Abstract
We report here on the isolation and primary characterization of the yohM gene of Escherichia coli. We show that yohM encodes a membrane-bound polypeptide conferring increased nickel and cobalt resistance in E. coli. yohM was specifically induced by nickel or cobalt but not by cadmium, zinc, or copper. Mutation of yohM increased the accumulation of nickel inside the cell, whereas cells harboring yohM in multicopy displayed reduced intracellular nickel content. Our data support the hypothesis that YohM is the first described efflux system for nickel and cobalt in E. coli. We propose rcnA (resistance to cobalt and nickel) as the new denomination of yohM.
Nickel and cobalt are both required as trace elements in prokaryotes to fulfill a variety of metabolic functions, but high intracellular concentrations of these transition metals are toxic. One of the strategies evolved by bacteria to prevent damage is to export excess metal by efflux systems. Plasmid-borne determinants responsible for nickel and/or cobalt resistance have been described for the heavy-metal-resistant bacterium Ralstonia metallidurans (11, 15), among which are members of the resistance-nodulation-cell division superfamily: the best-characterized CzcCBA (cobalt-zinc-cadmium) three-component cation antiporter (14) and the homologous CnrCBA (cobalt-nickel resistance) (10) and NccCBA (nickel-cobalt-cadmium resistance) (18) efflux systems. Moreover, cobalt can be extruded from the cytoplasm by the cation diffusion facilitator CzcD of R. metallidurans at the expense of the proton motive force or a potassium gradient (15). Cobalt may also be a substrate of Zn-CPx-type ATPases, as in Helicobacter pylori (8). There is no evidence for the transport of nickel by one of these two modes of efflux. Instead, this metal can be transported outside the cytoplasm by NreB from R. metallidurans (7) or NrsD from Synechocystis sp. strain PCC 6803 (6), which are members of the major facilitator superfamily and which each exhibit 12 putative transmembrane helices and a histidine-rich carboxy terminus contributing to nickel resistance.
In Escherichia coli, anaerobic hydrogenase isoenzymes and urease (in ureolytic strains) require incorporation of nickel to become active (12). Complex assembly processes involve accessory proteins, namely, HypB, implicated in nickel insertion into hydrogenase, and UreE, which delivers nickel to urease. HypB and UreE are well conserved among bacteria apart from a terminal histidine-rich stretch whose function would be nickel storage and which is absent in E. coli proteins (3, 5). In order to gain insights into nickel trafficking and, more precisely, to find proteins that would be involved in nickel resistance, we searched the E. coli genome database with a query based on a consensus alignment of the UreE and HypB histidine-rich variants. The best returned hit was yohM, whose product bears a histidine-rich domain in its center. The aim of the present work is to demonstrate the implication of yohM in nickel and cobalt trafficking in E. coli.
Inactivation of yohM confers sensitivity to nickel and cobalt.
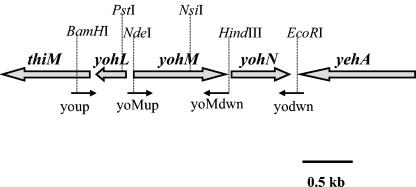
The yohM gene was identified because its product contains a remarkable histidine-rich loop (see below and Fig. 4, top). The yohM gene is surrounded upstream by yohL, which is divergently transcribed, and downstream by yohN (Fig. 1). The whole region was amplified from MC4100 chromosomal DNA by using the youp and yodwn primers, and the resulting 2,003-bp BamHI-EcoRI fragment was cloned into a pUC18 vector (19), resulting in plasmid pAR123. A yohM insertional mutant, in which a uidA-Kanr cassette derived from pUIDK11 (2) was inserted into yohM at the NsiI site, was then constructed. The transcriptional yohM-uidA fusion was recombined back to the chromosome of recBC sbcBC strain JC7623 (17) and was further moved into the wild-type (wt) strain MC4100 (laboratory collection) via P1 phage transduction to obtain strain ARY023. To perform complementation studies, yohM alone was cloned. For that purpose, a DNA fragment amplified with the youp and yoMdwn primers was digested by PstI and HindIII and then introduced into pUC18, resulting in pAR020.
FIG. 4.
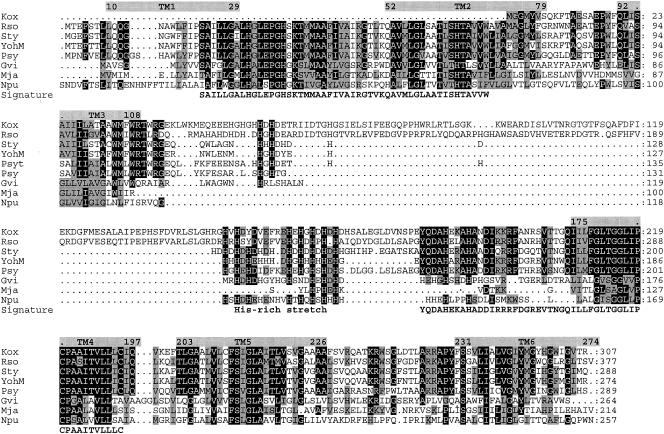
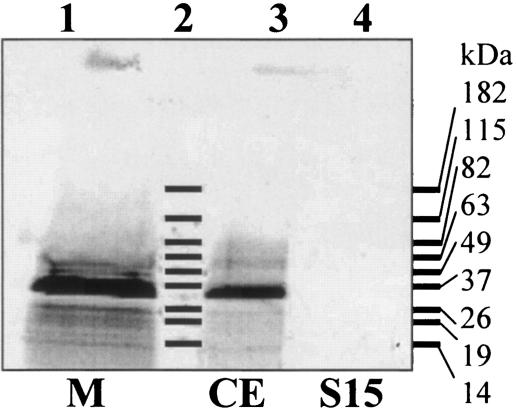
Sequence alignment and topology of YohM and putative open reading frames. (Top) YohM, E. coli (accession number P76425); Sty, Salmonella enterica serovar Typhimurium LT2 (accession number NP_461941.1); Psyt, Pseudomonas syringae pv. tomato strain DC3000 (AAO57731.1); Psy, Pseudomonas syringae pv. syringae B728a (ZP_00127113.2); Kox, Klebsiella oxytoca, NirC (AAR82965.1); Rso, Ralstonia solanacearum GMI1000 (NP_522113.1); Gvi, Gloeobacter violaceus PCC 7421 (NP_926273.1); Npu, Nostoc punctiforme (ZP_00108926.1); and Mja, Methanocaldococcus jannaschii DSM 2661 (NP_248085.1). Amino acid residues at any position that are identical in at least 60% of the sequences throughout the alignment are shaded in black, and residues exhibiting similarity in at least 60% or more of the sequences are shaded in grey. Hydropathy profile analysis predicted six transmembrane helices (grey rectangles above sequences). Numbers indicate coordinates of the amino acids flanking each transmembrane segment (TM). TM3 and TM4 are separated by a 70-amino-acid-long domain encompassing the histidine-rich region of YohM. Signature line indicates the two motifs which are signatures of the YohM family; these motifs were generated by using the MEME and MAST programs (1). (Bottom) In vivo specific labeling of YohM. Strain B834 harboring pAR02T (yohM in pET30 [Novagen]) was grown in the presence of [35S]Met. Cellular fractions were analyzed by SDS-15% PAGE and autoradiography. Lane 1, membrane fraction (M); lane 2, molecular weight; lane 3, crude extract (CE); and lane 4, soluble fraction (S15).
FIG. 1.
Genetic organization of the yohLMN cluster of E. coli. Large grey arrows indicate the transcriptional orientation of the respective genes. Thin black arrows represent the primers used for gene amplification. Restriction sites relevant to this study are indicated by dashed lines. thiM (SwissProt accession number P76423), hydroxyethyl thiazole kinase; yohL (P76424), yohM (P76425), yohN (P76426), and yehA (P33340), unknown function.
To assess whether yohM could be responsible for some metal resistance in E. coli and to determine the nature of the metals to which yohM would be sensitive, a plate sensitivity assay was carried out. Wild-type strain MC4100 was sensitive to all of the tested metals except manganese (Table 1). Among them, only nickel and cobalt promoted increased growth inhibition for the yohM mutant ARY023, since the zone of inhibition was increased by 38% for nickel and 30% for cobalt in comparison with wt strain MC4100. Furthermore, when expressed in trans from the multicopy plasmid pAR020, yohM conferred a marked enhancement of nickel and cobalt resistance to the host mutant strain (two- to threefold). The presence of plasmid-borne yohM did not affect the response to the other tested metals. Thus, yohM is shown to be a nickel and cobalt resistance gene in E. coli.
TABLE 1.
Metal sensitivity of the yohM mutant of E. coli
| Metal | Zone of inhibition (mm)a
|
||
|---|---|---|---|
| MC4100 (wt) | ARY023 (yohM) | ARY023/pAR020 (yohM/yohM+) | |
| Ni | 16 | 22 | 7 |
| Co | 17 | 22 | 5 |
| Cu | 5 | 5 | 5 |
| Zn | 4 | 4 | 4 |
| Mn | 0 | 0 | 0 |
| Cd | 14 | 14.5 | 14 |
Cells (104) were spread on N medium glucose agar plates (16). The zone of inhibition (in millimeters) around a filter disk containing 20 μl of a 5 mM solution of either NiCl2, CoCl2, CuSO4, ZnSO4, MnSO4, or CdCl2 was measured after a 24-h aerobic incubation at 37°C. Similar results were obtained after 24 h at 37°C under anaerobic conditions (GasPak jar).
Determination of nickel and cobalt MICs.
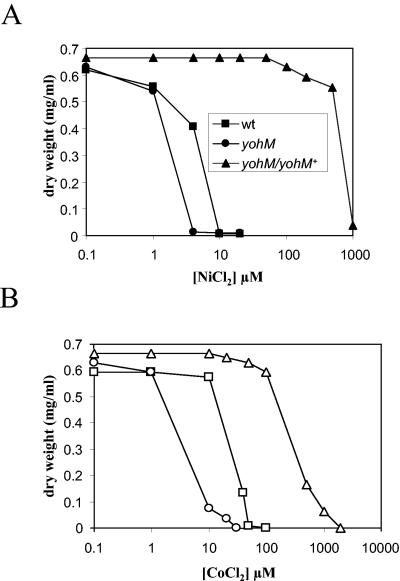
The toxic effect of nickel and cobalt on the wt and mutant strains was further assayed by measuring the final optical density at 600 nm (OD600) of a 12-h culture in minimal medium supplemented with NiCl2 or CoCl2. In agreement with the results of the metal sensitivity plate assay, the yohM strain displayed higher nickel and cobalt sensitivities (Fig. 2). More precisely, growth arrest occurred at 4 μM NiCl2 and 30 μM CoCl2 for ARY023 (yohM) compared with 10 μM NiCl2 and 50 μM CoCl2 for MC4100 (wt). Interestingly, in the mutant transformed with pAR020 (yohM+), the wild-type resistance levels were not only recovered but greatly enhanced, as in this case, ARY023/pAR020 was able to resist nickel or cobalt concentrations 100-fold higher. This result might be explained by two parameters, the high number of copies of vector pUC18 harboring yohM and the participation of the pUC18 lac promoter in yohM transcription, given that in the recombinant plasmid, yohM and Plac are in the same orientation.
FIG. 2.
Effect of nickel and cobalt on growth. Overnight cultures of MC4100 (wt) (squares), ARY023 (yohM::uidA) (circles), and ARY023/pAR020 (yohM::uidA/yohM+) (triangles) in minimal medium supplemented with 0.4% glucose were diluted into 1:100 fresh identical medium with the indicated concentrations of NiCl2 (A) or CoCl2 (B). Cell growth was monitored as the OD600 after 12 h of incubation at 37°C, with shaking, and converted to BDW. Experiments were performed in triplicate, and the values given are averages.
The yohM gene is induced by nickel and cobalt.
To analyze the metal-dependent expression of yohM, the transcriptional yohM-uidA fusion carried by ARY023 was used. After 6 h of growth in rich medium in the absence or in the presence of 0.5 mM CuSO4, ZnCl2, CdCl2, CoCl2, or NiCl2, β-glucuronidase activity was assayed. In the absence of added metals, there was almost no expression of the yohM::uidA fusion, as an activity of 7 (±2) nmol of paranitrophenol (PNP) · min−1 · mg−1 of bacterial dry weight (BDW) was measured. Addition of Co2+ or Ni2+ strongly induced the expression of the fusion, as activities of 220 (±20) and 240 (±25) nmol of PNP · min−1 · mg−1 of BDW, respectively, were recorded. Interestingly, the addition of either Cd2+, Cu2+, or Zn2+ had no effect on the transcription of yohM. Thus, the expression of yohM is specifically induced by nickel or cobalt and yohM is solely expressed when these metals are present, strongly suggesting that the function of yohM is to detoxify the cell with regard to nickel or cobalt.
yohM encodes a nickel-cobalt efflux system in E. coli.
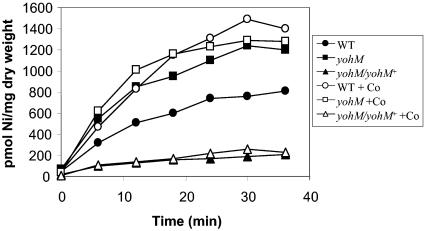
The preceding results have clearly shown that yohM confers increased resistance to nickel or cobalt in E. coli. This suggests that YohM can function as an efflux system. In such a case, the concentration of cytosolic metal ions should be higher in the sensitive cells (yohM) than in the resistant cells (wt). Alternatively, resistance could be the result of a storage mechanism, i.e., binding of the metal by the histidine-rich loop, and this would result in an increased concentration of metal ions in the resistant cells. The in vivo activity of YohM was monitored by using a 63Ni2+ uptake assay as described previously (13). In favor of the first hypothesis, the yohM mutant accumulated nearly twice the level of nickel as the wild type (Fig. 3, filled symbols). This finding was strongly reinforced by the measurements recorded from the sensitive strain complemented by plasmid-borne yohM, which contained less than one-fifth of the wild-type content. To assess whether YohM was also a cobalt efflux system, a nickel uptake assay was performed in the presence of 5 μM 63Ni and a large excess of cold cobalt (50 μM). If both cobalt and nickel compete for the same efflux system, then the concentration of nickel in the wild-type strain should be significantly higher when cobalt is present than when it is absent, whereas no change is expected for the yohM mutant. Indeed, the curve obtained for the wild-type strain in the competition assay reflected an increased intracellular nickel accumulation, which reached a level similar to that found in the yohM mutant (Fig. 3, open symbols). The latter displayed no significant change between the two assays. Likewise, for the yohM mutant harboring the plasmid-borne yohM gene, the curves obtained in the presence and absence of cobalt are superimposable, indicating the huge efflux capacity of the overexpressed system.
FIG. 3.
63Ni uptake. MC4100 (wt) (circles), ARY023 (yohM::uidA) (squares), and ARY023/pAR020 (yohM::uidA/yohM+) (triangles) cells were grown in Luria-Bertani medium in the presence of 0.5 mM NiCl2 at 37°C under aerobic conditions until mid-log phase (OD600 = 0.6), harvested, and then washed once with buffer consisting of 66 mM KH2PO4-K2HPO4 (pH 6.8) and 0.4% glucose. Cells were resuspended in the same buffer with a fivefold concentration factor. The uptake assay was performed in the presence of 5 μM 63NiCl2 (filled symbols) or 5 μM 63NiCl2 and 50 μM CoCl2 (open symbols). Aliquots (100 μl) were filtered at the indicated times. The intracellular concentration of 63Ni per milligram of bacterial dry weight was determined.
YohM is a histidine-rich membrane protein.
yohM is predicted to encode a 274-amino-acid protein with a molecular mass of 30,419 Da and to possess a remarkable histidine-rich region (amino acids 121 to 146) containing 17 histidines, 3 aspartates, and 3 glutamates out of 26 residues (Fig. 4, top panel). Using yoMup and yoMdwn primers, yohM was cloned into the expression vector pET30 at the NdeI and HindIII sites, resulting in plasmid pAR02T. YohM was specifically labeled with [35S]methionine in vivo by use of the T7 RNA polymerase system and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). One major band with an estimated mass of 32 kDa was observed in the crude extracts, in close agreement with the calculated mass (Fig. 4, bottom panel, lane 3). The separation of crude extract into membrane and soluble fractions allowed us to assign [35S]Met-labeled YohM to the membrane fraction (Fig. 4, bottom panel, lanes 1 and 4). Topology prediction, using the TM-Pred program (9), predicted YohM to be an inner membrane protein encompassing six transmembrane domains (Fig. 4, top panel). However, the proper orientation of the protein remains to be elucidated, as in silico analysis provided no clear indication of the orientation of the histidine-rich loop, which could be either periplasmic or cytoplasmic. When used as a probe for a BLAST query against the nonredundant database, YohM showed highest similarities with uncharacterized putative proteins from different species of alpha, beta, and gamma proteobacteria, cyanobacteria, and archaea (Fig. 4, top panel). A striking feature is the existence of the histidine-rich loop in all of them, as the BLAST default settings exclude this form of repeated residues from the query because they are considered low-complexity segments; these sequences were not retrieved because they possess a histidine stretch. When looking for conserved regions which could serve as signatures of the YohM family, two motifs were defined (Fig. 4, top panel). Now considering the residues known to be putative nickel or cobalt ligands and located outside the histidine-rich region, nine of them can be highlighted because of their strict or strong conservation. With regard to the YohM sequence, these residues are H27, H33, H63, H121, H123, H153, H157, D160, and C187. Interestingly, all of these residues are present in one or the other signature motif.
From a functional point of view, YohM resembles NrsD from Synechocystis sp. strain PCC 6803 (6) and NreB from R. metallidurans (7). Indeed, all are membrane-bound proteins which possess a histidine-rich domain. They belong to the major facilitator superfamily and are strongly suggested to be responsible for nickel resistance by an efflux mechanism. However, in contrast to NreB and NrsD, which are predicted to contain 12 transmembrane helices and histidine-rich C termini, YohM is supposed to contain 6 transmembrane segments and a histidine-rich domain located in the center of the polypeptide. Moreover, YohM transports cobalt in addition to nickel, which is the sole metal transported by NreB and NrsD. YohM and similar proteins can be very partially aligned with members of the nickel cobalt transporter family (NiCoT), which are nickel uptake permeases (4) (data not shown). Nevertheless, YohM does not harbor the NiCoT signature present in the second transmembrane helix of these eight-helix permeases; YohM thus does not belong to the NiCoT family.
In conclusion, YohM seems to be the first reported member of a new class of nickel and cobalt exporters, and we propose to assign it the new designation rcnA, corresponding to resistance to cobalt and nickel.
Acknowledgments
This work was supported by a BQR grant from the National Institute of Applied Sciences Lyon and by an Environmental Nuclear Toxicology grant from the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 2.Bardonnet, N., and C. Blanco. 1992. uidA-antibiotic resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 93:243-248. [DOI] [PubMed] [Google Scholar]
- 3.Brayman, T. G., and R. P. Hausinger. 1996. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J. Bacteriol. 178:5410-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eitinger, T., and M. A. Mandrand-Berthelot. 2000. Nickel transport systems in microorganisms. Arch. Microbiol. 173:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Fu, C., J. W. Olson, and R. J. Maier. 1995. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc. Natl. Acad. Sci. USA 92:2333-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Dominguez, M., L. Lopez-Maury, F. Florencio, and J. Reyes. 2000. A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grass, G., B. Fan, B. Rosen, K. Lemke, H. Schlegel, and C. Rensing. 2001. NreB from Achromobacter xylosoxidans 31A is a nickel-induced transporter conferring nickel resistance. J. Bacteriol. 183:2803-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrmann, L., D. Schwan, R. Garner, H. L. Mobley, R. Haas, K. P. Schafer, and K. Melchers. 1999. Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol. Microbiol. 33:524-536. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. 373:166. [Google Scholar]
- 10.Liesegang, H., K. Lemke, R. Siddiqui, and H. Schlegel. 1993. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 175:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mergeay, M., S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie, and R. Wattiez. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 12.Mulrooney, S. B., and R. P. Hausinger. 2003. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27:239-261. [DOI] [PubMed] [Google Scholar]
- 13.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 14.Nies, D. 1995. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 177:2707-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nies, D. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 16.Park, M. H., B. B. Wong, and J. E. Lusk. 1976. Mutants in three genes affecting transport of magnesium in Escherichia coli: genetics and physiology. J. Bacteriol. 126:1096-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandler, S. J., and A. J. Clark. 1994. RecOR suppression of recF mutant phenotype in Escherichia coli K-12. J. Bacteriol. 176:3661-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt, T., and H. G. Schlegel. 1994. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J. Bacteriol. 176:7045-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]