in DicB interaction as it showed no interaction in the yeast two-hybrid assay. Furthermore, E156A could not cooperate with DicB to inhibit cell division, nor could it be directed to the septal rings by DicB. In contrast, E156A interacted weakly with MinD in the yeast two-hybrid system but could still be targeted to the septum. Importantly, E156A could be activated efficiently by MinD to inhibit division and functioned normally with MinD and MinE to spatially regulate division when it was present in a single copy. Altogether, the data suggest that E156 is important for the DicB interaction. On the other hand, the residues in the RSGQ motif that we examined are not required for DicB binding and activation. Thus, the binding sites for DicB and MinD within MinC are somewhat distinct, but they may have some overlap, and that is why we observed that the interaction between MinD and E156A was reduced in the yeast two-hybrid assay.
MinC-septal component interaction.
MinD activates MinC to inhibit division by recruiting it to the membrane and ultimately the septum. However, these targeting steps can be bypassed if MinC is expressed at levels that are 25- to 50-fold above the physiological level (6). As expected, overexpression of R172A and R133A inhibited division as effectively as MinC, confirming that these mutations do not affect the untargeted aspect of MinC's inhibitory activity. It was previously estimated that by recruiting MinC to the membrane, MinD increases MinC's concentration at the membrane about 25-fold (13). Recent studies performed by two groups demonstrated that placing MinC directly on the cytoplasmic membrane enhanced its inhibitory activity. In one case this was done by adding the C-terminal amphipathic helix of Bacillus subtilis MinD to the C terminus of MinC (37). The resultant activation of MinC led to the suggestion that the septal targeting by MinD was not important. However, the level of MinC fusion was not measured, and it may have been above the physiological level. In the second approach the transmembrane domain of ZipA was attached to the N terminus of MinC (18). In this case the division-inhibiting activity of the MinC fusion was enhanced, but it could be further enhanced by adding MinD, indicating that MinD was doing more than placing MinD on the membrane. Our demonstration that R172A can be recruited to the membrane but cannot be activated to inhibit division and is therefore primarily deficient in targeting to the septum provides strong support for the importance of the targeting step in MinD's activation of MinC.
R172A interacted with MinD and DicB in the yeast two-hybrid system but could not efficiently inhibit division or be targeted to the septum by either protein. Although the interaction with these activators was less than the interaction with wild-type MinC in the yeast two-hybrid system, additional results indicated that the interaction with MinD should be sufficient for targeting. R172A was recruited to the membrane by MinD and oscillated normally in the presence of MinD and MinE. These results demonstrate that the interaction with MinD was sufficient for membrane targeting, which we were able to confirm in vitro. Also the interaction between R172A and DicB was stronger than the interaction observed with R170A, which was activated and targeted efficiently by DicB. Thus, it is unlikely that the defect of R172A lies in the interaction with the activators. Instead, it is likely that the defect is in the interaction with the septal target. We speculate that there may be a common septal component involved in the recruitment of the MinCD and MinC-DicB complexes.
The septal targeting of MinC by DicB, but not that of MinD, is ZipA dependent, indicating that the two complexes may target different components in the septum (19). Although targeting of the MinC-DicB complex requires ZipA, it only appears to recognize ZipA when it is present at the septum. A MinC-DicB fusion interacts weakly with ZipA in the yeast two-hybrid system; however, DicB is unable to target MinC to ZipA at the membrane in cells that lack Z rings. This result implies that ZipA alone is not sufficient to recruit the MinC-DicB complex or that ZipA undergoes a conformational change when it joins the Z ring, leading to recognition by the complex (19). Another possibility is that the target for the complex is bipartite (19). In this model DicB contacts ZipA and MinC contacts a second septal component, and both contacts are necessary for a stable association.
The MinC-MinD complex could also have a bipartite target, with MinD contacting one septal component and MinC contacting another. Septal association of MinD has not been observed in the absence of MinC in E. coli (18); however, MinD in B. subtilis is weakly associated with the septum. This association is observed in the absence of DivIVA, which is required for efficient localization of MinD (26). This association indicates that at least in B. subtilis MinD has a weak affinity for a septal component.
It is not clear in the bipartite target model if MinC in the MinCD complex contacts the same septal component that it contacts when it is in the DicB-MinC complex. If it does, the component cannot be ZipA. Also, FtsA and ZapA, the other two proteins recruited to the division site through direct interaction with FtsZ, do not play a role in the targeting of either complex (19). Therefore, an intriguing question is, what is the septal component that is recognized by MinC? Since the recruitment of all the known downstream proteins is dependent on FtsA and ZipA, these proteins are not likely to be the target. In that case, either FtsZ itself or some as-yet-unknown protein serves as the target of MinC when it is in the MinCD and MinC-DicB complexes. These possibilities require further investigation.
Acknowledgments
This work was supported by grant GM 29764 from the National Institutes of Health. This publication was also made possible by NIH grant 5P20RRO16443 from the COBRE Program of the NCRR.
REFERENCES
- 1.Bejar, S., F. Bouche, and J. P. Bouche. 1988. Cell division inhibition gene dicB is regulated by a locus similar to lambdoid bacteriophage immunity loci. Mol. Gen. Genet. 212:11-19. [DOI] [PubMed] [Google Scholar]
- 2.Cam, K., S. Bejar, D. Gil, and J. P. Bouche. 1988. Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 16:6327-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordell, S. C., R. E. Anderson, and J. Lowe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer, P. A., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 6.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1990. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. USA 87:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gil, D. 1990. Elaboration et caracterisation d'un nouveau type de vecteurs de clonage à nombre de copies regulable. Ph.D. thesis. Université de Toulouse, Toulouse, France.
- 10.Hale, C. A., H. Meinhardt, and P. A. J. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 12.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 13.Hu, Z., and J. Lutkenhaus. 2003. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 47:345-355. [DOI] [PubMed] [Google Scholar]
- 14.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, Z., C. Saez, and J. Lutkenhaus. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J. Bacteriol. 185:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. E., L. L. Lackner, C. A. Hale, and P. A. de Boer. 2004. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J. Bacteriol. 186:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse, K. 2002. A dynamic model for determining the middle of Escherichia coli. Biophys. J. 82:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labie, C., F. Bouche, and J. P. Bouche. 1990. Minicell-forming mutants of Escherichia coli: suppression of both DicB- and MinD-dependent division inhibition by inactivation of the minC gene product. J. Bacteriol. 172:5852-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lackner, L. L., D. M. Raskin, and P. A. de Boer. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185:735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutkenhaus, J., and M. Sundaramoorthy. 2003. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol. Microbiol. 48:295-303. [DOI] [PubMed] [Google Scholar]
- 24.Ma, L., G. F. King, and L. Rothfield. 2004. Positioning of the MinE binding site on the MinD surface suggests a plausible mechanism for activation of the Escherichia coli MinD ATPase during division site selection. Mol. Microbiol. 54:99-108. [DOI] [PubMed] [Google Scholar]
- 25.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 26.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84-96. [DOI] [PubMed] [Google Scholar]
- 27.Meinhardt, H., and P. A. J. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichoff, S., B. Vollrath, C. Touriol, and J. P. Bouche. 1995. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 18:321-329. [DOI] [PubMed] [Google Scholar]
- 29.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez-Arcos, S., V. Greco, H. Douglas, D. Tessier, D. Fan, J. Szeto, J. Wang, and J. R. Dillon. 2004. Conserved glycines in the C terminus of MinC proteins are implicated in their functionality as cell division inhibitors. J. Bacteriol. 186:2841-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin, D. M., and P. A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raskin, D. M., and P. A. J. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothfield, L. I., Y. L. Shih, and G. King. 2001. Polar explorers: membrane proteins that determine division site placement. Cell 106:13-16. [DOI] [PubMed] [Google Scholar]
- 34.Schindelin, H., C. Kisker, J. L. Schlessman, J. B. Howard, and D. C. Rees. 1997. Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature 387:370-376. [DOI] [PubMed] [Google Scholar]
- 35.Sen, M., and L. I. Rothfield. 1998. Stability of the Escherichia coli division inhibitor protein MinC requires determinants in the carboxy-terminal region of the protein. J. Bacteriol. 180:175-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szeto, T. H., S. L. Rowland, C. L. Habrukowich, and G. F. King. 2003. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J. Biol. Chem. 278:40050-40056. [DOI] [PubMed] [Google Scholar]
- 38.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, H., and J. Lutkenhaus. 2003. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J. Bacteriol. 185:4326-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, H., and J. Lutkenhaus. 2004. The switch I and II regions of MinD are required for binding and activating MinC. J. Bacteriol. 186:1546-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, H., R. Schulze, S. Cox, C. Saez, Z. Hu, and J. Lutkenhaus. 2004. Analysis of MinD mutations reveals residues required for MinE stimulation of the MinD ATPase and residues required for MinC interaction. J. Bacteriol. 187:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]