Abstract
The bacterial cell division protein FtsZ assembles into straight protofilaments, one subunit thick, in which subunits appear to be connected by identical bonds or interfaces. These bonds involve the top surface of one subunit making extensive contact with the bottom surface of the subunit above it. We have investigated this interface by site-directed mutagenesis. We found nine bottom and eight top mutants that were unable to function for cell division. We had expected that some of the mutants might poison cell division substoichiometrically, but this was not found for any mutant. Eight of the bottom mutants exhibited dominant negative effects (reduced colony size) and four completely blocked colony formation, but this required expression of the mutant protein at four to five times the wild-type FtsZ level. Remarkably, the top mutants were even weaker, most showing no effect at the highest expression level. This suggests a directional assembly or treadmilling, where subunit addition is primarily to the bottom end of the protofilament. Selected pairs of top and bottom mutants showed no GTPase activity up to 10 to 20 μM, in contrast to the high GTPase activity of wild-type FtsZ above 1 μM. Overall, these results suggest that in order for a subunit to bind a protofilament at the 1 μM Kd for elongation, it must have functional interfaces at both the top and bottom. This is inconsistent with the present model of the protofilament, as a simple stack of subunits one on top of the other, and may require a new structural model.
The bacterial cell division protein FtsZ assembles in vitro into protofilaments that are one subunit thick (30). These protofilaments can, under certain conditions, further associate into sheets and bundles (10, 12, 39), but the single protofilament seems to be the basic assembly unit. (While archaeal FtsZ forms two-stranded protofilaments and larger bundles, it also forms protofilaments that appear to be single stranded [26].) FtsZ is a prokaryotic homolog of eukaryotic tubulin (15, 24), which also forms protofilaments. The atomic structure of the tubulin protofilament, including the interface between subunits, is provided in the structure from electron crystallography (23, 24). The crystal structure of FtsZ did not show protofilaments, but electron microscopy suggests that the protofilament structure is very similar to tubulin. A recent crystal structure of a dimer of Methanococcus FtsZ showed contacts very similar to those in the tubulin protofilament (25). Both protofilaments are straight and have zero rotation of subunits and a similar subunit spacing of 4.3 to 4.1 nm (10, 16).
The tubulin protofilament is a linear strand of subunits connected by longitudinal bonds. Each longitudinal bond is an interface of the plus end of one subunit and the minus end of the subunit above it. The GTP bound to the plus end is sandwiched in this interface. Amino acids that make contact across the interface have been identified for α- and β-tubulin (23). Many of the contacting amino acids are conserved between tubulin and FtsZ, including the synergy or T7 loop on the minus end (NxDFxD in most FtsZ), which contacts the GTP of the subunit below it. A number of amino acids on the plus end are also conserved between tubulin and FtsZ (see Table 1, below, for the correspondence of amino acids in FtsZ and tubulin). Other FtsZ residues likely to be in the interface can be identified by their proximity to the GTP on the plus end or to the synergy loop on the minus end.
TABLE 1.
Complementation and dominant negative effects of longitudinal interface mutants
| FtsZ protein | Tubulin aaa | Compl. on plates | Compl. in liquid culture | Dom. neg. small coloniesb | Dom. neg. no coloniesc | GTP/FtsZd | GTPase Cc(μM)e | GTPase FtsZ −1 min−1f |
|---|---|---|---|---|---|---|---|---|
| Wild-type E. coli FtsZ | Yes | No | NA | 1.05 | 1.0 | 14.5 | ||
| Top mutants | ||||||||
| G21K | αβγ Q11 C | Yes | Yes | No | NA | 0.39 | ||
| N24K | αβγ Q15 | No | No | No | NA | 0.91 | ? | 0.17 |
| D45A | αβγ E71 | No | No | 5× | —g | |||
| Q47K | β G73 C | No | No | No | NA | 1.19 | ? | 0.5 |
| L68C | Not clear | Yes | Yes | No | NA | 1.3 | 2.0 | |
| L68D | Not clear | Yesh | Yes | No | NA | 0.56 | ? | 0.47 |
| L68W | Not clear | Yes | Yes | No | NA | 0.66 | ≈0.2 | 7.0 |
| G103K | αβγ S140 | No | No | No | NA | 0.20 | ? | 0.16 |
| G105S(Z84) | αβγ G142 | No (ts) | No (ts) | 0.23 | ? | 4.0 | ||
| E138K | β V/T177 C | No | No | No | NA | |||
| R142D | β D179 C | No | No | No | NA | |||
| F182C | αβγ F/Y224 C | Yes | Yes | No | NA | 0.37 | 5 | 5.5 |
| F182D | αβγ F/Y224 C | No | No | No | NA | 0.04 | ? | 0.14 |
| N186K | αβγ N228 | No | Yesi | No | NA | 0.66 | 1 | 3.7 |
| Bottom mutants | ||||||||
| D96A | Not clear | No | No | 4× | —g | 0.71 | ≈0.5 | 2.9 |
| N207C | αβγ N249 C | No | No | 3× | 5× | 0.74 | ? | 0.27 |
| D209A | αβγ D251 C | No | No | 4× | —g | 0.82 | ? | 0.17 |
| D209C | αβγ D251 C | No | No | No | NA | 1.06 | ? | 0.45 |
| D209K | αβγ D251 C | No | No | 3× | 4× | 0.01 | ? | 0.36 |
| F210A | αβγ L252 | No | No | 3× | 5× | 0.16 | 1 | 1.7 |
| D212A | α E254 C | No | No | 4× | —g | 0.88 | 1 | 0.16 |
| D212G(Z2) | No (ts) | No (ts) | 4× | —g | 0.78 | 2 | 0.20 | |
| D212C | No | No | 3× | —g | 0.27 | ? | 0.35 | |
| RLD271-3 | α VPK324-6 C | |||||||
| AKK | No | No | 1× | 4× | ||||
| D273K | α K326 C | Yes | No | NA | ||||
| Other species | ||||||||
| Synechocystis sp. FtsZ | No | 1× | —g | |||||
| B. subtilis FtsZ | No | 4× | —g | |||||
| M. pneumoniae FtsZ | No | No | NA |
The corresponding tubulin amino acid from the structure-based sequence alignment of Nogales et al. (22), with some adjustments to better match the structures. Tubulin amino acid numbers correspond to Nogales et al.'s numbering, which is correct for pig α-tubulin but counts two gaps in pig β-tubulin. αβγ means that this amino acid is found in α-, β-, and γ-tubulin. β or α indicates that this amino acid is found in β- or α-tubulin, which is the corresponding interdimer surface for top and bottom, respectively. C indicates that it was identified by Nogales et al. (23) as a contact residue within the heterodimer.
“Dom. neg. small colonies” indicates the expression level of mutant relative to the endogenous FtsZ (from Fig. 2) that first causes colonies to be visibly smaller than wild type. “No” means no negative effects were seen at any expression level. Note that there is some concern about these numbers, since significant effects were seen for small increases in expression level, over the range three to five times the wild type. Expression levels correspond to the following arabinose concentrations: 1×, 0.00015%; 3×, 0.0005%; 4×, 0.0015%; 5×, 0.005% and above.
“Dom. neg. no colonies” indicates the expression level that completely suppressed colony formation in the plate assay. NA, not applicable, indicates that no dominant negative effects were seen at any concentration.
The ratio of bound GTP to FtsZ is given.
Cc is the apparent critical concentration, below which GTP hydrolysis was negligible. “?” means that there was no evidence for a Cc. Either the line extrapolated close to zero, or the activity was weak overall and the extrapolation was not convincing.
This activity is the slope of the line above the Cc (Fig. 5).
Colonies became progressively smaller as expression increased from 0.0015% arabinose to 0.05% arabinose; colony number was unchanged.
L68D colonies appeared only after 2 days.
N186K grew for extended periods in liquid culture with smaller serial dilution, but would not grow when diluted more than 1/10,000.
The interface corresponding to the longitudinal bond has at least two important functions. First, it must provide free energy of association to keep the subunits bound to each other. This is achieved by multiple contacts across the interface. Second, it must carry out GTP hydrolysis. The synergy loop appears to provide key catalytic residues for hydrolysis, and hydrolysis is thought to require the formation of the interface to bring these residues in contact with the GTP of the subunit below (9, 18, 22, 34).
Mutations of amino acids in the protofilament bond interface could provide new insight into protofilament assembly and dynamics. We were especially interested in looking for mutants that had dominant negative effects. We expected that some of these might poison Z-ring assembly substoichiometrically, and this might give an indication of the size of the subassemblies in the Z-ring. For this we needed to develop a system for varying and quantitating the expression level of the mutated FtsZ relative to that of the endogenous wild-type FtsZ.
In the present study we prepared 11 mutants of the bottom interface and 13 mutants of the top interface and characterized them in vivo for complementation and dominant negative effects. We have determined the GTPase activity of each mutant and have examined the GTPase activity of selected pairs of bottom and top mutants. The results provide several novel insights into the interface of the protofilament bond and several surprises that will motivate new experiments and models for the function of the Z-ring.
MATERIALS AND METHODS
Strains and mutants.
Mutations in ftsZ were constructed using site-directed mutagenesis (QuikChange; Stratagene, La Jolla, Calif.) on pET11b-ftsZ. The complementation system used the JKD7-1/pKD3 conditional null strain as described by Dai and Lutkenhaus (7). Mutant alleles were cloned into pJSB2, a pBAD-based complementation vector, as described by Stricker and Erickson (36). All constructs were sequenced in pJSB2 to confirm the presence of the designed mutation and the absence of others within the ∼500 bp included in the sequence run. Only one inadvertent mutation was discovered in more than 100 sequencing runs. ftsZ genes from Synechocystis, Bacillus subtilis, and Mycoplasma pneumoniae were amplified by PCR from available plasmids and cloned into pJSB2.
Complementation and dominant negative assays.
Complementation tests were performed on plates and in liquid culture as described by Stricker and Erickson (36). Arabinose at 0.05% was used for induction, as expression at this level is near maximum for this construct (∼5 times the level of endogenous FtsZ) (see Fig. 2, below). Dominant negative tests were modified by the use of BW27783 as host strain, whose utility is explained in Results (14). BW27783 was a gift of Jay D. Keasling (University of California, Berkeley).
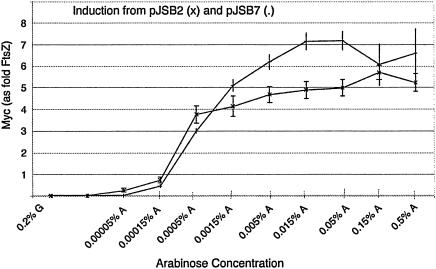
FIG. 2.
Expression level of FtsZ-myc relative to that of wild-type FtsZ as a function of arabinose concentration. The host strain was BW27783, and the expression plasmid was pJSB-ftsZ-myc, grown in LB medium at 37°C. Expression of FtsZ-myc is relative to the level of genomic wild-type FtsZ. The lower curve is from pJSB2 and the upper curve is from pJSB7, which has an improved Shine-Dalgarno sequence. Note that these expression levels are for strain BW27783, which has a constitutively expressed arabinose transporter. Comparable expression from other strains (e.g., the strain JKD7-1 used for complementation assays) probably requires higher arabinose levels.
Quantitation of pJSB induction.
We constructed two plasmids expressing FtsZ with a myc tag plus 10 additional amino acids appended to the C terminus (FtsZ-EQKLISEEDLQACKLGCFGG; myc tag underlined). Expression of this protein was driven by the pBAD promoter, using either a modified Shine-Dalgarno sequence of CGGAGT in the case of pJSB2-ftsZ-myc (36) or a consensus Shine-Dalgarno sequence of AGGAGG in the case of pJSB7-ftsZ-myc. BW27783 cells transformed with either FtsZ-myc vector were grown with various concentrations of arabinose to an optical density at 600 nm (OD600) of ∼0.5 to 0.7, harvested, and prepared for quantitative Western blotting as described by Lu et al. (17). Western blotting showed two closely spaced bands: a lower-molecular-weight band that comigrated with purified FtsZ protein and a larger band that corresponded to the FtsZ-myc protein. These bands were quantitated as described by Lu et al. (17) except that goat anti-rabbit immunoglobulin G conjugated to Alexa 488 (Molecular Probes, Eugene, Oreg.) was used as a secondary antibody, and a Molecular Dynamics Storm 860 fluorimager was used to collect data.
Immunofluorescence microscopy.
JKD7-1/pKD3 cells transformed with pJSB2-derived plasmids expressing mutant ftsZ alleles were grown as overnight cultures in Luria-Bertani (LB) plus 34 μg of chloramphenicol/ml, 100 μg of ampicillin/ml, and 0.2% (wt/vol) glucose at 30°C. Overnight cultures were diluted 1:2,000 in LB plus chloramphenicol and 0.05% (wt/vol) arabinose and grown at 42°C (complementation conditions) until the OD600 reached 0.5 to 0.7, at 240 to 300 min. This incubation time was sufficient for noncomplementing strains to stop growing due to loss of pKD3. Cultures were fixed, permeabilized, and stained as described previously (36). Images were captured using a Zeiss Axiophot microscope with 40× and 100× Plan-Neofluar objectives (numerical aperture, 1.4) and a Photometrics CoolSnap HQ cooled charge-coupled device camera and were processed for optimal contrast with Adobe Photoshop.
Protein purification.
Wild-type and mutant FtsZ proteins were purified using the protocol of Lu et al. (17), with some modifications. For some preparations we omitted Mg and DNase, instead using sonication to reduce the viscosity from DNA. For wild-type FtsZ, we continued to use the 20% ammonium sulfate cut followed by a 25% precipitation, but for most of the mutants we used a single precipitation at 25 to 35% saturated ammonium sulfate. We have added a step of column purification on an anion exchanger. The 25% ammonium sulfate pellet was resuspended in ∼10 ml of Z buffer (21) (50 mM Tris [pH 7.9], 1 mM EDTA, 10% [vol/vol] glycerol) plus 50 mM KCl. The resuspended pellet was prepared for chromatography by dialysis versus Z buffer plus 50 mM KCl and filtration through 0.2-μm filters. This filtered material was applied to a Resource Q 10/10 column (Amersham Biosciences, Piscataway, N.J.) and eluted with a linear gradient of 100 to 500 mM KCl in Z buffer. This chromatography was done at 4°C. The eluted protein was located by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and peak fractions were pooled. GTP was added to 100 μM, and the pools were aliquoted and stored at −80°C.
Assays for GTP binding and GTP hydrolysis.
To assay GTP binding, 0.2 mM GTP was added to 2.5 ml of FtsZ at 20 to 30 μM in HMK buffer (50 mM HEPES [pH 7.7], 5 mM Mg-acetate [MgAc], 350 mM KAc). The protein was loaded on a PD10 gel filtration column (Amersham) equilibrated in HMK buffer. After the sample was loaded onto the column, it was eluted with 3.5 ml of HMK. The first 1.0 ml was discarded, and the next 2.2 ml was collected in a single fraction. The protein concentration was determined by the bicinchoninic acid assay (Pierce, Rockford, Ill.). Bovine serum albumin was used as a standard, and FtsZ concentrations were adjusted to reflect the 0.75 color ratio of FtsZ and bovine serum albumin (18). Perchloric acid was added to 2.5% to the remainder of the sample, and after 30 min on ice the denatured protein was removed by centrifugation. The GTP concentration was determined by UV spectroscopy using an extinction coefficient of 12,700 at 256 nm.
GTPase activity was determined by measuring the concentration of evolved phosphate using an improved sensitivity malachite green assay (11). Proteins were diluted to the desired concentrations (ranging from 0.2 to 40 μM) in HMK buffer. The reaction was initiated by adding GTP to 1 mM at 37°C. Aliquots were taken at 0, 1, 2, 5, 10, and 20 min, transferred to tubes containing 2.3 volumes water and 3.3 volumes of 0.6 M perchloric acid, and placed on ice. After all time points, samples and monobasic potassium phosphate standards were aliquoted in triplicate to microtiter plates. An equal volume of a solution containing 0.78 mM malachite green, 8.3 mM sodium molybdate, 0.05% (vol/vol) Triton X-100, and 0.7 M HCl was added, the plates were incubated at room temperature for 30 min, and the A655 was measured using a Bio-Rad model 550 microplate reader. Free phosphate was plotted as a function of time, and the curves were examined to find a linear region (usually from 2 to 10 min or 5 to 20 min). The slope of the linear region was used to determine the hydrolysis rate. These rates were then plotted versus protein concentration to determine a critical concentration (Cc) and true hydrolysis rate above the Cc for each mutant.
RESULTS
Mutant design.
We mutated surface residues on the top and bottom of FtsZ judged to form the protofilament interface by reference to the structure of the tubulin protofilament (24) (Fig. 1; the corresponding amino acids in tubulin are listed in Table 1). In most cases we designed mutations to disrupt the interface. For example, an Asp-to-Lys mutation increases the size of the side chain and changes the charge from minus to plus; a Gly-to-Lys mutation changes the small glycine to a large side chain with a positive charge. We also investigated several less severe mutations.
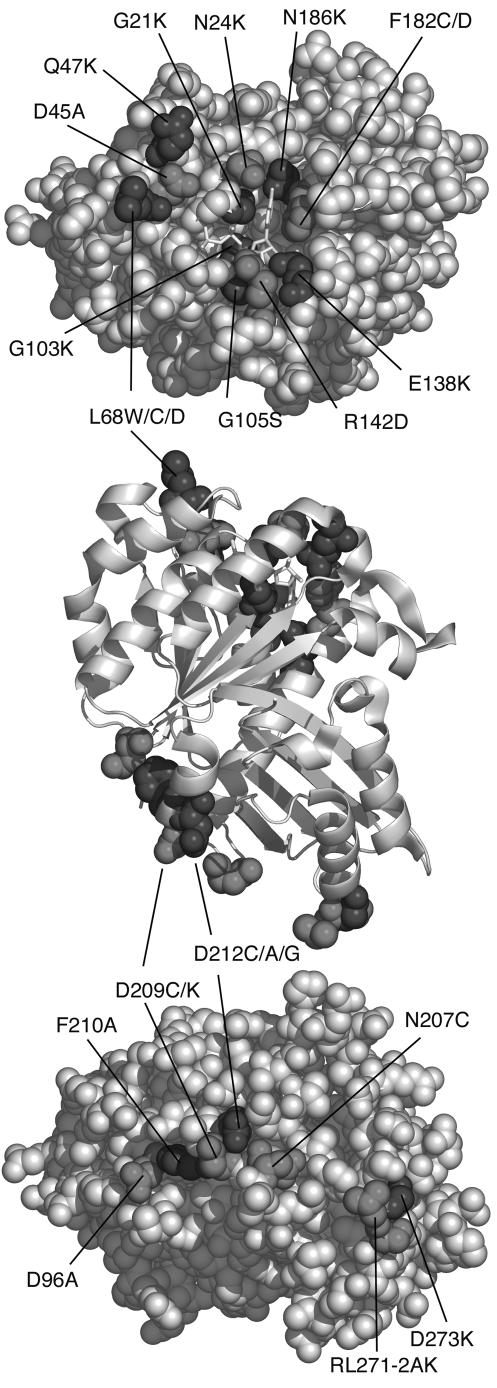
FIG. 1.
Map of the amino acid residues mutated on the top and bottom surfaces. The middle panel shows a front view in a ribbon diagram, with mutated amino acids in the spacefilling. The upper panel perspective is looking down on the top surface, and the bottom panel perspective is looking up at the bottom surface. GDP is shown in light sticks. Mutated amino acids are in dark grey and black. The figure was prepared using the program PyMol (http://pymol.sourceforge.net/).
Several of our bottom mutants lie in the synergy (T7) loop. N207 and D209 are completely conserved in FtsZ and all tubulins. D212 is conserved in all FtsZs, is E in α-tubulin (where it is involved in GTP hydrolysis of the underlying beta tubulin), and is K and G in β- and γ-tubulin, where it apparently does not participate in GTP hydrolysis. These synergy loop residues were previously mutated to Cys by Scheffers et al. (34), who kindly provided us with their expression plasmids. These Cys mutants are all blocked for GTPase, but some of them can assemble into protofilaments and/or coassemble with wild-type FtsZ (34) (our observations and data not shown). We therefore made more drastic mutations. D209 was replaced with K, changing the charge and increasing the length of the side chain. F210, which lies beside the synergy loop and appears to support it (Fig. 1), was changed to A. We expected that this might cause the synergy loop to collapse.
We also made mutations farther away from the synergy loop. In one case, we made a triple mutant, replacing R271, L272, and D273 with A, K, and K. These three amino acids are not highly conserved in FtsZ but were selected because they are homologous to SMK324-326 of β-tubulin. Nogales et al. (23) identified these residues as making a longitudinal contact. Finally, we included the mutant D96A in this analysis. We had thought that this might be in the longitudinal interface (Fig. 1), but the recent crystal structure of the FtsZ dimer (25) shows that this is actually about 10 Å above the interface, on a lateral surface.
We used two strategies to select top mutants. We mutated residues that make contact with the GTP molecule, with the goal of blocking GTP binding, and we mutated other nearby residues, with the goal of destabilizing the interface. We added or changed charge in some cases and added or removed bulky hydrophobic groups in other cases.
Test for complementation.
We used a complementation system that employs an ftsZ null host strain (JKD7-1) (7) carrying two plasmids. The rescue plasmid pKD3 expresses wild-type FtsZ but is temperature sensitive for replication and is lost at 42°C (7). The tester plasmid (pJSB2) expresses FtsZ (with the mutation of interest) under the control of the araBAD promoter (36). Each strain, carrying a different mutated ftsZ, was first grown overnight in liquid culture at 30°C and then plated on LB agarose containing different concentrations of arabinose. As in our previous study (36), complementing mutations and wild-type ftsZ gave similar response curves. Colonies began to appear at 0.005% arabinose, and their number plateaued at or above 0.05%. (Note that these arabinose levels are much higher than would be expected from Fig. 2, below. However, the experiment in Fig. 2 used the cell line BW27783, and the complementation cell line was JKD7-1. It is likely that JKD7-1 requires higher arabinose levels for induction of FtsZ at the wild-type level.) All of the mutants we tested either fully complemented, giving the same number and size of colonies as wild type, or gave no colonies at the highest arabinose.
All eight of the bottom mutants in the synergy loop failed to complement, indicating a loss of cell division function. The triple mutant RLD271-273AKK failed to complement, although the single mutant D273K did complement. Five of the 13 top mutants complemented the null. The remaining eight top mutants all failed to complement, and these were selected for further study.
It was important to confirm that the mutant proteins were expressed in the complementation system. To confirm this, we diluted overnight cultures of each of the strains and grew them for 3 h at 42°C with 0.05% arabinose to induce expression. We then pelleted the bacteria and loaded equal amounts of whole bacterial lysates (by total protein concentration) on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blotting confirmed that all mutants were expressed at a level of more than twice that of wild type (data not shown). The expression level is probably equal to the four- to fivefold overexpression seen in strain BW27783 (see below), but we used horseradish peroxidase staining here, which is not as quantitative as the Alexa Fluor used below.
Two of the mutants, E138K and R142D, were insoluble when overexpressed from pET. We investigated their solubility when expressed from pJSB2 by growing JKD7-1/pKD3 cells containing these mutations under maximal induction (0.05% arabinose) and fractionating cell lysates into soluble and insoluble fractions. Both E138K and R142D were found in the soluble and insoluble fractions at approximately equal levels (data not shown). When cells expressing wild-type FtsZ from pJSB2 were fractionated in this manner, approximately two-thirds of the FtsZ was soluble. We conclude that these mutants are expressed and soluble in the complementation test cells. Most of the other mutant proteins could be prepared as soluble proteins for in vitro studies, so we conclude these are also soluble under complementation conditions.
We also checked FtsZ with either an N- or C-terminal His tag. The N-terminal His tag protein assembled in vitro into protofilament pairs or small bundles, instead of single protofilaments. The C-terminal His tag sometimes formed large aggregates of spiral bundles, but after 1 min these resolved into normal protofilaments. In spite of these aberrations, both His tag proteins complemented the ftsZ null allele.
We checked for complementation in liquid culture by using a serial dilution log-phase growth assay, where the culture was diluted every 2 h to maintain log phase (36). Arabinose was added to 0.05%, and cells were grown at 42°C. Complementation was scored as the ability to maintain growth over several hours. The results agreed with the plate complementation assay with one exception. N186K, which failed to complement on plates, grew for at least 15 generations under serial dilution conditions. However, we found that when cultures were diluted more than 10,000 they failed to grow. This was not due to a titratable factor in the medium, as cultures that were diluted into medium filtered from growing log-phase cultures also failed to grow. We do not have an explanation for this phenomenon.
We note that the complementation test was performed at 42°C, and so temperature sensitive mutants would be scored as noncomplementing. Temperature-sensitive alleles cannot be detected when temperature is used to turn off the (essential) wild-type gene.
Dominant negative effects.
We next tested the ability of these mutated FtsZ proteins to interfere with wild-type FtsZ function. We were particularly interested in determining what level of mutant expression relative to wild type was needed to produce a dominant negative effect. This required regulating the expression of the mutant protein over known levels relative to wild type. A problem with using the pBAD vector for regulated expression is that Escherichia coli has a feedback system for arabinose transport that results in an all-or-nothing response. As the arabinose concentration is increased, an increasing number of cells turn on arabinose transport and expression from the araBAD promoter, but each activated cell is maximally induced. Khlebnikov et al. (14) created cell lines in which the arabinose transporter araE is constitutively expressed. In the BW27783 strain, individual cells respond more homogeneously by increasing expression levels as arabinose is increased. For our dominant negative experiments we used BW27783 transformed with our pBAD-based complementation plasmid (pJSB2) carrying ftsZ with the mutation of interest.
To quantitate the expression level of FtsZ from pJSB2, we constructed a reporter plasmid that had an extended myc tag on the C terminus of wild-type FtsZ. The addition of 20 amino acids raised the molecular mass of the expressed protein approximately 2,100 Da, allowing it to be resolved from wild-type FtsZ in Western blots stained with anti-FtsZ antibodies. We found that the expression level of FtsZ-myc varied from about 1/10 to five times the wild-type level over the range of arabinose tested (Fig. 2). Importantly, at 0.00015% arabinose the mutant protein was expressed at about the same level as wild-type FtsZ. Expression then jumped to four to five times the wild type at higher arabinose levels. We also constructed a version of this plasmid (pJSB7) by replacing our modified Shine-Dalgarno sequence with a sequence more closely matching the consensus sequence. This gave somewhat higher expression levels at the higher arabinose concentrations, up to seven times the wild-type level. In a separate experiment (data not shown), Western blotting showed that all mutant proteins were expressed at the same level as wild-type FtsZ (from pJSB2) at 0.05% arabinose, and we assume that this applies to all induction levels. We also determined the absolute expression levels per cell for endogenous FtsZ in BW27783. This FtsZ varied from 10,000 to 14,000 molecules per cell, somewhat lower than the 15,000 we determined for strain BL21 (a B/r A strain) (17), but significantly higher than the 3,200 determined for E. coli B/r K (31).
Each mutant was tested for dominant negative effects over a range of arabinose induction. Table 1 gives the level of FtsZ expression at which an effect was first seen (i.e., the level at which colonies were visibly smaller than wild type) and the expression level that resulted in complete inhibition of colony formation after an 18-h incubation. None of the mutants produced dominant negative effects at a substoichiometric level. Five of the bottom mutants (N207C, D209K, F210A, D212C, and RLD271-3AKK) reduced colony size at expression levels equal to or approximately fourfold higher than wild-type FtsZ. Three of these mutants (D209K, F210A, and RLD271-3AKK) were totally dominant negative, completely preventing cell division and colony growth. This lethality required expression at ∼4 to 5 times the wild-type level. Overexpression of wild-type E. coli FtsZ had no effect on colony number or size at the maximal expression from either pJSB2 or pJSB7.
Several studies have examined the effects of expressing FtsZ from foreign bacteria in E. coli (19, 32, 37). Foreign FtsZ can colocalize at the Z-ring when expressed at low levels and is toxic when expressed at higher levels. However, the level of expression that produced toxicity was not determined in these previous studies. We examined this question using our dominant negative expression system, to see if foreign FtsZ might poison substoichiometrically. FtsZ proteins from Synechocystis and B. subtilis were similar to bottom mutants in dominant negative effects. FtsZ from M. pneumoniae had no effect, probably due to its high degree of sequence divergence. We assumed that these foreign FtsZs were expressed at the same level as E. coli FtsZ at the same arabinose level, but we were not able to determine their expression levels because the antibody gives only partial cross-reactivity.
The effective level of arabinose might be different in liquid culture and agar plates, so we tested the bottom mutants for dominant negative effects in liquid culture. Overnight cultures were diluted 1/10,000 into medium with different levels of arabinose, and observed several hours later as their OD600 reached levels of 0.2 to 0.6. For D209K and F210A, turbidity was visibly reduced at 0.0005% arabinose (the same level that caused smaller colonies) and turbidity was eliminated at 0.0015 and 0.05% arabinose (the levels that completely inhibited colony formation) (Table 1). We conclude that the expression level is the same in liquid culture and plates. For the less deleterious mutants, which only showed reduced colony size in the plate assay, the liquid culture assay was less sensitive and did not demonstrate a clear dominant effect.
We should note one caveat to the quantitation. Several of the mutants showed major changes in phenotype from 0.0005 to 0.005 or 0.05% arabinose (Table 1), where Fig. 2 suggests the FtsZ level changes only from three to five times the wild-type level. This suggests that the effective level of FtsZ may be undergoing larger changes in this range than indicated by the graph.
Remarkably, none of the top mutants (except D45A) showed any dominant effects in the colony assay. Two top mutants were tested in the pJSB7 vector, which has an improved Shine-Dalgarno sequence and an expression level up to seven times the wild type. G103K and R142D both gave smaller colonies at the highest expression levels, while expression of wild-type FtsZ at this level did not affect colony size. We conclude that while the top mutants are not completely innocuous, they are significantly weaker in effect than the bottom mutants.
Structure of mutant Z-rings.
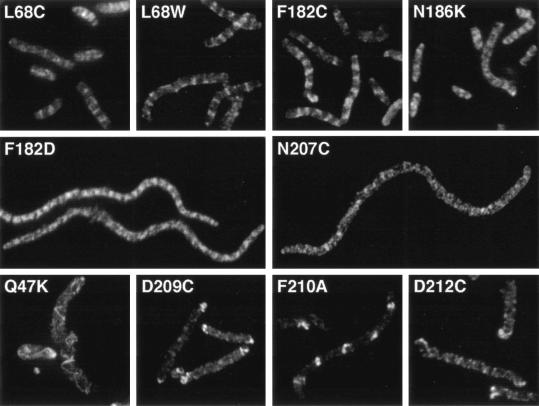
We investigated cell morphology and Z-ring structure under both complementation and dominant negative conditions. In both cases the cells were induced with 0.05% arabinose to give maximal induction of the mutant FtsZ. Mutant strains which complemented were typically longer than wild-type cells, but never more than four times as long. These mutants had some apparently normal Z-rings and also had FtsZ structures that appeared as partial rings or patches (Fig. 3, top row). We did not see spiral Z-rings in the complementing mutants. While the structures in these cells were not as sharp as the Z-rings in wild-type cells, they are apparently capable of performing cell division.
FIG. 3.
The Z-ring structure of selected mutants under complementation conditions, 240 to 300 min after shifting to 42°C and adding 0.05% arabinose.
Many of the noncomplementing mutants formed filamentous cells. These included the top mutants N24K, G103K, E138K, R142D, and F182D, as well as the bottom mutants N207C, D209K, and RLD271-3AKK. None of the filamentous cells had sharp Z-rings. Some had no organized FtsZ structures, such as N207C (Fig. 3). Others had various amounts of FtsZ structure between these two extremes. F182D had regular patches of strong FtsZ staining with a periodic variation in intensity (Fig. 3).
Several other noncomplementing mutants formed long cells that contained helical FtsZ structures. These included the previously described D212A and D212G (36), as well as Q47K, D209C, F210A, and D212C. Q47K formed the most dramatic helical structures, with a wide and variable pitch and often running throughout the cell (Fig. 3). D209C was unable to form any medial FtsZ structure, instead forming bright narrow-pitched spirals at the poles of the cell (Fig. 3). F210A formed rings, short two-turn spirals of narrow pitch, and bright FtsZ patches (Fig. 3). These structures appeared to be located at every other internucleoid space. D212C formed spirals of constant pitch and intensity that ran the length of the cell. These were similar to the spirals formed in D212A and D212G.
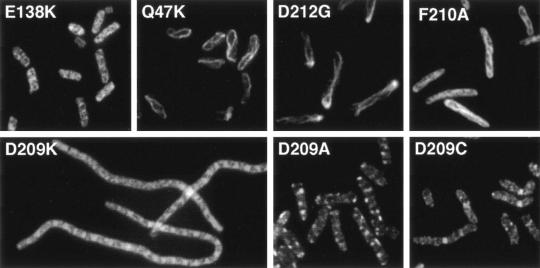
We also examined the structures formed under dominant negative conditions (Fig. 4). In most cases the structures were similar to those in the complementing cells, which is not surprising since the mutant protein is about five times the concentration of the wild-type FtsZ. Top mutants grown under these conditions had short cells, as would be expected since none of these mutants were dominant negative. (D45A has a dominant effect at high expression, but this effect seemed to be due to growth arrest of short cells.) Some of the top mutants formed essentially normal FtsZ structures, such as E138K, but there was a high background of diffuse patches (Fig. 4). Other top mutants formed spiral FtsZ structures. The Q47K spirals look very similar to the spirals formed in complementing conditions. We cannot tell if there are normal Z-rings present that are obscured by the spirals or if the spirals themselves can function and achieve division. The bottom mutants had a wider range of structural phenotypes under dominant negative conditions. D209C, the only bottom mutant that had no dominant effects, formed normal Z-rings in a patchy background. In contrast, D209A formed small bright patches that often failed to extend across the diameter of the cell and seemed to arrest cell growth. D209K, which had the strongest dominant negative effects, caused cell filamentation with bright patches and diffuse staining. D212G and F210A formed helical structures.
FIG. 4.
The Z-ring structure of selected mutants under dominant negative conditions. Strain BW27783 containing various pJSB2-FtsZ(mut) plasmids was fixed 240 to 300 min after addition of arabinose to 0.05%.
There were two classes of cell shape produced by the dominant negative bottom mutants. Some (N207C, D209K, and RLD271-3AKK) caused extensive filamentation. These were the strongest dominant mutants, producing smaller colonies at the lowest arabinose concentrations (Table 1). Others (D209A and D212A/C/G, F210A) produced cells that were only slightly elongated. These weaker mutants may have only begun to affect division in the later part of the 4- to 5-h induction period.
GTPase activity.
We measured the binding of GTP to wild-type and mutant proteins, as summarized in Table 1. Wild-type FtsZ bound 1 mol of GTP per mol of protein, as did several of the mutants. Other mutants showed a significant reduction in GTP binding.
The GTPase of wild-type FtsZ is known to exhibit a Cc around 1 μM, below which there is very low GTPase and above which the GTPase is directly proportional to FtsZ concentration (13, 30, 37). We confirmed that wild-type FtsZ in HMK buffer had a Cc for hydrolysis of 1.0 μM and an activity (slope) of 14 GTP per FtsZ per min above the Cc (Fig. 5; Table 1). This activity was found for several preparations of wild-type FtsZ, but some preparations gave an activity 20 to 30% lower. This may reflect the quality of protein preparations and suggests that activities of mutant proteins may also be variable within this range.
FIG. 5.
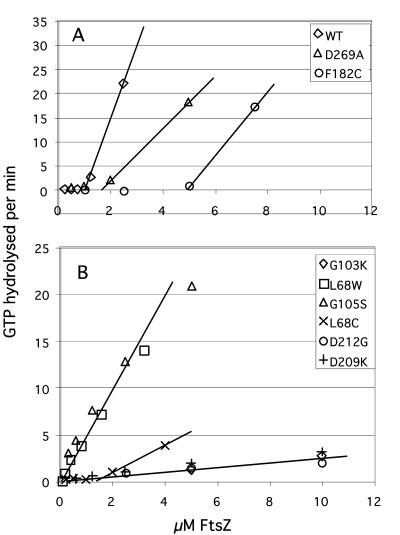
GTPase activity of selected FtsZ mutants as a function of protein concentration. Activity is expressed as micromolar GTP hydrolyzed per minute. (A) Wild-type FtsZ and two mutants with altered Cc. Wild-type FtsZ shows a linear increase in activity above a Cc of 1.0 μM. The slope of this line was 14.2 (GTP hydrolyzed per FtsZ per min). D269A is a complementing mutant characterized previously (18, 36) and was reexamined here. D269A and F182C show an activity (slope) above Cc of about 5 min−1. (B) Mutants that appear to have Cc near zero (and L68C, with Cc = 1.3 μM) and with various slopes. The four mutants with the lowest slopes are considered to have no GTPase activity.
In most past studies, FtsZ mutants have been assayed for GTPase at a single protein concentration, but we realized that a mutation could affect GTPase by altering either the Cc or the activity of the protein above Cc. We therefore assayed all mutants over a range of protein concentrations. The GTPase curves of several mutants are shown in Fig. 5, and in Table 1 we report the Cc and the activity above the Cc for wild type and most mutants. We found that some mutations substantially increased the Cc for GTPase, in particular F182C, L68C, and D269A. We had previously reported that D269A, a complementing mutant, had a GTPase activity only 10% of wild type, based on measurement at a single concentration (18). We now see that its Cc is increased to 2 μM, and above this Cc D269A has an activity about 30% of wild type, as did several other complementing mutants. F182C showed the largest increase in Cc and a similar activity of 30% of wild type above its Cc.
Several mutants showed an apparent reduction in Cc. L68W and G105S (FtsZ84) showed a Cc that was too close to zero to be measured, and an activity of about 4 to 5 GTP per FtsZ per min. We have confirmed by assembly studies that L68W has a Cc of 0.1 μM in HMK buffer, 10-fold lower than that of wild-type FtsZ (3). Several of the mutants showed a low hydrolysis activity of ∼0.15 to 0.2 GTP per FtsZ per min. We believe this activity is due to contaminating GTPase enzymes. We expressed and purified an unrelated inert protein by the same purification protocol used for FtsZ, and the end product showed a GTPase activity similar to or of higher level than the low GTPase FtsZ mutant proteins. We believe that the mutants that show activity higher than this level (for example, an activity of 0.35 to 0.5 GTP per FtsZ per min) have a low level of hydrolysis of their own.
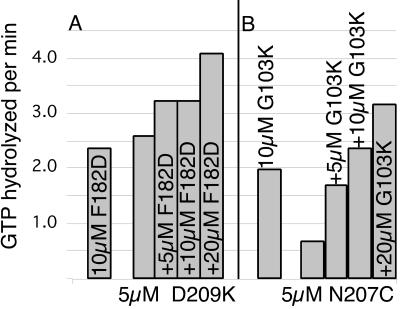
Since both top and bottom mutants presumably have a functional surface on the side opposite the mutation, we expected that a mixture of top and bottom mutants might form dimers with a normal protofilament interface and that these dimers might exhibit a GTPase activity. We tested the GTPase activity for selected pairs of top and bottom mutants (Fig. 6). The mixtures gave a GTPase that was no more than the sum of the two components, and in some cases somewhat less than additive. We thus found no evidence for GTPase activity that could be attributed to a top and bottom mutant dimer.
FIG. 6.
Pairs of top and bottom interface mutants were tested for GTPase activity, shown as GTP hydrolyzed per minute for the indicated concentrations. (A) D209K (bottom mutant) at 5 μM was tested with 0, 5, 10, and 20 μM F182D (top mutant). The left bar shows 10 μM F182D by itself. (B) N207C at 5 μM (bottom mutant) was mixed with increasing concentrations of G103K (top mutant).
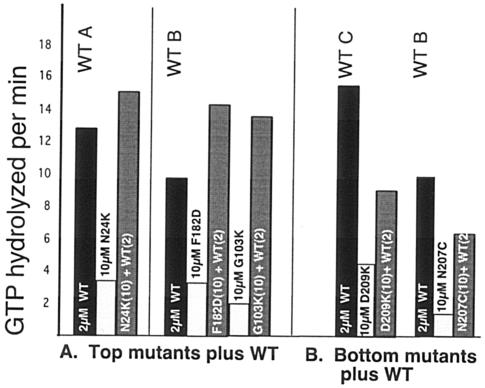
We also tested how top and bottom mutants affected the GTPase of wild-type FtsZ. We tested the effect of adding 10 μM mutant FtsZ to 2 μM wild-type FtsZ. Figure 7 shows that top mutants had almost no effect (the GTPase of the mixture was approximately the sum of the wild type plus mutant), while bottom mutants inhibited activity significantly, as already reported for some of them (34). Note, however, that the fivefold excess of bottom mutant reduced the GTPase of the wild-type FtsZ by only about half.
FIG. 7.
Effects of 10 μM concentrations of top and bottom mutants on the GTPase of 2 μM wild-type FtsZ. Numbers in parentheses indicate the concentration (micromolar). WT A, WT B, and WT C show the variation for different preparations (10 to 15 GTP per FtsZ per min). WT B was used for two experiments, showing the consistency of the assay. (A) Top mutants showed a small increase in GTPase that was approximately additive. (B) Bottom mutants decreased the GTPase of wild-type FtsZ.
DISCUSSION
One assumption implicit in our strategy is that the mutations of surface residues do not disrupt the folding of the protein and have only local effects on protein structure. In particular, the protein should be largely normal on the side opposite the mutation. This assumption is consistent with comprehensive studies of protein-protein binding, where amino acids in a protein-protein interface are systematically mutated to determine their contribution to the binding (4, 6, 8, 35). Mutations outside the interface rarely affect binding, and only those in the “hot spot” have a large effect on the binding affinity.
This assumption appears to be contradicted by our observation that several of the bottom mutants had substantially reduced binding of GTP. How could a point mutation of a surface residue on the bottom be transmitted to the GTP binding site on the top? We think it is unlikely that the bottom mutants generate a structural change that is transmitted all the way to the GTP binding site. We note that both D209K and F210A, which showed the strongest block of GTP binding, exhibited strong dominant negative effects, implying that their top surface is functional and likely bound to GTP in the cell. One possibility is that GTP binding requires a dimeric structure, perhaps related to the dimers of FtsZ-GDP found under conditions of molecular crowding (12). The full explanation awaits further studies.
We attempted negative-stain electron microscopy (EM) to characterize the assembly of mutant proteins and their coassembly with wild-type FtsZ, but the results were not quantitative and often ambiguous. We then used our recently developed tryptophan fluorescence assay (3) to characterize selected mutants (Y. Chen and H. P. Erickson, unpublished observations). A double mutant, L68W/D209K, showed no fluorescence change upon GTP addition, either by itself at >10 μM or with wild-type FtsZ. Thus, it cannot assemble. D209A and D209C coassembled with L68W, consistent with previous studies (18, 33). The top mutant Q47K also coassembled with L68W, consistent with the prominent spiral polymers seen in bacteria by immunofluorescence (Fig. 3 and 4). This limited survey confirms that some of our mutants are coassembly mutants and some are cap mutants, as discussed next.
There are two ways that a mutant protein could poison wild-type FtsZ and produce dominant negative effects. The first is by coassembly to produce an aberrant polymer, and the second is by capping polymers and/or sequestering monomers. A precedent for the first is the study of Anders et al. (1), who tested the synergy loop mutant E254A of α-tubulin (E255A in yeast), which corresponds to our FtsZ mutants D212A/C/G. The mutant tubulin coassembled with wild-type tubulin, and these microtubules were much more resistant to disassembly than normal. The tubulin E254A mutant substoichiometrically poisoned yeast microtubules when expressed at a level of only 1/20 that of wild-type α-tubulin.
The second way to produce a dominant negative effect is by capping. An example of a microtubule capping protein is N-ethyl maleimide-modified tubulin. This modified tubulin was found to block growth of the minus end, while not affecting the plus end. It thus seems to act as a minus end cap. It also was effective at substoichiometric levels, in a range from 0.1 to 1.0 relative to wild-type tubulin (27). While this covalent modification results in a substoichiometric cap, mutation of the proteins might not necessarily have this effect. In comprehensive studies of mutants in yeast α- and β-tubulin, only 2 out of 49 α-tubulin (29) and 2 out of 46 β-tubulin (28) mutant clusters were dominant negative. The tubulin mutants were only tested at a 1:1 stoichiometry, a ratio where we saw no dominant effects for any of our FtsZ mutants. α-Tubulin E254A may be a rare mutant with substoichiometric dominant negative effects.
Bottom mutants.
Several of our bottom mutants (D209A/C and D212A/C/G) are likely to be coassembly mutants, since they have been reported to assemble on their own or to coassemble with wild-type FtsZ in vitro (18, 34). However, in contrast to the substoichiometric poisoning seen in α-tubulin E254A, these mutants only inhibited cell growth when expressed at four to five times the level of wild-type FtsZ. This suggests that FtsZ protofilaments can function for cell division with a majority of the interfaces being mutated and incapable of GTP hydrolysis. These putative coassembly mutants were the least deleterious of the bottom mutants. They reduced colony size but not colony number at the highest expression level, ∼5 times wild type. Several bottom mutants are likely cap mutants: N207C, D209K, F210A, and RLD271-3AKK. These mutants appear to be more toxic, since they completely block colony formation at the higher expression levels.
It is probably more meaningful to compare the level of the mutants to that of the wild-type monomer, rather than to total FtsZ. Total FtsZ was measured to be 11 μM in strain BL21 (17) and 7 to 10 μM in strain BW27783 (present study). If the Cc in the cell is 1 μM, which was determined in vitro under physiological buffer conditions (3, 12), the wild-type FtsZ should have 6 to 9 μM subunits in protofilaments and 1 μM free subunits. (This assumes that the 70% of the FtsZ that is in the cytoplasm is also mostly assembled into protofilaments, which we think is likely but have not yet demonstrated.) A bottom cap mutant present at 4 to 5 times the total FtsZ would actually have an effective concentration 25 to 50 times the concentration of free wild-type monomers. It seems that these bottom cap mutants are more than an order of magnitude weaker than wild-type FtsZ in competing for protofilament assembly.
The cell division inhibitor SulA binds to the bottom interface of FtsZ and probably blocks assembly of FtsZ by acting as a steric block of that interface (5). It has been suggested that SulA may act substoichiometrically by capping FtsZ protofilaments (5). There is also one report that SulA does indeed poison E. coli substoichiometrically (20). However, we would suggest that the FtsZ-SulA complex should act like a bottom cap mutant. If so it would interact only weakly with protofilaments and would have to act stoichiometrically by binding and sequestering FtsZ subunits. It should be interesting to reinvestigate the stoichiometry of SulA to FtsZ in vivo and in vitro.
Top cap mutants are even weaker, possible treadmilling.
Even more surprising than the weak activity of the bottom mutants are the top mutants. They showed little or no dominant negative effect even at the highest expression level. This is 5 times the level of wild-type FtsZ, or 50 times the 1 μM Cc. A sevenfold overexpression using pJSB7 caused reduced colony size for some mutants but did not completely block growth. Consistent with the lack of activity in vivo, a fivefold excess of top mutant had no effect on wild-type GTPase activity. Several of the top mutants produced aberrant Z-ring morphology, even though they did not reduce colony size. In a related study, the top mutant T108A (FtsZ3) caused Z-ring aberrations when overexpressed (2). Therefore, the top mutants are not completely innocuous but are significantly weaker in effect than the bottom mutants.
The weak activity of the top mutants suggests the possibility that assembly of protofilaments may be directional. If subunits add primarily at the bottom end, and dissociate primarily from the top, top mutants may be inactive because there is no significant assembly at the top end. Assembly at one end and disassembly at the other requires an irreversible reaction step like GTP hydrolysis (38) and is termed treadmilling.
Interface mutants and cooperativity.
Although the N- and C-terminal globular domains of FtsZ appear to be independently folding domains (25), the present study suggests that their assembly surfaces do not act independently. If one interface surface is missing or damaged, the other has a much weaker interaction than predicted by the ∼1 μM critical concentration (which is equal to the Kd for elongation). Our results suggest that the subunit needs a functional interface at both the top and bottom in order to interact with protofilaments at the ∼1 μM Kd. At the present time we cannot propose a structural basis for this speculation, but it may be related to the enigma of the apparent cooperativity of FtsZ assembly (3). We may require a model of the protofilament structure that is very different from the simple stacking of subunits on top of each other.
Acknowledgments
We thank Jay D. Keasling (University of California, Berkeley) for strain BW27783 and Joe Lutkenhaus (University of Kansas) for strain JKD7.
The work was supported by NIH grant GM66014.
REFERENCES
- 1.Anders, K. R., and D. Botstein. 2001. Dominant-lethal alpha-tubulin mutants defective in microtubule depolymerization in yeast. Mol. Biol. Cell 12:3973-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, E., and J. Lutkenhaus. 1990. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J. Bacteriol. 172:5602-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Y., K. Bjornson, S. D. Redick, and H. P. Erickson. 2005. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 88:505-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clackson, T., and J. A. Wells. 1995. A hot spot of binding energy in a hormone-receptor interface. Science 267:383-386. [DOI] [PubMed] [Google Scholar]
- 5.Cordell, S. C., E. J. Robinson, and J. Lowe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. USA 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham, B. C., and J. A. Wells. 1993. Comparison of a structural and a functional epitope. J. Mol. Biol. 234:554-563. [DOI] [PubMed] [Google Scholar]
- 7.Dai, K., and J. Lutkenhaus. 1991. ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173:3500-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLano, W. L. 2002. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 12:14-20. [DOI] [PubMed] [Google Scholar]
- 9.Erickson, H. P. 1998. Atomic structures of tubulin and FtsZ. Trends Cell Biol. 8:133-137. [DOI] [PubMed] [Google Scholar]
- 10.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geladopoulos, T. P., T. G. Sotiroudis, and A. E. Evangelopoulos. 1991. A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 192:112-116. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, J. M., M. Jimenez, M. Velez, J. Mingorance, J. M. Andreu, M. Vicente, and G. Rivas. 2003. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 278:37664-37671. [DOI] [PubMed] [Google Scholar]
- 13.Huecas, S., and J. M. Andreu. 2003. Energetics of the cooperative assembly of cell division protein FtsZ and the nucleotide hydrolysis switch. J. Biol. Chem. 278:46146-46154. [DOI] [PubMed] [Google Scholar]
- 14.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241-3247. [DOI] [PubMed] [Google Scholar]
- 15.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 16.Löwe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motility Cytoskel. 40:71-86. [DOI] [PubMed] [Google Scholar]
- 18.Lu, C., J. Stricker, and H. P. Erickson. 2001. Site-specific mutations of FtsZ—effects on GTPase and in vitro assembly. BMC Microbiol. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, X. L., Q. Sun, R. Wang, G. Singh, E. L. Jonietz, and W. Margolin. 1997. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J. Bacteriol. 179:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee, A., and J. Lutkenhaus. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17:462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogales, E., K. H. Downing, L. A. Amos, and J. Lowe. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5:451-458. [DOI] [PubMed] [Google Scholar]
- 23.Nogales, E., M. Whittaker, R. A. Milligan, and K. H. Downing. 1999. High-resolution model of the microtubule. Cell 96:79-88. [DOI] [PubMed] [Google Scholar]
- 24.Nogales, E., S. G. Wolf, and K. H. Downing. 1998. Structure of the αβ tubulin dimer by electron crystallography. Nature 391:199-203. [DOI] [PubMed] [Google Scholar]
- 25.Oliva, M. A., S. C. Cordell, and J. Lowe. 2004. Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11:1243-1250. [DOI] [PubMed] [Google Scholar]
- 26.Oliva, M. A., S. Huecas, J. M. Palacios, J. Martin-Benito, J. M. Valpuesta, and J. M. Andreu. 2003. Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J. Biol. Chem. 278:33562-33570. [DOI] [PubMed] [Google Scholar]
- 27.Phelps, K. K., and R. A. Walker. 2000. NEM tubulin inhibits microtubule minus end assembly by a reversible capping mechanism. Biochemistry 39:3877-3885. [DOI] [PubMed] [Google Scholar]
- 28.Reijo, R. A., E. M. Cooper, G. J. Beagle, and T. C. Huffaker. 1994. Systematic mutational analysis of the yeast β-tubulin gene. Mol. Biol. Cell 5:29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards, K. L., K. R. Anders, E. Nogales, K. Schwartz, K. H. Downing, and D. Botstein. 2000. Structure-function relationships in yeast tubulins. Mol. Biol. Cell. 11:1887-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romberg, L., M. Simon, and H. P. Erickson. 2001. Polymerization of FtsZ, a bacterial homolog of tubulin. Is assembly cooperative? J. Biol. Chem. 276:11743-11753. [DOI] [PubMed] [Google Scholar]
- 31.Rueda, S., M. Vicente, and J. Mingorance. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salimnia, H., A. Radia, S. Bernatchez, T. J. Beveridge, and J. R. Dillon. 2000. Characterization of the ftsZ cell division gene of Neisseria gonorrhoeae: expression in Escherichia coli and N. gonorrhoeae. Arch. Microbiol. 173:10-20. [DOI] [PubMed] [Google Scholar]
- 33.Scheffers, D., J. G. de Wit, T. den Blaauwen, and A. J. Driessen. 2001. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 494:34-37. [DOI] [PubMed] [Google Scholar]
- 34.Scheffers, D. J., J. G. de Wit, T. den Blaauwen, and A. J. Driessen. 2002. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41:521-529. [DOI] [PubMed] [Google Scholar]
- 35.Schreiber, G., and A. R. Fersht. 1995. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248:478-486. [DOI] [PubMed] [Google Scholar]
- 36.Stricker, J., and H. P. Erickson. 2003. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J. Bacteriol. 185:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, X., and J. Lutkenhaus. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9:435-442. [DOI] [PubMed] [Google Scholar]
- 38.Wegner, A. 1976. Head to tail polymerization of actin. J. Mol. Biol. 108:139-150. [DOI] [PubMed] [Google Scholar]
- 39.Yu, X. C., and W. Margolin. 1997. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 16:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]