Abstract
Low-copy-number plasmids all encode multiple systems to ensure their propagation, including replication, partition (active segregation), and postsegregational killing (PSK) systems. PSK systems kill those rare cells that lose the plasmid due to replication or segregation errors. PSK systems should not be used as the principle means of maintaining the plasmid. The metabolic cost of killing the many cured cells that would arise from random plasmid segregation is far too high. Here we describe an interesting exception to this rule. Maintenance of the large virulence plasmid of Shigella flexneri is highly dependent on one of its PSK systems, mvp, at 37°C, the temperature experienced during pathogenesis. At 37°C, the plasmid is very unstable and mvp efficiently kills the resulting cured bacterial cells. This imposes a major growth disadvantage on the virulent bacterial population. The systems that normally ensure accurate plasmid replication and segregation are attenuated or overridden at 37°C. At 30°C, a temperature encountered by Shigella in the outside environment, the maintenance systems function normally and the plasmid is no longer dependent on mvp. We discuss why the virulent pathogen tolerates this self-destructive method of propagation at the temperature of infection.
Shigella flexneri is an etiologic agent of bacillary dysentery, a serious invasive disease of the human colonic mucosa. Virulence is dependent, in large part, on the products of a 230-kb virulence plasmid referred to by a number of names, including pMYSH6000 and pWR100, according to the particular isolate of the bacterium studied. The virulence phenotype is correlated with the ability of the cells to bind the dye Congo Red and form red colonies on Congo Red agar (the Crb+ phenotype). Strains that have lost the virulence plasmid or are otherwise unable to express the virulence genes, form white colonies on this medium (Crb−) (16). Cells express virulence at 37°C, but not at 30°C, due to the temperature regulation of virulence genes under the control of the virB regulon (8). Expression of virulence puts the cells at a selective disadvantage in comparison to avirulent variants. Thus, at 37°C, some white colonies appear after prolonged growth of wild-type cells. These are the progeny of rare cells that have lost virulence by mutation or by loss of the plasmid (16).
The complete sequence of pWR100 has been determined (4). It reveals several plasmid maintenance functions, including an IncFII replication system, two plasmid partition systems, and two postsegregational killing (PSK) systems: the mvp system and a locus resembling the ccd PSK system of plasmid F (4). The presence of multiple plasmid maintenance systems is typical of large, low-copy-number plasmids found in gram-negative bacteria. They invariably encode partition systems that actively distribute copies of the plasmid to daughter cells during cell division and also one or more PSK systems (10). PSK systems consist of genes for the production of a toxin and its specific antidote. Should a cell lose the plasmid, the antidote rapidly decays but the toxin persists and kills the cell or inhibits its growth. In this way, cured cells are effectively eliminated from the population, thus ensuring the continued maintenance of the plasmid (20). When the core plasmid maintenance systems, including the partition system, function efficiently, the PSK system should rarely come into play, as very few cured cells are produced (10). Thus, PSK systems can be regarded as back-up plasmid maintenance systems that deal with occasional errors. This makes sense, because frequent use of the system would impose a major disadvantage to the host due to frequent cell killing.
This functional relationship between PSK and the plasmid partition system has recently been demonstrated directly in Escherichia coli (3). A model plasmid was constructed with just a low-copy-number replicon and the PSK system mvp. It was accurately maintained, but at a cost to the host growth rate and viability. Its maintenance was highly dependent on mvp function. When a partition system (P1par) was added, accurate maintenance was retained but the normal cell growth rate was restored. The plasmid was no longer dependent on mvp function for maintenance (3). Thus, plasmids with both PSK and partition systems are stably maintained and do not measurably disadvantage the host, because the PSK system rarely comes into play.
MATERIALS AND METHODS
Media, chemicals, and DNA manipulations.
Media, reagents, enzymes, buffers, and chemicals were as previously described (1). Cells were grown in Luria-Bertani (LB) broth supplemented when needed with kanamycin (12.5 μg/ml), chloramphenicol (25 μg/ml), or ampicillin (50 μg/ml). Strains were tested for Congo Red binding on tryptic soy broth (TSB) agar plates (1.5% agar) containing 0.025% Congo Red (Sigma Chemical Co., St. Louis, Mo.). Plasmid DNA was prepared using the Wizard Plus Miniprep/Maxiprep DNA purification system (Promega Corp., Madison, Wis.). E. coli strain DH5α was used for plasmid growth and DNA manipulations. Plasmid DNA was introduced into S. flexneri strains by electroporation (18).
Bacterial strains and plasmids.
E. coli strain DH5α is a φ80dlacZΔM15 Δ(lacZYA-argF)U169 derivative of strain DH5 (7). The wild-type, virulent S. flexneri strain 2a 2457T was as described elsewhere (6). BS547 is an avirulent, kanamycin-resistant derivative of S. flexneri with a mxiM1::aphA-3 insertion in the virulence plasmid (15). The bulk of the studies used this strain, because the plasmid can be followed by kanamycin resistance and the strain is not subject to the selective pressure that favors loss of virulence in wild-type cells (16). MxiM is required for the assembly of the type III secretory apparatus that is essential for virulence (15).
Plasmid pALA33 contains the pBR322 origin of replication, the P1 replication region, and a gene for ampicillin resistance, as described previously (12). Plasmid pALA136 was derived from pALA33 by introduction of a chloramphenicol resistance cassette in place of the ampicillin resistance gene, as described elsewhere (12). Plasmid pALA1196 consists of the 1,127-bp mvp region from the S. flexneri virulence plasmid pMYSH6000, inserted in plasmid pALA136 (12). Plasmid pALA2515 contains the same 1,127-bp region as a BamHI fragment inserted into the unique BamHI site of pALA136 (12). Plasmids pALA2522, pALA2523, pALA2525, pALA2526, pALA2529, and pALA2546 are deletion derivatives of pALA2515 as previously described (14). Plasmid pALA2558 consists of the BamHI-SalI fragment of pALA2529 containing the mvp locus, inserted between the equivalent sites in the vector pGB2.cat. Plasmid pGB2.cat has the spectinomycin resistance gene of pGB2 (5) replaced by the Tn9 chloramphenicol resistance gene (21).
Blocking the maintenance of the wild-type virulence plasmid by extra copies of the mvp locus.
S. flexneri strain 2a 2457T was grown in LB broth, and plasmid pALA136 or its mvp-containing derivative pALA1196 were introduced by electroporation as described previously (18). Suitable dilutions were plated on LB-chloramphenicol plates at 30 or 37°C. The resulting colonies were purified by restreaking under the same conditions, and three of the resulting colonies were separately resuspended in 5 ml of LB-chloramphenicol broth. Suitable dilutions were immediately plated on TSB-Congo Red plates (16) to give separate colonies at 37°C, and the proportion of red to white colonies was determined after overnight growth. The broth cultures were grown for 8 h at 30 or 37°C, diluted 104-fold, and grown for a further 8 h at 30 or 37°C. Suitable dilutions of the resulting cultures were plated to give separate colonies on TSB-Congo Red plates at 37°C, and the proportion of red to white colonies was determined after overnight growth. The rate of accumulation of Crb− cells (i.e., the presumed rate of loss of the virulence plasmid) during the approximately 20 generations of growth in liquid medium was calculated from the initial and final ratios of red to white colonies according to the formula of Roberts et al. (13).
Loss of the virulence plasmid in the mxi mutant, kanamycin-resistant S. flexneri strain BS547.
S. flexneri strain BS547 was grown in LB broth plus kanamycin at 30°C, and plasmid pALA136 or its derivatives were introduced by electroporation. Chloramphenicol-resistant colonies were selected on LB-ampicillin or LB-chloramphenicol agar at 30 or 37°C and purified by restreaking under the same conditions, and two single colonies from each test were picked and resuspended in 5 ml of LB broth with chloramphenicol. The cultures were incubated at 30 or 37°C for 5 h, and suitable dilutions were plated to test for kanamycin resistance. The cultures were diluted 5 × 106-fold in the same medium and incubated at 30 or 37°C for 12 h. Samples were plated again to test for kanamycin resistance. The cultures were subject to two further rounds of 5 × 106-fold dilution and incubation, and samples were plated after each round of growth. From each plating, 80 separate colonies were picked with a sterile toothpick and stabbed sequentially to LB plates without and with kanamycin. The proportion of kanamycin-sensitive colonies was taken as a measure of the proportion of cells that had lost the kanamycin resistance virulence plasmid. Loss of the plasmid from sample colonies was confirmed by PCR tests as described below.
Competitive growth tests.
S. flexneri strain BS547 was grown in LB broth plus kanamycin at 30°C, and plasmid pALA33 or its derivative, plasmid pALA2515, was introduced by electroporation. The resulting purified strains were grown at 30°C, mixed together in LB broth, diluted, and grown at 37°C using the protocol for measuring virulence plasmid loss. Aliquots were removed periodically and plated for viable counts on LB agar (total viable cells), LB agar with chloramphenicol (viable BS547 pALA2515 cells), or LB agar with ampicillin (viable BS547 pALA33 cells).
PCR tests for loss of the virulence plasmid.
Presence of the virulence plasmid was assayed by PCR amplification of the virF, virB, and repA genes. Amplification of the chromosomal gene rpoS was used to control for the efficiency of the PCRs. Primer sequences were taken from GenBank sequence file accession numbers AL391753 (plasmid sequences) and U00119 (rpoS sequence). The primer pairs were as follows: virF forward (complementary bases 39288 to 39255), virF reverse (bases 38589 to 38608), virB forward (complementary bases 102154 to 102130), virB reverse (bases 101430 to 101454), repA forward (bases 203064 to 203082), repA reverse (complementary bases 203880 to 203862), rpoS forward (bases 102 to 121), and rpoS reverse (complementary bases 1375 to 1356).
Template DNA was prepared directly from single subcultured colonies by the protocol described by Powell et al. (11), modified for use with the Expand high-fidelity PCR system (Roche Applied Sciences, Indianapolis, Ind.). The reaction products were analyzed on 1% (wt/vol) agarose gels. The products were of the correct size as judged by comparison with 1-kb DNA ladder from Invitrogen (Carlsbad, Calif.).
Reverse transcription-PCR tests for the induction of the virB regulon.
Cultures of S. flexneri strain BS547 were transformed with plasmid pATM324, which carries an l-arabinose-inducible virB gene (17). The cells were grown overnight, diluted 1:100, and grown to an optical density at 600 nm of 0.4 at 30 or 37°C in LB broth with and without 0.2% (wt/vol) l-arabinose. Total RNA was prepared with the RNeasy mini kit from QIAGEN (Valencia, Calif.) using the QIAshredder homogenization protocol for the isolation of total RNA from gram-negative bacteria. A second round of DNase I treatment was then carried out, using RNase-free DNase I from Amersham Biosciences (Piscataway, N.J.) according to the manufacturer's recommendations. This DNase I was removed using the RNeasy mini kit protocol for RNA cleanup (QIAGEN).
The isolated RNA was assayed for ipaB, spa33, and repA sequences by reverse transcription using 0.1 ng of RNA template per assay mixture for ipaB and spa33 primer reactions and 10 ng of RNA template per assay mixture for the repA primer reaction. The resulting cDNA was amplified by 30 cycles of PCR amplification using the same primer pairs. Primer sequences (GenBank sequence file accession number AL391753) were as follows: ipaB forward (complementary bases 107837 to 107809), ipaB reverse (bases 106787 to 106812), spa33 forward (bases 127194 to 127215), spa33 reverse (complementary bases 127964 to 127937), repA forward (bases 203229 to 203249), and repA reverse (complementary bases 203894 to 203873). The reaction products were analyzed on 1.5% (wt/vol) agarose gel. The product sizes for the ipaB, spa33, and repA sequences were predicted to be 1,051, 771, and 666 bp, respectively. These appear correct by comparison with the bands of the PCR size markers from Sigma-Aldrich, Inc. (St. Louis, Mo.).
RESULTS
Blocking the plasmid mvp function leads to rapid loss of a virulence marker in the wild-type S. flexneri population. S. flexneri strain 2a 2457T is a virulent, invasive strain and gives the Crb+ (red colony) phenotype on Congo Red plates at 37°C. Any cells that lose the virulence plasmid are avirulent and give the Crb− (white colony) phenotype (9). The virulence plasmid of strain 2a 2457T appears to be essentially the same as pWR100 from strain 2a M90T that was recently sequenced (4). It encodes an mvp PSK system.
The PSK function of mvp can be blocked by introducing an extra source of the MvpA antidote on an additional plasmid (14). Now, if the mvp-containing plasmid is lost, MvpA continues to be produced and the MvpT can no longer kill the cell. We blocked mvp activity in strain 2457T in this way by introducing a pBR322-based plasmid carrying the entire mvp locus (pALA1196) into the strain by electroporation. We did not expect this to have any measurable effect on virulence plasmid maintenance. The virulence plasmid encodes replication and partition systems closely resembling those that promote faithful plasmid replication and segregation to daughter cells in other plasmid types (4). Thus, very few plasmid-free cells should ever arise, and PSK by mvp should hardly ever come into play.
At 30°C, this expectation was justified; introduction of pALA1196 had no measurable effect on virulence plasmid stability as measured by retention of the Crb+ phenotype (Table 1). However, at 37°C, 30% of cells containing pALA1196 formed Crb− (white) colonies after 20 generations of growth (Table 1). Virulent cells containing the vector without mvp (pALA136) produced no Crb− colonies under the same conditions. This suggested that the virulence plasmid is an exception to the rule and relies heavily on the action of the mvp PSK system for its continued maintenance at 37°C.
TABLE 1.
Loss of Crb+ phenotype on introduction of a plasmid carrying the mvp locus
| Incoming plasmida | Growth temp (°C) | Loss of Crb+ phenotypeb |
|---|---|---|
| pALA136 | 30 | <1 |
| pALA1196 | 30 | 0.6 |
| pALA136 | 37 | <1c |
| pALA1196 | 37 | 34 |
The relevant plasmid was introduced into S. flexneri 2a 2457T, selecting for chloramphenicol resistance.
The rate of loss (as a percentage) of the Crb+ phenotype assayed during 20 generations of unselected growth at the specified temperature, as described in Materials and Methods.
Loss was measured through 20 generations. This growth period was insufficient to detect the low level of loss of the Crb+ phenotype normally seen when S. flexneri 2a 2457T cells are grown at 37°C (16).
Extra copies of the mvp locus lead to complete loss of an mxiM mutant virulence plasmid at 37°C.
We showed above that expression of additional copies of mvp leads to loss of the Crb+ phenotype in cells carrying the wild-type virulence plasmid. Is this due to loss of the entire plasmid, as was predicted, or to loss of a virulence function? This was investigated using S. flexneri strain BS547. This is an avirulent derivative of strain 2457T with an aphA-3 insertion mutation in the mxiM virulence gene of the virulence plasmid (15). Here, retention of the plasmid can be monitored by following the kanamycin resistance encoded by the plasmid.
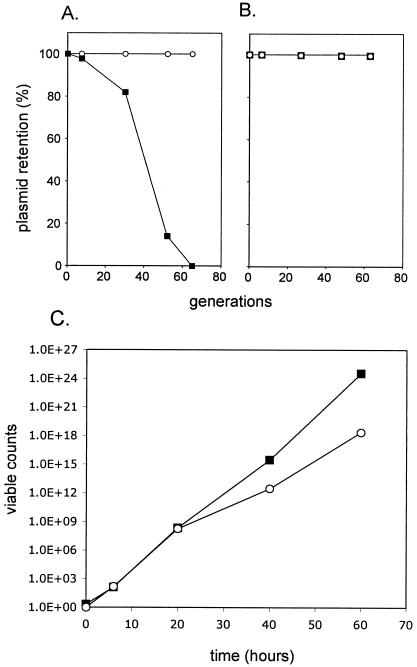
We blocked mvp function by introducing a multicopy, pBR322-based plasmid carrying an additional copy of the mvp genes (pALA2515). The kanamycin resistance (Kmr) of the cells in the culture was progressively lost at 37°C. After an initial lag, the loss rate was 2 to 3% per generation. Virtually no Kmr cells remained after 65 generations of unselected growth (Fig. 1A). Control strains containing no additional plasmid or containing the pALA136 vector produced no measurable loss.
FIG. 1.
Blocking effect of an additional cloned mvp region at 37°C. (A and B) Retention of the kanamycin resistance marker carried on the virulence plasmid in strain BS547 was assayed after the stated number of generations of growth without selection in LB broth at 37°C (A) or 30°C (B). Circles, BS547 cells containing the vector, pALA136; squares, BS547 cells containing the plasmid pALA2515, a derivative of pALA136 that contains the intact mvp locus. Both strains retained the virulence plasmid kanamycin resistance marker throughout the experiment when grown at 30°C. After the initial lag, the slope of the response to pALA2515 at 37°C corresponded to a virulence plasmid loss rate of 2 to 3% per generation (2). (C) Viable counts of strain BS547 during competitive growth in LB broth at 37°C. Squares, BS547 cells containing the plasmid pALA2515 containing the mvp locus; circles, BS547 cells containing the vector, pALA33. The two variants were grown together in competition and were distinguished by the different antibiotic resistance markers that they carry (see Materials and Methods).
The cells containing pALA2515 appeared to grow faster at 37°C than cells containing the vector in the above experiment (data not shown). To check this, we carried out a growth competition experiment (Fig. 1C). Cells containing pALA2515 (chloramphenicol resistance) were mixed in equal amounts with cells containing a variant of pALA136 (pALA33, ampicillin resistance). The mixture was grown at 37°C under the same conditions as the above experiment, and samples were plated periodically for chloramphenicol or ampicillin resistance on LB plates at 30°C. As shown in Fig. 1C, the cells containing pALA2515 had a considerable growth advantage over cells containing the vector during long-term growth.
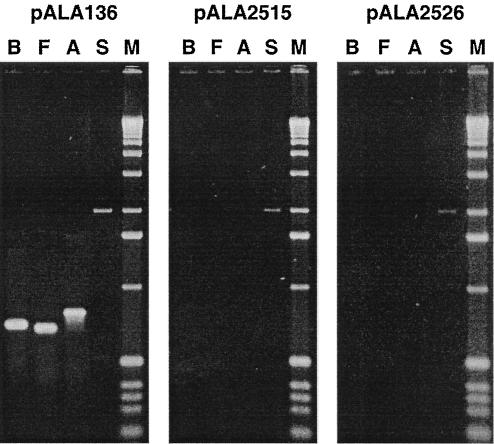
The kanamycin-sensitive colonies produced in response to pALA2515 were screened for loss of other plasmid loci by PCR analysis. In addition to kanamycin resistance, the cells had lost sequences in the virulence plasmid replicon (repA) and the plasmid-encoded virulence genes virB and virF, while retaining the host chromosomal locus rpoS (Fig. 2). We conclude that introduction of the mvp region present in plasmid pALA2515 leads to the loss of the entire virulence plasmid at 37°C. No loss of the plasmid markers was seen in cells growing at 30°C (Fig. 1B).
FIG. 2.
The entire virulence plasmid is lost when mvp activity is blocked at 37°C. Lanes correspond to amplified products for the virulence plasmid genes virB (lane B), virF (lane F), or repA (lane A), the chromosomal gene rpoS (lane S), and size markers (lane M). The template DNA was derived from colonies of strain BS547 recovered from the experiments shown in Fig. 1 and 3. Panel 1, typical kanamycin-resistant line recovered from cells containing the plasmid vector pALA136; panel 2, kanamycin-sensitive line recovered from cells containing pALA2515, which has the entire mvp region; panel 3, kanamycin-sensitive line recovered from cells containing pALA2526 (mvpA gene only). The products of the virF, virB, repA, and rpoS primers were predicted to be 700, 725, 816, and 1,274 bp, respectively, and appear to be correct as estimated by comparison with the 1,636-, 1,018-, and 517-bp bands of the 1-kb DNA ladder (central three bands in the size marker lanes).
Expression of the mvpA gene is sufficient to promote plasmid loss from the 37°C population.
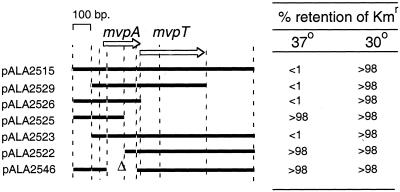
We reasoned that the virulence plasmid must be inherently unstable at 37°C and that killing of the cured cells by mvp must ensure that the cells retain the plasmid. This causes the growth of the population to slow down at 37°C. When killing is blocked by the MvpA antidote encoded by the extra copies of mvp on pALA2515, the cured cells survive and rapidly accumulate in the population. In order to confirm that MvpA synthesis from pALA2515 was responsible for the effect, we carried out a deletion analysis of pALA2515. As predicted, the critical determinant for the accumulation of cells lacking the virulence plasmid at 37°C mapped to the mvpA gene (Fig. 3).
FIG. 3.
The determinant for blocking virulence plasmid stability at 37°C maps to the mvpA gene. The extents of the mvp region carried by derivatives of the plasmid vector pALA136 are shown on the left. These plasmids were introduced into strain BS547 by electroporation, selecting for chloramphenicol resistance. After approximately 65 generations of unselected growth at 30 or 37°C, retention of the kanamycin resistance marker of the virulence plasmid was assayed as described in Materials and Methods. The intact mvp locus promoted loss of the virulence plasmid (pALA215). An in-frame deletion within mvpA in an otherwise-intact 1,127-bp mvp region abolished the effect (pALA2546). Plasmid pALA2526 gave the full effect. It carries only the mvpA gene and its upstream control region, showing that the mvpA gene is necessary and sufficient to promote loss of the virulence plasmid at 37°C.
Induction of the virB regulon is not the cause of rapid plasmid loss at 37°C.
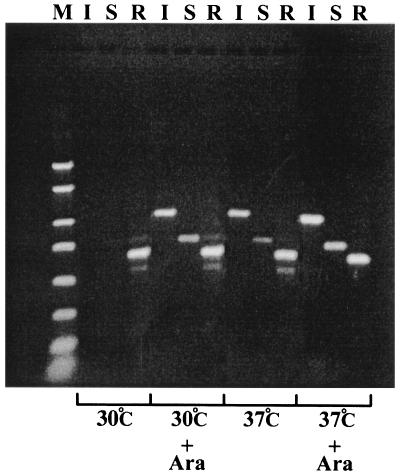
We have shown that the maintenance of virulence plasmid is inherently unstable at 37°C but not at 30°C. This implies that some key plasmid maintenance function or functions are attenuated or overridden at the higher temperature. The plasmid has a known temperature-dependent regulatory system, the virB regulon. A number of virulence-associated plasmid genes are controlled by virB and become induced only at the higher temperature (19). It seemed possible that some key plasmid maintenance genes might be under negative regulation within the virB regulon. Alternatively, plasmid loss at 37°C might be due to some unknown regulon product, such as a site-specific endonuclease, that damages the plasmid. In either case, induction of virB at 30°C should also make the plasmid unstable. We expressed virB in strain BS547 at 30 or 37°C by using an arabinose-inducible expression system (see Materials and Methods). Despite the fact that the virB regulon was demonstrably induced at both temperatures (Fig. 4), maintenance of the virulence plasmid continued to be inherently unstable only at 37°C (Table 2). Thus, temperature regulation of plasmid maintenance is not part of the virB regulon but is due to an independent temperature effect on plasmid maintenance, either involving the replication and partition systems or some function that actively damages the plasmid at 37°C.
FIG. 4.
l-Arabinose induction of the virB regulon as detected by reverse transcription-PCR. Total RNA was prepared from strain BS547 transformed with pATM324, which has the virB gene under the control of l-arabinose induction (see Materials and Methods). Reverse transcription was used to synthesize cDNA for virulence plasmid genes ipaB (lane I), spa33 (lane S), and repA (lane R). The cDNA was amplified by PCR. The gel shows the PCR products from cells grown at 30 or 37°C with and without l-arabinose. Lane M, DNA size markers. The product sizes for the ipaB, spa33, and repA sequences (1,051, 771, and 666 bp, respectively) appear correct in comparison with the 500-, 750-, 1,000-, and 1,500-bp bands of the size markers (second through fifth bands from the top of the marker lane). The ipaB and spa33 genes, members of the virB regulon, are induced by arabinose at 30°C, whereas the constitutively expressed replication gene repA is expressed whether arabinose is present or not.
TABLE 2.
Instability of the virulence plasmid as a function of growth temperature and virB gene expression
| Incoming plasmida | Growth temp (°C) | Resident virB plasmidb % | Loss of virulence plasmid (Kms) in 100 generationsc |
|---|---|---|---|
| pALA2558 | 30 | pATM278 | <2 |
| pALA2558 | 30 | pATM324 virB | <2 |
| pALA2558 | 37 | pATM278 | 95 |
| pALA2558 | 37 | pATM324 virB | >98 |
The incoming plasmid was a chloramphenicol-resistant derivative of plasmid pGB2, which carries the mvp locus.
pATM324 has the virB gene cloned into the plasmid vector pBAD18, which has an l-arabinose-inducible promoter under the control of araC. pATM278 is similar but lacks the virB gene.
The loss of kanamycin resistance carried by the virulence plasmid was measured after 100 generations of growth at the stated temperature in LB broth supplemented with 0.2% l-arabinose.
DISCUSSION
Inherent instability of the virulence plasmid at 37°C.
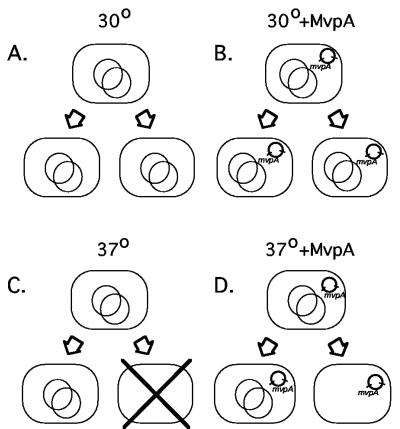
Our interpretation of the data presented here is shown in Fig. 5. It is based on the principles that have been established experimentally using a model mvp-containing plasmid in E. coli (3). At 30°C, the core plasmid maintenance systems of the virulence plasmid are efficient. Very few plasmid-free cells are produced, and the mvp PSK system rarely comes into play (Fig. 5A). When the mvp system is effectively switched off by supplying an additional copy of the mvpA gene in trans, the maintenance of the virulence plasmid is not measurably compromised (Fig. 5B). At 37°C, the core maintenance systems of the virulence plasmid become ineffective, due to down-regulation or temperature sensitivity of some key component or to induction of some function that damages the plasmid. Many cured cells are produced. These are killed by the MvpT toxin, which persists after the plasmid is lost (Fig. 5C). This causes a considerable growth disadvantage to the population (Fig. 1C). When the mvp system is switched off by supplying an additional copy of mvpA, plasmid-free cells rapidly accumulate in the 37°C population because they are not killed (Fig. 5D). This population can grow normally (Fig. 1C).
FIG. 5.
Basis for reliance on mvp for virulence plasmid maintenance at 37°C. The mvp PSK system of the large virulence plasmid (large circles) can effectively be switched off by supplying the antidote protein MvpA from a second plasmid (small bold circles). (A) At 30°C, maintenance of the virulence plasmid (large circles) is good, and very few cured cells are formed. (B) Switching off the mvp system has little effect at 30°C, because there are very few cured cells to kill. (C) At 37°C, virulence plasmid maintenance is poor, cured cells arise frequently, and these are killed by the mvp PSK system. (D) When the second plasmid expressing MvpA is introduced, the mvp system can no longer kill the cured cells that arise at 37°C. Cured cells rapidly accumulate in the population.
The identity of the component or components that compromise plasmid maintenance at 37°C is unknown. However, they do not appear to involve the virB regulon or inherent temperature sensitivity of one of the two plasmid partition systems, pWR100par (K. Sergueev and S. Austin, unpublished observations).
The virulence-dependent genetic instability previously described.
Many of the virulence genes of the plasmid are located in a 31-kb invasion region containing the ipa mxi and spa genes. These are positively regulated by the factor virB. Genes under the control of virB are expressed at 37°C but are repressed at 30°C (8). During growth at 37°C, some spontaneous variants arise on long-term culture that have lost the ability to express the virulence genes, either by plasmid mutation or by complete loss of the plasmid (16). This genetic instability reflects a selective disadvantage of virulence gene expression. We avoided this selective effect in our studies by limiting growth of cells with the wild-type plasmid to 20 generations at 37°C, or by using mxiM mutant plasmid that is not subject to selection because the cells are avirulent (see Materials and Methods). However, we can now extend the interpretation of this genetic instability effect, given our findings. The virulence plasmid is inherently unstable at 37°C. This effect is independent of the virB regulon. The cells lose the plasmid at a rate of 2 to 3% per generation. However, these cured cells are efficiently killed by the action of mvp, which remains active at 37°C. Very few cured cells survive. These remain virtually undetectable unless the cell population is virulent. In this case, the few cured cells have a growth advantage and eventually outgrow the population after extended growth (16). Because the original surviving cells are very rare, the outgrowing, avirulent population includes a high proportion of cells that retain the plasmid but have lost virulence by spontaneous mutation. Although the inherent instability of the plasmid is independent of virB control, the selection and eventual outgrowth of the rare cured cells that survive from the virulent population do depend on virB, because virB controls virulence.
Implications for the pathogenic lifestyle.
The use of PSK as a major component for the maintenance of a naturally occurring plasmid is a completely unexpected phenomenon. Low-copy-number plasmids encode highly efficient replication and partition systems that ensure that PSK rarely comes into play. This is necessary because, although effective, PSK is an expensive strategy for preventing the accumulation of cured cells in the population. This is well illustrated in the growth competition experiment described in Fig. 1C: mvp is effective in preventing the accumulation of cured cells, but the cells are at a major growth disadvantage to cells in which mvp is disabled.
Virulent Shigella cells infect their human host at 37°C. If we assume that the properties of the plasmid are not grossly altered in the environment of the human host, virulence plasmid retention presumably depends on mvp killing during infection. Why would the virulence plasmid become unstable at the very temperature at which the functions that it carries are needed, and why should the bacterial cells tolerate a system that continuously kills a portion of the bacterial cell population during virulent infection? Perhaps the loss of the plasmid and death of part of the infecting bacterial population is not deleterious to the infection process or the transmission of the organism to other hosts. It may even be advantageous. Unrestricted exponential growth of the pathogen might kill the host prematurely and limit transmission. Also, it is possible that dead cells provide some advantage to the pathogen. For example, an accumulation of dead bacterial cells might help to encapsulate the infection, providing a barrier to infiltration by the immune system, or it might facilitate release of toxins into the host tissues that aid the infection process.
Acknowledgments
We thank Tony Maurelli for guidance and use of facilities during the performance of the Congo Red assays and for his generous gifts of avirulent S. flexneri strain BS547 and plasmids pATM278 and pATM324.
REFERENCES
- 1.Abeles, A. 1986. P1 plasmid replication. Purification and DNA-binding activity of the replication protein RepA. J. Biol. Chem. 261:3548-3555. [PubMed] [Google Scholar]
- 2.Boe, L., K. Gerdes, and S. Molin. 1987. Effects of genes exerting growth inhibition and plasmid stability on plasmid maintenance. J. Bacteriol. 169:4646-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brendler, T., L. Reaves, and S. Austin. 2004. Interplay between plasmid partition and postsegregational killing systems. J. Bacteriol. 186:2504-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 5.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 6.Formal, S. B., G. J. Dammin, E. H. LaBrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 8.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect. Immun. 43:397-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 11.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radnedge, L., M. A. Davis, B. Youngren, and S. J. Austin. 1997. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 179:3670-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts, R. C., and D. R. Helinski. 1992. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J. Bacteriol. 174:8119-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayeed, S., L. Reaves, L. Radnedge, and S. Austin. 2000. The stability region of the large virulence plasmid of Shigella flexneri encodes an efficient post-segregational killing system. J. Bacteriol. 182:2416-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuch, R., and A. T. Maurelli. 1999. The mxi-Spa type III secretory pathway of Shigella flexneri requires an outer membrane lipoprotein, MxiM, for invasin translocation. Infect. Immun. 67:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuch, R., and A. T. Maurelli. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect. Immun. 65:3686-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuch, R., R. C. Sandlin, and A. T. Maurelli. 1999. A system for identifying post-invasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway of Shigella flexneri in intercellular dissemination. Mol. Microbiol. 34:675-689. [DOI] [PubMed] [Google Scholar]
- 18.Sheen, J. 1997. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 19.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887-893. [DOI] [PubMed] [Google Scholar]
- 20.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267:836-837. [DOI] [PubMed] [Google Scholar]
- 21.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]