Graphical abstract
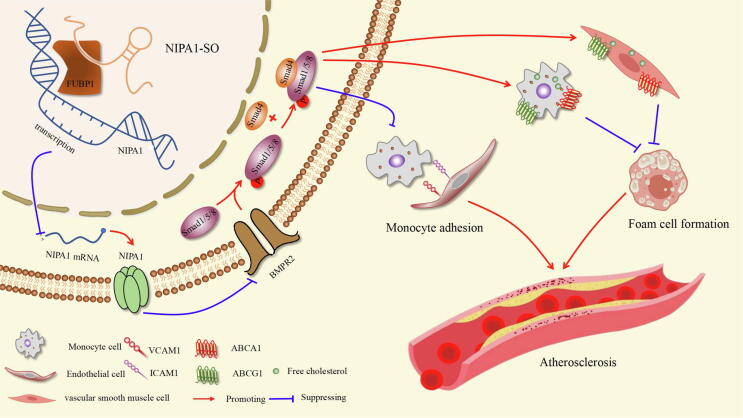
Schematic diagram of the signalling pathways regulated by NIPA1-SO. Our study has uncovered an athero-protective role of the lncRNA NIPA1-SO, which, by interacting with a single transcription factor, is capable of inhibiting monocyte adhesion and foam cell formation, two fundamental processes in atherosclerosis. The transcription factor FUBP1 negatively regulates NIPA1 expression; this inhibitory effect is increased by the interaction of NIPA1-SO with FUBP1, resulting in lower transcription of NIPA1. A reduction in NIPA1 protein results in increased BMPR2 protein due to a reduction in NIPA1-mediated BMPR2 endocytosis and degradation, leading to higher levels of Smad1/5/8 phosphorylation (pSmad1/5/8), which, through complexation with Smad4, inhibit transcription of the adhesion molecules VCAM1 and ICAM1, reducing monocyte adhesion to endothelial cells. Further, the pSmad1/5/8:Smad4 complex promotes ABCA1 and ABCG1 transcription, both of which promote cholesterol efflux via high-density lipoprotein (HDL) particles, thereby inhibiting foam cell formation.
Keywords: Atherosclerosis, LncRNA, NIPA1, Vascular adhesion molecule
Highlights
-
•
Human atherosclerotic plaques have reduced expression of the long non-coding RNA NIPA1-SO and increased levels of NIPA1.
-
•
NIPA1-SO inhibits monocyte adhesion to endothelial cells and decreases cholesterol accumulation.
-
•
Increasing the expression of NIPAI-SO or knockout of NIPA1 reduces atherosclerosis in an animal model.
Abstract
Long non-coding RNAs (lncRNAs) are emerging as important players in gene regulation and cardiovascular diseases. However, the roles of lncRNAs in atherosclerosis are poorly understood. In the present study, we found that the levels of NIPA1-SO were decreased while those of NIPA1 were increased in human atherosclerotic plaques. Furthermore, NIPA1-SO negatively regulated NIPA1 expression in human umbilical vein endothelial cells (HUVECs). Mechanistically, NIPA1-SO interacted with the transcription factor FUBP1 and the NIPA1 gene. The effect of NIPA1-SO on NIPA1 protein levels was reversed by the knockdown of FUBP1. NIPA1-SO overexpression increased, whilst NIPA1-SO knockdown decreased BMPR2 levels; these effects were enhanced by the knockdown of NIPA1. The overexpression of NIPA1-SO reduced while NIPA1-SO knockdown increased monocyte adhesion to HUVECs; these effects were diminished by the knockdown of BMPR2. The lentivirus-mediated-overexpression of NIPA1-SO or gene-targeted knockout of NIPA1 in low-density lipoprotein receptor-deficient mice reduced monocyte-endothelium adhesion and atherosclerotic lesion formation. Collectively, these findings revealed a novel anti-atherosclerotic role for the lncRNA NIPA1-SO and highlighted its inhibitory effects on vascular inflammation and intracellular cholesterol accumulation by binding to FUBP1 and consequently repressing NIPA1 expression.
Introduction
Long non-coding RNAs (lncRNAs), defined as gene transcripts of ≥200 nucleotides in length that are not translated into proteins, can modulate the expression of protein-coding genes and consequently affect cell behavior [1]. Several lncRNAs have been implicated in the pathogenesis of atherosclerosis [2]. The biological functions of many lncRNAs, however, are still unknown.
The non-imprinted in Prader-Willi/Angelman Syndrome region protein 1 (NIPA1) gene is highly expressed in the brain, and mutations in this gene have been implicated in the development of an autosomal dominant form of hereditary spastic paraplegia (HSP) [3], [4], [5]. Transmembrane protein NIPA1 can function as a Mg2+ transporter and control Mg2+ uptake; this is the basis for HSP [6]. The deficiency of four highly-conserved and non-imprinted genes, namely NIPA1, NIPA2, CYFIP1, TUBGCP5, was previously associated with congenital heart disease [7]. Recently, emerging evidence showed a significant upregulation of NIPA1 gene expression in pulmonary arterial smooth muscle cells (PASMCs) during chronic hypoxia and monocrotaline-induced pulmonary hypertension, a life-threatening pulmonary vascular disease in humans [8]. However, whether and how NIPA1 plays a role in atherosclerosis remains to be elucidated.
Here, we report that the lncRNA NIPA1-SO, whose function was previously unknown, modulated the expression of the transmembrane magnesium transporter protein NIPA1, and that NIPA1-SO affected monocyte adhesion to vascular endothelial cells and intracellular cholesterol level in a BMPR2-dependent manner. Furthermore, using a well-established mouse model of atherosclerosis, we demonstrated that augmenting NIPA1-SO expression or abolishing NIPA1 could inhibit atherogenesis.
Results
Human atherosclerotic plaques exhibit reduced NIPA1-SO and increased NIPA1 expression
We previously performed differential expression analysis of human atherosclerotic tissues in comparison with normal arterial specimens using Arraystar LncRNA Expression Microarray V3.0, which contained probes for 24,748 lncRNAs and 24,420 protein-coding transcripts. This analysis detected a number of differentially expressed lncRNAs (Appendix Fig. S1A and data file S1) and mRNAs (Appendix Fig. S2A and data file S2), some of which have been described recently [9], [10], [11]. One of the most significantly downregulated lncRNA in atherosclerotic plaques was ENSG00000274253 (LOC283683, referred to here as NIPA1-SO standing for NIPA1 sense overlapping); the expression level of this gene was 43.29 ± 0.06-fold lower in human atherosclerotic plaques than in normal intimal tissues (P < 0.001) (Appendix Fig. S1A and B, and data file S1). The gene encoding NIPA1-SO was located at nucleotide position 23,094,330 to 23,115,254 on chromosome 15 (GRCh37/hg19) and overlapped with the NIPA1 gene which was located at position 23,043,278–23,086,843. Interestingly, the level of NIPA1 expression was significantly higher in atherosclerotic plaques than in normal intimal tissues (a 13.67 ± 0.08-fold difference, P < 0.001) (Appendix Fig. S2A-C, and data file S2). Subsequent analyses using quantitative reverse transcription polymerase chain reaction (RT-PCR), fluorescence in situ hybridization, Western blotting, and immunohistochemistry, confirmed that NIPA1-SO expression was decreased, whilst NIPA1 was increased, in atherosclerotic plaques (Fig. 1A-D).
Fig. 1.
NIPA1-SO and NIPA1 are differentially expressed in human normal and atherosclerotic tissues. (A) Expression of NIPA1-SO and NIPA1 were assessed via RT-PCR in both human normal and atherosclerotic plaque tissues. Data are presented following normalization to either U6 RNA (NIPA1-SO) or GAPDH (NIPA1) and to the normal carotid tissue samples using the ΔΔCt method (n = 5). (B) FISH analysis of human normal and atherosclerotic tissue samples. Green fluorescence shows expression of NIPA1-SO whilst blue fluorescence indicates nuclei stained with the DNA stain DAPI (n = 5). Scale bar = 200 µm. (C) Western blotting of NIPA1 protein levels in human normal and plaque tissues (n = 5). Protein levels were normalized to β-actin and to the levels of expression in normal tissue. (D) Immunohistochemistry of human normal and atherosclerotic carotid artery tissue samples to examine NIPA1 expression (brown). Data shown are normalized to normal intima tissue samples (n = 5). (E) Fluorescence in situ hybridization was employed to determine the intracellular locations of NIPA1-SO in cultured human umbilical vein endothelial cells (HUVECs). NIPA1-SO was stained green with RNA probes, while 18S (a cytoplasmic marker) and U6 (a nuclear marker) were stained red with fluorescent secondary antibody (n = 3). *P < 0.05 by unpaired 2-tailed Student’s t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Quantitative RT-PCR analyses showed that the expression level of NIPA1-SO in human atherosclerotic plaques was approximately 0.05% relative to the expression level of the housekeeping gene GAPDH (Appendix Fig. S3A) and that cultured human umbilical vein endothelial cells (HUVECs), THP-1 cells, and human aortic smooth muscle cells (HASMCs) contained approximately 15, 8, and 7.5 copies of NIPA1-SO per cell, respectively (Appendix Fig. S3B). Our experiments further indicated that NIPA1-SO did not contain a poly-A tail (Appendix Fig. S3C) and that NIPA1-SO was present in both the nucleus and cytoplasm of HUVECs (Fig. 1E).
NIPA1-SO downregulates NIPA1 expression
Double immunostaining studies of atherosclerotic arterial tissues showed that NIPA1 was expressed in endothelial cells (ECs, identified by positive staining for the endothelial cell marker CD34), monocytes/macrophages (MΦs, positive staining for LGALS3), and vascular smooth muscle cells (VSMCs, positive staining for α-smooth muscle actin) (Fig. 2A).
Fig. 2.
NIPA1-SO negatively regulates NIPA1 expression. (A) Immunofluorescence staining of human atherosclerotic arterial tissue samples was used to determine cell-type specific NIPA1 expression. The left-hand images show expression of the cell type-specific markers used to determine cell identity; CD34 for endothelial cells (ECs), LGALS3 for macrophages (MΦs) and α-smooth muscle actin for smooth muscle cells (SMCs). Green fluorescence indicates expression of the indicated markers and thus cell type identity. The central images show NIPA1 staining (red) for the same images as those shown on the left. The merged images on the right demonstrate co-localization of NIPA1 with the indicated cell type markers (example regions of co-localization are indicated using arrows). For all images, nuclei were stained with DAPI (blue). (B and C) Cultured human umbilical vein endothelial cells (HUVECs), THP-1 cells, and human aortic smooth muscle cells (HASMCs) were transduced with lentiviral vectors encoding NIPA1-SO (LV-NIPA1-SO) or a mock control lentivirus (LV-Mock). (B) The level of NIPA1 mRNA expression was quantified using RT-PCR. Data is presented following normalization to the endogenous control gene GAPDH and normalization to the LV-Mock samples (n = 5). (C) The level of NIPA1 protein in response to lentiviral-mediated overexpression of NIPA1-SO in endothelial, monocyte and vascular smooth muscle cells was also assessed using Western blotting, normalizing to β-actin and the LV-mock samples (n = 5). (D) The level of NIPA1 protein was examined by Western blotting in response to NIPA1-SO knockdown via shRNA (n = 5). Data were normalized to β-actin and control shRNA samples. *P < 0.05 by unpaired 2-tailed Student’s t-test, n = 5 per group. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Because lncRNAs can regulate the expression of nearby genes [1], we investigated whether NIPA1-SO modulated NIPA1 expression. We transduced cultured HUVECs, THP-1 cells, and HASMCs with either an shRNA to knockdown NIPA1-SO or a lentivirus to increase NIPA1-SO expression. Cells with lentivirus-mediated overexpression of NIPA1-SO had reduced NIPA1 expression compared to cells transduced with a control lentivirus vector (Fig. 2B and C) while cells with shRNA-mediated knockdown of NIPA1-SO showed increased NIPA1 expression when compared to transduction with a control shRNA (Fig. 2D), thus suggesting a down-regulatory effect of NIPA1-SO on NIPA1 expression. In addition, RNA-sequencing of cultured HUVECs transduced with either a NIPA1-SO expressing lentivirus or the lentivirus vector (control) confirmed that NIPA1-SO could inhibit NIPA1 expression (Appendix Fig. S4A, B, and data file S5).
To investigate whether NIPA1-SO interacted with the NIPA1 gene, we performed chromatin isolation by RNA purification (ChIRP) analysis [12] using NIPA1-SO RNA probes to pull down chromatin in cultured HUVECs. Next, we performed PCR using the pulled-down DNA as template and primers annealing to the NIPA1 gene promoter. Analysis detected NIPA1 in the chromatin DNA pulled down by the NIPA1-SO RNA probes, thus suggesting that NIPA1-SO interacted with the NIPA1 gene (Appendix Fig. S6). In comparison, the negative controls GAPDH and ACTB were barely detectable in the pulldown by the NIPA1-SO RNA probes; NIPA1 was barely detectable in chromatin DNA pulled down by a LacZ probe serving as a negative control (Appendix Fig S6).
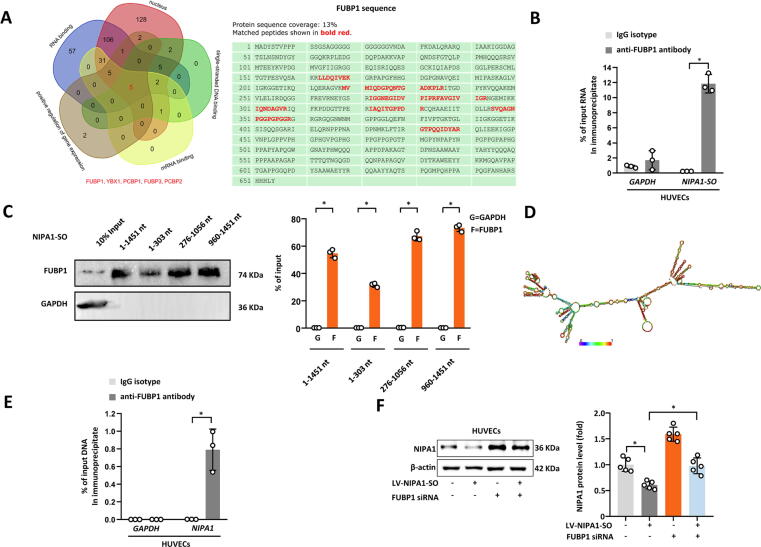
Mass spectrometry analysis of the chromatin pulled down by the NIPA1-SO RNA probes detected the presence of a number of proteins, five of which have reported properties: (1) nuclear location, (2) single-stranded DNA binding, (3) mRNA binding, (4) positive regulation of gene regulation, and (5) RNA binding (Fig. 3A). Of these five proteins, FUBP1 (far upstream element binding protein 1) protein was most abundantly detectable in the NIPA1-SO pulldown (Fig. 3A and Appendix Fig. S5). To verify that NIPA1-SO could interact with FUBP1, an RNA immunoprecipitation (RIP) assay was performed with an anti-FUBP1 antibody, followed by RT-PCR using the precipitated RNA sample as a template and primers complementary to the NIPA1-SO lncRNA sequence. Analysis showed the presence of NIPA1-SO in the RNA sample precipitated by the anti-FUBP1 antibody (Fig. 3B), suggesting that NIPA1-SO interacts with FUBP1. To investigate which parts of NIPA1-SO were likely to be required for the interaction with FUBP1, we performed FUBP1 immunoblotting on different chromatin samples pulled-down using different regions of NIPA1-SO as specific probes. The experiment indicated the involvement of the 5́-region (1–303 nt), the middle (276–1056 nt), and 3́-region (960–1451 nt) of NIPA1-SO (Appendix Fig. S6 and Fig. 3C and D).
Fig. 3.
Negative regulation of NIPA1 expression by NIPA1-SO involves FUBP1. (A) Chromatin of cultured HUVECs were pulled down using NIPA1-SO RNA probes and subjected to protein mass spectrometry analysis. (B) RNA immunoprecipitation was used to determine if the transcription factor FUBP1, identified by mass spectrometry analysis of ChIP samples, interacts with NIPA1-SO (n = 3). An anti-FUBP1 antibody was used to immunoprecipitate cellular RNA in HUVECs, and RT-PCR was used to measure the quantity of immunoprecipitated NIPA1-SO RNA as a percentage of the input RNA. Primers amplifying the GAPDH gene were used as a control for the RT-PCR and an IgG isotype antibody was used as a control for the RNA immunoprecipitation. (C) Biotin-labeled full-length (1–1451 nt) and truncated (1–303 nt, 276–1056 nt, and 960–1451 nt) NIPA1-SO sequences were prepared by the in vitro transcription method. The labeled sequences were incubated with cell lysate at 4 °C for 3 h and then with streptavidin-conjugated magnetic beads to isolate NIPA1-SO-protein complexes, followed by Western blot analysis of FUBP1. n = 3. (D) The tertiary structure of NIPA1-SO was analyzed by RNAfold, which indicates that NIPA1-SO has three functional structure areas: 1–303 nt, 276–1056 nt, and 960–1451 nt, respectively. (E) Chromatin immunoprecipitation using an anti-FUBP1 antibody, followed by PCR analysis of NIPA1, was used to determine if FUBP1 interacts with the NIPA1 gene in HUVECs (n = 3). Data are presented as a percentage of input DNA with IgG isotype immunoprecipitation and GAPDH PCR used as controls for the immunoprecipitation and PCR, respectively. (F) Western blotting was used to determine if NIPA1-SO-mediated down-regulation of NIPA1 is dependent upon FUBP1 using lentiviral overexpression with and without FUBP1 siRNA in HUVECs (n = 5). Data are presented normalized to β-actin and the untreated control (without lentiviral transduction or siRNA transfection). *P < 0.05 by unpaired 2-tailed Student’s t-test or one-way ANOVA with Tukey’s post hoc tests.
Furthermore, we performed chromatin immunoprecipitation (ChIP) using an anti-FUBP1 antibody, followed by PCR amplification of the precipitated chromatin DNA sample with primers complementary to the NIPA1 gene. This assay showed enrichment of NIPA1 in the chromatin precipitated by the anti-FUBP1 antibody (Fig. 3E), suggesting that FUBP1 interacted with NIPA1.
To investigate whether the effect of NIPA1-SO on NIPA1 expression was dependent upon FUBP1, we transfected HUVECs with a lentivirus that expressed NIPA1-SO and a siRNA to knockdown FUBP1. Then, we performed Western blot analysis of NIPA1. This experiment showed that the knockdown of FUBP1 abolished the suppressive effect of NIPA1-SO on NIPA1 expression (Fig. 3F), suggesting that NIPA1-SO influences NIPA1 expression in a FUBP1-dependent manner. Furthermore, cells that were transfected with the FUBP1 siRNA alone exhibited increased NIPA1 protein levels (Fig. 3F), indicating that FUBP1 negatively regulated NIPA1; taken together with our earlier findings, these data implied that the functionality of FUBP1 was enhanced by NIPA1-SO.
NIPA1-SO increases the levels of BMPR2 via NIPA1
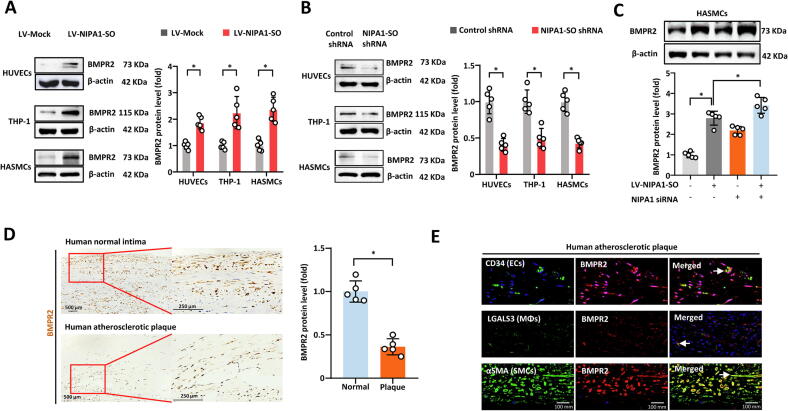
Previous studies have shown that NIPA1 interacted with BMPR2 and thereby promoted endocytosis and lysosomal degradation of BMPR2 [13]. Having found that NIPA1-SO suppressed NIPA1 expression, we next investigated whether NIPA1-SO could influence BMPR2 levels via NIPA1. Our experiments showed that the lentivirus-mediated overexpression of NIPA1-SO in HUVECs, THP-1 cells and HASMCs resulted in an increase in BMPR2 level (Fig. 4A) while the knockdown of NIPA1-SO resulted in a decrease in BMPR2 (Fig. 4B). The effect of NIPA1-SO overexpression on BMPR2 level was augmented by the knockdown of NIPA1 (Fig. 4C), suggesting that NIPA1-SO increased BMPR2 levels by reducing NIPA1.
Fig. 4.
NIPA1-SO increases BMPR2 by reducing NIPA1 expression. (A) HUVECs, THP-1 cells, and HASMCs were transduced with NIPA1-SO-expressing lentiviral vectors or a control lentivirus (LV-Mock) (n = 5). Western blotting was used to assess BMPR2 protein levels in response to transduction. Data shown are normalized to the mock control samples. (B) Knockdown of NIPA1-SO in HUVECs, THP-1 cell, and HASMCs, using shRNA, was also used to assess the role of NIPA1-SO in BMPR2 expression via Western blot analysis (n = 5). Data were normalized to the control shRNA samples following normalization to β-actin. (C) To demonstrate that the regulatory effect of NIPA1-SO on BMPR2 is mediated by NIPA1, NIPA1-targeting siRNA and overexpression of NIPA1-SO was used to assess BMPR2 expression using Western blotting (n = 5). Data for the treatment combinations shown were normalized to β-actin and the control sample (no NIPA1-SO overexpression or NIPA1 knockdown). (D) Protein levels of BMPR2 (brown staining) in atherosclerotic plaque tissues or normal tissue samples was assessed via immunohistochemistry (n = 5). Data shown are normalized to the values obtained from normal intimal tissue samples. (E) Immunofluroescence staining of atherosclerotic plaque tissue was performed to assess the expression of BMPR2 in different cell types. The left-most images show immunofluorescence staining with cell-type specific markers for endothelial cells (EC; CD34), monocytes (MΦ; LGALS3L) and vascular smooth muscle cells (VSMC; α-smooth muscle actin) with green fluorescence. The center images show corresponding BMPR2 immunofluorescence staining (red) in the three cell types. The images on the right show merged images, demonstrating regions of co-localization between the cell type markers and BMPR2 (as indicated by an arrow). In all immunofluorescence images, staining with DAPI (blue) was used to visualize cell nuclei. (A-E) *P < 0.05 by unpaired 2-tailed Student’s t-test, n = 5 per group. Data are expressed as the mean (±SD) fold differences between the cells transfected with NIPA1-SO-expressing lentivirus and cells transfected with the lentivirus vector in three independent experiments. *P < 0.05 by unpaired 2-tailed Student’s t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In addition, we investigated whether there was a difference in BMPR2 level between atherosclerotic plaques and normal intimal tissue. Immunohistochemical analysis showed that BMPR2 levels were lower in atherosclerotic plaques than in normal intimal tissues (Fig. 4D). Furthermore, co-localization of BMPR2 with the endothelial cell marker CD34, macrophage marker LGALS3 and the vascular smooth muscle cell marker αSMA was observed (Fig. 4E). Together, these results demonstrated that NIPA1-SO upregulated BMPR2 in a NIPA1-dependent manner.
NIPA1-SO and NIPA1 influence THP-1 cell adhesion to HUVECs
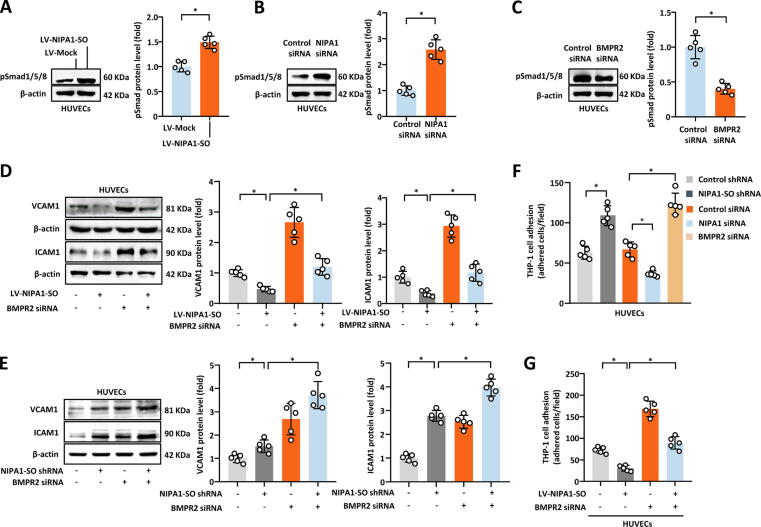
It has previously been reported that BMPR2 can inhibit the expression of intercellular adhesion molecule-1 (ICAM1) and vascular cell adhesion molecule-1 (VCAM1) in endothelial cells, thus leading to the reduced adhesion of monocytes to endothelial cells; this is thought to be mediated by the Smad1/5/8-mediated signaling pathway [14], [15]. We hypothesized that NIPA1-SO, BMPR2 and NIPA1 may influence these events and performed NIPA1-SO overexpression and knockdown studies. Our experiments showed that overexpressing NIPA1-SO in HUVECs increased the level of pSmad1/5/8 (Fig. 5A) whereas the knockdown of NIPA1 or BMPR2 had the opposite effect (Fig. 5B and C). Additionally, we found that the overexpression of NIPA1-SO in vascular endothelial cells resulted in the reduced expression of VCAM1 and ICAM1 (Fig. 5D), while the knockdown of NIPA1-SO had the opposite effect (Fig. 5E). Furthermore, the effect of NIPA1-SO on VCAM1 and ICAM1 expression was attenuated by BMPR2 knockdown (Fig. 5D and E), suggesting that NIPA1-SO inhibits ICAM1 and VCAM1 via the BMPR2-mediated pathway.
Fig. 5.
NIPA1-SO, via NIPA1 signaling, alters expression of adhesion molecules by HUVECs, influencing THP-1 cell adhesion. (A to C) The effects of NIPA1-SO, NIPA1 and BMPR2 upon pSmad1/5/8 expression were examined via Western blotting and either lentiviral-mediated overexpression (A) or knockdown with NIPA1 siRNA (B) or BMPR2 siRNA (C). Data were normalized to β-actin and to control samples (n = 5). (D) VCAM1 and ICAM1 expression in response to overexpression of NIPA1-SO and/or siRNA-mediated knockdown of BMPR2 was examined via Western blotting. Data were normalized to β-actin and to control samples (n = 5). (E) Knockdown of NIPA1-SO and BMPR2, both separately and in combination, was used to assess VCAM1 and ICAM1 expression via Western blotting (n = 5). Data are presented following normalization to β-actin and untreated control cells. (F) Monocyte adhesion in response to endothelial cell knockdown of NIPA1-SO, NIPA1 and BMPR2 was performed. Data are presented as a count of adhered cells per field of the acquired images (n = 5). (G) The effect of NIPA1-SO overexpression and BMPR2 knockdown upon monocyte adhesion was also assessed (n = 5). Data shown are a count of the number of adhered cells per image field. *P < 0.05 by unpaired 2-tailed Student’s t-test (A to C) or one-way ANOVA with Tukey’s post hoc tests (D to G).
Next, we investigated whether NIPA1-SO, BMPR2 and NIPA1 could affect monocyte adhesion to endothelial cells. Monocyte adhesion assays showed that the knockdown of NIPA1-SO or BMPR2 in HUVECs resulted in increased THP-1 cell adhesion to HUVECs, while the knockdown of NIPA1 had the opposite effect (Fig. 5F). In contrast, the augmented expression of NIPA1-SO caused a reduction in THP-1 cell adhesion to HUVECs and this effect was attenuated by BMPR2 knockdown (Fig. 5G), suggesting that NIPA1-SO inhibits THP-1 cell adhesion to HUVECs in a BMPR2-dependent manner.
NIPA1-SO affects lipid levels in THP-1 cells and HASMCs
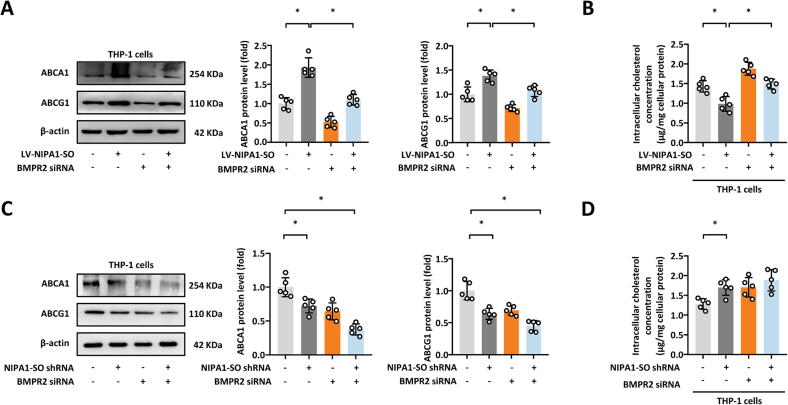
There is evidence to suggest that the BMPR2 gene influences lipid levels [16]. Therefore, we investigated whether NIPA1-SO and NIPA1 could affect lipid metabolism and found that in both THP-1 cells (Fig. 6) and HASMCs (Appendix Fig. S7), lentivirus-mediated NIPA1-SO overexpression resulted in the increased expression of the cholesterol efflux transporters ATP binding cassette subfamily A member 1 (ABCA1) and ABCG1 (Fig. 6A and Appendix Fig. S7A) and decreased the levels of intracellular cholesterol (Fig. 6B and Appendix Fig. S7B). The shRNA-mediated knockdown of NIPA1-SO had the opposite effect (Fig. 6C and D, Appendix Fig. S7C and D). The effect of NIPA1-SO on the protein levels of ABCA1 and ABCG1 on intracellular cholesterol level was attenuated by the siRNA-mediated knockdown of BMPR2 (Fig. 6B and D, Appendix Fig. S7B and D), suggesting that NIPA1-SO could influence cellular lipid metabolism in a BMPR2-dependent manner.
Fig. 6.
NIPA1-SO, via BMPR2, increases ABCA1 and ABCG1 levels in THP-1 cells. (A and B) Western blotting of the cholesterol efflux-mediators ABCA1 and ABCG1 (A) and intracellular cholesterol concentration (B), in human monocytes (THP-1 cells) in response to lentiviral overexpression of NIPA1-SO and BMPR2 siRNA knockdown (n = 3). (C and D) Western blotting of ABCA1 and ABCG1 (C) and intracellular cholesterol concentration (D), in human THP-1 cells with NIPA1-SO shRNA knockdown and BMPR2 siRNA transfection (n = 3). Data in (A) and (C) are presented following normalization to β-actin and untreated control cells. *P < 0.05 by one-way ANOVA with Tukey’s post hoc tests (A and C) or unpaired 2-tailed Student’s t-test (B and D).
NIPA1-SO and NIPA1 affect atherosclerotic plaque formation
Having found that NIPA1-SO and NIPA1 were differentially expressed in atherosclerotic plaques (compared to the normal arterial intima) and that the NIPA1-SO-FUBP1-NIPA1-BMPR2 pathway influenced adhesion molecule expression, monocyte adhesion to vascular endothelial cells and lipid metabolism, we next investigated whether NIPA1-SO and NIPA1 were involved in the development of atherosclerosis.
To test if NIPA1-SO influenced atherogenesis, LDLR-/- mice were injected with either an NIPA1-SO-expressing lentivirus or a control lentivirus and then fed an atherogenic Western diet for 12 weeks. After this, the atherosclerotic lesions in the two groups were compared. This experiment showed that aortic atherosclerotic lesions in the NIPA1-SO-expressing group were smaller, contained less lipid, and had a thicker fibrous cap, than aortic atherosclerotic lesions in the control group (Fig. 7A). An ex vivo assay showed that there was a reduction in monocyte adhesion to the aortic endothelium in the NIPA1-SO-expressing group when compared with the control group (Fig. 7B).
Fig. 7.
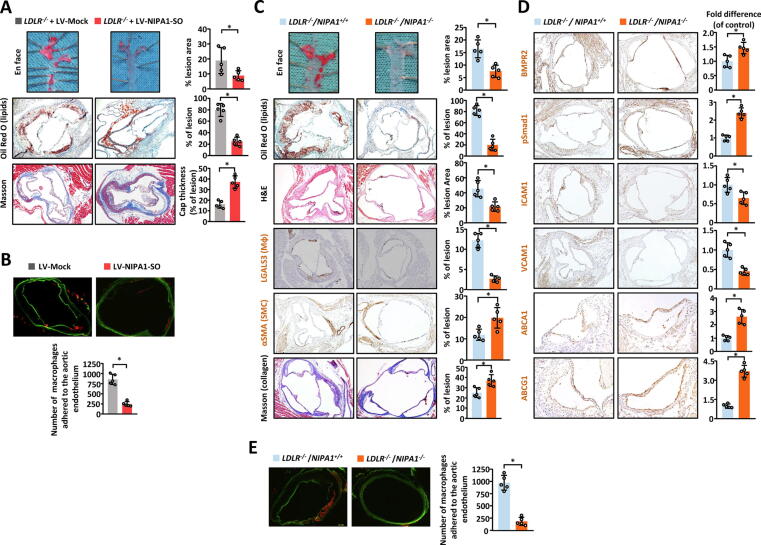
NIPA1-SO and NIPA1 influence atherosclerotic plaque formation in mice in vivo. (A) En face analysis of aortas from LDLR-/-/NIPA1+/+ mice injected with lentivirus to overexpress NIPA1-SO or a control lentivirus (LV-Mock) was performed to demonstrate if NIPA1-SO is involved in atherogenesis in vivo (n = 5). Tissue samples were stained with Oil Red O and the lesion area quantified as a percentage of total en face area. Immunohistochemistry analysis of lesions in LDLR-/-/NIPA1+/+ mice overexpressing NIPA1-SO was performed to determine if the composition of atherosclerotic lesions in such mice differs from control mice. Data shown is the % of lesion area positive for Oil Red O (lipid) staining or Masson staining. (B) Aortic ring monocyte adhesion assays were performed on LDLR-/-/NIPA1+/+ mice injected with NIPA1-SO lentiviral particles or a control lentivirus as described in C (n = 5). (C) En-face dissection and subsequent Oil Red O lipid staining and immunohistochemical analysis of LDLR-/-/NIPA1+/+ and LDLR-/-/NIPA1-/- mice fed an atherogenic Western diet for 12 weeks was performed (n = 5). En face aorta samples were stained with Oil Red O (red) to visualize atherosclerotic plaques. Lesion areas, as a percentage of total en face area, are shown. Further analysis of atherosclerotic lesions from the two mouse strains was also performed using Oil Red O staining (quantifying the level of lipid as percentage of the lesion area). Hematoxylin (blue) and eosin (pink) (H&E) staining was also performed to visualize the nuclei and cytoplasm/extracellular matrix (respectively). H&E staining was quantified as a percentage of total lesion area. Immunohistochemistry was used to stain for monocytes (using the marker LGALS3) and smooth muscle cells (using α-smooth muscle actin) as indicated by brown staining. The abundance of these cell types as a percentage of total lesion area is shown. Staining with Masson’s trichrome stain was used to determine the quantity of collagen, as a percentage of total lesion area. (D) Immunohistochemistry of LDLR-/-/NIPA1+/+ and LDLR-/-/NIPA1-/- mice was performed for BMPR2, pSmad1, ICAM1, VCAM1, ABCA1 and ABCG1, with brown staining indicating the presence of the protein of interest. For all immunohistochemistry experiments, the level of each protein, relative to the control (LDLR-/-/NIPA1+/+) mice, is presented (n = 5). (E) An aortic ring monocyte adhesion assay was performed using LDLR-/-/NIPA1+/+ and LDLR-/-/NIPA1-/- mouse aorta samples to determine if NIPA1 knock-out alters monocyte adhesion to endothelial cells (n = 5). The aortic rings were labeled with calcein-AM and incubated with Dil–labeled mouse peritoneal monocytes. Data presented are the total number of monocytes adhered to the aortic ring for the two groups. Data are presented as the total number of cells adhering to the aortic endothelium. *P < 0.05 by unpaired 2-tailed Student’s t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To test if NIPA1 played a role in atherogenesis, we generated NIPA1 knock-out mice (on a LDLR-/- background) and fed both LDLR-/-/NIPA1+/+ control mice and LDLR-/-/NIPA1-/- mice an atherogenic Western diet for 12 weeks, followed by analyses of atherosclerotic plaque development in the two groups. Analysis found that aortic atherosclerotic lesions were smaller in LDLR-/-/NIPA1-/- mice than LDLR-/-/NIPA1+/+ control mice (Fig. 7C). Immunohistochemical analyses of plaque composition showed that atherosclerotic lesions in LDLR-/-/NIPA1-/- mice contained less lipid and fewer monocytes/macrophages but more VSMCs and collagen (indicative of more stable plaques) than atherosclerotic lesions in LDLR-/-/NIPA1+/+ control mice (Fig. 7C). Additionally, immunohistochemical analyses of BMPR2, pSmad1/5/8, ICAM1, VCAM1, ABCA1, and ABCG1 showed that compared with atherosclerotic lesions in LDLR-/-/NIPA1+/+ control mice, atherosclerotic lesions in LDLR-/-/NIPA1-/- mice had higher levels of BMPR2, pSmad1, ABCA1, and ABCG1, but lower levels of ICAM1 and VCAM1 (Fig. 7D). An ex vivo assay demonstrated that there was a reduction in monocyte adhesion to the aortic endothelium in the LDLR-/-/NIPA1-/- group when compared with the LDLR-/-/NIPA1+/+ control group (Fig. 7E).
Furthermore, LDLR-/-/NIPA1-/- mice had lower blood levels of ICAM1, VCAM1, triglycerides, total cholesterol, low density lipoprotein cholesterol (LDLc) but higher levels of high-density lipoprotein cholesterol (HDLc) when compared with LDLR-/-/NIPA1+/+ control mice (Table S1).
Discussion
In this study, we found that NIPA1-SO, a previously uncharacterized lncRNA, played a protective role in the development of atherosclerosis. We found that NIPA1-SO down-regulated the expression of the transmembrane protein NIPA1 by interacting with the transcription factor FUBP1 and that there was a decrease of NIPA1-SO and an increase of NIPA1 in atherosclerotic plaques. Furthermore, we found that NIPA1-SO increased the cellular level of BMPR2 by inhibiting BMPR2 endocytosis by NIPA1, and that NIPA1-SO suppressed monocyte adhesion to endothelial cells and intracellular cholesterol accumulation via a BMPR2-dependent pathway. Using a well-established mouse model of atherosclerosis, we demonstrated that augmenting NIPA1-SO expression or abolishing NIPA1 resulted in a reduction in the development of atherosclerotic lesions. The pathways uncovered in this study are summarized in Fig. 8.
Fig. 8.
Schematic diagram of the signalling pathways regulated by NIPA1-SO. Our study has uncovered an athero-protective role of the lncRNA NIPA1-SO, which, by interacting with a single transcription factor, is capable of inhibiting monocyte adhesion and foam cell formation, two fundamental processes in atherosclerosis. The transcription factor FUBP1 negatively regulates NIPA1 expression; this inhibitory effect is increased by the interaction of NIPA1-SO with FUBP1, resulting in lower transcription of NIPA1. A reduction in NIPA1 protein results in increased BMPR2 protein due to a reduction in NIPA1-mediated BMPR2 endocytosis and degradation, leading to higher levels of Smad1/5/8 phosphorylation (pSmad1/5/8), which, through complexation with Smad4, inhibit transcription of the adhesion molecules VCAM1 and ICAM1, reducing monocyte adhesion to endothelial cells. Further, the pSmad1/5/8:Smad4 complex promotes ABCA1 and ABCG1 transcription, both of which promote cholesterol efflux via high-density lipoprotein (HDL) particles, thereby inhibiting foam cell formation.
These findings added to the growing body of evidence demonstrating the importance of lncRNAs in atherosclerosis. Several lncRNAs have been implicated in the pathogenesis of atherosclerosis. For instance, CDKN2B-AS (also known as ANRIL), is located on chromosome 9p21, the best-known and most widely-replicated coronary artery disease risk locus identified by genome-wide association studies [17]. CDKN2B-AS has been shown to modulate the expression of the cell cycle regulators CDKN2B and CDKN2A, thereby influencing vascular smooth muscle cell proliferation and thus contributing to the development of atherosclerotic plaques [18]. LncRNA CARMN deficiency has been reported to promote atherosclerosis in mice in vivo [19]. Mechanistically, the loss of CARMN promotes VSMC proliferation and phenotypic switching [19]. Apart from CDKN2B-AS and CARMN, the lncRNAs MeXis [20], MALAT1 [21], and NEXN-AS1 [10] have also been shown to be important players in the development of atherosclerosis. To our knowledge, our present study is the first to reveal that NIPA1-SO also plays a (protective) role in atherogenesis.
Non-coding RNAs, including lncRNAs, can be encapsulated into extracellular vesicles and released into the blood and other biofluids in mammals [22]. The non-coding RNAs included in extracellular vesicles become stable because they cannot be degraded by RNA degradation enzyme ribonucleases (RNases), which commonly exist in extracellular fluids [23]. Thus, extracellular lncRNAs represent promising diagnostic and prognostic biomarkers for human diseases. Several lncRNAs have been suggested to serve as diagnostic markers of coronary heart disease. For example, the levels of lncRNA H19 were found to be dramatically elevated in the plasma of patients with coronary heart disease [24]. Furthermore, hyperhomocysteinemia, an independent risk factor for atherosclerosis, induced H19 expression in the aorta of adult C57BL/6 mice [25]. Although we found that NIPA1-SO was significantly downregulated in human atherosclerotic plaques, examining the plasma levels of NIPA1-SO is of significance in future studies.
The transmembrane protein NIPA1 has been implicated in nervous system development, and mutations in the NIPA1 gene have been associated with autosomal dominant spastic paraplegia [4], [5]. Our study revealed a previously unknown role for this protein in the development of atherosclerosis. Mechanistically, our study showed that NIPA1 enhanced adhesion molecule expression and monocyte adhesion to vascular endothelial cells and indicated that it also promoted a pro-atherogenic lipid profile, with high levels of total cholesterol, LDLc and triglycerides but reduced HDLc. Of relevance, there is reported evidence that NIPA1 physically interacts with BMPR2 and induces the internalization and degradation of BMPR2 [13], [15]. We have also shown that NIPA1 negatively regulated the protein levels of BMPR2. Bone morphogenetic protein 2/4 (BMP2/4) was known to play a role in pulmonary arterial smooth muscle cells via BMP2/4-BMPR2-Smad1/5/8 axis [26]. Our results identified that the knockdown of BMPR2 attenuated pSmad1/5/8 signaling in HUVECs. Furthermore, genome-wide association studies have shown a link between genetic variants in the BMPR2 gene and levels of total cholesterol, LDLc, and HDLc [27], [28].
There is little information in the literature about how NIPA1 expression is regulated. Our study indicated that NIPA1-SO physically interacted with both the transcription regulator FUBP1 and the NIPA1 gene, and down-regulated NIPA1 expression in an FUBP1-dependent manner. Interestingly, FUBP1 has been reported to be capable of binding to RNA and DNA molecules and regulating the expression of several other genes [29]. Our study identified NIPA1-SO as a novel RNA that FUBP1 could interact with and revealed that the NIPA1 gene was negatively regulated by FUBP1.
In summary, our study revealed the previously unknown involvement of NIPA1-SO and NIPA1 in atherosclerosis, such that NIPA1-SO played a protective role against the development of atherosclerotic lesions, whilst NIPA1, which was down-regulated by NIPA1-SO, promoted atherogenesis. The findings from our study were pertinent to a better understanding of the pathogenesis of atherosclerosis and suggested that further investigations of NIPA1-SO and NIPA1 as potential therapeutic targets for atherosclerosis are warranted.
Materials and methods
Sources and analyses of human tissue samples
The study was conducted following the principles in the Declaration of Helsinki. Twenty human atherosclerotic plaque samples (classified as type V or VI lesions according to the Committee on Vascular Lesions of the Council on Atherosclerosis, American Heart Association [30]), were taken from patients undergoing carotid endarterectomy between May 2013 and May 2014 in the Department of Vascular Surgery, Nanfang Hospital, Southern Medical University, China. None of the patients had received previous treatment for atherosclerosis before surgery. Arteries without macroscopic evidence of atherosclerosis, collected from individuals who died either in a road traffic accident or due to cerebral edema, were used as controls. The study was approved by the Institutional Review Board of Nanfang Hospital, Southern Medical University (Guangzhou, China) and written, informed consent was obtained from participants or relatives of the deceased.
Animals
All animal experiments in this study were approved by the Experimental Animal Center of Guangzhou Medical University and performed following the NIH Guide for the Care and Use of Laboratory Animals. LDLR-/-/NIPA1-/- mice were generated by crossing NIPA1-/- mice (purchased from the Laboratory Animal Center of Peking University (Beijing, China) with LDLR-/- mice with a C57BL/6 background (purchased from the Laboratory Animal Center of Peking University (Beijing, China)). Genotype segregation in the offspring followed the expected Mendelian frequency, and we did not observe any developmental/morphological abnormalities. For atherosclerosis development, 8-week-old male LDLR-/-/NIPA1+/+ and LDLR-/-/NIPA1-/- mice were placed on an atherogenic Western diet for 12 weeks. After 12 weeks, mice were inhalationally anesthetized with 2% isoflurane and sacrificed by cervical dislocation. 1 ml of blood was collected by cardiac puncture before the mice were sacrificed, and tissues were collected for further analyses.
8-week-old male LDLR-/- mice infected with lentivirus to overexpress NIPA1-SO (LV-NIPA1-SO), or with a negative control lentivirus via tail vein injection, were fed an atherogenic Western diet for 12 weeks. After 12 weeks, the mice in each group were anesthetized as described above and were sacrificed by cervical dislocation. Blood and tissues were collected for further analyses.
Cell culture
Human THP-1 cells, HASMCs and HUVECs were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). THP-1 monocytes were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% fetal calf serum (FCS). Human vascular endothelial cells and vascular smooth muscle cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. All cells were incubated at 37 °C in an atmosphere of 5% CO2. Cells were seeded in 6- or 12-well plates or 60-mm dishes and grown to 60%-80% confluence before use.
En face analysis
Immediately after the mice were sacrificed, the aortas were excised and fixed in 4% paraformaldehyde for quantification of plaque formation using the en face technique. Briefly, after the adventitial tissue was carefully removed, the aorta was opened longitudinally, stained with Oil Red O (0.5% in 60% isopropanol) (Sigma) for 15 min, and pinned to a blue wax surface. En face images were obtained using a stereomicroscope (SZX12; Olympus) equipped with a digital camera (Dxm1200, Nikon) and analyzed using Adobe Photoshop version 7.0 and Scion Image software 4.0. The percentage of the luminal surface area stained by Oil Red O was determined.
Quantification of NIPA1-SO in human atherosclerotic plaques
Total RNA from human atherosclerotic plaques was extracted using TRIzol reagent (Invitrogen). After reverse transcription, real-time PCR was performed on the ABI 7500 Real-Time PCR System (Applied Biosystems) with SYBR Green detection chemistry (Takara Bio). All samples were assayed in triplicate. Data were analyzed using the ΔΔCt method, with GAPDH RNA as a reference.
NIPA1-SO copy number analysis
Total RNA was isolated from HUVECs, THP-1 cells, and HASMCs, respectively, and reversed transcribed into cDNA, followed by quantitative PCR of NIPA1-SO. The copy number of NIPA1-SO RNA per cell was calculated using a standard curve generated by quantitative PCR using known amounts of NIPA1-SO cDNA as templates, the numbers of cell from which RNA samples were extracted, and the Ct values of the NIPA1-SO RT-PCR.
Lentivirus production and cell line infection
Human THP-1 cells, HASMCs and HUVECs were cultured as described above. Lentiviral (LV) particles were packaged with constructs expressing either green fluorescent protein only (GFP; LV-Mock), NIPA1-SO and GFP (LV-NIPA1-SO; for NIPA1-SO overexpression), or a shRNA targeting NIPA1-SO and GFP (LV-shRNA-NIPA1-SO; for NIPA1-SO knockdown). GFP expression from all vectors was used to monitor transduction efficiency. The cells were infected with the LV stock at a multiplicity of infection of 100 (THP-1 cells) or 20 (HASMCs and HUVECs) transducing units per cell in the presence of 8 mg/mL of polybrene. The cells were washed with fresh, complete media after 24 h of incubation and GFP-positive cells were counted 96 h post-transduction.
Transfection with small interfering RNA
Small interfering RNAs (siRNAs) targeting FUBP1, NIPA1 and BMPR2, and a non-targeting, negative control siRNA, were purchased from Ribo Biotechnology (China). Cells (2 × 106/well) were transfected using Lipofectamine 2000 (ThermoFisher) according to the manufacturer’s instructions. After 24- or 48-hours post-transfection, RT-PCR and Western blotting were performed respectively.
Construction of recombinant plasmids
The pIRES2-EGFP and PCR-XL-TOPO vectors (containing NIPA1, which were assembled from chemically-synthesized oligonucleotides by PCR) were purchased from Invitrogen. The EcoRI-NIPA1-IRES-EGFP-XhoI fragment was amplified using the PCR-XL TOPO and pIRES2-EGFP vectors as templates for overlap-extension PCR. Gel electrophoresis was performed, and the relevant band was excised from the gel, digested with EcoRI andXhoI in a double-digest, and, incorporated into the pcDNA3.1 (+) vector, and then transformed into competent Escherichia coli DH5α cells for further amplification and use. The recombinant plasmids were verified by sequencing and named pcDNA3.1-NIPA1. The plasmid was transfected as indicated using Lipofectamine 2000 (ThermoFisher), according to the manufacturer’s instructions.
LncRNA and coding RNA microarray analysis
Total RNA was extracted from either human atherosclerotic plaques (from 3 subjects) or human healthy artery specimens (from 3 subjects) with TRIzol reagent (Invitrogen). The purity and integrity of the total RNA samples were verified by spectrophotometry and gel electrophoresis (Appendix Fig. S8A and B). mRNA was purified from total RNA using mRNA-ONLY Eukaryotic mRNA Isolation Kit (Epicentre) and each mRNA sample transcribed into fluorescent cRNA using a random priming method. Labeled cRNAs were hybridized with Human LncRNA Expression Microarray v3.0 (8 × 60 K, Arraystar) and scanned using Agilent Scanner G2505C. Data were normalized and analyzed with the use of Agilent Feature Extraction software (version 11.0.1.1) and GeneSpring GX v11.5.1 (Agilent). The expression microarray data have been deposited in the NCBI Gene Expression Omnibus database (GSE97210).
Fluorescent in situ hybridization
FISH was used to investigate NIPA1-SO copy number using the PathVysion NIPA1-SO RNA probe kit (Vysis) and the Dako Histology FISH Accessory kit. For the Dako Histology kit, the manufacturer's instructions were modified, in order to optimize the technique and decrease the time required for sample processing). Sections were incubated at 56 °C overnight, deparaffinized in two series of xylene, and rehydrated with an ethanol series. Slides were pre-treated with Pre-treatment Solution in a water bath at 97 °C for 10 min. Enzymatic digestion was carried out with Ready-to-Use Pepsin for 3 min at room temperature (endoscopic biopsies) or 6 min at 37 °C (surgical specimens). After dehydration with a graded ethanol series, 10 μl of NIPA1-SO probe was applied to each tissue section. The slides and probe were denatured at 80 °C for 5 min and hybridized at 37 °C overnight in a Dako Hybridizer. The next day, the sections were washed with Stringent Wash Buffer at 65 °C for 10 min in a water bath. Then, the slides were dehydrated with a graded ethanol series and mounted using 10 μl of mounting medium containing 4′, 6-diamino-2-phenylindole (DAPI).
Analysis of nuclear versus cytoplasmic ratio of NIPA1-SO
The nuclear and cytoplasmic samples of HUVECs were prepared by nucleocytoplasmic separation kit. Total RNA from nuclear or cytoplasmic was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Real-time reverse transcription quantitative PCR (RT-PCR), using SYBR Green detection chemistry, was performed on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). Finally, the nuclear versus cytoplasmic ratio of NIPA1-SO were analyzed.
NIPA1-SO polyadenylation assay
Total RNA from HUVECs were extracted using TRIzol reagent and divided into two groups. In the plus PloyA group, a ploy A tail was added. Subsequently, reverse transcription using oligo dT as primer was carried out in both groups, followed by quantitative PCR using a gene specific upstream primer and oligo dA as a downstream primer.
RNA sequencing
Total RNA was extracted from cultured endothelial cells transduced with either a NIPA1-SO expressing lentivirus or the lentivirus vector (n = 3/group), with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol, followed by RNA sequencing. The RNA samples were amplified by RT-PCR to libraries with a concentration of >2 nM. The library was sequenced using Illumina NovaSeq 6000 at 6 GigaBases (150 bp ends) per sample. We aligned reads of six samples to the <research species> reference genome using HISAT2 (https://daehwankimlab.github.io/hisat2/, version: hisat2-2.0.4) package [31]. Next, we analyzed the differential expression of lncRNAs using the DESeq2 [32] between two groups. The lncRNAs with a false discovery rate (FDR) below 0.05 and absolute fold change ≥ 2 were considered as differentially expressed lncRNAs.
RNA isolation and real-time quantitative PCR analysis
Total RNA from cultured cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Real-time quantitative PCR (RT-PCR), using SYBR Green detection chemistry, was performed on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). The expression of GAPDH was used as the internal control for analysis of mRNA level. The expression of U6 RNA was used as the endogenous control for analysis of lncRNA expression from tissues and cells. The PCR primer sequences are shown in Table S2. Melt curve analysis of all RT-PCR products was performed and to confirm production of a single DNA duplex. All samples were measured in triplicate, and the mean Ct value was considered for comparative analysis. Quantitative measurements were determined using the ΔΔCt method, with data normalized to the aforementioned control genes.
Comprehensive identification of RNA binding proteins by mass spectrometry (ChIRP-MS)
Cell harvesting (30 × 108), lysis, disruption, and ChIRP were essentially performed as described by Chu et al. [12] with the following modifications: 1) human vascular endothelial cells were crosslinked in 3% formaldehyde for 30 min, followed by 0.125 M glycine quenching for 5 min; 2) hybridization of ChIRP probes (described in Table S3) was initiated late in the day and left to continue overnight; 3) for mass spectrometry experiments, lysates were pre-cleared by incubation with 30 µl washed beads per ml of lysate at 37 °C for 30 min with shaking. Prior to hybridization, beads were removed from the lysates twice using a magnetic stand; 4) for RNase control, lysates were pooled first and aliquoted into two equal amounts. 1/1000 vol of 10 mg/ml RNase A (Sigma) was added to the RNase control sample and both control and non-treated samples were incubated at 37 °C for 30 min with mixing prior to the hybridization steps. RNA extraction was performed from a small aliquot of post-ChIRP beads as described [12]. For protein elution, beads were collected with a magnetic stand, resuspended in biotin elution buffer (12.5 mM biotin (Invitrogen), 7.5 mM HEPES pH 7.5, 75 mM NaCl, 1.5 mM EDTA, 0.15% SDS, 0.075% sarkosyl, and 0.02% sodium deoxycholate), mixed at room temperature for 20 min, and then at 65 °C for 10 min. The eluate was transferred to a fresh tube and the beads were eluted again. The two eluates were pooled, and residual beads were removed again using the magnetic stand. ¼ total volume of 10% trichloroacetic acid was added to the eluate, and after thorough mixing, proteins were precipitated at 4 °C overnight. The next day, proteins were pelleted at 16,000 g at 4 °C for 30 min. Supernatant was carefully removed from the tubes and protein pellets were washed once with cold acetone, and pelleted again at 16,000 g at 4 °C for 5 min. The acetone was removed and the pellets were briefly centrifuged again and, after removal of residual acetone, left to air-dry for 1 min at room temperature. The protein pellet was then immediately dissolved in appropriate volumes of 1 × Laemmli buffer and boiled at 95 °C for 30 min with occasional mixing to reverse the cross-links. Final protein samples were size-separated in bis-tris SDS-PAGE gels (Invitrogen) for Western blots or mass spectrometry.
Chromatin immunoprecipitation (ChIP) assays
ChIP analysis of human vascular endothelial cells using anti-FUBP1 antibody (Abcam, ab213525) was performed using the protocol provided by the ChIP kit manufacturer (Pierce Agarose ChIP Kit, Thermo Scientific). Immunoprecipitated DNA was analyzed on a LightCycler 480 (Roche) using LightCycler 480 SYBR Green I Master Mix (Roche). PCR primer sequences are shown in Table S4.
RNA immunoprecipitation (RIP)
Nuclear extracts from cultured human vascular endothelial cells (HUVECs) were prepared and incubated with anti-FUBP1 antibody (Abcam, ab213525) or a non-specific mouse IgG, at 4 °C overnight. Protein G Plus Protein A agarose beads (Calbiochem) were then added and the mix was incubated at 4 °C for 2 h, follow by centrifugation to pellet the beads. After washing and elution, immunoprecipitated RNA was extracted with the use of TRIzol reagent (Takara Bio) and RT-PCR was the performed to detect NIPA1-SO binding to immunoprecipitated FUBP1, followed by agarose gel electrophoresis. The PCR primer sequences are shown in Table S5.
RNA pulldown
Biotin-labeled full-length (1–1451 nt) and truncated (1–303 nt, 276–1056 nt, and 960–1451 nt) NIPA1-SO sequences were prepared by the in vitro transcription method. The labeled sequences were incubated with cell lysate at 4 °C for 3 h and then with streptavidin-conjugated magnetic beads to isolate NIPA1-SO-protein complexes, followed by Western blot analysis of FUBP1.
Intracellular cholesterol concentration assay
Intracellular cholesterol concentration was extracted using a mixture of chloroform, isopropanol and NP-40 in the 7:11:0.1 ratio and measured with the use of Cholesterol/Cholesteryl Ester Quantitation Assay kit (Abcam, ab65359). The results were expressed as microgram cholesterol/mg cellular protein.
Analyses of aortic sinus atherosclerotic lesions
The upper portion of the heart and proximal aorta were obtained from sacrificed mice, embedded in Optimal Cutting Temperature compound (Fisher), and stored at −80 °C. Serial 10 μm thick cryosections of aorta, beginning at the aortic root, were collected for a distance of 400 μm. Sections were stained with either Oil Red O, hematoxylin and eosin, or Masson's trichrome, or were immunostained for either LGALS3, αSMA, BMPR2, pSmad1/5/8, ICAM1, VCAM1, ABCA1, or ABCG1. Digitized color images of stained sections were analyzed using Image-Pro Plus image analysis software (Media Cybernetics, Rockville, MD, USA).
Immunohistochemistry
Sections (4 μm thick) of formalin-fixed, paraffin-embedded human atherosclerotic plaques and normal arterial wall specimens, mouse atherosclerotic plaque specimens from mice overexpressing LV-NIPA1-SO or arterial wall specimens from mice transduced with a negative control lentivirus, were subjected to immunohistochemistry, comprising gradient alcohol dehydration, antigenic retrieval, 3% hydrogen peroxide blocking and non-immune sheep serum blocking. The human sections were incubated with an anti-NIPA1 antibody (Abcam, ab121744). The mouse-derived sections were incubated with anti-galectin 3 (LGALS3) antibody (Abcam, ab2785), anti-alpha smooth muscle actin (αSMA) antibody (Abcam, ab32575), anti-anti-ABCA1 antibody (Abcam, ab18180), anti-ABCG1 antibody (Abcam, ab217023), anti-ICAM1 antibody (Abcam, ab119871), anti-VCAM1 antibody (Abcam, ab134047), and anti-BMPR2 antibody (Abcam, ab130206), anti-pSmad1 (S187) antibody (Abcam, ab73211), respectively. Negative controls were performed by incubating samples with non-immune sheep serum. The immunohistochemical staining was visualised using a colorimetric kit (Gene Tech, GK500705) and images were acquired using an Olympus BX50 microscope with a digital color camera and analyzed using Image J (version 1.8.0).
Immunohistofluorescence analysis
Sections (4 μm thick) formalin-fixed, paraffin-embedded human atherosclerotic plaques and normal arterial wall specimens, respectively, were incubated with an anti-NIPA1 antibody (Abcam, ab121744) together with a mouse antibody for either human CD34 (Abcam, ab8536), human αSMA (Abcam, ab7817), or human CD68 (Abcam, ab955), followed by incubation with a Texas red–conjugated anti-rabbit secondary antibody (Abcam, ab6719) and an FITC-conjugated anti-mouse secondary antibody (Abcam, ab6785). Nuclei were counterstained with DAPI. The fluorescent images were acquired using an Olympus BX50 microscope with a digital color camera and analyzed using Image J (version 1.8.0).
Fluorescence confocal microscopy
Human vascular endothelial cells (HUVECs) were seeded on coverslips, fixed with paraformaldehyde, permeabilized with 0.25% Triton X-100, and then incubated with a digoxin-labeled probe for detecting NIPA1-SO. Thereafter, cells were incubated with a DyLight 594-conjugated IgG fraction (Abcam, ab96873) coupled with a monoclonal mouse anti-digoxin antibody (Abcam, ab116590). In a separate experiment, fixed and permeabilized HUVECs were incubated with an anti-NIPA1 antibody (Abcam, ab121744) and subsequently with a fluorophore-conjugated goat anti-rabbit antibody (Abcam, ab6717). Nuclei were counterstained with DAPI. Cells were imaged using an Olympus FV1000 confocal laser scanning microscope.
Monocyte adhesion to aortic rings
Aortas were isolated from LDLR -/-/NIPA1+/+ and LDLR-/-/NIPA1-/- mice and placed into a Petri dish filled with cold, sterile PBS to keep the tissue moist. The aorta samples were dissected to remove extraneous tissue (such as fat tissue) and then sliced into 1 mm rings. The aortic rings were labeled with calcein-AM, then washed with PBS and then incubated with Dil–labeled mouse peritoneal monocytes (isolated from C57BL/6 mice) for 2 h at 37 °C with agitation at 90 rpm. Subsequently, the aortic rings were washed with PBS and embedded cross-wise in optimum cutting temperature medium, snap-frozen, and stored at − 80 °C. Multiple 10 μm sections were cut using a cryotome, fixed in formalin, and analyzed using fluorescence microscopy. Images were captured and analyzed to determine the number of macrophages that had adhered to the luminal endothelium of the aortic rings.
Western blot analyses
Protein extracts from cultured cells and tissue samples were prepared in accordance with established methods. Protein extracts were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then subjected to Western blot analyses using rabbit polyclonal anti-NIPA1, -FUBP1, -BMPR2 antibodies (Proteintech Group), rabbit polyclonal anti-pSmad1/5/8 antibodies (Abcam) and rabbit polyclonal anti-ICAM-1, anti-VCAM-1, and anti-β-actin antibodies (Santa Cruz Biotechnologies). Chemiluminescent visualization was performed using the ECL Plus Western Blot Detection System (Amersham Biosciences).
Cell adhesion assay
HUVECs were plated into 6-well plates (1 × 105/well) to and cultured at 37◦C/5% CO2 to form a confluent monolayer. The cells were washed with medium and incubated with fresh growth medium in the presence or absence of TNF-α (10 ng/ml) for 8 h. THP-1 monocytes (at 2 × 105 cells/ml) stained with 5 μM carboxyfluorescein succinimidyl ester (Genecopoeia) was then added to each well and incubated for 30 min under standard culture conditions (37 °C, 5% CO2). After 4–6 h of incubation, non-adherent monocytes were gently removed by washing twice with PBS. Monocyte adhesion was quantified by counting the number of adhered monocytes with green fluorescence using a fluorescence microscope. Experiments were repeated at least three times.
Measurement of serum biochemical parameters
The serum concentrations of ICAM-1 and VCAM-1 in mice were measured by ELISA kits with an anti-ICAM1 antibody (Abcam ab100688) and an anti-VCAM1 antibody (Abcam, ab201278), respectively. The total cholesterol (TC), triglyceride (TG), high density lipoprotein cholesterol (HDL-C) and low density lipoprotein cholesterol (LDL-C) levels in the serum were measured using a Roche automatic biochemical analyzer (Roche, C7500).
Statistical analyses
Data were analyzed using Statistical Package for the Social Sciences (SPSS) software (version 13.0). Normally distributed data are presented as mean ± SD or median (interquartile range in skewed-distributed data), unless otherwise indicated. Continuous variables were analyzed by one-way ANOVA with Bonferroni’s correction applied for multiple comparisons, or by unpaired 2-tailed Student’s t-test, with P < 0.05 considered significant. The correlations between selected protein levels and biochemical parameters were analyzed using Pearson’s or Spearman’s correlation, with P < 0.05 considered significant.
Data and materials availability
The data, analytic methods, and study materials will be/have been made available to other researchers for purposes of reproducing the results or replicating the procedure. The supplementary materials have been uploaded to Nutstore. The web hyperlink is provided below. https://www.jianguoyun.com/p/DZrlZS4QsPHhChiZqMsEIAA.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (Grant numbers: 82072335, 81871701, 82070466, 81800364, and 81974046), the Natural Science Fund of Guangdong (Grant numbers: 2020B1515020013, 2018A030313533, and 2017A030313535), Guangzhou Women and Children' s Medical Center (Grant number: GWCMC2020-6-010), and Fujian Joint Funds for the Innovation of Science and Technology (Grant number: 2018Y9099). SY thanks the support of the British Heart Foundation (Grant numbers: RG/16/13/32609, RG/19/9/34655, SP/19/2/344612, PG/16/9/31995, and PG/18/73/34059). This work falls under the portfolio of research conducted within the NIHR Leicester Biomedical Research Centre.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.01.017.
Contributor Information
Xiao-Yan Dai, Email: xdai@gzhmu.edu.cn.
Shu Ye, Email: sy127@leicester.ac.uk.
Yan-Wei Hu, Email: ywhu0618@163.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchida S., Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 3.Rudenskaya GE, Kadnikova VA, Bessonova LA, Sparber PA, Kurbatov SA, Mironovich OL, et al. Autosomal dominant spastic paraplegias. Zh Nevrol Psikhiatr Im S S Korsakova. 2021;121:75–87. [DOI] [PubMed]
- 4.Rainier S., Chai J.H., Tokarz D., Nicholls R.D., Fink J.K. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am J Hum Genet. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed J.A., Wilkinson P.A., Patel H., Simpson M.A., Chatonnet A., Robay D., et al. A novel NIPA1 mutation associated with a pure form of autosomal dominant hereditary spastic paraplegia. Neurogenetics. 2005;6:79–84. doi: 10.1007/s10048-004-0209-9. [DOI] [PubMed] [Google Scholar]
- 6.Goytain A., Hines R.M., El-Husseini A., Quamme G.A. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J Biol Chem. 2007;282:8060–8068. doi: 10.1074/jbc.M610314200. [DOI] [PubMed] [Google Scholar]
- 7.Vanlerberghe C., Petit F., Malan V., Vincent-Delorme C., Bouquillon S., Boute O., et al. 15q11.2 microdeletion (BP1-BP2) and developmental delay, behaviour issues, epilepsy and congenital heart disease: a series of 52 patients. Eur J Med Genet. 2015;58:140–147. doi: 10.1016/j.ejmg.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Wang D., Zhu Z.L., Lin D.C., Zheng S.Y., Chuang K.H., Gui L.X., et al. Magnesium supplementation attenuates pulmonary hypertension via regulation of magnesium transporters. Hypertension. 2021;77:617–631. doi: 10.1161/HYPERTENSIONAHA.120.14909. [DOI] [PubMed] [Google Scholar]
- 9.Bai H.L., Lu Z.F., Zhao J.J., Ma X., Li X.H., Xu H., et al. Microarray profiling analysis and validation of novel long noncoding RNAs and mRNAs as potential biomarkers and their functions in atherosclerosis. Physiol Genomics. 2019;51:644–656. doi: 10.1152/physiolgenomics.00077.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y.W., Guo F.X., Xu Y.J., Li P., Lu Z.F., McVey D.G., et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J Clin Invest. 2019;129:1115–1128. doi: 10.1172/JCI98230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X.H., Lu Z.F., Kang C.M., Li X.H., Haworth K.E., Ma X., et al. The long noncoding RNA RP11-728F11.4 promotes atherosclerosis. Arterioscler Thromb Vasc Biol. 2021;41:1191–1204. doi: 10.1161/ATVBAHA.120.315114. [DOI] [PubMed] [Google Scholar]
- 12.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Shaw W.R., Tsang H.T., Reid E., O'Kane C.J. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat Neurosci. 2007;10:177–185. doi: 10.1038/nn1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim C.W., Song H., Kumar S., Nam D., Kwon H.S., Chang K.H., et al. Anti-inflammatory and antiatherogenic role of BMP receptor II in endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:1350–1359. doi: 10.1161/ATVBAHA.112.300287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang H.T., Edwards T.L., Wang X., Connell J.W., Davies R.J., Durrington H.J., et al. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signalling. Hum Mol Genet. 2009;18:3805–3821. doi: 10.1093/hmg/ddp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng J., Gao J., Li Y., Yang Y., Dang L., Ye Y., et al. BMP4 enhances foam cell formation by BMPR-2/Smad1/5/8 signaling. Int J Mol Sci. 2014;15:5536–5552. doi: 10.3390/ijms15045536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo Sardo V., Chubukov P., Ferguson W., Kumar A., Teng E.L., Duran M., et al. Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell. 2018;175(1796–1810):e1720. doi: 10.1016/j.cell.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacante F., Rodor J., Lalwani M.K., Mahmoud A.D., Bennett M., De Pace A.L., et al. CARMN loss regulates smooth muscle cells and accelerates atherosclerosis in mice. Circ Res. 2021;128:1258–1275. doi: 10.1161/CIRCRESAHA.120.318688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sallam T., Jones M., Thomas B.J., Wu X., Gilliland T., Qian K., et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24:304–312. doi: 10.1038/nm.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cremer S., Michalik K.M., Fischer A., Pfisterer L., Jae N., Winter C., et al. Hematopoietic deficiency of the long noncoding RNA MALAT1 promotes atherosclerosis and plaque inflammation. Circulation. 2019;139:1320–1334. doi: 10.1161/CIRCULATIONAHA.117.029015. [DOI] [PubMed] [Google Scholar]
- 22.Gruner H.N., McManus M.T. Examining the evidence for extracellular RNA function in mammals. Nat Rev Genet. 2021;22:448–458. doi: 10.1038/s41576-021-00346-8. [DOI] [PubMed] [Google Scholar]
- 23.Sorrentino S. Human extracellular ribonucleases: multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell Mol Life Sci. 1998;54:785–794. doi: 10.1007/s000180050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong G., Jiang X., Song T. The overexpression of lncRNA H19 as a diagnostic marker for coronary artery disease. Rev Assoc Med Bras. 1992;2019(65):110–117. doi: 10.1590/1806-9282.65.2.110. [DOI] [PubMed] [Google Scholar]
- 25.Glier M.B., Ngai Y.F., Sulistyoningrum D.C., Aleliunas R.E., Bottiglieri T., Devlin A.M. Tissue-specific relationship of S-adenosylhomocysteine with allele-specific H19/Igf2 methylation and imprinting in mice with hyperhomocysteinemia. Epigenetics. 2013;8:44–53. doi: 10.4161/epi.23063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng L., Liu X., Teng X., Yuan W., Duan L., Meng J., et al. DAN plays important compensatory roles in systemic-to-pulmonary shunt associated pulmonary arterial hypertension. Acta Physiol (Oxf) 2019;226:e13263. doi: 10.1111/apha.13263. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann T.J., Theusch E., Haldar T., Ranatunga D.K., Jorgenson E., Medina M.W., et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat Genet. 2018;50:401–413. doi: 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet 2018;50:1514–1523. [DOI] [PMC free article] [PubMed]
- 29.Zhang J., Chen Q.M. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013;32:2907–2916. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stary HC, Chandler AB, Dinsmore RE, Fuster V, Glagov S, Insull W, Jr., et al. A definition of advanced types of atherosclerotic lesions and a histological classification of atherosclerosis. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation 1995;92:1355–1374. [DOI] [PubMed]
- 31.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.