Abstract
FliN is a component of the bacterial flagellum that is present at levels of more than 100 copies and forms the bulk of the C ring, a drum-shaped structure at the inner end of the basal body. FliN interacts with FliG and FliM to form the rotor-mounted switch complex that controls clockwise-counterclockwise switching of the motor. In addition to its functions in motor rotation and switching, FliN is thought to have a role in the export of proteins that form the exterior structures of the flagellum (the rod, hook, and filament). Here, we describe the crystal structure of most of the FliN protein of Thermotoga maritima. FliN is a tightly intertwined dimer composed mostly of β sheet. Several well-conserved hydrophobic residues form a nonpolar patch on the surface of the molecule. A mutation in the hydrophobic patch affected both flagellar assembly and switching, showing that this surface feature is important for FliN function. The association state of FliN in solution was studied by analytical ultracentrifugation, which provided clues to the higher-level organization of the protein. T. maritima FliN is primarily a dimer in solution, and T. maritima FliN and FliM together form a stable FliM1-FliN4 complex. Escherichia coli FliN forms a stable tetramer in solution. The arrangement of FliN subunits in the tetramer was modeled by reference to the crystal structure of tetrameric HrcQBC, a related protein that functions in virulence factor secretion in Pseudomonas syringae. The modeled tetramer is elongated, with approximate dimensions of 110 by 40 by 35Å, and it has a large hydrophobic cleft formed from the hydrophobic patches on the dimers. On the basis of the present data and available electron microscopic images, we propose a model for the organization of FliN subunits in the C ring.
The bacterial flagellum is a complex assembly formed from about two dozen proteins having copy numbers ranging from a few to more than 10,000 (1, 6, 35, 41, 42). The FliN protein is a major component of the C ring, a drum-shaped structure at the bottom of the basal body that is approximately 45 nm in diameter and 15 nm high (19, 31, 68, 79, 81) (Fig. 1). FliN interacts with FliG and FliM to form the switch complex, which is essential for assembly of the flagellum and also for rotation and clockwise (CW)-counterclockwise (CCW) switching (27, 40, 59, 67, 71, 77, 78). The switch complex attaches to the MS ring, which is in the cytoplasmic membrane and is formed from a single protein, FliF (24, 72). The precise locations of FliG, FliM, and FliN in the switch complex are unknown, but FliG and FliM are likely to be near the upper (membrane-proximal) part of the C ring, because FliG binds to FliF (18, 21, 33, 68) and also makes multiple contacts with FliM (9, 44-46, 70). The C-terminal domain of FliM is independently stable and forms the binding site for FliN (46, 70).
FIG. 1.
Simplified flagellar assembly scheme. The arrows represent many assembly steps that are omitted for simplicity. The switch complex and the export apparatus are assembled at about the same time, relatively early in flagellar assembly. The export apparatus transports the components that form the exterior axial structures (the rod, hook, hook-associated proteins, and filament [cross-hatched]) into a central channel that traverses the length of the flagellum, indicated by the dashed lines. IM, inner membrane; PG, peptidoglycan; OM, outer membrane; HAPs, hook-associated proteins.
The sequence of events in flagellar assembly has been deduced from studies of mutants arrested at various steps (28, 37, 42, 62, 63). The MS ring is formed first (Fig. 1). The proteins that form the rod, hook, and filament are actively transported to their destinations by way of a central channel through the structure (41, 51). Export is driven by an apparatus at the base of the flagellum, which is evolutionarily related to the type III secretion system used by pathogenic bacteria for the export of virulence factors (8, 36, 41). This export apparatus is centered within the MS ring and is formed from six membrane proteins and at least three cytosolic proteins (Fig. 1) (42, 47). It assembles at about the same time as the switch complex, but the sequence of events is not known exactly. The proteins to be exported arrive in a partially folded state in association with specialized chaperones (2, 4, 5, 20). Energy for transport comes from the hydrolysis of ATP, which is catalyzed by the protein FliI (14, 17) and is regulated by the protein FliH (48). Although the C ring appears to be well separated from the membrane-bound parts of the export apparatus (30), FliN has been implicated in flagellar export. Proteins with sequence similarity to FliN are found in the virulence factor export apparatus of various species, where they presumably carry out functions related to export rather than motility (42, 65). A temperature-sensitive FliN mutant was unable to regrow flagellar filaments after shearing at the restrictive temperature, indicating that there was a failure in the export of flagellin or the flagellar cap protein (75).
Other mutations in FliN have a range of effects that depend on the expression level of the protein. Certain fliN mutations prevent rotation while allowing flagellar assembly when the protein is expressed at normal levels (27) but allow both assembly and rotation when the protein is overexpressed (40). When wild-type FliN is underexpressed, flagella are still assembled, but their rotation is slow and irregular (65). These observations suggest that incomplete C rings are sufficient for flagellar assembly but cannot support normal motor rotation. The C-terminal parts of FliN (residue ∼58 to the end) are best conserved and most important for function; a FliN fragment lacking 57 N-terminal residues supported swarming motility at about one-third the wild-type rate (65).
The molecular mechanisms of motor rotation, CW-CCW switching, and flagellar export are not understood, mainly due to a lack of structural information. Structures are known for relatively few flagellar proteins and for none of the export apparatus proteins except the chaperone FliS (15). To provide a structural basis for understanding the functions of FliN, we solved its structure by X-ray crystallography and studied its state of association by analytical ultracentrifugation. A stable complex of FliN with FliM was also characterized. The data obtained lead to a model for the structure of the C ring and highlight the functional importance of a prominent hydrophobic patch on the surface of FliN.
MATERIALS AND METHODS
Cloning and protein expression.
The Thermotoga maritima homolog of fliN was identified first by a BLAST search of the unpublished genome data and then from the published genome sequence. In certain (mainly gram-positive) species the FliN homolog has additional N-terminal parts that make it about twice as large as it is in other species, and it is designated FliY (7). The T. maritima genome encodes two proteins (designated FliY-1 and FliY-2 in the annotated sequence) that together resemble FliY. The second of these is the homolog of FliN and was the subject of the present study. Constructs encoding residues 23 to 154 of T. maritima FliN and full-length T. maritima FliM were obtained by PCR amplification of genomic DNA (a gift from R. Huber, University of Regensburg). Primer sequences for PCR were based on the T. maritima genome sequence (53). T. maritima FliN is predicted to have 154 residues on the basis of the genome sequence (53). The T. maritima FliN protein studied here is a 132-residue construct that lacks 22 residues from the N terminus but encompasses the parts found to be essential in Escherichia coli (65), and its size is similar to that of the E. coli protein (137 residues). The constructs were cloned into the NdeI and BamHI sites of the vector pSBETa for fliN (58) or pAED4 for fliM (13).
Either the FliN protein alone or FliN and FliM together were overexpressed in BL21(DE3) cells (61). The cells were grown overnight at 37°C to an optical density at 600 nm of approximately 1.5 and then induced with 0.6 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. The cells were collected by centrifugation, frozen in liquid nitrogen, and stored at −70°C.
Protein purification.
Frozen cells were thawed and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA), and protease inhibitors were added at the following concentrations: phenylmethylsulfonyl fluoride, 170 μg/ml; pepstatin A, 0.7 μg/ml; leupeptin, 0.5 μg/ml; and aprotinin, 2 μg/ml. The cells were sonicated, passed through a French pressure cell, sonicated again, and then centrifuged at 20,000 × g for 30 min at 4°C. The cold supernatant was mixed with 3 volumes of boiling lysis buffer and kept at 85°C for 15 min. The lysate was cooled in an ice-water bath, and the denatured proteins were pelleted by centrifugation at 100,000 × g for 1 h at 4°C.
For purification of FliN, a saturated (NH4)2SO4 solution was added to the clarified lysate (∼200 ml) to obtain a final (NH4)2SO4 concentration of 0.5 M, and the solution was loaded onto a phenyl-Sepharose hydrophobic affinity column (Pharmacia) equilibrated in 50 mM Tris-HCl (pH 8.0)-0.5 M (NH4)2SO4-5 mM EDTA. The column was washed with the same buffer until the UV trace returned to the baseline (∼500 ml). Proteins were eluted from the column in 50 mM Tris-HCl (pH 8.0)-0.15 M (NH4)2SO4-5 mM EDTA. Fractions containing FliN were pooled, dialyzed, concentrated by ultrafiltration, and loaded onto a Superdex-200 size exclusion column (Pharmacia). The column was run in 50 mM Tris-HCl (pH 8.0)-200 mM NaCl. Fractions containing FliN were pooled and concentrated by ultrafiltration.
For purification of FliM plus FliN, clarified lysate (typically ∼200 ml) was loaded directly onto a Q-Sepharose column (Pharmacia), which was washed with ∼500 ml of 50 mM Tris-HCl (pH 8.0) and then developed with a 0 to 1 M NaCl gradient in the same buffer. The FliM and FliN proteins eluted together; fractions containing the proteins were pooled, and saturated (NH4)2SO4 was added to obtain a final (NH4)2SO4 concentration of 0.5 M. The solution was loaded onto a phenyl-Sepharose column equilibrated in 50 mM Tris-HCl (pH 8.0)-0.5 M (NH4)2SO4, and proteins were eluted with a 0.5 to 0 M (NH4)2SO4 gradient. Fractions containing FliM plus FliN were pooled, concentrated by ultrafiltration, and loaded onto a Superdex-200 column. The column was run in 50 mM Tris-HCl (pH 8.0)-200 mM NaCl. Fractions containing FliM plus FliN were identified by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, and concentrated by ultrafiltration.
The E. coli FliN studied here was the full-length protein and was purified as described previously (54).
Analytical ultracentrifugation.
For analytical ultracentrifugation experiments we used the proteins described above (full-length E. coli FliN, residues 23 to 154 of T. maritima FliN, and full-length T. maritima FliM). Sedimentation equilibrium experiments were conducted at 20°C with a Beckman Optima XL-A analytical ultracentrifuge by using an AnTi60 rotor with six-channel, 12-mm-thick, charcoal-Epon centerpieces. Three channels contained protein samples at different concentrations, and three channels contained buffer in dialysis equilibrium with the protein solution for use as optical references. The buffer was 50 mM Tris (pH 8.0)-200 mM NaCl. The FliN concentration ranged from 0.5 to 2 μM (on a monomer basis), and the concentration of FliM plus FliN ranged from 0.05 to 0.2 μM (total subunit concentration).
Samples were centrifuged until sedimentation equilibrium and chemical equilibrium were attained. Absorbance data were taken at 0.001-cm intervals and were averages of 10 measurements. Absorption measurements were obtained at 230 nm. The approach to equilibrium was monitored by comparison of scans taken at 4-h intervals. Partial specific volumes were calculated for each protein from the amino acid sequence, resulting in the following values: E. coli FliN, 0.7368 ml/g; T. maritima FliN, 0.7578 ml/g; and T. maritima FliM, 0.7443 ml/g. The buffer density was calculated and corrected for temperature by using the method of Laue et al. (39), which resulted in a value of 1.0079 g/ml at 20°C. A 360-nm scan was used to check for anomalous light bending in the 230-nm scan and to define the range of usable data.
Plots of absorbance versus radial distance were fitted by nonlinear regression by using the ORIGIN software package (Microcal Software) and an additional module supplied by Beckman Instruments. Data obtained from the different loading concentrations and from different rotational speeds (when applicable) were analyzed simultaneously. The fits incorporated a baseline offset to account for the zero offset of the optical system or the presence of any absorbing, nonsedimenting components. Initial estimates of the baseline offset term were obtained by overspeeding the sample at the completion of the run to deplete the meniscus of solute.
Velocity-sedimentation experiments were carried out with the XL-A ultracentrifuge with an AnTi60 rotor at 20°C by using double-sector centerpieces. A protein sample (300 to 400 μl) was added to one channel, and buffer was added to the reference compartment. Absorbance data were acquired at 280 nm with radial increments of 0.002 cm in continuous-scanning mode. The rotational velocity for all three proteins was between 35,000 and 45,000 rpm. Scans were taken at 2-min intervals. The sedimenting boundaries were fitted to a model incorporating a distribution of sedimentation coefficients by using the software package DCDT (60).
FliN/FliM ratio in purified complex.
The purified FliM-FliN complex was dissociated in SDS-PAGE loading buffer, and the subunits were resolved on 15% polyacrylamide gels. The gels were stained with Coomassie blue G-250, and bands were quantified by using video densitometry and the program NIH-IMAGE. Proteins were loaded at a range of concentrations, and plots of absorbance versus loading were used to estimate relative protein levels. As a further check, purified FliM and FliN were resolved on SDS—12% PAGE gels and visualized by silver staining by using a protocol that reportedly has been optimized for uniformity and linearity (49, 80). Bands were quantified by video densitometry by using the same method that was used for Coomassie blue-stained gels, except that a green filter was used.
Crystallization and data collection.
Following sizing chromatography, FliN was concentrated to 3 mg/ml. Crystals were grown at room temperature in sitting drop trays with a well solution containing 18% (vol/vol) 2-methyl-2,4-pentanediol and 100 mM morpholineethanesulfonic acid (MES) (pH 5.9). The drops were set up with a ratio of well solution to protein of 1:1. The crystals grew to full size in about 12 h. The crystals used for data collection typically were 200 by 50 by 50 μm.
FliN crystals were cooled by rapid immersion in liquid nitrogen after a brief (ca. 10-s) soak in 25% 2-methyl-2,4-pentanediol-100 mM MES (pH 5.9). X-ray diffraction data were collected at 100K by using a charge-coupled device Quantum 4 detector (ADSC) at ALS beamline 5.0.2. Data were processed (Table 1) by using DENZO and SCALEPACK (55). The crystals belonged to space group P3121 with the following unit cell dimensions: a = 100.2 Å and c = 87.9 Å. The asymmetric unit contained two FliN molecules, and the solvent content was approximately 70%.
TABLE 1.
Data collection statistics
| Parameter | Value |
|---|---|
| Wavelength (Å) | 0.979277 |
| Resolution (Å) | 87.7-3.4 (3.6-3.4)a |
| Highest-resolution shell (Å) | 3.52-3.4 |
| No. of observed reflections | 80,998 |
| No. of unique reflections | 7,246 |
| Completeness (%) | 95.9 (93.4) |
| Rsym (%)b | 7.0 (28.9) |
| Avg I/σ(I) | 7.3 (1.8) |
| Mosaicity (°) | 0.351 |
The values in parentheses are data for the highest-resolution shell.
Rsym = 100(Σ|I−〈I〉|/ΣI), where I is the intensity of the individual measurement and 〈I〉 is the average intensity from multiple observations.
Structure determination and refinement.
The crystal structure of a similar but slightly smaller T. maritima FliN construct was recently solved by the Joint Center for Structural Genomics (PDB accession code 1O6A). We were able to obtain phases for our data by performing molecular replacement using the programs AMORE (52) and MOLREP (73) and the coordinates for 1O6A. The resulting maps were clear and showed continuous density characteristic of α-helices and β sheets throughout the model region. The program O (29) was used for model building, and refinement calculations were performed with REFMAC5 (50). Refinement statistics are shown in Table 2. Figures were prepared by using the programs PYMOL (12) and RasMol (57).
TABLE 2.
Refinement statistics
| Parameter | Value |
|---|---|
| Resolution range (Å) | 87.7-3.4 (3.6-3.4)a |
| No. of reflections in working set | 6,906 |
| No. of reflections in free-R set | 340 |
| No. of protein atomsb | 1,352 |
| R factor (%)c | 22.4 (33.7) |
| Free-R factor (%)d | 28.6 (41.7) |
| RMSD (bond lengths) (Å) | 0.014 |
| RMSD (bond angles) (°) | 1.76 |
| % of φ and ψ anglese | |
| Most favored | 77.3 |
| Additional allowed | 20.0 |
| Generously allowed | 1.3 |
| Disallowed | 1.3 |
The values in parentheses are data for the highest resolution shell.
Nonhydrogen atoms only.
R factor = 100[∑(||Fobs| − |Fcalc||)/∑|Fobs|].
Rfree is the R factor for a selected subset of reflections (5%) that were not included in the refinement calculations.
Stereochemistry was assessed with PROCHECK (38).
Hydrophobic-patch mutant.
Residue Val113 in the hydrophobic patch of FliN was mutationally replaced with Asp by using the Altered Sites (Promega) procedure with fliN cloned in plasmid pLS4 (40). The mutation was confirmed by DNA sequencing. The mutated gene was then transferred into plasmid pHT39, which allowed controlled expression of FliN from the lac promoter (65). Assays for swarming in soft agar, swimming in liquid, and flagellation were performed as described previously (66) by using the fliN deletion strain DFB223 transformed with wild-type or mutant plasmids. The swarm plates contained tryptone broth and 0.28% agar. Flagella were stained by a wet mount procedure (23).
Protein structure accession number.
Coordinates and diffraction data for the FliN crystal structure have been deposited in the RCSB Protein Data Bank under accession number 1YAB.
RESULTS
FliN from T. maritima functions in E. coli.
For the structural study and certain other experiments below we used the FliN protein from T. maritima, whereas previous information on FliN function and localization has come mainly from studies of Salmonella enterica serovar Typhimurium (19, 27, 81) or E. coli (65, 67). T. maritima is a hyperthermophile with a single polar flagellum (25). The amino acid sequences of T. maritima and E. coli FliN are 39% identical overall and 55% identical in the segment that is most important for function and whose structure is described below. To further validate the use of the T. maritima protein, we used a soft-agar swarming assay to test the ability of the T. maritima fliN gene to complement an E. coli fliN-null strain. fliN-null cells are nonflagellate, immotile, and unable to swarm in soft agar (65). When transformed with a plasmid expressing the T. maritima fliN construct, the cells swarmed at a rate that was about 20% of the wild-type rate (Fig. 2). When examined under the microscope, cells expressing T. maritima FliN showed fair motility but were more tumbly than wild-type cells. This functional complementation indicates that T. maritima FliN provides a valid structural model for the protein from E. coli.
FIG. 2.
Complementation of the fliN-null strain DFB223 by the fliN gene from T. maritima. This strain has an in-frame chromosomal deletion of fliN (65). Plasmid pPNB10 encodes residues 23 to 154 of the T. maritima FliN protein. A tryptone plate containing 0.28% Bacto agar (Difco) was spotted with 1 μl of an overnight culture of each strain and incubated at 32°C for 24 h.
Crystal structure of FliN.
A construct encompassing residues 23 to 154 of T. maritima FliN was overexpressed in E. coli and purified, and it formed crystals that diffracted to a resolution of about 4 Å. Selenomethionine-substituted protein was similarly purified and crystallized, and data were collected at three wavelengths near the selenium edge, at a resolution of 3.4 Å. Anomalous scattering from selenium was detectable, but the quality of the data was not sufficient to allow phase determination. Phases for the best 3.4-Å data set (Table 1) were eventually obtained by molecular replacement using coordinates for a T. maritima FliN structure determined by the Joint Center for Structural Genomics and released to the Protein Data Bank in November 2003 (accession code 1O6A). The FliN molecule is a dimer. Simulated-annealing omit maps gave interpretable electron density for residues 68 to 152 of chain A and residues 68 to 154 of chain B. Forty-five residues at the N terminus of each chain were not visible, presumably because they were less ordered. The structure was rebuilt and refined at a 3.4-Å resolution to an R factor (Rfree) of 22.4% (28.6%) (Table 2), with good geometry. The resulting structure is very similar to the structure of 1O6A overall, with a root mean square deviation (RMSD) of 0.9 Å for main-chain atoms in the central portion of the molecule (residues 80 to 150). Like the present structure, 1O6A did not show clear electron density for parts N terminal of residue 68.
The FliN dimer is roughly the shape of a saddle, and the overall dimensions are 60 by 35 by 30 Å (Fig. 3). The two chains in the crystallographic asymmetric unit are similarly folded (RMSD for main-chain atoms, 0.4 Å) and are related by an approximate twofold axis. Each chain contains three α helices and five β strands. The two chains intertwine to form two β barrels near the middle of the molecule, each containing strands from both subunits. The subunits are held together by extensive β-strand interactions, chiefly between the β1 strands of the two chains and between β2 of one chain and β5 of the other. Each end of the molecule is capped by a group of three helices, α2 from one subunit and α1 and α3 from the other. Helices α2 and α3 are packed against each other and against the rest of the molecule. Helix α1 is directed away from the main body of the molecule and is positioned differently in our structure than in 1O6A, implying that its location is influenced by crystal packing forces (Fig. 3B).
FIG. 3.
(A) Two views of the FliN dimer, with one chain gold and the other white. Beta strands are labeled β1 to β5, and alpha helicesare labeled α1 to α3. The dimer twofold axis is vertical in the top diagram, and the view in the lower diagram is along the twofold axis, from the top of the dimer as viewed in the top diagram. (B) FliN structure solved in this study (left), 1O6A FliN structure (right), and overlay of the two structures (middle). The view is along the twofold dimer axis, but from the direction opposite that in the lower diagram in panel A. The major difference is in the position of helix α1, as indicated. (C) Simulated annealing omit map computed by using CNS (10), with the following residues omitted: residues 103 to 105 (top strand), 126 to 129 (bottom strand), and 134 to 138 (middle strand). NCS restraints were not applied during the simulated annealing refinement. The Fo − Fc map is contoured at 3.5× RMSD.
Conserved hydrophobic residues are found in the buried core of the protein and also in a patch on the protein surface. Conservation of buried hydrophobic residues is not unusual and indicates that the various FliN homologs shown in Fig. 4 have similar folds. Several conserved hydrophobic side chains (primarily Leu85, Thr110, Val128, Val130, and Phe135) form a patch on the protein surface that is centered on the twofold axis (Fig. 5). These hydrophobic residues do not appear to have critical structural roles, and so their conservation might indicate a specific functional role for this part of the protein surface.
FIG. 4.
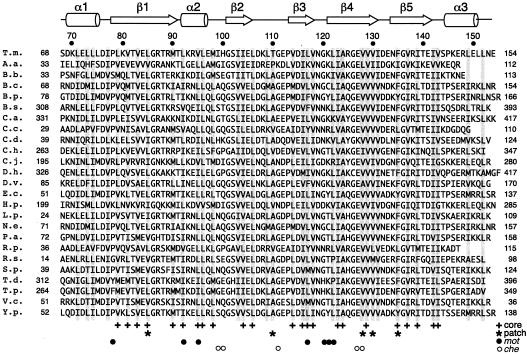
FliN sequence alignment for the part of FliN whose structure is reported here (residues 68 to the end for the T. maritima protein). The N-terminal part of the protein is more variable and is partially dispensable for function in E. coli (65). The secondary structure is indicated at the top. Gray bars in the alignment indicate positions where the hydrophobic character is conserved. At the bottom plus signs indicate residues that are mostly buried and contribute to the core of the FliN dimer, asterisks indicate residues that make major contributions to the surface hydrophobic patch, solid circles indicate the positions of mutations that give an immotile but flagellate phenotype, and open circles indicate the positions of mutations that give a motile but nonchemotactic (switching-impaired) phenotype. T.m., Thermotoga maritima; A.a., Aquifex aeolicus; B.b., Borrelia burgdorferi; B.c., Burkholderia cepacia; B.p., Bordetella pertussis; B.s., Bacillus subtilis; C.a., Clostridium acetobutylicum; C.c., Caulobacter crescentus; C.d., Clostridium difficile; C.h., Carboxydothermus hydrogenoformans; C.j., Campylobacter jejuni; D.h., Desulfitobacterium hafniense; D.v., Desulfovibrio vulgaris; E.c., Escherichia coli; H.p., Helicobacter pylori; L.p., Legionella pneumophila; N.e., Nitrosomonas europaea; P.a., Pseudomonas aeruginosa; R.p., Rhodopseudomonas palustris; R.s., Rhodobacter sphaeroides; S.p., Shewenella putrefaciens; T.d., Treponema denticola; T.p., Treponema pallidum; V.c., Vibrio cholerae; Y.p., Yersinia pestis.
FIG. 5.
Two surface representations of the FliN dimer, with the surface hydrophobic patch highlighted in yellow. Residues contributing to the patch are indicated. The views are similar to those in Fig. 3, except that they are rotated by approximately 20° about the twofold axis to reveal the concave shape of the hydrophobic patch (in the upper diagram).
Association state of FliN and a stable FliM-FliN complex.
The crystal structure shows that FliN is likely to be a dimer or a larger molecule in solution. To study the association state of FliN in solution, we performed analytical ultracentrifugation experiments with purified FliN proteins from both T. maritima and E. coli. T. maritima FliN behaved like a single species in sedimentation equilibrium experiments, with a molecular mass (30.2 ± 2 kDa) indicating a dimer (calculated dimer molecular mass, 30.2 kDa) (Fig. 6). A dimer-tetramer equilibrium model was also tested, but the fits were not significantly better and only trace amounts of tetramer were predicted (data not shown). The E. coli protein also behaved like a single species in sedimentation equilibrium experiments, with a molecular mass (59.6 ± 6 kDa) indicating a tetramer (calculated molecular mass, 59.4 kDa) (Fig. 6). In velocity-sedimentation experiments, E. coli FliN behaved like a single species, with a sedimentation coefficient of 3.1 S, which is smaller than expected for a globular tetramer but too large for a dimer (Fig. 7). Assuming a typical level of hydration (0.5 g of water per g of protein), the shape factor for the FliN tetramer is ∼1.4, indicating an elongated shape.
FIG. 6.
Equilibrium sedimentation of FliN and the FliM-FliN complex. (Top panel) Data for T. maritima FliN. The fitted lines correspond to a molecular mass of 30.2 ± 2 kDa (calculated dimer molecular mass, 30.2 kDa). The three graphs give data obtained at different radii, as indicated at the bottom. (Middle panel) Data for E. coli FliN. The fitted lines correspond to a molecular mass of 59.6 ± 6 kDa (calculated tetramer molecular mass, 59.4 kDa). (Bottom panel) Data for the T. maritima FliM-FliN complex. The fitted lines correspond to a molecular mass of 98.6 ± 3.5 kDa (calculated molecular mass for FliM1-FliN4 complex, 98.5 kDa).
FIG. 7.
Velocity sedimentation of E. coli FliN (top panel) and the T. maritima FliM-FliN complex (bottom panel). Representative absorbance traces are shown on the left, at ∼7-min intervals for FliN and at ∼11-min intervals for the FliM-FliN complex. The scans were analyzed by the method of Stafford (60) to obtain the distributions of apparent sedimentation coefficients [g(s*)] shown on the right.
FliN and FliM coassemble into nascent flagella during the early step of C-ring formation (37, 42), and the proteins were shown to bind to each other in blot overlay experiments (70) and pull-down assays (67). To examine the state of purified FliM and FliN in solution, we coexpressed T. maritima FliN and FliM in cells and purified them by column chromatography. FliM and FliN remained together through ion-exchange, hydrophobic affinity, and gel filtration steps, which resulted in a stable, soluble complex that contained only the two proteins. The good solubility of FliM and FliN together is in contrast with the solubility of FliM alone (from either T. maritima or E. coli), which forms inclusion bodies when it is overexpressed (54, 76; unpublished results with T. maritima FliM). The FliM-FliN complex had an apparent molecular mass of 140 kDa as determined by gel filtration. In sedimentation equilibrium experiments the complex behaved like a single species having a molecular mass of 98.6 ± 3.5 kDa (Fig. 6). The complex appears to be quite stable, as it did not dissociate even at protein concentrations in the nanomolar range.
The experimentally estimated molecular mass (98.6 kDa) is very close to the value expected for a FliM1-FliN4 complex (calculated molecular mass, 98.5 kDa), but it might also fit a FliM2-FliN2 complex (106 kDa), given the uncertainties. To estimate the relative levels of FliM and FliN in the complex, we quantified the proteins on Coomassie blue-stained gels (Fig. 8). The FliN/FliM ratio estimated in this way was 3.2:1. A similar experiment in which silver staining was used gave a FliN/FliM ratio of 4.2:1 (data not shown). The measured FliN/FliM ratio is thus consistent with the subunit composition FliM1-FliN4 and rules out the composition FliM2-FliN2.
FIG. 8.
Quantification of FliM and FliN proteins in the FliM-FliN complex. The gel was stained with Coomassie blue. Relative protein levels were estimated by assuming that the staining intensity of a band is proportional to the mass of protein in the band. The ratio of the slopes of the fitted lines (and thus the ratio of staining intensities for the FliN and FliM bands) is 1.3:1. Given the molecular masses of FliM and FliN (37.2 and 15.1 kDa, respectively), this corresponds to an estimated FliN/FliM ratio of 3.2:1.
The FliM-FliN complex behaved like a single species in velocity-sedimentation experiments, with a sedimentation coefficient of 5.3 S (Fig. 7). When we used the mass determined by sedimentation equilibrium and hydration like that assumed for isolated FliN, the calculated shape factor was ∼1.1, which indicates that the shape of the FliM-FliN complex is less eccentric than the shape of FliN alone.
Comparison to HrcQB: a model for the FliN tetramer.
HrcQB is a paralog of FliN that functions in the type III secretion apparatus of the phytopathogen Pseudomonas syringae (3, 16, 26). HrcQB and FliN show significant sequence similarity (see Fig. S1 in the supplemental material), and the recently reported crystal structure of the C-terminal domain of HrcQB (HrcQBC) has a fold very similar to that of FliN (16) (PDB accession code 109Y). Like FliN, the subunits of HrcQBC intertwine to form dimers, but the HrcQBC dimers are further associated into tetramers in the crystal. The association between HrcQBC dimers buries more than 1,200 Å2 of surface and is stabilized by hydrophobic interactions, hydrogen bonds between backbone segments in an antiparallel β-strand arrangement, and hydrogen bonds between side chains. The sedimentation experiments with FliN showed that it also forms tetramers, either by itself (the E. coli protein) or in a complex with FliM (the T. maritima proteins). The large shape factor of the FliN tetramer suggests that it has an elongated shape, as does the HrcQBC tetramer. The dimer-dimer interface in HrcQBC is formed in part from the hydrophobic residues Ile85, Val111, and Val113 (the residue numbers are the numbers for the full HrcQBC sequence; the numbers used in PDB entry 109Y are 44 lower). Hydrophobic character is conserved at the corresponding positions in FliN (Ile103, Val129, and Ile131 in T. maritima FliN) (Fig. 4). These correspondences suggest that the FliN tetramer may have a subunit arrangement similar to that of HrcQBC. Accordingly, we used the HrcQBC structure as a guide in docking two FliN dimers together to form a model for the tetramer.
As noted above, the dimer-dimer interface in HrcQBC is stabilized by four hydrogen bonds between backbone atoms, as well as by hydrophobic interactions. The initial FliN tetramer model was constructed by manually aligning the dimers to bring together the backbone hydrogen bonding groups (Val130 O to Asp132′ N and Ile103 O to Ile103′ N, and their symmetry-related counterparts). The structure was then energy minimized by using a utility in Swiss-PDB viewer (22). Following energy minimization, the four backbone H bonds were retained, and more than 1,600 Å2 of surface was buried, including the hydrophobic residues Ile103, Val129, and Ile131 (Fig. 9). The dimer-dimer interface of the energy-minimized structure was also stabilized by hydrogen bonds between the side chains of residues Glu105, Asp132, Arg138, and Glu105. The sequence alignment shows that these residues retain hydrogen bond donor or acceptor potential in most FliN proteins, and the corresponding residues in HrcQBC (Glu87, Glu114, and Gln120) contribute most of the interdimer hydrogen bonds in that molecule. Thus, while the details should be considered speculative, the model shows that the main features of the dimer-dimer interface in HrcQBC can be reproduced with FliN. The modeled FliN tetramer is elongated, and the approximate dimensions are 110 by 40 by 35 Å.
FIG. 9.
Model of the FliN tetramer. (A) View along the twofold axis of the tetramer. The backbone of one dimer is turquoise, and the backbone of the other dimer is pink. Hydrophobic residues at the dimer-dimer interface (and also their dimer-related counterparts away from the interface) are yellow; residues of the hydrophobic patches are yellow and space filling. (B) View from a direction perpendicular to the tetramer twofold axis (from the right side of panel A). (C) Close-up of the dimer-dimer interface and comparison to the interface observed in the structure of the HrcQBC tetramer. The direction of view is similar to that in panel B. The yellow residues are conserved hydrophobic residues in both FliN and HrcQB. The dashed lines indicate hydrogen bonds between backbone atoms that are observed in the HrcQBC crystal structure and predicted in the FliN tetramer model.
Functional importance of the hydrophobic patch.
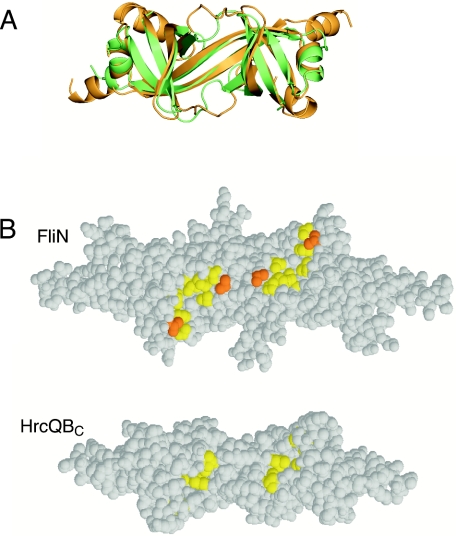
The hydrophobic patches on the dimers (Fig. 5) come together in the tetramer to form a hydrophobic cleft that is approximately 50 Å long (Fig. 10B). Although FliN and HrcQB have similar folds overall, there are clear differences in the segment that frames the hydrophobic patch (residues 107 to 114 in T. maritima FliN) (Fig. 10A). The conformation of this segment appears to be well determined by the crystal structures, because it is the same in our structure as in 1O6A, which involved different crystallization conditions and a different space group. The 107 to 114 segments in the FliN dimer are farther apart than the corresponding segments in HrcQBC, and together with the more hydrophobic character of residues 110 and 130 in FliN, this makes the hydrophobic patch larger in FliN than in HrcQB (Fig. 10B and C).
FIG. 10.
Comparison of FliN and HrcQBC structures in the region of the hydrophobic patch. (A) Ribbon diagram showing the FliN dimer (gold) superimposed on half of the HrcQBC tetramer (green). The view is along the twofold dimer axis, looking onto the hydrophobic patch. The largest differences between FliN and HrcQBC occur in the loops connecting β2 and β3 (residues 107 to 114 of FliN), which in FliN frame the hydrophobic patch. (B) The hydrophobic patch is larger in FliN than in HrcQBC. The modeled FliN tetramer and the crystal structure of the HrcQBC tetramer are shown, and the hydrophobic residues of the patch are yellow or orange. The view is along the twofold axis of the tetramer (as in Fig. 9A). Orange indicates a valine residue (Val130 in T. maritima, corresponding to Val113 in E. coli) that was mutated to aspartic acid to test the functional importance of the patch.
The strong conservation of residues that form the hydrophobic patch suggests that it is important for function. To test this proposal, we mutated the E. coli fliN gene to replace a well-conserved hydrophobic residue of the patch, Val113, with Asp. This residue corresponds to Val130 in the T. maritima protein, as indicated in Fig. 10B (orange). A plasmid encoding the fliN mutation was transformed into the fliN-null strain DFB223 (63), and motility and flagellation were examined. The mutation caused a severe reduction in swarming in soft agar at 32°C (Fig. 11). Cells taken from the edges of the swarms were highly motile when they were viewed with a microscope but swam smoothly, indicating that there was a CCW bias of the flagellar motors. Cells cultured in liquid medium (tryptone broth, 32°C) showed a delay in the onset of motility compared to wild-type cells, and staining showed that there were fewer flagella. At later stages of growth in liquid, mutant cells were highly motile but aberrantly smooth. The motility impairment was more severe when cells were grown in tryptone broth at a higher temperature. Wild-type cells cultured at 38°C exhibited fair motility at the mid-log phase, whereas most cells of the mutant were immotile and nonflagellate.
FIG. 11.

Mutation of a residue in the hydrophobic patch eliminates swarming in soft-agar tryptone plates. E. coli strain DFB223, null for fliN, was transformed with plasmids that encode either wild-type E. coli FliN (w.t.) or FliN with the mutation V113D. The plate was inoculated with 2 μl of saturated overnight cultures and incubated at 32°C for 8 h.
DISCUSSION
The flagellar C ring is a fairly large structure (in Salmonella, it is 45 nm in diameter and 15 nm high) (19, 68, 79) and contains more than 100 subunits of FliN (31). Sedimentation experiments give clues to the arrangement of FliN dimers in the flagellum. Because E. coli FliN forms a stable tetramer and T. maritima FliN forms a stable FliM1-FliN4 complex, we propose that an FliM1-FliN4 complex is the building block of the C ring. The copy number of FliM in the flagellum has been estimated to be 34 ± 6 (80, 81), and en face electron micrographs of the C ring show subunit structure with a rotational symmetry of about 34-fold, varying slightly from specimen to specimen (69, 79). A typical C ring should then contain about 34 copies of the FliM1-FliN4 unit and therefore 136 copies of FliN, in fair agreement with an experimental estimate of 111 ± 13 (81).
The crystal structure of HrcQBC provides a basis for modeling the arrangement of subunits in the FliN tetramer. Patterns of sequence conservation and similar patterns of hydrogen bonding seen in HrcQBC and the modeled FliN tetramer support the view that the FliN tetramer has an organization similar to that of HrcQBC. An elongated shape for the FliN tetramer is also supported by the large shape factor (1.4) determined in the velocity-sedimentation experiment. Although the residues at the dimer-dimer interface have dimer-related counterparts that are exposed at the ends of the tetramer, we did not see evidence of any FliN complexes larger than a tetramer. Further end-to-end association of the tetramers might be prevented by the N-terminal parts of the protein, which are sizable (more than 50 residues) but were not sufficiently ordered to be seen in either crystal structure.
The shorter dimensions of the FliN tetramer are 3 to 4 nm, which is comparable to the subunit spacing seen in end views of the C ring (4 nm). The 11-nm dimension of the tetramer is sufficient to span about three-fourths the height of the C ring (19, 31, 68, 79). We therefore propose that the FliN tetramers are arranged in the C ring with their long axes approximately parallel to the axis of the flagellum (Fig. 12). Given such a subunit arrangement, the C ring could be viewed as a fusion of two rings, an upper ring and a lower ring, each formed from a circular array of FliN dimers. Thin-metal replica images of the C rings of Salmonella have a two-layer appearance consistent with such an architecture, and partially disrupted C rings sometimes appear to lack portions of the lower layer (32). The C-ring architecture proposed here is probably applicable to a wide range of bacterial species but may not be universal. Certain species (e.g., Bacillus subtilis) use the much larger FliY protein in place of FliN (7), and their C rings might be constructed differently.
FIG. 12.
Model for the arrangement of FliN tetramers in the C ring. The orientation shown for the FliN tetramers is suggested by en face electron micrographs of the C ring that showed a ∼34-fold subunit structure and ∼4-nm subunit spacing (69, 79).
The present structural study of FliN has some bearing on FliM, because a segment in the C-terminal domain of FliM shows sequence similarity to FliN (46). Several residues are conserved as hydrophobic residues in both proteins, and the structure shows that these residues form much of the core of FliN (Fig. 4; also see Fig. S2 in the supplemental material). The secondary-structure predictions for this part of FliM agree well with the secondary structures seen in the structure of FliN, except that β1 is predicted to be interrupted by a loop in FliM (see Fig. S2 in the supplemental material). Based on the correspondence between the proteins, we propose that the C-terminal domain of FliM is folded like the C-terminal domain of FliN, except that β1 folds back onto itself and β2 and β3 are joined by a reverse turn, which allows the β barrel to be formed from a single chain rather than two chains. This segment of FliM is also the part that binds to FliN (46, 70), and its structural resemblance to FliN may be important for this binding. A similar correspondence has been noted between HrcQB and its binding partner, HrcQA (16).
FliN is essential for flagellar assembly, because deletion of the fliN gene results in a nonflagellate phenotype (65). Its functions in assembly are not easily disrupted by point mutations. Irikura et al. (27) analyzed a number of fliN mutations and found that flagellar assembly was prevented only by frameshifts or premature termination. Most missense mutations give a mot phenotype, in which flagella are assembled but do not rotate. Some rotation was restored when the mot mutant proteins were overexpressed, which suggests that the mutations decreased the level of the protein or diminished its ability to form fully functional C rings. The positions of mot mutations in the structure appear to be consistent with this suggestion (see Fig. S3 in the supplemental material). One mot mutation replaced a fairly well-conserved Gly residue (Gly120 in the T. maritima protein) whose φ and ψ angles (80° and −10°, respectively) are not as favorable for a larger side chain, one mutation introduced Pro in place of a residue (Leu117) whose φ angle (−115°) was incompatible with Pro, and another mutation replaced a fairly well-conserved Pro residue (Pro 78) in the segment that separates α1 from β1. Two mot mutations replaced nonpolar residues that are largely buried (positions 92 and 95). The remaining positions that gave a mot phenotype (positions 121 and 122) were together on the side surfaces of the tetramer, where they might participate in interactions that stabilize the C ring. One of these mutations (at position 122) was found to cause a reduction in the length of the flagellar hook (43). This might indicate that FliN has a role in binding to hook subunits, as proposed by Makishima and coworkers (43), but it is also consistent with a more general role for FliN in flagellar assembly.
Mutations that affect the CW-CCW bias of the motor (designated che, for nonchemotactic) have been reported for five positions in FliN (27). These mutations occur at the hypothesized dimer-dimer interface (residues 99 and 100 in T. maritima FliN, corresponding to residues 82 and 83 in E. coli or Salmonella) and also in the neighborhood of the hydrophobic patch (positions 111, 127, and 128 in T. maritima) (see Fig. S3 in the supplemental material). One of the mutations near the hydrophobic patch (at position 111) appears to affect not only switching but also flagellar assembly, as shown by a reduction in the length of the flagellar hooks (43). The relative scarcity of motile but nonchemotactic mutants has been taken to indicate that FliN has only a small role in switching (27). Alternatively, FliN might play a critical role in switching, but with only a small part of the protein participating directly. The positions of the che mutations on the structure suggest that the dimer-dimer interface and the hydrophobic patch could have roles in switching. A site-directed mutation in the hydrophobic patch, described below, has a phenotype consistent with this.
In a study to identify important protonatable residues, Zhou et al. replaced several conserved acidic residues in FliN with alanine and found that none were critical for flagellar assembly or function. The positions mutated were positions 72, 76, 107, 112, 115, 127, 133, and 141 (T. maritima positions), most of which are on the sides of the saddle (the faces perpendicular to the hydrophobic patch) (see Fig. S3 in the supplemental material). Although none of the alanine replacements of acidic residues prevented FliN function, many of them resulted in an increase in the level of FliN needed for optimal swarming (82). This might indicate that these residues have a role in stabilizing the protein or facilitating its incorporation into the C ring. Glu127 is near the hydrophobic patch and might have other roles; replacement of this residue with lysine gives a nonchemotactic phenotype (27).
The surface hydrophobic patch is the most conspicuous feature of FliN. In the modeled FliN tetramer, the hydrophobic patches come together to form a ca. 50-Å-long hydrophobic cleft. This hydrophobic surface feature appears to have a function specific to FliN, because it is much smaller on HrcQBC, which is otherwise fairly similar. Sequence alignments show that nonpolar character is well conserved in the residues that form the hydrophobic patch on FliN, and a Val-to-Asp mutation in the patch affected both motor switching (at 30°C) and flagellar assembly (at 38°C), so that swarming was prevented (Fig. 11). We cannot yet assign a particular function to the hydrophobic patch. The reduced flagellation and the CCW motor bias might indicate that FliN functions in both flagellar assembly and CW-CCW switching. While the switching defect is most dramatic, the hydrophobic patch is unlikely to function exclusively in switching because the residues that form it are conserved as hydrophobic residues even in Aquifex aeolicus and Buchnera, organisms that lack CheY and other proteins of the chemotactic signaling pathway (11, 64).
In the fliN mutant characterized by Vogler et al. (75), a temperature-sensitive defect in assembly was shown to result from a block in flagellar export. Further characterization of hydrophobic patch mutants should reveal whether they are defective in flagellar export or in other steps of assembly. Hydrophobic surface features that look similar are found on some small heat shock proteins that function as chaperones (34, 74), and one possibility is that FliN functions as a cochaperone for flagellar export by providing docking sites for chaperone-cargo complexes. The large structural differences between FliN and HrcQBC in the region of the hydrophobic patch (Fig. 10) are consistent with such an export function, because the virulence factor export apparatus acts on a different set of substrates and utilizes different chaperones (15).
Supplementary Material
Acknowledgments
We thank F. Whitby, H. Schubert, N.-L. Chan, and B. Howard for assistance and advice at many stages of this project, D. Mackay and G. Cortez for assistance with cloning and protein purification, J. Harmon and K. Paul for assistance with mutagenesis and swarming assays, and J. S. Parkinson for helpful comments.
This work was supported by grant R01-GM61145 and training grant 5T32-GM08537 from the National Institutes of Health. The protein-DNA core facility at the University of Utah receives support from the National Cancer Institute (grant 5P30 CA42014). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by the National Institutes of Health National Center for Research Resources Biomedical Technology Program and the National Institute of General Medical Sciences.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aizawa, S.-I. 2001. Flagella, p. 155-175. In Bacterial structure. Academic Press, New York, N.Y.
- 2.Aldridge, P., J. E. Karlinsey, and K. T. Hughes. 2003. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 49:1333-1345. [DOI] [PubMed] [Google Scholar]
- 3.Alfano, J. R., and H. S. Kim. 1997. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in an hrp-dependent manner within tobacco cells. Mol. Plant-Microbe Interact. 10:580-588. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, J. C. Q., and C. Hughes. 2000. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8:202-204. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, J. C. Q., and C. Hughes. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol. Microbiol. 39:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, D. S., and G. W. Ordal. 1992. Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol. Microbiol. 6:2715-2723. [DOI] [PubMed] [Google Scholar]
- 8.Blocker, A., K. Komoriya, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 11.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olson, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 12.Delano, W. L. 2002. The PYMOL molecular graphics system. [Online.] http://www.pymol.org.
- 13.Doering, D. S. 1992. Functional and structural studies of a small F-actin binding domain. Ph.D. thesis. Massachusetts Institute of Technology, Cambridge, Mass.
- 14.Dreyfus, G., A. W. Williams, I. Kawagishi, and R. M. Macnab. 1993. Genetic and biochemical analysis of Salmonella typhimurium FliI, a flagellar protein related to the catalytic subunit of the FoF1 ATPase and to virulence proteins of mammalian and plant pathogens. J. Bacteriol. 175:3131-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evdokimov, A. G., J. Phan, J. E. Tropea, K. M. Routzahn, H. K. Peters, M. Pokross, and D. S. Waugh. 2003. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat. Struct. Biol. 10:789-793. [DOI] [PubMed] [Google Scholar]
- 16.Fadouloglou, V. E., A. P. Tampakaki, N. M. Glykos, M. N. Bastaki, J. M. Hadden, S. E. Phillips, N. J. Panopoulos, and M. Kokkinidis. 2004. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. USA 101:70-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, F., and R. M. Macnab. 1996. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271:31981-31988. [DOI] [PubMed] [Google Scholar]
- 18.Francis, N. R., V. M. Irikura, S. Yamaguchi, D. J. DeRosier, and R. M. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 20.Fraser, G. M., J. C. Q. Bennett, and C. Hughes. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569-580. [DOI] [PubMed] [Google Scholar]
- 21.Grunenfelder, B., S. Gehrig, and U. Jenal. 2003. Role of the cytoplasmic C terminus of the FliF motor protein in flagellar assembly and rotation. J. Bacteriol. 185:1624-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PDB viewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 23.Heimbrook, M. E., W. L. L. Wang, and G. Campbell. 1986. Easily made flagella stains, abstr. R-22, p. 240. Abstr. 86th Annu. Meet. Am. Soc. Microbiol. 1986. American Society for Microbiology, Washington, D.C.
- 24.Homma, M., S.-I. Aizawa, G. E. Dean, and R. M. Macnab. 1987. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 84:7483-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber, R., T. A. Langworthy, H. Koenig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 26.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, C. J., and R. M. Macnab. 1990. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J. Bacteriol. 172:1327-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 30.Katayama, E., T. Shiraishi, K. Oosawa, N. Baba, and S.-I. Aizawa. 1996. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J. Mol. Biol. 255:458-475. [DOI] [PubMed] [Google Scholar]
- 31.Khan, I. H., T. S. Reese, and S. Khan. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 89:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan, S., R. Zhao, and T. S. Reese. 1998. Architectural features of the Salmonella typhimurium flagellar motor switch revealed by disrupted C-rings J. Struct. Biol. 122:311-319. [DOI] [PubMed] [Google Scholar]
- 33.Kihara, M., G. U. Miller, and R. M. Macnab. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, K. K., R. Kim, and S. H. Kim. 1998. Crystal structure of a small heat-shock protein. Nature 394:595-599. [DOI] [PubMed] [Google Scholar]
- 35.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93-134. [DOI] [PubMed] [Google Scholar]
- 36.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S.-I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 37.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S.-I. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049-1067. [DOI] [PubMed] [Google Scholar]
- 39.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. D. Harding, A. J. Rowe, and J. C. Horton (ed.), Ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 40.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propeller and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 43.Makishima, S., K. Komoriya, S. Yamaguchi, and S.-I. Aizawa. 2001. Length of the flagellar hook and the capacity of the type III export apparatus. Science 291:2411-2413. [DOI] [PubMed] [Google Scholar]
- 44.Marykwas, D. L., and H. C. Berg. 1996. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol. 178:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564-576. [DOI] [PubMed] [Google Scholar]
- 46.Mathews, M. A. A., H. L. Tang, and D. F. Blair. 1998. Domain analysis of the FliM protein of Escherichia coli. J. Bacteriol. 180:5580-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minamino, T., and R. M. Macnab. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37:1494-1503. [DOI] [PubMed] [Google Scholar]
- 49.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov, G. N., A. A. Vagin, A. Lebedev, K. S. Wilson, and E. J. Dodson. 1999. Efficient anisotropic refinement of macromolecuular structures using FFT. Acta Crystallogr. Sect. D 55:247-255. [DOI] [PubMed] [Google Scholar]
- 51.Namba, K., I. Yamashita, and F. Vonderviszt. 1989. Structure of the core and central channel of bacterial flagella. Nature 342:648-654. [DOI] [PubMed] [Google Scholar]
- 52.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. Sect. D 57:1367-1372. [DOI] [PubMed] [Google Scholar]
- 53.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelbert, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 54.Oosawa, K., T. Ueno, and S.-I. Aizawa. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otwinowski, Z. 1993. Oscillation data reduction program. SERC Daresbury Laboratory, Warrington, United Kingdom.
- 56.Rost, B., and C. Sander. 1993. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 57.Sayle, R., and E. J. Milner-White. 1995. RasMol: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 58.Schenk, P. M., S. Baumann, R. Mattes, and H. H. Steinbiss. 1995. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare Arg tRNAs. BioTechniques 19:196-198:200. [PubMed] [Google Scholar]
- 59.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stafford, W. F. 1992. Boundary analysis in sedimentation transport experiments—a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 203:295. [DOI] [PubMed] [Google Scholar]
- 61.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki, T., T. Iino, T. Horiguchi, and S. Yamaguchi. 1978. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J. Bacteriol. 133:904-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki, T., and Y. Komeda. 1981. Incomplete flagellar structures in Escherichia coli mutants. J. Bacteriol. 145:1036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endsymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 65.Tang, H., S. Billings, X. Wang, L. Sharp, and D. F. Blair. 1995. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J. Bacteriol. 177:3496-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang, H., and D. F. Blair. 1995. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J. Bacteriol. 177:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang, H., T. F. Braun, and D. F. Blair. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209-221. [DOI] [PubMed] [Google Scholar]
- 68.Thomas, D., D. G. Morgan, and D. J. DeRosier. 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J. Bacteriol. 183:6404-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toker, A. S., and R. M. Macnab. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273:623-634. [DOI] [PubMed] [Google Scholar]
- 72.Ueno, T., K. Oosawa, and S.-I. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 73.Vagin, A., and A. Teplyakov. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. Sect. D 56:1622-1624. [DOI] [PubMed] [Google Scholar]
- 74.van Montfort, R. L., E. Basha, K. L. Friedrich, C. Slingsby, and E. Vierling. 2001. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 8:1025-1030. [DOI] [PubMed] [Google Scholar]
- 75.Vogler, A. P., M. Homma, V. M. Irikura, and R. M. Macnab. 1991. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J. Bacteriol. 173:3564-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Welch, M., K. Oosawa, S.-I. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaguchi, S., H. Fujita, A. Ishihara, S.-I. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young, H. S., H. Dang, Y. Lai, D. J. DeRosier, and S. Khan. 2003. Variable symmetry in Salmonella typhimurium flagellar motors. Biophys. J. 84:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao, R., C. D. Amsler, P. Matsumura, and S. Khan. 1996. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J. Bacteriol. 178:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao, R., N. Pathak, H. Jaffe, T. S. Reese, and S. Khan. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195-208. [DOI] [PubMed] [Google Scholar]
- 82.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.