Abstract
The purple sulfur photosynthetic bacterium Thiocapsa roseopersicina BBS contains a heat-stable membrane-associated hydrogenase encoded by the hyn operon. Expression from the hyn operon regulatory region is up-regulated under anaerobic conditions. cis elements were mapped between positions −602 and −514 upstream from the hynS gene. Within this region two sequences that resemble DNA sites for FNR were recognized. The gene of an FNR homologue, FnrT, was identified in the genome of T. roseopersicina, and an fnrT knockout mutant was constructed. Anaerobic induction of hynS expression was abolished in the fnrT mutant, suggesting that FnrT is an activator of the hynS promoter. The T. roseopersicina hynS promoter could be activated in Escherichia coli, and this regulation was dependent on E. coli FNR. In vitro experiments with purified E. coli Ala154 FNR protein and purified E. coli RNA polymerase showed that FNR bound to two sites in the hyn regulatory region, that FNR could activate transcription initiation at the hynS promoter, and that FNR bound at the two target sites activated to different extents.
Thiocapsa roseopersicina BBS is a purple sulfur photosynthetic γ proteobacterium belonging to the Chromatiaceae family. It can be cultivated under photosynthetic anaerobic conditions and requires reduced sulfur compounds for growth. Like certain other members of Chromatiaceae, T. roseopersicina can be propagated chemolithoautotrophically in dark, aerobic conditions (21). Thus, the most important environmental factors affecting T. roseopersicina growth are light, oxygen, carbon, nitrogen, and sulfur compounds.
Hydrogenases catalyze the reversible oxidation of molecular H2. Three major classes of hydrogenases are known: iron-sulfur-cluster-free hydrogenases, Fe hydrogenases, and [NiFe] hydrogenases (41). Hydrogenases can have different roles; they can serve as redox safety valves to dispose of excess reducing power formed during fermentation or as generators of chemical energy by oxidation of H2 (10). In some organisms the same enzyme can have both functions, but more frequently each hydrogenase has a specialized physiological role in the cell (10, 41). Biosynthesis of hydrogenases can be regulated through various mechanisms. For example, the presence of the substrate molecule, H2, triggers the expression of some hydrogenases through a hydrogen-sensing hydrogenase (HupUV and HoxBC in Rhodobacter capsulatus and Ralstonia eutropha, respectively) and a two-component system (HupT/HoxJ and HupR/HoxA) (14, 27). Other hydrogenase-encoding operons are regulated by environmental factors, like oxygen, nitrate in Escherichia coli (32), or nickel in Nostoc species (2).
Very little is known about how T. roseopersicina senses its environment, and nothing is known about transcriptional regulation in this organsim. In previous work, we focused on the hydrogenases of T. roseopersicina. We cloned and characterized two sets of membrane-bound [NiFe] hydrogenase genes, the hynS-isp1-isp2-hynL set (formerly hydS and hydL) (30) and the hupSLCDHIR set (13), and a third, soluble hydrogenase set (hoxEFUYH) (31) together with other components that are necessary for hydrogenase maturation (15, 28). T. roseopersicina provides an attractive model system for comparative studies of the structure-function-stability relationships of different hydrogenase isoenzymes (25). The membrane-associated dimeric hydrogenase HynSL of T. roseopersicina was reported to be stable in the presence of heat and some proteases (24). Also, the organization of the hyn operon is unusual compared to the organization of the hydrogenase-encoding operons (41).
The expression of hydrogenases in many different microorganisms is induced in anaerobic conditions. In many instances, hydrogen is produced only under anoxic conditions (10), and many hydrogenases are inactivated by oxygen (reference 35 and references therein). The availability of O2 is one of the most important regulatory signals in bacteria (38); intricate signal transduction mechanisms for responding to this factor have evolved (40), and oxygen can affect gene expression through various mechanisms (reviewed in reference 4). For example, transcription of the E. coli hya (hydrogenase 1) and hyb (hydrogenase 2) operons, encoding membrane-bound hydrogenases, is controlled by the FNR and ArcA regulators along with the NarL/NarX and NarP/NarQ systems and the AppY regulator (8, 32). In E. coli, FNR and ArcA are the two major global regulators that control gene expression in response to the availability of oxygen. Both FNR and ArcA have been intensively studied, and homologues are found in a wide range of microorganisms (reviewed in reference 38). FnrN has been reported to regulate the Rhizobium leguminosarum hyp genes involved in the maturation of HupSL hydrogenase (17).
In this paper, we report that T. roseopersicina contains an FNR homologue and that this homologue is involved in regulation of the hyn operon in response to anaerobiosis. We also report that this homologue is similar to E. coli FNR, and we discuss the organization of the T. roseopersicina hynS promoter and regulatory region.
MATERIALS AND METHODS
Bacterial strains and growth media.
The strains used are listed in Table 1. T. roseopersicina strains were grown in liquid cultures for 3 to 4 days in Pfennig's mineral medium supplemented with 0.1% NH4Cl (23). Plates were solidified with 7 g of Phytagel (Sigma) per liter and were supplemented with acetate (2 g/liter) when selection for transconjugants was performed, and they were incubated for 2 weeks in anaerobic jars by using the GasPack (BBL) or AnaeroCult (Merck) system. Cultures were illuminated with continuous light at 27 to 30°C (15). In the presence of oxygen the medium was supplemented with 5 g of d-glucose per liter, and the cells were cultivated in the dark under air (23). E. coli strains were grown in Luria-Bertani medium. Antibiotics were used at the following concentrations: for E. coli, 100 μg of ampicillin per ml, 50 μg of kanamycin per ml, 20 μg of gentamicin per ml, and 50 μg of erythromycin per ml; and for T. roseopersicina, 5 μg of gentamicin per ml, 50 μg of erythromycin per ml, and 20 μg of kanamycin per ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Thiocapsa roseopersicina strains | ||
| BBS | Wild type | 7 |
| FNRTM | fnrT::Emr | This study |
| Escherichia coli strains | ||
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]c | Stratagene |
| M182 | E. coli K-12 Δlac | 11 |
| M182fnr | Like M182 but fnr | 6 |
| MC4100 | F−araD139 Δ(argF-lac)U169 ptsF25 deoC1 relA1 flbB350 rpsL150 λ− | 12 |
| RM313 | Like MC4100 but arcA1 zjj::Tn10 | 37 |
| RM315 | Like MC4100 but arcA1 zjj::Tn10, Δfnr | 37 |
| S17-1 (λpir) | 294 (recA pro res mod) Tpr Smr (pRP4-2-Tc::Mu-Km::Tn7), λ pir | 18 |
| Plasmids | ||
| pBluescript SK(+) | Ampr, cloning vector | Stratagene |
| pTSH2/8 | 4,631-bp BamHI fragment of the hyn operon in pBluescribe19+ | 30 |
| pGEM T-Easy | Ampr, cloning vector | Promega |
| pFNR1 | 262-bp fragment of fnrT of T. roseopersicina in pGEM T-Easy | This study |
| pFNR7 | 1,906-bp BamHI fragment of fnrT of T. roseopersicina in pBluescript SK(+) | This study |
| pPR9TT | RK2 vector with the promoterless lacZ gene, Ampr Cmr | 34 |
| pBBRMCS5 | Gmr, broad-host-range vector | 22 |
| pFLAC | Broad-host-range ′lacZ vector, Gmr | This study |
| pHYDPRO1 | 1,214-bp hynS promoter region in pBluescript SK(+) | This study |
| pHYDR1 | BamHI-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR2 | trhydo11-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR3 | trhydo12-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR4 | SphI-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR5 | PstI-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR6 | FspI-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR7 | Eco47III-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR8 | EcoRI-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR9 | hynpo12-HindIII fragment of pHYDPRO1 in pFLAC | This study |
| pHYDR17 | pHYDR9, but FNR site II mutated | This study |
| pHYDR18 | pHYDR9, but FNR site I mutated | This study |
| pK18mobsacB | Kmr, mob+sacB+ | 39 |
| pRL271 | Cloning vector carrying sacB, Emr (ermC) Cmr | GenBank accession no. L05081 |
| pFNRTM1 | SmaI-BamHI fragment of pFNR7 in SmaI-BamHI-digested pK18mobsacB | This study |
| pFNRTM2 | SalI-EcoRI fragment of pRL271 cloned in XhoI site of pFNRTM1 | This study |
| pSR | Ampr, pBR322 derivative for cloning EcoRI-HindIII promoter fragments, with an oop terminator sequence downstream of the HindIII site; also encodes a control RNA I | 20 |
| pHYNP1 | 132-bp hynpo7-hynpo8 PCR fragment in pBluescript SK(+) | This study |
| pHYNP3 | 148-bp EcoRI-HindIII fragment of pHYNP1 in pSR | This study |
| pHYNP5 | pHYNP3 but FNR site II mutated | This study |
| pHYNP6 | pHYNP3 but FNR site I mutated | This study |
Conjugation.
Plasmids were transferred into T. roseopersicina BBS recipient strains by using conjugation conditions described previously (15).
DNA manipulation.
DNA manipulation was performed by using standard techniques (1, 33) or the specifications of the manufacturers.
Construction of plasmids. (i) Reporter fusion constructs.
The plasmids used are listed in Table 1. Plasmid pFLAC was created as follows. A blunted 3,161-bp NotI-KpnI fragment, containing lacZ from pPR9TT, was ligated with the 4,064-bp SspI fragment of pBBRMC5. The promoter region of the hynS gene was amplified from pTSH2/8 with primers T7 (5′-GTAATACGACTCACTATAGGGC-3′) and trhydo10 (5′-AAGCTTAGGCTCTCGCCGAGTGTT-3′), which contained an artificially introduced HindIII site (underlined). The BamHI-digested 1,214-bp product was cloned into the EcoRV-BamHI site of pBluescript SK(+), yielding pHYDPRO1. Fragments of the pHYDPRO1 insert that were different lengths were ligated into the XhoI (polished)-HindIII site of pFLAC, resulting in pHYDR1 and pHYDR4-8 (Table 1 and Fig. 1). pHYDR2 and pHYDR3 were constructed by inserting the SphI-digested trhydo11 (5′-TTCAGGCGATGGAGCAGGAG-3′)-trhydo10 PCR product (293 bp) or trhydo12 (5′-ACCGAGGCGCTCGACATCTT-3′)-trhydo10 PCR product (108 bp) into the ApaI (polished)-SphI site of pHYDR1. pHYDR9 was constructed by inserting the NcoI-PstI-digested hynpo12 (5′-AGCCCATGGAGCGTTCAGGTCTTCCAGAG-3′)-hynpo8 (5′-TCTGCACACCTCGGCCTACT-3′) PCR fragment into the corresponding sites of pHYDR1.
FIG. 1.
β-Galactosidase activities in T. roseopersicina cells (Tr) harboring various hynS upstream region-lacZ constructs. The data are expressed in Miller units (see Materials and Methods), and the values in parentheses are standard deviations. The upstream end of each hynS fragment with respect to the hynS translational initiation site is indicated.
(ii) Plasmids for in vitro transcription and DNA binding.
The 132-bp hynpo7 (5′-AGCGTTCAGGTCTTCCAGAG-3′)-hynpo8 PCR fragment was cloned into the EcoRV site of the pBluescript SK(+)vector, resulting in pHYNP1. pHYNP3 was constructed by inserting the 148-bp EcoRI-HindIII fragment of pHYNP1 into the corresponding site of plasmid pSR. Thus, pHYNP3 carrying the FNR-regulated promoter was cloned upstream of the lambda oop transcription terminator.
(iii) Mutagenesis of the FNR binding half-sites.
We used primers hynpo10 (5′-CCAGAGGACGCGGTTTGGATAATTCGATCTCGCTAACG-3′) and hynpo11 (5′-GAGCTGCGCTGCATAAAATCAGGGCTGCAGC-3′) to mutagenize the template pHYNP3, generating pHYNP5 (FNRI+/FNRII−) and pHYNP6 (FNRI−/FNRI+), respectively. The mutations were recloned into the lacZ reporter plasmid after amplification of the region of interest with primers hynpo12 and hynpo8. The PCR fragment was digested with NcoI and PstI and cloned into the corresponding sites of pHYDR9, yielding pHYDR17 (FNRI+/FNRII−) and pHYDR18 (FNRI−/FNRI+). In all cases where PCR was involved, the sequences were checked.
Identification, cloning, and sequencing of the fnrT gene.
A multiple alignment of the known FNR proteins was constructed, and conserved domains were chosen for designing PCR primers corresponding to the MVCEIPF region (amino acids 120 to 126) and the DIGNYLGL region (amino acids 199 to 206) of the E. coli FNR protein. PCR was carried out with primers FNRo2 (5′-AGICCSAGRTARTTICCGATRTC-3′) and FNRo3 (5′-ATGGTITGYGARATCCCSTT-3′) (where S is C or G, R is A or G, and Y is C or T) and T. roseopersicina genomic DNA. The isolated PCR product of the correct size (262 bp) was cloned into the pGEM T-Easy vector and sequenced. A Southern analysis was performed with digested genomic DNA by using the FNRo2-FNRo3 PCR fragment as a probe. A BamHI partial genomic library was created in pBluescript SK(+), and a clone containing a 1.9-kbp insert, designated pFNR7, was selected after colony hybridization. Plasmid pFNR7 was subcloned, and both strands were sequenced by primer walking.
Mutagenesis of the fnrT gene.
The 1,492-bp SmaI-BamHI region from pFNR7 was cloned into the corresponding sites of pK18mobsacB, resulting in pFNRTM1. After digestion of pFNRTM1 with XhoI and polishing, the truncated SalI-EcoRI fragment (918 bp) of pRL271 (GenBank accession no. L05081) containing the erythromycin resistance gene was inserted (pFNRTM2). The construct was transformed into the E. coli S-17(λpir) strain and then conjugated into T. roseopersicina BBS. Screening for mutant strains was based on erythromycin resistance, and then double-recombinant clones that were resistant to erythromycin and sensitive to kanamycin were selected, resulting in FNRTM.
Enzyme assays.
Hydrogenase uptake activities of membrane fractions were determined by using benzyl viologen (28). The β-galactosidase activity of the toluene-permeabilized cell extracts was assayed as described previously for T. roseopersicina (23, 29). Cells were assayed at the late logarithmic growth stage, as we found no significant difference due to the growth phase. One Miller unit corresponded to 1 μmol of o-nitrophenyl-β-galactoside (Sigma-Aldrich) hydrolyzed per min normalized to the optical density at 600 nm for E. coli and the optical density at 650 nm for T. roseopersicina.
RNA isolation and quantitative reverse transcription-PCR.
RNA was isolated with the TRI reagent (Sigma-Aldrich) by following the manufacturer's recommendations. Prior to reverse transcription, the RNA was treated with DNase I as previously described (15). Reverse transcription was performed by using an iScript cDNA synthesis kit (Bio-Rad). Quantification of cDNA was performed with the iCycler iQ real-time PCR detection system (Bio-Rad) by using IQ SYBR Green Supermix. The following primers were used: for hynS, hydrtR (5′-CGACAGCCAGATGACC-3′) and hydrtF (5′-CCACTGATAAAACACTCGG-3′); and for the crtD gene of T. roseopersicina (23), caroR (5′-CTCCTCGTCGGCGAAGAG-3′) and caroF (5′-CGCATCCCTGACCGACTATC-3′). The amount hynS cDNA was normalized to the level of crtD cDNA.
Preparation of FNR protein.
Overexpression and purification of the FNR protein containing the Ala154 substitution were described by Wing et al. (42).
Gel retardation assays.
Purified EcoRI-HindIII promoter fragments of pHYNP3 were end labeled with [γ-32P]ATP, and 0.5 ng of fragment was incubated with 0 to 3 μM purified Ala154 FNR. The sample buffer mixture (final volume, 10-μl) contained 0.1 M potassium glutamate, 1 mM EDTA, 10 mM potassium phosphate buffer (pH 7.5), 50 μM dithiothreitol, 5% glycerol, 0.5 mg of bovine serum albumin ml−1, and 25 ng of herring sperm DNA (Gibco) μl−1. Following incubation at 37°C for 20 min, samples were electrophoresed in 0.25× Tris-borate-EDTA on a 6% polyacrylamide gel at 12 V cm−1 and were analyzed by using a Bio-Rad Molecular Imager FX and Quantity One software (Bio-Rad).
DNase I footprinting experiments.
DNase I footprinting experiments were performed with a 32P-end-labeled AatII-HindIII fragment of pHYNP3 by using the protocols of Savery et al. (36). Each reaction mixture (20 μl) contained 4 nM (final concentration) template DNA along with 0 to 3 μM purified Ala154 FNR in a solution containing 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM dithiothreitol, 500 μg of bovine serum albumin ml−1, and 25 μg of herring sperm DNA ml−1. All samples were analyzed by denaturing polyacrylamide gel electrophoresis. Gels were calibrated with Maxam-Gilbert G+A sequencing reactions for the labeled fragment, and the results were quantified by using a phosphorimager.
In vitro transcription assays.
Plasmids pHYNP3, pHYNP5, and pHYNP6 were used as templates for in vitro transcription. Plasmid DNA (final concentration, 10 nM) was incubated at 37°C for 20 min with various concentrations of Ala154 FNR (0 to 1 μM). Each 20-μl reaction mixture contained 40 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM KCl, 0.1 mM dithiothreitol, 0.2 μg of bovine serum albumin μl−1, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.05 mM UTP, and 5 μCi of [α-32P]UTP. Following incubation, E. coli RNA polymerase holoenzyme (RNAP) (Epicentre) was added to a final concentration of 50 nM, and the mixture was incubated at 37°C for a further 20 min. The reactions were stopped by addition of 10 μl of formamide buffer (95% [vol/vol] deionized formamide, 20 mM EDTA, 0.05% [wt/vol] bromophenol blue, 0.05% [wt/vol] xylene cyanol FF). Samples were run on 6% denaturing polyacrylamide gels and were analyzed by using a phosphorimager. FNR-dependent transcripts were quantified with reference to the FNR-independent RNA I transcript encoded by the pSR vector.
Permanganate footprint analysis.
Reaction mixtures contained AatII-HindIII promoter fragments that had been labeled at the HindIII site with [γ-32P]ATP, 0 to 0.5 μM purified D154A FNR, and 0 to 50 nM RNAP. The reaction was performed in DNase I footprinting buffer, but herring sperm DNA was omitted. After treatment with potassium permanganate (10 mM [final concentration] for 4 min) to modify single-stranded T residues, the reactions were quenched by addition of 2.5 volumes of stop solution (3 M ammonium acetate, 0.1 mM EDTA, 1.5 M β-mercaptoethanol). Following phenol-chloroform extraction, ethanol precipitation, and treatment with 1 M piperidine (90°C for 30 min), samples were resuspended in formamide buffer. Permanganate cleavage patterns were analyzed by using 6% polyacrylamide sequencing gels and were visualized with a phosphorimager.
Bioinformatics tools.
Protein sequence comparisons with the various databases were done with the BLAST (P, X) programs (www.ncbi.nih.nlm.gov). Multiple alignments were constructed with the CLUSTAL X program.
Nucleotide sequence accession number.
The 1,906-bp sequence of pFNR7 has been deposited in the GenBank database under accession number: AY629341.
RESULTS
Regulation of hynS expression by anaerobiosis: mapping of cis-acting elements.
In order to study the regulation of expression of the T. roseopersicina hyn operon, a 1,220-bp DNA fragment containing the first 45-bp hynS coding sequence, together with upstream sequences, was cloned into the broad-host-range lac expression vector pFLAC to create an in-frame hynS::lacZ gene fusion. The resulting recombinant plasmid, pHYDR1, was introduced into T. roseopersicina BBS, and β-galactosidase activities were measured during growth in different conditions. The measurements revealed significantly lower expression when cells were propagated aerobically in the dark (25 ± 3.4 Miller units) than under anaerobic conditions (46 ± 13 Miller units). Similar β-galactosidase activities were observed when we used a construct recombined with the bacterial chromosome (17.9 ± 2.5 Miller units under aerobic conditions and 43.2 ± 9.4 Miller units under anaerobic conditions). Other factors, such as hydrogen or nitrogen-fixing conditions, had no effect on β-galactosidase expression (data not shown).
To map cis-acting elements involved in the regulation of hynS::lacZ fusion expression, a series of pHYDR1 derivatives containing nested deletions of upstream sequences were constructed (pHYDR2 to pHYDR9) (Table 1 and Fig. 1). In this series, the upstream end of the hynS fragment varied from position −1171 to −167 with respect to the hynS translational initiation site, while the downstream end was fixed at position 45. β-Galactosidase expression was not significantly changed by deletion of sequences from position −1171 to position −602 (pHYDR9). Further deletion of sequences from positions −602 to −514 (pHYDR5) significantly reduced the anaerobic enhancement of expression, and thus, the 89-bp region defined by these deletions is likely to contain a cis regulatory sequence involved in anaerobic activation in T. roseopersicina. Inspection of this region revealed the presence of two sequence elements that resemble the DNA binding site for E. coli FNR protein (Fig. 2). These two elements are centered at position −529 (FNR I) and position −568 (FNR site II) relative to the start codon of hynS (see below).
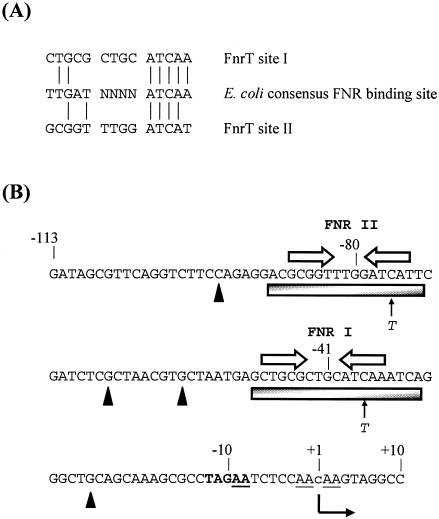
FIG. 2.
hyn regulatory region and FNR binding sites. (A) Sequences of T. roseopersicina FNR site I and FNR site II identified in this work aligned with the known consensus E. coli FNR binding site sequence. Vertical lines indicate residue identity. (B) Sequence of the FNR-regulated hyn promoter region from position −113 to position 10 relative to the putative transcription start site (position 1). The putative transcription start site is located 492 bp upstream of the hynS start codon. The initiation site, detected in an in vitro reaction, is indicated by a lowercase letter, and the direction of transcription is indicated by a bent arrow. Open arrows indicate the locations of DNA sites for FNR binding (FNR I and FNR II), while the shaded boxes show the extent of protection afforded by FNR in DNase I footprint experiments. The solid triangles indicate hypersensitive sites. The cleavage sites produced by potassium permanganate footprint analysis are underlined. The positions of single-base-pair substitutions are indicated. The −10 promoter element is indicated by boldface type. Selected positions are indicated.
Identification and knockout of a T. roseopersicina fnrT gene.
The presence of possible DNA sites for FNR in the segment of the hynS regulatory region involved in anaerobic control prompted us to investigate if the genome of T. roseopersicina contains an fnr homologue. Thus, degenerate primers were designed on the basis of the conserved regions of the known FNR proteins to amplify the FNR-like sequences from the genome of T. roseopersicina (see Materials and Methods). This generated a 262-bp fragment that was cloned, and sequence analysis confirmed that it was likely part of a gene related to the fnr gene family. This fragment was then used to isolate a 1.9-kbp fragment carrying the full-length fnr gene (designated fnrT to indicate its origin) and flanking sequences from a T. roseopersicina genomic library. Sequence analysis of this fragment showed that fnrT encodes a 254-amino-acid protein with 69% similarity and 46% identity to the E. coli FNR protein (GenBank accession number AY629341). The sequence comparison showed that the amino acid sequences of the DNA binding helix-turn-helix motif are very similar in the two proteins, strongly suggesting that they recognize similar binding motifs. The FnrT protein contains four cysteine residues, Cys-16, -19, -25, and -118, that correspond to Cys-20, -23, -29, and -122 of E. coli FNR. Note that in E. coli FNR these cysteine residues contribute to the formation of an iron-sulfur cluster that is essential for activation of FNR in anaerobic conditions (16).
The cloned fnrT gene was disrupted with an erythromycin cassette to generate plasmid pFNRTM2, which was conjugated into T. roseopersicina BBS. Double-recombinant colonies were isolated to obtain an fnrT mutant, FNRTM. Southern blot analyses were performed with genomic DNA from FNRTM to confirm the inactivation of the chromosomal fnrT gene by a double-crossover event. In the ongoing T. roseopersicina genome project, no open reading frame with the same direction of transcription has been identified downstream of the fnrT gene; therefore, we supposed that the cassette did not cause downstream polarity (data not shown). We observed phenotypic differences between the wild-type and fnrT-deficient strains when they were growing in the presence of different sulfur and carbon sources. For example, the fnrT-deficient cells contained sulfur globules when they were grown in the presence of acetate, which was not observed in wild-type T. roseopersicina cells (Á. T. Kovács et al., unpublished observations). This suggests that FnrT has a global role in T. roseopersicina. These phenotypic changes could be complemented with the cloned fnrT gene. Plasmid pHYDR1 carrying the hynS::lacZ fusion was transformed into the wild-type and FNRTM mutant T. roseopersicina BBS strains, and transformants were grown anaerobically or aerobically and assayed for β-galactosidase activity. The results showed that there was no anaerobic induction of hynS::lacZ expression in the FNRTM mutant (27 ± 5.1 Miller units under anaerobic conditions; 23 ± 3.2 Miller units under aerobic conditions). This confirmed the requirement for FnrT for anaerobic induction of hyn expression. We also assayed HynSL hydrogenase enzyme activity directly and measured hynS mRNA levels using real-time quantitative PCR in the wild-type and FNRTM T. roseopersicina strains. The level of hynS mRNA was normalized to the level of crtD that is regulated by a CrtJ/PpsR-like protein in T. roseopersicina (23). As hydrogenase enzymes are inactivated in the presence of oxygen, the activity was assayed only in cells growing under anaerobic conditions. As determined by both assays, hyn operon expression was greatly decreased in the FNRTM mutant strain; the level of hyn operon mRNA decreased to 4.9% ± 0.6% compared to the mRNA level in the wild type, while the HynSL hydrogenase activity decreased to 25% ± 8.8% of the wild-type HynSL activity. The relative messenger levels in cells grown under aerobic conditions were 35.1% ± 1.3% and 4.1% ± 0.3% in the wild-type and FNRTM mutant strains, respectively. The hydrogenase activities of the HoxYH and HupSL proteins were not affected in the T. roseopersicina FNRTM mutant strain (data not shown).
These results suggest that the anaerobic induction of hynS expression is mainly due to the action of FnrT in the hynS regulatory region. A sequence analysis (Fig. 2B) revealed a possible −10 hexamer promoter element, TAGAAT, downstream of FNR site I. If this element was functional, it would place the likely site for transcription initiation at position −492 from the hynS start codon. As we could not identify the genuine start site using primer extension or 5′ random amplification of cDNA ends, we mapped the end of the transcript using reverse transcription-PCR and showed that it was located between positions 50 and −40 relative to the proposed putative site (data not shown). In this case, the two DNA sites for FnrT were centered at positions −41.5 and −80.5 relative to the putative transcription start site. Recall that E. coli FNR protein binds as a dimer to sequences that resemble TTGAT-N4-ATCAA (6), and note that the downstream half-site was more conserved than the upstream half of the FNR binding site (Fig. 2A).
In order to confirm the importance of the two putative DNA sites for FNR identified after our nested deletion analysis (Fig. 1), point mutations were introduced into pHYDR9 to inactivate either FNR site I (pHYDR18) or FNR site II (pHYDR17) (Fig. 2A). To do this, we replaced the downstream FNR half-sites, ATCA(A/T), with ATTA(A/T), so that they were no longer recognized by a native FNR DNA binding helix (5). Table 2 shows that anaerobic induction of the hynS::lacZ fusion was reduced by mutation of either DNA site for FNR, suggesting that FnrT binding at both sites plays a role in activation.
TABLE 2.
β-Galactosidase activities in a wild-type T. roseopersicina strain containing hynS-lacZ fusions with a wild-type or mutated FNR binding site
| Plasmid | β-Galactosidase activity in T. roseopersicina BBS (Miller units)a
|
|
|---|---|---|
| Without O2 | With O2 | |
| pHYDR1 (full length, wild type) | 45.6 ± 13.1 | 25.1 ± 4.5 |
| pHYDR9 (wild type)b | 46.5 ± 8.7 | 27.1 ± 4.3 |
| pHYDR18 (FNR site I mutant) | 28.8 ± 7.9 | 23.4 ± 2.5 |
| pHYDR17 (FNR site II mutant) | 34.8 ± 10.1 | 27.2 ± 6.7 |
The values are means ± standard deviations.
See Fig. 1.
Activation of the hynS promoter in a heterologous E. coli system.
Our results obtained with the T. roseopersicina fnrT gene and hynS cis elements suggest striking similarities with E. coli fnr and regulatory elements of fnr-regulated genes. In particular, the organization of the hynS promoter suggested in Fig. 2B with the principal DNA site for FnrT centered at position −41.5 is identical to the organization of many FNR-dependent promoters in E. coli. Because of the technical problems of studying activation in vitro by using T. roseopersicina FnrT and RNA polymerase (including a lack of purified and active proteins), we investigated whether we could measure hynS promoter function in a heterologous E. coli system. Thus, first, pHYDR1, containing the hynS::lacZ reporter fusion, was introduced into E. coli. Table 3 clearly shows that there was enhanced expression under anaerobic conditions. Furthermore, induction was completely lost in an fnr mutant but was not affected in an arcA mutant.
TABLE 3.
β-Galactosidase activities in wild-type or global anaerobic regulator mutant E. coli strains containing hynS-lacZ fusions
| Strain | Genotype | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|---|
| Without O2 | With O2 | ||
| M182 | Wild type | 41.2 ± 9.1 | 3.4 ± 0.9 |
| M182fnr | fnr | 3.3 ± 1.1 | 2.6 ± 0.6 |
| MC4100 | Wild type | 51.1 ± 1.2 | 4.7 ± 1.2 |
| RM313 | arcA | 55.4 ± 0.8 | 5.3 ± 2.4 |
| RM315 | fnr arcA | 11.5 ± 2.5 | 7.2 ± 1.8 |
The values are means ± standard deviations.
To confirm that anaerobic induction in E. coli is dependent on the same cis elements as those in T. roseopersicina, we used a deletion fusion construct series, pHYDR2 to pHYDR9 (Table 1). These plasmids were introduced into E. coli Δlac strain M182, and the β-galactosidase values in Table 4 confirm that the same upstream region which was involved in anaerobic activation in T. roseopersicina mediated the activation of transcription in E. coli. To confirm the importance of the two putative DNA sites for FNR, assays were carried out with pHYDR18 and pHYDR17, in which FNR site I and FNR site II, respectively, were mutated. Table 4 shows that anaerobic induction of the hynS::lacZ fusion was reduced by mutation of either DNA site for FNR, suggesting that FNR binding to each site plays a role in induction and both sites are needed for full activation. However, a mutation in FNR site I had a more significant effect on transcription than a mutation in FNR site II.
TABLE 4.
β-Galactosidase activities in a wild-type E. coli strain carring a deletion fusion construct series or FNR binding site mutations
| Plasmid | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|
| Without O2 | With O2 | |
| pHYDR1 | 41.2 ± 9.1 | 3.4 ± 0.9 |
| pHYDR2 | 32.9 ± 8.4 | 2.5 ± 0.3 |
| pHYDR3 | 40.1 ± 7.9 | 2.9 ± 0.8 |
| pHYDR4 | 38.5 ± 4.6 | 4.1 ± 0.6 |
| pHYDR9 | 45.3 ± 9.1 | 4.5 ± 0.4 |
| pHYDR5 | 1.9 ± 0.9 | 1.5 ± 0.2 |
| pHYDR6 | 0.9 ± 0.5 | 0.9 ± 0.4 |
| pHYDR7 | 1.1 ± 0.5 | 1.1 ± 0.3 |
| pHYDR8 | 0.5 ± 0.4 | 0.6 ± 0.4 |
| pHYDR18 (FNR site I mutant) | 12.2 ± 4.4 | 2.5 ± 0.9 |
| pHYDR17 (FNR site II mutant) | 26.1 ± 4.7 | 2.1 ± 0.3 |
The values are means ± standard deviations.
In vitro studies of E. coli FNR and RNA polymerase binding to the T. roseopersicina hyn regulatory region.
Taken together, the results described above strongly suggest that activation of the T. roseopersicina hynS promoter by E. coli FNR in E. coli is very similar to activation by T. roseopersicina FnrT in T. roseopersicina. We therefore used previously developed in vitro systems to study the T. roseopersicina hynS promoter using purified E. coli RNAP and purified FNR. To facilitate in vitro studies with the FNR protein under aerobic conditions, we used Ala154 FNR, which carried the D154A substitution that stabilized active FNR in the presence of O2 (19, 26).
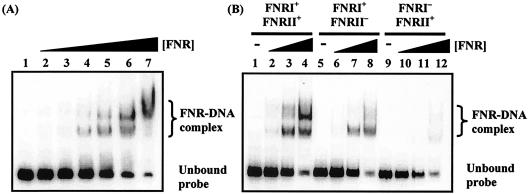
First, electrophoretic mobility shift assays were used to investigate FNR binding to the hyn regulatory region. The EcoRI-HindIII fragment of pHYNP3 carried hyn regulatory region sequences from position −113 to position 24 relative to the putative transcriptional initiation site of hynS (Fig. 2B). Different concentrations of purified Ala154 FNR protein were incubated with 32P-end-labeled hyn regulatory region DNA and separated by polyacrylamide gel electrophoresis (Fig. 3A). Two retarded FNR-DNA complexes were detected, which was consistent with the binding of FNR to two sites. In line with this, Fig. 3B shows that mutations in FNR site II greatly reduced the amount of the less mobile FNR-DNA complex. Interestingly, a mutation in FNR site I almost totally abolished the bandshifts, suggesting that FNR was unable to bind to FNR site II in the absence of prior binding to FNR site I.
FIG. 3.
Binding of E. coli FNR to the hyn regulatory region. (A) End-labeled hyn regulatory region fragment (carrying the sequence from position −113 to position 10) incubated with different concentrations of purified Ala154 FNR. The concentrations of FNR in the reaction mixtures were as follows: lane 1, no protein; lanes 2 to 7, 0.125, 0.25, 0.5, 1, 2, and 3 μM, respectively. (B) End-labeled hyn regulatory region fragments from pHYNP3 (FNRI+/FNRII+), pHYNP5 (FNRI+/FNRII−), and pHYNP6 (FNRI−/FNRII+) incubated with different concentrations of purified FNR. DNA was incubated with no protein (lanes 1, 5, and 9), 0.3 μM FNR (lanes 2, 6, and 10), 1 μM FNR (lanes 3, 7, and 11), and 3 μM FNR (lanes 4, 8, and 12).
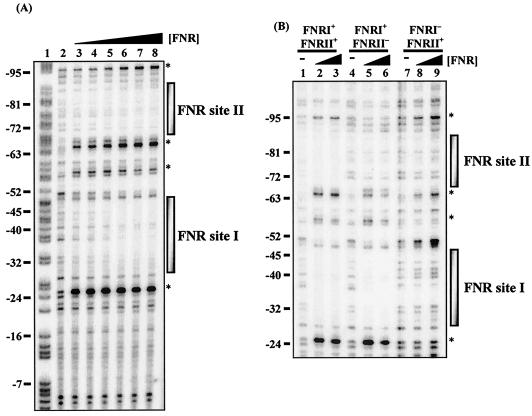
The binding of FNR to the hyn regulatory region was also investigated by using DNase I footprinting. Data in Fig. 4A are consistent with FNR binding to two DNA sites centered at positions −41.5 and −80.5 with respect to the proposed transcription start site (position 1). Further experiments (Fig. 4B) showed that mutations that inactivated FNR site II prevented binding of FNR to site II but did not affect binding to site I. In contrast, mutations that inactivated FNR site I prevented binding of FNR to both site I and site II.
FIG. 4.
DNase I footprint analysis with E. coli FNR for the hyn regulatory region. (A) End-labeled pHYNP3 AatII-HindIII fragment (containing the sequence from position −113 to position 10) incubated with different concentrations of the E. coli Ala154 FNR protein and subjected to DNase I footprinting. The concentrations of FNR in the reaction mixtures were as follows: lane 2, no protein; lanes 3 to 8, 0.125, 0.25, 0.5, 1, 2, and 3 μM, respectively. (B) DNase I footprint analysis for end-labeled pHYNP3 (FNRI+/FNRII+), pHYNP5 (FNRI+/FNRII−), and pHYNP6 (FNRI−/FNRII+) AatII-HindIII fragments. DNA was incubated with no protein (lanes 1, 4, and 7), 1 μM FNR (lanes 2, 5, and 8), and 3 μM FNR (lanes 3, 6, and 9). The gel was calibrated with a Maxam-Gilbert G+A sequencing reaction for the labeled fragment (lane 1), and selected positions are indicated on the left. The shaded boxes indicate the extent of protection afforded by Ala154 FNR binding. The asterisks indicate hypersensitive sites.
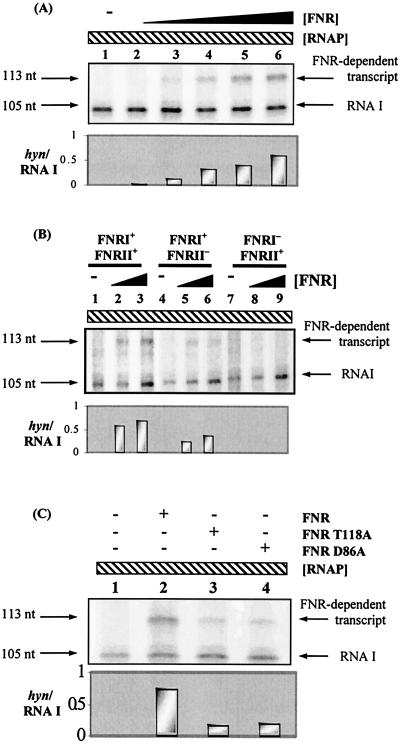
Figure 5 shows that E. coli Ala154 FNR could activate transcription initiation by RNAP at the hynS promoter. In this experiment the hynS promoter fragment was cloned upstream of a ρ-independent transcription terminator in plasmid vector pSR. In the absence of FNR, the 105-nucleotide control RNA I transcript was observed. In the presence of FNR, an additional 113-nucleotide transcript appeared (Fig. 5A). The size of this transcript indicated that FNR-dependent transcripts initiated at the predicted location (i.e., position 1 in Fig. 2B, which corresponded to position −492 upstream of the hynS start codon). Figure 5B shows that mutations in FNR site I abolished the FNR-dependent transcription activation, while mutations in FNR site II reduced activation by 40 to 50%. The experimental data shown in Fig. 5C indicate that activation at the hynS promoter by Ala154 FNR, carrying a second alanine substitution at either T118 (which inactivated activating region 1) or D86 (which inactivated activating region 3), was significantly reduced.
FIG. 5.
E. coli FNR-dependent transcription from the hyn regulatory region. (A) pHYNP3 construct used as the template for in vitro transcription. Ten nanomolar template plasmid DNA was incubated with 50 nM RNAP, nucleoside triphosphates, and [α-32P]UTP along with different concentrations of FNR (0 to 1 μM). Transcripts were analyzed on a denaturing polyacrylamide gel. The positions of the control RNA I transcript (105 nucleotides [nt]) and the FNR-dependent transcript (113 nucleotides) are indicated. Lanes were loaded as follows: lane 1, no FNR protein, lanes 2 to 6, 0.0625, 0.125, 0.25, 0.5, and 1 μM FNR, respectively. The size of the FNR-dependent transcript was determined by using a G+A ladder (data not shown). (B) In vitro transcription analysis of the pHYNP3 (FNRI+/FNRII+), pHYNP5 (FNRI+/FNRII−), and pHYNP6 (FNRI−/FNRII+) vectors. Lanes were loaded as follows: lanes 1, 4, and 7, no FNR protein; lanes 2, 5, and 8, 0.3 μM FNR; lanes 3, 6, and 9, 1 μM FNR. (C) In vitro transcription analysis with AR1 (T118A) or AR3 (D86A) mutant FNR proteins. The RNAP concentration was 50 nM in each reaction. Lane 1 contained no FNR protein, while lanes 2 to 4 contained 1 μM wild-type, T118A, and D86A FNR proteins, respectively. The histograms show the level of hyn transcript from each transcription in relation to the RNA I transcript.
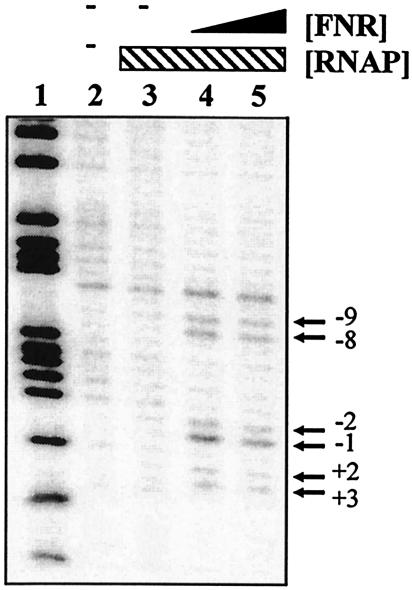
Finally, we checked for promoter opening by using potassium permanganate footprinting. Figure 6 shows that Ala154 FNR directed open-complex formation by RNAP between nucleotides −9 and 3 relative to the transcription start site, a region that overlapped the proposed hynS −10 promoter element, TAGAAT.
FIG. 6.
Potassium permanganate footprint analysis of complexes formed in the hyn promoter region: cleavage products produced when the end-labeled pHYNP3 AatII-HindIII fragment was incubated with purified RNAP and FNR and then subjected to potassium permanganate footprint analysis. Lane 2 contained no protein. Lanes 3 to 5 contained 50 nM RNAP polymerase and the following concentrations of FNR: lane 3, no FNR; lane 4, 250 nM; lane 5, 500 nM. The gel was calibrated with a Maxam-Gilbert G+A sequencing reaction for the labeled fragment (lane 1). The locations of the permanganate-induced cleavage sites are indicated on the right.
DISCUSSION
Many hydrogenases are regulated by oxygen and are optimally expressed only in anoxic conditions (10). However, in most cases the regulatory cascade transducing the signal from the environment to the initiation complex is not known. Transcriptional regulation of the T. roseopersicina hyn operon was studied by using in-frame lacZ fusion constructs, and it was found that the expression was affected by oxygen. An upstream activating sequence, located 602 to 514 bp from the start codon of the translational start site of hynS, was found to be essential for the elevated activity observed under anaerobic conditions. This region contained two DNA sequences that resemble consensus DNA sites for FNR, a transcription factor known to be responsible for anaerobic induction of many genes in many bacteria. For both sequences, the 3′ part of the palindromic binding site is more similar to the E. coli FNR consensus binding site than the 5′ part is. The presence of these sequence elements prompted us to search for an FNR homologue in T. roseopersicina BBS, and this resulted in isolation of the fnrT gene. This gene encodes an FNR homologue that is most similar to the ANR protein of Pseudomonas aeruginosa (73% similarity and 52% identity) (43). Alignment of FnrT with other sequences revealed striking similarity to E. coli FNR, notably in the helix-turn-helix sequence responsible for DNA binding. The presence of FNR in a member of the Chromatiaceae is fascinating since members of this family grow mainly under anaerobic conditions. T. roseopersicina is one of the few Chromatiaceae strains that are able to grow under aerobic conditions (21), which may explain its need for an oxygen-sensing system. However, until now, nothing was known about this, and the only other reported transcription factor was PpsR, which is responsible for aerobic repression of genes involved in carotenoid biosynthesis (23).
The role of FnrT in the anaerobic regulation of hyn expression was tested by creating a T. roseopersicina fnrT mutant. FnrT was essential for elevated hyn promoter activity under oxygen-free conditions. However, by using the hynS::lacZ fusion reporter system relatively high basic expression was measured in the presence of air or in the fnrT mutant strain. The simplest way to explain our data is to suppose that the hynS regulatory region contains an FnrT-dependent promoter which requires FnrT binding to the two putative DNA sites for FNR plus at least one other FnrT-independent promoter that drives expression in aerobic conditions. The discrepancy between the effect of the fnrT knockout on expression of the hynS::lacZ fusion and hynS messenger levels may be due to differences in the translational efficiencies of the two transcripts.
Examination of the base sequence of the hynS regulatory region suggested that the FnrT-dependent promoter may be organized like many FNR-dependent promoters of E. coli. This, together with the extensive similarities between the T. roseopersicina FnrT and E. coli FNR proteins, prompted us to examine the activity of the hynS::lac fusion in E. coli. We found that E. coli FNR is fully able to activate the hynS promoter in vivo and that activation requires both of the proposed DNA sites for FNR. Strikingly, in E. coli, expression is more dependent on FNR and better coupled to anaerobiosis. The simplest explanation for this is that the FnrT-independent promoter(s) responsible for aerobic promoter activity in T. roseopersicina is not functional in E. coli.
Our observation that the T. roseopersicina hynS promoter could be served by E. coli FNR and RNAP in vivo led us to perform a series of in vitro studies with a view to understanding its organization better. We are aware of the shortcomings of this approach as we still lack rigorous proof that the same promoter elements are used in both T. roseopersicina and E. coli. In vitro transcription experiments confirmed the transcription start point located 41.5 bp downstream of FNR site I and 80.5 bp downstream of FNR site II and identified a likely −10 hexamer element. These experiments showed that while FNR binding at upstream site II contributes to activation, FNR binding at site I is essential. This is consistent with our observation that activation requires both activating region 1 and activating region 3 of FNR. Recall that activating region 3 is functional only at promoters where the FNR binds to a site that overlaps the promoter −35 element (so-called class II promoters), while activating region 1 is functional in activation irrespective of the FNR binding location (9). Most FNR-dependent promoters have a single DNA site for FNR centered near position −41.5. However, many such promoters have a second DNA site for FNR located further upstream, and a recent systematic study by Barnard et al. (3) showed that upstream bound FNR at certain positions (positions −71.5, −81.5, −91.5, and −101.5) could act as an enhancer of transcription initiation, while at other locations (positions −85.5 and −95.5) it functioned as a repressor. Our results are consistent with these findings; thus, in the hyn regulatory region, the center-to-center distance between the two FNR sites is 39 bp, suggesting that the upstream bound FNR has a synergistic role. Interestingly, binding to the upstream DNA site for FNR depended on the presence of a functional downstream binding site. So far there is no other example where binding of one FNR enhances the binding of a second FNR to a lower-affinity site.
ADDENDUM
Following the suggestion of one of our reviewers, we searched for an open reading frame between the hynS start codon and the putative FNR-dependent transcription initiation site. We identified an open reading frame which starts 14 bp downstream of the putative initiation site and terminates 18 bp upstream of the start codon of the hynS gene. Based on a homology search alone, it is not possible to implicate the putative product of this open reading frame in hydrogenase enzyme formation.
Acknowledgments
This work was supported by EU 5th Framework Programme projects QLK5-1999-01267, QLK3-2000-01528, QLK3-2001-01676, and ICA1-CT-2000-70026, by OM KFHÁT grants OMFB-00525/02, OMFB-00242/02, OMFB-00768/03, and NKFP OM-00072/01 to K.L.K., and by Wellcome Trust grant WT066685 to S.J.W.B and D.F.B, as well as by an FEBS short-term fellowship to A.T.K.
We thank Gary Sawers (John Innes Centre, Norwich, United Kingdom) for E. coli RM313 and RM315 and Svein Valla (Norwegian University of Science and Technology, Trondheim, Norway) for PR9TT. We thank Anne Barnard for many helpful discussions. We also thank Bio-Rad Hungary for the opportunity to use iCycler real-time PCR. We gratefully acknowledge Rózsa Verebély for excellent technical assistance.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology. Wiley, New York, N.Y.
- 2.Axelsson, R., and P. Lindblad. 2002. Transcriptional regulation of Nostoc hydrogenases: effects of oxygen, hydrogen, and nickel. Appl. Environ. Microbiol. 68:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, A. M., J. Green, and S. J. Busby. 2003. Transcription regulation by tandem-bound FNR at Escherichia coli promoters. J. Bacteriol. 185:5993-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, C. E., S. Elsen, and T. H. Bird. 1998. Mechanisms for the redox control of gene expression. Annu. Rev. Microbiol. 53:495-523. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. I., K. L. Gaston, J. A. Cole, and S. J. Busby. 1989. Cloning of binding sequences for the Escherichia coli transcription activators, FNR and CRP: location of bases involved in discrimination between FNR and CRP. Nucleic Acids Res. 17:3865-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, A., and S. Busby. 1994. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol. Microbiol. 11:383-390. [DOI] [PubMed] [Google Scholar]
- 7.Bogorov, L. V. 1974. About the properties of Thiocapsa roseopersicina BBS, isolated from estuaria of White Sea. Mikrobiologija 43:326-332. [PubMed] [Google Scholar]
- 8.Brondsted, L., and T. Atlung. 1994. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J. Bacteriol. 176:5423-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning, D., D. Lee, J. Green, and S. J. W. Busby. 2002. Secrets of bacterial transcription initiation taught by the Escherichia coli FNR protein, p. 127-142. In D. Hodgson and C. Thomas (ed.), Signals, switches, regulons, and cascades. Cambridge University Press, Cambridge, United Kingdom.
- 10.Cammack, R., M. Frey, and R. Robson (ed.) 2001. Hydrogen as a fuel: learning from nature. Taylor & Francis, London, United Kingdom.
- 11.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casadaban, M., and S. Cohen. 1980. Analysis of gene control signal by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 13.Colbeau, A., K. L. Kovács, J. Chabert, and P. M. Vignais. 1994. Cloning and sequencing of the structural (hupSLC) and accessory (hupDHI) genes for hydrogenase biosynthesis in Thiocapsa roseopersicina. Gene 140:25-31. [DOI] [PubMed] [Google Scholar]
- 14.Dischert, W., P. M. Vignais, and A. Colbeau. 1999. The synthesis of Rhodobacter capsulatus HupSL hydrogenase is regulated by the two-component HupT/HupR system. Mol. Microbiol. 34:995-1006. [DOI] [PubMed] [Google Scholar]
- 15.Fodor, B., G. Rákhely, Á. T. Kovács, and K. L. Kovács. 2001. Transposon mutagenesis in purple sulfur photosynthetic bacteria: identification of hypF, encoding a protein capable of processing [NiFe] hydrogenases in α, β, and γ subdivisions of the proteobacteria. Appl. Environ. Microbiol. 67:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, J., B. Bennett, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez, D., Y. Hernando, J.-M. Palacios, J. Imperial, and T. Ruiz-Argueso. 1997. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum biovar viciae UPM791. J. Bacteriol. 179:5264-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero, M., V. Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing nonantibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiley, P. J., and W. S. Reznikoff. 1991. Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 173:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondrateva, E. N., I. P. Petushkova, and V. G. Zhukov. 1975. Growth and oxidation of sulfur compounds by Thiocapsa rosoepersicina in darkness. Mikrobiologija 44:389-394. [PubMed] [Google Scholar]
- 22.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 23.Kovács, Á. T., G. Rákhely, and K. L. Kovács. 2003. Genes involved in the biosynthesis of photosynthetic pigments in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 69:3093-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovács, K. L., C. Bagyinka, and G. Tigyi. 1988. Proteolytic resistance and its utilization in purification of hydrogenase from Thiocapsa roseopersicina. Biochim. Biophys. Acta 935:166-172. [Google Scholar]
- 25.Kovács, K. L., B. Fodor, Á. T. Kovács, G. Csanádi, G. Maróti, J. Balogh, S. Arvani, and G. Rákhely. 2002. Hydrogenases, accessory genes and the regulation of [NiFe] hydrogenase biosynthesis in Thiocapsa roseopersicina. Int. J. Hydrogen Energy 27:1463-1469. [Google Scholar]
- 26.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 27.Lenz, O., M. Bernhard, T. Buhrke, E. Schwartz, and B. Friedrich. 2002. The hydrogen-sensing apparatus in Ralstonia eutropha. J. Mol. Microbiol. Biotechnol. 4:255-262. [PubMed] [Google Scholar]
- 28.Maróti, G., B. D. Fodor, G. Rákhely, Á. T. Kovács, S. Arvani, and K. L. Kovács. 2003. Selectivity and cooperativity of accessory proteins in the biosynthesis of [NiFe] hydrogenases. Eur. J. Biochem. 270:2218-2227. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Rákhely, G., A. Colbeau, J. Garin, P. M. Vignais, and K. L. Kovács. 1998. Unusual organization of the genes coding for HydSL, the stable [NiFe] hydrogenase in the photosynthetic bacterium Thiocapsa roseopersicina BBS. J. Bacteriol. 180:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rákhely, G., Á. T. Kovács, G. Maróti, B. D. Fodor, G. Csanádi, D. Latinovics, and K. L. Kovács. 2004. Cyanobacterial-type, heteropentameric, NAD+-reducing NiFe hydrogenase in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 70:722-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard, D. J., G. Sawers, F. Sargent, L. McWalter, and D. H. Boxer. 1999. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology 145:2903-2912. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Santos, P. M., L. Di Bartolo, J. M. Blatny, E. Zennaro, and S. Valla. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 35.Sasikala, K., C. V. Ramana, P. R. Rao, and K. L. Kovács. 1993. Anoxygenic phototrophic bacteria: physiology and advances in hydrogen production technology. Adv. Appl. Microbiol. 38:211-295. [Google Scholar]
- 36.Savery, N., T. Belyaeva, and S. Busby. 1996. Protein DNA interactions: introduction and protein DNA interactions: DNase I footprinting, hydroxyl radical footprinting, potassium permanganate footprinting, preparation of G ladder by DMS treatment and piperidine cleavage, p. 21-33. In K. Docherty (ed.), Essential techniques: gene transcription. BIOS Scientific Publishers, Oxford, United Kingdom.
- 37.Sawers, G., and B. Suppmann. 1992. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J. Bacteriol. 174:3474-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawers, G. 1999. The aerobic/anaerobic interface. Curr. Opin. Microbiol. 2:181-187. [DOI] [PubMed] [Google Scholar]
- 39.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 40.Unden, G., S. Becker, J. Bongaerts, G. Hollighaus, J. Schirawsky, and S. Six. 1995. O2-sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch. Microbiol. 164:81-90. [PubMed] [Google Scholar]
- 41.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 42.Wing, H. J., J. Green, J. R. Guest, and S. J. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 272:29061-29065. [DOI] [PubMed] [Google Scholar]
- 43.Zimmermann, A., C. Reimmann, M. Galimand, and D. Haas. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5:1483-1490. [DOI] [PubMed] [Google Scholar]