Abstract
Mycobacterium tuberculosis is the etiological agent of tuberculosis (TB), which kills approximately 2 million people a year despite current treatment options. A greater understanding of the biology of this bacterium is needed to better combat TB disease. The M. tuberculosis genome encodes as many as 15 adenylate cyclases, suggesting that cyclic AMP (cAMP) has an important, yet overlooked, role in mycobacteria. This study examined the effect of exogenous cAMP on protein expression in Mycobacterium bovis BCG grown under hypoxic versus ambient conditions. Both shaking and shallow standing cultures were examined for each atmospheric condition. Different cAMP-dependent changes in protein expression were observed in each condition by two-dimensional gel electrophoresis. Shaking low-oxygen cultures produced the most changes (12), while standing ambient conditions showed the fewest (2). Five upregulated proteins, Rv1265, Rv2971, GroEL2, PE_PGRS6a, and malate dehydrogenase, were identified from BCG by mass spectrometry and were shown to also be regulated by cAMP at the mRNA level in both M. tuberculosis H37Rv and BCG. To our knowledge, these data provide the first direct evidence for cAMP-mediated gene regulation in TB complex mycobacteria.
Tuberculosis (TB) is a leading cause of human death by an infectious agent, killing approximately 2 million people each year. Nearly one-third of the world's population is latently infected with the Mycobacterium tuberculosis bacillus, and 7 to 8 million new TB cases occur annually (49). A deadly synergy with the human immunodeficiency virus epidemic and an increasing proportion of drug-resistant cases contribute to TB's recent resurgence and complicate its treatment (7, 9). A better understanding of M. tuberculosis biology is needed to devise more effective treatments against TB. Gene regulation is critical for the interaction of the tubercle bacilli with their environment, and deciphering these key regulatory pathways remains a major challenge in the field. Sequencing of the M. tuberculosis genome extended TB research onto a global scale, allowing researchers to take advantage of techniques in the realm of functional genomics, primarily those involving proteomics and transcriptomics.
Global gene regulation studies in M. tuberculosis to date have focused almost exclusively on the regulatory roles of two-component systems and sigma factors (10, 23, 31, 33, 34, 44, 51, 59). Two-component systems, consisting of a membrane-bound sensor protein and a cytoplasmic effector protein, represent a conserved mechanism among bacteria for regulating gene responses to external stimuli (3). The mycobacterial genome encodes 11 complete two-component systems, four isolated regulators, and three isolated sensors, some of which are necessary for virulence and for early phases of intracellular growth (8, 23, 31, 44, 51). The M. tuberculosis genome also encodes 13 alternative sigma factors (8). Mutations in some of these sigma factors cause increased sensitivity to a variety of environmental conditions and also attenuated virulence (8, 10, 33, 34).
Cyclic AMP (cAMP) is an important signaling molecule that controls a wide variety of cellular functions in many organisms, including virulence factors from a diverse range of pathogens (4, 12, 18, 46, 55, 58). cAMP is produced in the cell by adenylate cyclases (ACs) (45). A recent study used Bayesian computational methods to identify 15 putative cyclases in the M. tuberculosis genome (35). All candidate open reading frames contained a cyclase homology domain associated with one of a variety of cytoplasmic, receptor-type, and integral membrane structural motifs (35). Two of these genes (Rv1625c and Rv2435c) belong to the mammalian type adenylyl cyclase grouping, and both enzymes are active (19, 28, 35, 50, 54). Functionality has also been shown for the mycobacterial cyclases Rv1264, Rv1318c, Rv1319c, Rv1320c, and Rv3645 (6, 27).
Little is known about the role of cAMP in mycobacteria, although it is found in both fast- and slow-growing, as well as pathogenic and nonpathogenic, species (42, 43). A correlation between intracellular cAMP levels and phospholipid synthesis in Mycobacterium smegmatis has been reported (22). Elevated cAMP levels cause significant increases in cardiolipin and phosphatidylinositol mannosides (22). Additionally, it has been reported that cAMP levels in macrophages increase upon uptake of live Mycobacterium microti and Mycobacterium bovis BCG. This increase in cAMP correlated with a decrease in phagosome lysosome fusion (30). cAMP-mediated gene expression in mycobacteria has not been examined. The present study explores the possible role of cAMP signaling as a third global regulatory system in TB complex mycobacteria.
We have examined the proteome of M. bovis BCG, with and without exposure to exogenous cAMP, to identify cAMP-regulated proteins. We report here that cAMP causes increased expression of at least 15 proteins, including Rv1265, Rv2971, GroEL2, PE_PGRS6a, and malate dehydrogenase (MDH). This regulation occurs more often under low-oxygen, CO2-enriched growth conditions than in ambient air, indicating a possible physiological niche for cAMP regulation during host infection.
MATERIALS AND METHODS
Bacterial cultures.
M. bovis BCG (Pasteur strain; Trudeau Institute) and M. tuberculosis H37Rv (ATCC 25618) were grown in 7H9 liquid medium (Difco) supplemented with 0.5% glycerol, 10% oleic acid-dextrose-albumin-catalase, and 0.05% Tween 80. Cultures were grown in a controlled atmospheric incubator (series II; Forma Scientific, Inc.) as described elsewhere (14) at a maximum depth of 2 mm under conditions of ambient air (20% O2, 0% CO2), low oxygen (1.3% O2, 5% CO2), or ambient oxygen plus CO2 (20% O2, 5% CO2) in 25-cm2 tissue culture flasks, either left standing undisturbed or shaking with constant agitation (15, 47). Where appropriate, 10 mM dibutyryl cAMP (dbcAMP; Sigma) or 5 to 20 mM butyrate (Sigma) was added on day 3 of culture during early log-phase growth.
Sample preparation and one-dimensional (1-D) SDS-PAGE.
Whole-protein extracts were obtained from 7-day late log-phase cultures. Cells were harvested and washed with Dulbecco's phosphate-buffered saline plus 1% (vol/vol) Tween 80. Cells were lysed in 0.3% sodium dodecyl sulfate (SDS), 200 mM dithiothreitol (DTT), and 30 mM Tris-HCl (pH 8.0) plus 1% protease inhibitor (Sigma) by heating at 80°C for 20 min. Cellular debris was pelleted, and protein was quantitated with the NanoOrange protein quantitation kit (Molecular Probes) following the manufacturer specifications. Fifty micrograms of protein was loaded onto SDS-12% polyacrylamide gel electrophoresis (PAGE) gels and electrophoresed at 50 mA for 2 h and then at 75 mA for 2.5 h. Gels were silver stained (38), and the banding patterns were compared visually.
Sample preparation and 2-D gel electrophoresis of M. bovis BCG proteins.
Bacteria were harvested by centrifugation and washed three times with Dulbecco's phosphate-buffered saline plus 0.2% (wt/vol) EDTA and 1% (vol/vol) protease inhibitor cocktail (Sigma). Cells were resuspended in Tris-SDS buffer (0.3% [wt/vol] SDS, 50 mM Tris-HCl [pH 8.0]) and lysed as described previously (14). Cellular debris was pelleted at 50,000 × g, and protein samples were precipitated with 10% trichloroacetic acid.
Protein samples were separated by 2-D SDS-PAGE as described previously (15). Briefly, ≈200 μg of protein was separated by isoelectric focusing in polyacrylamide tube gels of 1.5 mm (inner diameter [i.d.]), 15 cm (length). Focusing was performed using a constant voltage of 667 V over 18 h. The second dimension was run on SDS-12% PAGE gels 16 by 20 cm and 1.5 mm thick. Gels were stained either by standard silver staining methods or SYPRO Ruby fluorescent dye (Molecular Probes), and spot patterns were compared visually.
For mass spectrometry (MS) samples, preparative 2-D gels were run with 500 μg of protein focused on isoelectric focusing tubes 3 mm thick, 13 cm long at 667 V over 18 h and separated in the second dimension on 3-mm-thick 16- by 20-cm 12% polyacrylamide gels. Gels were stained with Coomassie brilliant blue stain (Bio-Rad) or SYPRO Ruby (Molecular Probes).
Protein spot quantitation.
Protein spots of interest were quantitated from SYPRO Ruby stained 2-D gels run as described above. Only spots that differed reproducibly in three biologically independent repeats were analyzed. Gels were visualized using a FluorImager 595 (Molecular Dynamics) at 488 nm with excitation filter 610RG and analyzed with the ImageQuant software package (Molecular Dynamics). Gels were normalized on the basis of total fluorescence to correct for differences in staining intensity and/or protein loading between gels.
MS analysis.
Protein spots of interest were cut from 2-D PAGE gels and prepared as described previously (15) for analysis by ion trap nano-liquid chromatography (LC)-nanoelectrospray MS. Peptides obtained from trypsin digestion were loaded onto a Finnigan LCQ Deca MS machine fitted with a Dionex microautosampler coupled to a 75-μm (i.d.) reverse-phase chromatography system (PicoFrit). Tandem MS (MS/MS) spectrum data obtained from automatically selected ions were used to search the NCBI nonredundant database using the SEQUEST database search program with a species-limited search filter for Mycobacterium species.
Alternatively, protein spots were prepared for analysis by tandem quadrupole time-of-flight MS by dehydrating with 50% CH3CN-200 mM NH4HCO3 followed by in-gel reduction and alkylation by incubation with 10 mM DTT-100 mM NH4HCO3 at 56°C for 30 min and then 55 mM iodoacetamide-100 mM NH4HCO3 for 20 min. Gel pieces were rehydrated and digested overnight in 50 mM NH4HCO3 and either 12.5 ng of trypsin or chymotrypsin (Boehringer Mannheim sequence grade)/μl. Peptides were extracted with one wash of 20 mM NH4HCO3, followed by three washes of 5% formic acid-50% acetonitrile.
These peptides were analyzed on a nanoelectrospray LC quadrupole time-of flight 2 tandem MS machine equipped with a desalting column packed with R2 stationary phase (Applied Biosystems), in series with a nanoscale C18 column (75 μm [i.d.] by 110 mm) packed with Beta basic C18 resin (TermoHypersil Keystone). MS/MS data were obtained from automatically selected ions and used to search the NCBI nonredundant database using the Mascot search program (Matrix Science).
RNA extraction.
M. bovis BCG or M. tuberculosis H37Rv cultures were pelleted, washed with 0.5% Tween 80, and resuspended in RNase-free water. RNA was extracted as described previously (32). Briefly, cells were disrupted mechanically using a bead beater (BioSpec Products) with two rounds of beating on high for 100 s in a mixture of 0.1-mm zirconia-silica beads (BioSpec Products), 45% Divolab no. 1 (Diversey), 45% acid phenol, and 10% chloroform-isoamyl alcohol (24:1). RNA was reextracted with an equal volume of chloroform-isoamyl alcohol and precipitated with isopropanol and 3 M sodium acetate (pH 5.2). RNA was resuspended in RNase-free water, and DNA contamination was removed using the RNeasy Mini kit following manufacturer specifications (QIAGEN).
RT-PCR.
cDNA was prepared for use in reverse transcriptase PCRs (RT-PCRs). A 0.5-μg aliquot of RNA was incubated with 0.125 μg of random primers RPA00, RPT00, RPC00, and RPG00 (Table 1) at 100°C for 1 min, followed by 65°C for 5 min. A mixture of 10 mM dNTP, 0.1 M DTT, RNase Out (Invitrogen), 5× first-strand buffer (Invitrogen), and RT Superscript III (Invitrogen) was added to the RNA-random primer mixture and incubated at 25°C for 10 min, 42°C for 1 h, and 70°C for 15 min. RNA levels for genes of interest were examined through a series of cDNA dilutions (0 to 1:1,000) run in PCRs for 10, 20, and 30 cycles at 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min. This ensured that the conditions chosen for quantitation were in the linear range of the PCR. Five microliters of a 1:100 dilution for BCG, 1:30 dilution for H37Rv, was used for semiquantitative PCRs run for 30 cycles each. Control reactions were performed against 16S RNA using cDNA diluted 10−5 or 10−6. The 16S RNA PCR products from all growth conditions were normalized to one another before quantitation of individual genes, to ensure equal levels of starting RNA in each reaction. 16S RNA PCRs were also performed using total RNA without RT, to ensure the absence of DNA contamination.
TABLE 1.
Primers used for RT-PCRb
| Gene | Primer sequencea |
|---|---|
| Rv1265c | F, ATG TAA TCA TGT AAT TAT GAG GC |
| R, GCG ATA GTC ATT CGC TCCG | |
| fixB | F, CGA CAA ATA CCT GAT CAC CC |
| R, GAA GAT GCT GTG GAC ACC C | |
| GroEL2 3′ | F, CAT CGC CGG ACG AGT GGC |
| R, CGA TCT GCT TCA GCG GGG | |
| GroEL2 5′ | F, GTA AAG GTG ACA TTG GGC |
| R, CGT TTG AGA CCG AGC G | |
| PPE39- | F, CAC CGG ATT TTG GAA CGG CG |
| R, GCG TGC CCG GAT TAA ACC C | |
| tuf | F, GTG CGG AAG TAG AAC TGC GG |
| R, AGG AAG TTG AGA TCG TCG GC | |
| Rv2971 | F, GCG CAA TTG CAG CCT CCG |
| R, TTG TCC AGC CTG CC | |
| mdh | F, GGA TGA CGT TCG CGT CGG G |
| R, GAA TTC ATC CTC GAT CCA GGC | |
| PE_PGRS6 | F, GGA TTT GGC GGG TAT TGG G |
| R, CTG CTG CTC CAC GTT GGC | |
| PE_PGRS3 | F, TCA ATG TGG TGA ACG AGC CC |
| R, CGA CGC CTA TGG AGT TCC C | |
| PE_PGRS43 | F, GAC GGC GAT CTT CCT GGG |
| R, GAG CAC AGC ATT GAA ACCG | |
| 16S rRNA | F, GCG AAC GGG TGA GTA ACA CG |
| R, TGA AAG AGG TTT ACA ACC CG |
F, forward primer; R, reverse primer.
Random primers for cDNA synthesis were the following: RPA00, NNN NNA; RPG00, NNN NNG; RPC00, NNN NNC; RPT00, NNN NNT.
Western blotting.
2-D gels of interest were transferred to polyvinylidene fluoride membranes by semidry transfer using a TransBlot semidry transfer cell (Bio-Rad) at 20 V for 1 h. Membranes were blocked with 5% nonfat dry milk in 0.5% Tween 20, Tris-buffered saline (50 mM Tris-HCl [pH 7.6], 150 mM NaCl; TTBS). Monoclonal antibodies to GroEL, MC 4220 and MC 5205 (TB Research Materials and Vaccine Testing Contract), were used at a 1:2,500 dilution followed by horseradish peroxidase-conjugated goat anti-mouse secondary antibody at 1:5,000 in TTBS. The ECL detection kit (Amersham) was used to detect antibody bound to the membrane, and blots were exposed to Kodak BioMax MS-1 film (Sigma).
RESULTS
BCG culture conditions affect cAMP-mediated changes in protein expression.
We hypothesized that the presence of up to 15 ACs indicates an important role for cAMP in M. tuberculosis. An excess-cAMP model was established to explore this possibility, using methods adapted from a system developed to study cAMP virulence genes in fungal pathogens (1, 2). BCG was grown shaking under ambient conditions either in the presence or absence of exogenous cAMP for various lengths of time to optimize the system. 1-D SDS-PAGE was used to analyze total protein lysates from these bacteria. These optimization studies evaluated cAMP type (cAMP or dbcAMP), concentration (100 μm to 10 mM), and exposure times (1 h to 4 days). Conditions of 10 mM dbcAMP over 4 days were chosen as a result of these studies (data not shown). This 10 mM concentration is similar to exogenous cAMP concentrations reported in other models (1, 2, 11, 29).
We previously reported differences in M. tuberculosis and BCG gene expression between cultures grown under shallow standing versus shaking conditions (14, 15, 47). We reasoned that cAMP could have differing effects depending on the particular environmental conditions faced by the bacilli. Changes in protein expression caused by exogenous dbcAMP were examined when BCG was grown under a variety of culture conditions including standing in either ambient oxygen (20% O2, 0% CO2) or low oxygen (1.3% O2, 5% CO2) or shaking under either oxygen condition.
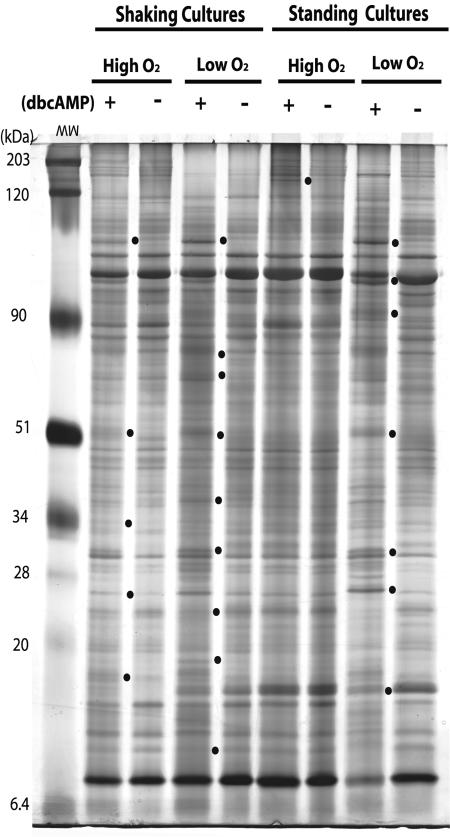
SDS-PAGE gels were used to identify banding pattern changes between extracts obtained from BCG grown either in the presence or absence of 10 mM dbcAMP under each condition. The number of banding pattern differences varied depending on culture conditions. At least nine differences were identified upon addition of dbcAMP to cultures grown shaking in low oxygen (Fig. 1). In contrast, cells grown standing in ambient oxygen had very few pattern changes in response to exogenous cAMP. One cAMP-dependent difference was observed in standing ambient cultures, five under shaking ambient air, and seven under standing low oxygen (Fig. 1). These results indicate that cAMP regulates gene expression in mycobacteria and that environmental factors such as hypoxia and/or CO2 can play a role.
FIG. 1.
1-D SDS-PAGE gel showing protein changes after addition of 10 mM dbcAMP to BCG cultures grown shaking or standing in 1.3% O2, 5% CO2 (low O2) or 20% O2, 0% CO2 (high O2). Banding pattern changes between cultures grown in the presence (+) or absence (−) of exogenous cAMP (10 mM) are marked by dots.
Identification of differentially expressed cAMP-dependent proteins in BCG.
2-D GE was performed to further characterize the differentially expressed proteins observed by 1-D SDS-PAGE analysis. Visual comparison of silver-stained 2-D gels was consistent with the 1-D SDS-PAGE results (Fig. 1), in that more proteins were regulated by dbcAMP in shaking low oxygen than under any other condition (Table 2). Expression of at least 12 proteins was upregulated by cAMP in shaking low oxygen, six under shaking ambient air, five under standing low oxygen, and two under standing ambient air. No proteins were detected at reduced levels in the presence of exogenous dbcAMP. Only protein spots that were observed on three biologically independent 2-D gel profiles were counted.
TABLE 2.
Proteins found to be differentially regulatedf by exogenous cAMP in BCG grown under various conditions
| Protein spot no.a | Shaking
|
Standing
|
||||||
|---|---|---|---|---|---|---|---|---|
| High O2d
|
Low O2e
|
High O2
|
Low O2
|
|||||
| +b cAMP | −b cAMP | + cAMP | − cAMP | + cAMP | − cAMP | + cAMP | − cAMP | |
| 1 | +c | −c | + | − | − | − | + | − |
| 2 | − | − | + | − | − | − | − | − |
| 3 | − | − | + | − | − | − | + | + |
| 4 | − | − | + | − | − | − | − | − |
| 5 | + | − | + | − | − | − | + | − |
| 6 | + | − | + | − | + | − | + | − |
| 7 | + | − | + | − | − | − | − | − |
| 8 | + | − | + | − | − | − | − | − |
| 9 | − | − | + | − | + | + | + | + |
| 10 | − | − | + | − | − | − | − | − |
| 11 | + | − | + | − | − | − | − | − |
| 12 | − | − | + | − | − | − | − | − |
| 13 | + | + | + | + | + | + | + | − |
| 14 | ? | ? | + | + | + | − | + | + |
| 15 | − | − | − | − | − | − | + | − |
Numbers correspond to protein spots from shaking low-oxygen 2-D gel profiles (Fig 2).
+, 10 mM exogenous cAMP added to cultures; −, no exogenous cAMP added.
+, presence of protein spot under given condition; −, absence; ?, could not be determined.
High-oxygen conditions: 20% O2, 0% CO2.
Low-oxygen conditions: 1.3% O2, 5% CO2.
Results in boldface indicate proteins differentially regulated by exogenous cAMP.
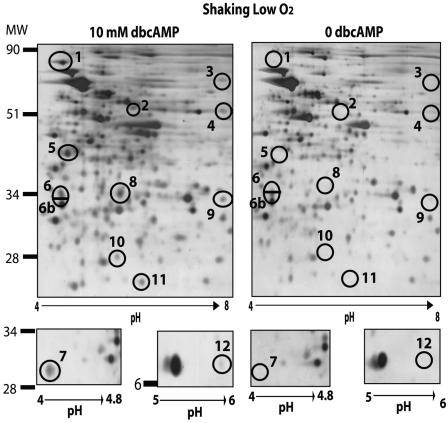
Shaking low-oxygen conditions were chosen for further studies on the identification and quantitation of cAMP-responsive, differentially expressed proteins (Fig. 2). Seven spots representing cAMP-regulated proteins were excised from Coomassie brilliant blue- or SYPRO Ruby-stained gels for identification by MS using either an ion trap nanoelectrospray mass spectrometer or an LC-quadrupole time-of-flight apparatus (Tables 3 and 4). Putative identities were successfully assigned to five of these protein spots (Table 5), as discussed below. A sixth protein, FixB, was also identified from constitutive spot 6b.
FIG. 2.
Representative 2-D gel profile of BCG grown shaking in low oxygen (1.3% O2, 5% CO2) in the presence or absence of 10 mM dbcAMP. Circled protein spots represent protein changes consistently seen upon addition of dbcAMP (10 mM) in three biologically independent experiments. Numbers correspond to protein spot numbers in Tables 2 to 5.
TABLE 3.
MS data, obtained from analysis on an ion trap nano LC-nanospray mass spectrometer, for identified protein spots from 2-D GE
| Spot no.a | Protein ID | Ion strengthb | MS/MS peptide matches |
|---|---|---|---|
| 5 | GroEL2 | 1.1e6 | gi|13879960 +7 (K)DETTIVEGAGDTDAIAGR |
| 2.0e6 | gi|13879960 +4 (K)QIAFNSGLEPGVVAEK | ||
| 1.5e6 | gi|13879960 +22 (P)DLLPLLEK | ||
| 6 | Rv2971 | 9.9e5 | gi|13882834 +2 (R)LIDTAYAYGNEAAVGR |
| 2.9e6 | gi|13882834 +4 (K)YVDAWGGMIQSR | ||
| 2.0e6 | gi|13882834 +4 (K)YVDAWGGM*IQSR | ||
| 6.5e6 | gi|13882834 +2 (R)LLDNPTVTSIASEYVK | ||
| 3.1e6 | gi|13882834 +2 (R)WNLQLGNAVVVR | ||
| 7.6e6 | gi|13882834 +4 (K)YVDAWGGM*IQSR | ||
| 8.6e5 | gi|13882834 +4 (K)YVDAWGGMIQSR | ||
| 4.5e6 | gi|13882834 +2 (R)VREDPLTYAGT | ||
| 6b | fixB | gi|31619801 +3 (R)GVGSAEN#FSVVEALADSLGAAVGASR | |
| 5.7e6 | gi|31619801 +3 (R)AGAVEAEPAAGAGEQVSVEVPAAAEN#AAR | ||
| 1.1e7 | gi|31619801 +3 (R)AAVDSGYYPGQFQVGQTGK | ||
| 6.5e6 | gi|31619801 +3 (M)AEVLVLVEHAEGALK | ||
| 3.1e6 | gi|31619801 +3 (R)AAVDSGYYPGQFQVGQTGK | ||
| 1.6e5 | gi|31619801 +3 (R)AGAVEAEPAAGAGEQVSVEVPAAAEN#AAR | ||
| 1.0e7 | gi|31619801 +3 (R)AAVDSGYYPGQFQVGQTGK | ||
| 2.8e7 | gi|31619801 +3 (R)ALGEPAAVVVGVPGTAAPLVDGLK | ||
| 5.7e6 | gi|31619801 +5 (K)VSAELITAAR | ||
| 8.1e6 | gi|31619801 +3 (K)IYVAESDLVDK | ||
| 4.0e6 | gi|31619801 +5 (R)IGSGLLVDVVDVR | ||
| 1.7e6 | gi|31619801 +3 (K)TIVAVNKDEEAPIFEIADYGVVGDLFK | ||
| 1.0e6 | gi|31619801 +3 (R)EPAVAGDRPELTEATIVVAGGR | ||
| 1.1e5 | gi|31619801 +3 (R)AGAVEAEPAAGAGEQVSVEVPAAAENAAR | ||
| 1.5e6 | gi|31619801 +3 (R)EPAVAGDRPELTEATIVVAGGR | ||
| 2.8e6 | gi|31619801 +3 (R)EPAVAGDRPELTEATIVVAGGR | ||
| 1.0e6 | gi|31619801 +3 (R)ALGEPAAVVVGVPGTAAPLVDGLK | ||
| 1.3e7 | gi|31619801 (K)VAPQLTEVIK |
Numbers correspond to protein spots from shaking low-oxygen 2-D gel profiles (Fig 2).
Ion scores of ≥106 are considered significant.
TABLE 4.
Mass spectrometry data, obtained from analysis by tandem quadrupole time-of-flight MS, for identified protein spots from 2-D gel electrophoresis
| Spot no.a | Protein ID | Ion scoreb | MS/MS peptide data |
|---|---|---|---|
| 5 | GroEL2 | 39 | QIAFNSGLEPGVVAEK |
| 78 | DETTIVEGAGDTDAIAGR | ||
| 27 | SALQNAASIAGLFLRREAVVADKPEK | ||
| 32 | VIGAGKPLLIIAEDVEGEALSTLVVNK | ||
| 7 | PE_PGRS6a | 14 | AGAAGG |
| 2 | AGGNGG | ||
| 11 | GVGGVGG | ||
| 9 | DSGGDGG | ||
| 17 | LFGNGG | ||
| 13 | DSGGDGG | ||
| 9 | LFGNGG | ||
| 20 | GAGGFGQ | ||
| 12 | GAGGFGQ | ||
| 12 | GAGGFGQ | ||
| 12 | GAGGFGQ | ||
| 20 | GAGGFGQ | ||
| 14 | AQQYQ | ||
| 8 | mdh | 96 | VAVTGAAGQIGYSLLFR |
| 11 | Rv1265 | 53 | QVLDTLVDAGVAR |
| 27 | LPGLESPEEESAAR |
Numbers correspond to protein spots from shaking low-oxygen 2-D gel profiles (Fig 2).
Extensive homology indicated by ion score: GroEL2 > 26; PE_PGRS6a (total score) > 48; mdh > 25; Rv1265 > 43.
TABLE 5.
Quantitation of cAMP-induced proteins from BCG cultures grown shaking in 1.3% O2, 5% CO2
| Spot no.a | Rv ID | Protein ID | Protein function/category | Fold induction
|
||
|---|---|---|---|---|---|---|
| Protein | BCG mRNAb | Mtb mRNAc | ||||
| 1 | PE_PGRS | PE/PE_PGRS superfamily | 7.9 ± 1.3 | |||
| 2 | 3.1 ± 0.3 | |||||
| 3 | 4.3d | |||||
| 4 | 5.5d | |||||
| 5 | Rv0440 | GroEL2 | Heat shock protein/chaperone | 10.7 ± 0.7 | 2.3 | 3.2 |
| 6 | Rv2971 | Rv2971 | 2,5 Diketo-d-gluconic acid reductase | 3.4 ± 0.1 | 4.4 | 2.6 |
| 7 | Rv0532 | PE_PGRS6a | PE/PE_PGRS superfamily | 24.4 ± 2.6 | 3.0 | 2.0 |
| 8 | Rv1220 | Mdh | MDH | 6.1 ± 0.2 | 5.7 | 3.6 |
| Rv0440 | GroEL2 | Heat shock protein/chaperone | 6.1 ± 0.2 | 2.3 | 3.2 | |
| 9 | 3.0 ± 0.4 | |||||
| 10 | 4.4 ± 1.3 | |||||
| 11 | Rv1265 | Rv1265 | Unknown function/upregulated in macrophages | 8.4 ± 0.7 | 8.3 | 3.8 |
| 12 | 5.8d | |||||
Numbers correspond to protein spots from shaking low-oxygen 2-D gel profiles (Fig 2).
Fold levels determined by semiquantitative RT-PCR on BCG RNA.
Fold levels determined by semiquantitative RT-PCR on Mtb H37Rv RNA.
Only one round of quantitation was done for these spots, due to localized streaking on 2-D gel profiles.
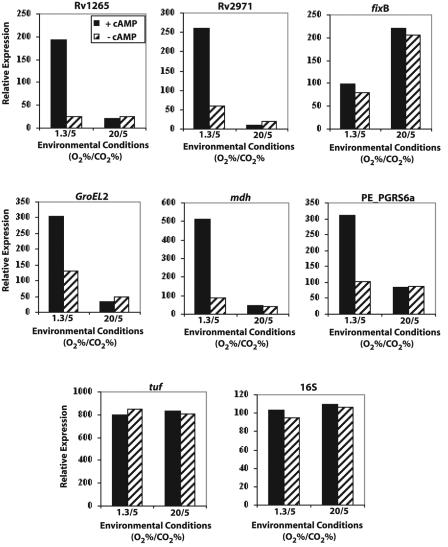
Semiquantitative RT-PCR was performed to determine the extent of transcriptional regulation for the gene corresponding to each cAMP-induced protein. All five genes whose expression was regulated at the protein level were also upregulated at the mRNA level, while fixB expression remained constitutive (Table 5; Fig. 3). Transcription of these genes was also examined when cultures were grown shaking in 20% O2, 5% CO2 to determine whether CO2 supplementation, rather than hypoxia, was causing the cAMP regulation that we observed. No cAMP-induced transcription was observed for these genes under high-O2 conditions, indicating that hypoxia is the important environmental condition in these studies (Fig. 3).
FIG. 3.
RT-PCR quantitation data indicating cAMP-mediated transcriptional regulation of differentially expressed proteins identified from 2-D GE studies. BCG cultures were grown with (solid bars) or without (hatched bars) exogenous cAMP (10 mM) shaking in 1.3% O2, 5% CO2 (1.3/5) or 20% O2, 5% CO2 (20/5). Genes examined included Rv1265, Rv2971, fixB, GroEL3, mdh, and PE_PGRS6a. Controls included tuf and normalized 16S RNA.
The results were as follows.
(i) Rv1265.
Protein spot 11 (Fig. 2) was identified as containing Rv1265, a putative open reading frame with unknown function (NCBI gi 7476706 ). Two peptide sequences from the trypsin-digested spot matched the Rv1265 sequence, providing approximately 12% coverage of the protein (Table 4). The predicted mass of this protein is 25.2 kDa and the pI is 7.11, which correspond to the migration of spot 11 on the 2-D gel (Fig. 2). A BLASTP search of the NCBI database found only two proteins with significant homology, and both were hypotheticals. Streptomyces avermitilins MA-4680 (gi 29834027 ) had 57% identity (81 of 142) and 68% positives (99 of 142), while Chloroflexus aurantiacus (gi 229729281 ) had 41% identity (42 of 101) and 61% positives (62 of 101). Neither match is informative with regards to the possible function of Rv1265.
RT-PCR data showed an 8.3-fold cAMP-dependent induction in the Rv1265 mRNA transcripts when cells were cultured shaking in low oxygen, but not under shaking 20% O2, 5% CO2 conditions (Fig. 3). mRNA levels correlated well with the observed protein change (Table 5) and indicated that cAMP regulation of Rv1265 expression is at the transcriptional level.
(ii) Rv2971.
Spot number 6 (Fig. 2) was identified as containing Rv2971, a putative oxidoreductase of the aldo/keto reductase family (NCBI gi 13882834 ). Eight trypsin-digested peptides matched the Rv2971 sequence, giving 23.8% protein coverage (Table 3). Rv2971 is a 282-amino-acid protein with a predicted mass of 30.3 kDa and pI of 4.7, both of which correspond to the location of spot 6 on the 2-D gel (Fig. 2). A search of the NCBI nonredundant database found significant homology to 2,5 diketo-d-gluconic acid reductase, which catalyzes the reduction of 2,5- diketo-d-gluconic acid to 2-keto-l-gulonic acid from a variety of bacteria (39). Rv2971 is an essential gene for M. tuberculosis laboratory growth, although its physiological role is unknown (53).
The close proximity of spot 6 to 6b, which was constitutively present, made it difficult to determine whether spot 6 contained a distinct cAMP-induced protein or a modified version of the protein in spot 6b. Protein spot 6b was identified as FixB (NCBI gi 31619801 ), a probable electron transfer flavoprotein subunit. Eighteen trypsin-digested peptides matched the FixB sequence, giving 65% protein coverage (Table 3). The size, 32 kDa, and pI, 4.5, of FixB were consistent with its location on the 2-D gel profile. The RT-PCR data also supported separate gene identifications for the proteins in each of these spots. Rv2971 was induced 4.4-fold after cAMP addition to shaking low-oxygen cultures (Fig. 3), while there was no change in the level of fixB (Fig. 3).
(iii) GroEL2.
Spot number 5 (Fig. 2) was identified as containing the 60-kDa chaperonin GroEL2 (NCBI gi 31617204 ). The predicted mass for GroEL2 is 56.7 kDa, with a pI of 4.85. Protein spot 5 on the 2-D gel migrated at the expected pI, but its apparent molecular weight (MW) of 35,000 is lower than that predicted for GroEL2. The identification of the protein in this spot was independently confirmed by a second MS laboratory because of this MW anomaly. In one case, three peptides were identified that matched the GroEL2 sequence with a protein coverage of 7.8% (Table 3). A second laboratory identified the same protein with four peptides matching the sequence and covering 16% of the sequence (Table 4). RT-PCR data on groEL2 expression further supported the MS identification, as groEL2 mRNA levels increased 2.3-fold in response to cAMP under shaking low-oxygen conditions (Fig. 3).
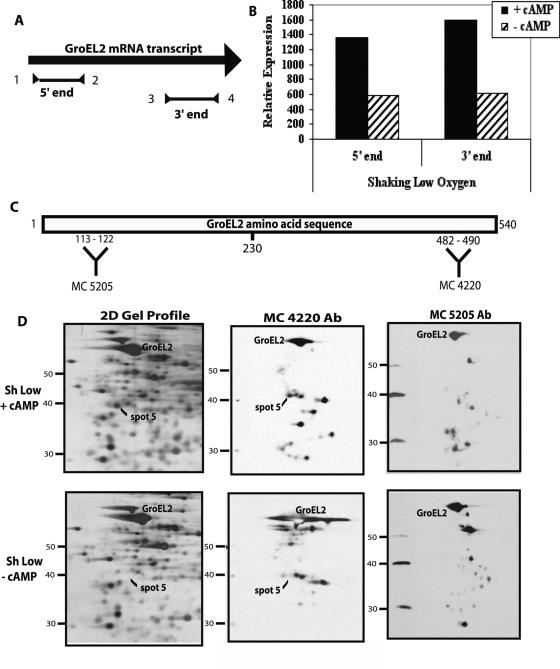
All of the trypsin-cleaved peptides that matched GroEL2 in the MS analyses were clustered toward the carboxy terminus of the protein, and the first matched peptide began at amino acid 229 (data not shown). Removal of the first 230 amino acids would result in a protein of 32.2 kDa with a pI of 4.94, as predicted from the ExPASY protein analysis website (http://www.expasy.org). A protein of this size correlates with the migration of protein spot 5. We reasoned that protein spot 5 could contain a cleavage product of the GroEL2 protein or a product from a smaller mRNA made from an alternative start site.
RT-PCR was used to compare groEL2 mRNA levels using primers targeted to either the 5′ or 3′ ends of the transcript (Fig. 4A; Table 1). More transcription from the 3′ end would suggest an alternate start site, leading to higher levels of RNA at the 3′ end. However, results were similar with both sets of primers, ruling out the alternate start site possibility (Fig. 4B).
FIG. 4.
Testing GroEL2 alternate transcriptional start site versus protein cleavage models. (A) Schematic of the GroEL2 mRNA transcript indicating the RT-PCR products obtained from the 5′ end (primers 1 and 2) or 3′ end (primers 3 and 4). (B) RT-PCR quantitation data indicating cAMP-mediated transcriptional upregulation from both the 5′ and 3′ ends of the GroEL2 transcript. BCG cultures were grown with (solid bars) or without (hatched bars) exogenous cAMP (10 mM) under shaking low-oxygen (1.3% O2, 5% CO2) conditions. (C) Schematic of the GroEL2 amino acid sequence, indicating the region of protein recognized by antibodies MC 4220 and MC 5205. (D) Western blot of shaking low-oxygen 2-D gel profiles (left panel) with (+) or without (−) exogenous cAMP (10 mM) and probed with either MC 4220 (middle panel) or MC 5205 (right panel) to detect possible cleavage products of GroEL2.
The cleavage product hypothesis was further explored by Western analysis. Western blot assays were performed using monoclonal antibodies that recognize the amino (MC 5205) or carboxy (MC 4220) terminus of GroEL. The amino acid sequences recognized by each antibody are also present in the GroEL2 protein sequence. Therefore, the antibodies recognize both GroEL and GroEL2 (Fig. 4C). MC 4220 detected protein spot 5 under conditions of exogenous cAMP, but MC5205 did not. Full-length GroEL2 was detected by both antibodies. The failure of MC5205 to detect protein spot 5 indicates that this spot contains a cleavage product of GroEL2 from which the amino terminal has been lost. As expected, neither antibody detected protein in spot 5 under conditions without exogenous cAMP (Fig. 4D). Both antibodies also recognized additional spots, consistent with the identification of GroEL and GroEL2 from a number of protein spots observed on various 2-D gel profiles (40, 52). In particular, a specific 48-kDa GroEL2 cleavage product has been reported under hypoxic conditions; however, this differs from the fragment we observed in the presence of cAMP (56).
(iv) PE_PGRS6a.
Protein spot 7 (Fig. 2) was identified as containing PE_PGRS6a (NCBI gi 31791714 ), a member of the PE multigene family, which includes nearly 100 genes with significant homology to one another (5). In approximately 65 of these genes the PE domain is linked to a polymorphic GC-rich repetitive sequence (PGRS) which is rich in glycine (40%) and alanine (25%) residues (5). Protein spot 7 did not digest well with trypsin, so chymotrypsin was used for MS analysis. Thirteen peptides matched the PE_PGRS6a sequence, with a coverage of 7% by amino acid count (Table 4). The location of spot 7 on the gel corresponded to a predicted mass of 40 kDa and a pI of 4.2, which are close to the 48 kDa, pI 4.0 of PE_PGRS6a.
PE_PGRS3, Rv3345, and PE_PGRS43 were also identified with significant scores, but none of these proteins' predicted MW or pI matched the location of protein spot 7 on the 2-D gel. RT-PCR data showed cAMP-responsive regulation only for PE_PGRS6a and not for any of the other three genes, confirming the PE_PGRS6a identification (data not shown). PE_PGRS6a mRNA levels were induced threefold by exogenous cAMP under shaking low-oxygen conditions (Fig. 3). This mRNA induction is approximately ninefold lower than the protein induction (Table 5), suggesting that additional posttranscriptional levels of regulation mediate expression of this protein.
(v) MDH.
Protein spot 8 (Fig. 2) was identified as containing MDH (NCBI gi 15608380 ), an enzyme of the tricarboxylic acid and glyoxylate shunt pathways (13). One trypsin-digested peptide matched the MDH protein sequence, giving 5% protein coverage with a highly significant ion score (Table 4). MDH is a 329-amino-acid protein with a predicted mass of 34.4 kDa and pI of 4.7, both of which correspond to the location of spot 8 on the 2-D gel (Fig. 2). The MDH identification was also supported by RT-PCR data showing a 5.7-fold increase in mdh mRNA levels under shaking hypoxic conditions (Fig. 3).
Regulatory effects are a result of cAMP, not butyrate.
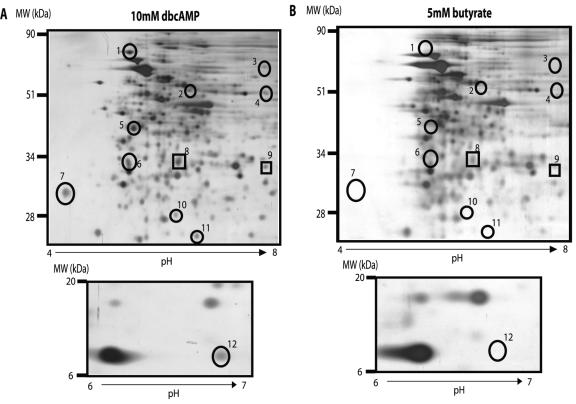
dbcAMP was used throughout this study because its increased hydrophobicity over cAMP allows it to more efficiently cross through the lipid cell wall. Butyrate is released from the dbcAMP molecule after it enters the cell, and free butyrate has been associated with gene regulatory effects (16, 41, 57). 2-D gel profiles of BCG grown shaking under low-oxygen conditions were examined after addition of 5 mM butyrate in place of dbcAMP to determine whether any of the protein changes we had observed were due to butyrate rather than cAMP. Expression of two proteins, numbers 8 (MDH) and 9, were partially upregulated by the addition of butyrate, showing approximately one-third of the cAMP-induced expression (Fig. 5). RT-PCR was also performed for all identified dbcAMP-regulated genes after the addition of 20 mM butyrate, in place of dbcAMP, to the BCG cultures. Butyrate at 20 mM did not affect the mRNA level of any genes tested, including mdh (data not shown). These data support the conclusion that the majority of observed regulatory effects are a result of the exogenous cAMP, and not of butyrate.
FIG. 5.
Representative 2-D gel of BCG cultures grown shaking in low oxygen (1.3% O2, 5% CO2) after a 4-day incubation with either 10 mM dbcAMP (A) or 5 mM butyrate (B). Circled protein spots indicate proteins upregulated upon addition of exogenous dbcAMP. Boxed spots indicate proteins where regulation is partially affected by the addition of exogenous butyrate, as well as by the addition of exogenous dbcAMP.
cAMP-regulated transcription in virulent M. tuberculosis H37Rv.
We were interested to determine whether the genes regulated by cAMP in BCG behaved similarly in the virulent M. tuberculosis strain H37Rv. RT-PCR was performed using mRNA obtained from cultures of H37Rv grown either shaking in low oxygen or standing in ambient air, with or without exogenous cAMP, as was done for BCG. All five cAMP-responsive genes identified from BCG also showed similar transcriptional regulation in H37Rv. Four genes, groEL2, mdh, Rv2971, and PE_PGRS6, showed very similar levels of induction for BCG and H37Rv. Rv1265 showed an induction level that was approximately one-half as strong in H37Rv as it was in BCG (Table 5). fixB expression was constitutive in M. tuberculosis, as it was in BCG.
DISCUSSION
Overview of the cAMP-regulated network of genes in BCG.
The goal of this study was to determine whether cAMP regulates gene expression in TB complex mycobacteria. While the physiological role of cAMP in mycobacteria is still unclear, this study provides the first evidence that cAMP regulates gene expression in mycobacteria and that this regulation is affected by bacterial growth conditions. On both 1-D and 2-D gels, protein patterns changed most noticeably under shaking low-oxygen conditions. Bacteria in these cultures were growing logarithmically under all conditions except standing low oxygen (15), so the observed gene expression changes were likely to be associated with specific adaptations to a CO2-rich, hypoxic environment rather than a growth arrest or generalized stress response. Based on RT-PCR data from 20% O2, 5% CO2 conditions, it is most likely the hypoxic component of this environment which is important for cAMP regulation. This suggests that cAMP may be used by the bacterium as a signaling molecule in response to the low-oxygen environments that the TB bacterium will face during its infection of a mammalian host.
The differences in cAMP regulation observed between ambient standing and low-oxygen shaking cultures were notable and were surprising to us. Previously we showed that genes of the ACG family regulate similarly in cultures grown shaking under hypoxic conditions or standing in ambient air (14, 15, 47). Standing cultures are only 2 to 3 mm deep, and we have suggested that a hypoxic microenvironment forms as the cells use up available oxygen faster than it is replenished by diffusion (14, 15, 47). Although it is likely that standing cultures contain other environmental components, these have been difficult to identify, as all genes studied to date are regulated similarly under hypoxic and standing conditions (14, 15). The difference between cAMP-dependent gene regulation in shaking low-oxygen and standing ambient cultures is the first evidence that shallow standing conditions are distinct from shaking hypoxic cultures and that factors besides hypoxia affect gene expression in the standing microenvironment. This cAMP regulation model will allow us to identify additional environmental factors that affect mycobacterial gene regulation in standing cultures that may be relevant to infection.
It is likely that the 15 proteins that were consistently upregulated in this study represent only a small subset of cAMP-regulated genes, because bacteria grown without exogenous cAMP still contained normal levels of endogenous cAMP. This native cAMP could render many genes insensitive to the effects of the added exogenous cAMP. This limitation might be overcome in future studies involving the manipulation of M. tuberculosis phosphodiesterase expression to modulate cAMP levels within the bacterium. An additional possibility is that the cAMP response occurs during a short-lived window and that the peak response was missed at the 4-day time point. This would also result in underestimation of the total number of cAMP-regulated genes. Lastly, the relative insensitivity of analyzing total proteins, rather than de novo-synthesized proteins, would reduce the number of proteins identified and could explain the absence of downregulated proteins in this study. Downregulated proteins would not be identified until existing protein was sufficiently degraded. It is also possible that genes downregulated by cAMP may already be shut off by native cAMP.
cAMP regulation in macrophages.
Two out of the five genes identified in this study have previously been reported to be upregulated during intracellular growth in macrophages. Hobson et al. observed an 8-fold transcriptional upregulation for Rv1265, while Li et al. showed a 3.9-fold increase in groEL2 transcription after 24-h macrophage infection (21, 24). In both cases, these macrophage induction levels are similar to the levels of cAMP-induced transcription that we observed in this study (8.3- and 2.3-fold, respectively). Additionally, two homologous members of the PE_PGRS superfamily have been identified as necessary for Mycobacterium marinum strains to replicate in macrophages (48). PE_PGRS6a, also a member of the PE_PGRS superfamily, was identified in this study as being upregulated by cAMP, although its function is unknown. Together, these data suggest a correlation between cAMP regulation and intracellular growth, which will be explored in future studies.
In other infectious organisms this correlation has already been established. Mutants of the airborne fungal pathogen Aspergillus fumigatus deficient in AC or G-protein subunits show decreased survival after ingestion by human monocyte-derived macrophages (26). These mutants also show reduced expression of a virulence factor, pksP, known to decrease phagolysosome fusion and protect against reactive oxygen species (26). This is consistent with a previous report that elevated cAMP levels correlated with reduced phagolysosome fusion during mycobacterial infection of macrophages (30).
M. tuberculosis bacteria are expected to face low-oxygen, high-CO2 conditions similar to those used in this study after ingestion by macrophages, as the intracellular environment of the macrophage is approximately 2.6% O2, 6.6% CO2 (20). These conditions may provide a regulatory signal for the bacterium, which is relayed by cAMP as a second messenger, to effect regulation of genes that are needed to survive during intracellular growth or later in infection.
cAMP and persistence.
A second low-oxygen environment the bacteria encounter during infection is in a granuloma during latency. The conditions within the granuloma are not well characterized, but it is thought to provide a low-oxygen, low-nutrient environment where the bacteria exist in an undefined persistent state (25, 37). M. tuberculosis's ability to establish a chronic, persistent infection involves a metabolic shift to C2 substrates, which are obtained readily from the host by β-oxidation of fatty acids (55). This metabolic shift occurs by bypassing steps of the tricarboxylic acid cycle via the glyoxylate shunt.
Isocitrate lyase, the key enzyme in the glyoxylate shunt, is upregulated during macrophage infections, and disruption of the icl gene in M. tuberculosis attenuated bacterial persistence in immune-competent mice (17, 36). No change in growth during acute infection was observed (36). Isocitrate lyase produces glyoxylate from isocitrate, which is then acted on by malate synthase to produce malate (13). MDH converts malate to oxaloacetate, which is then able to enter gluconeogenesis, allowing the cell to build six-carbon sugars from two-carbon sources (13). Increased levels of MDH may be necessary to maintain a stoichiometry of reactants to products to keep the glyoxylate shunt enzyme reactions going forward. We found that mdh expression levels were increased by cAMP in shaking low-oxygen cultures. This cAMP link with mdh regulation indicates a possible role for cAMP in persistence of M. tuberculosis infection by regulation of the glyoxylate shunt metabolism and is consistent with the increased effects of cAMP under hypoxic conditions.
Conclusions and future directions.
To our knowledge, this report presents the first evidence that cAMP has a gene regulatory role in TB complex mycobacteria. This regulation occurs at both the transcriptional and posttranscriptional levels in virulent M. tuberculosis and BCG. cAMP-regulated genes include several associated with biologically relevant conditions, from growth essentiality (Rv2971) to intramacrophage regulation (Rv1265, groEL2) and stress response (groEL2). This study also raises many questions regarding the potentially complex nature of cAMP-mediated signal transduction in M. tuberculosis. Additional work is needed to determine the full extent of cAMP signaling in M. tuberculosis, as it is likely that the genes identified in this study represent just a small portion of M. tuberculosis's cAMP-regulated network (M. A. Gazdik and K. A. McDonough, unpublished data).
Both the number and diversity of functional ACs in M. tuberculosis are extraordinary compared with other microorganisms (27, 35), and we think it likely that each cyclase is associated with a distinct signaling pathway. An exciting future challenge will be to determine how M. tuberculosis maintains discrete regulatory circuits using a single signal molecule and such a large number of ACs. This might be achieved by a combination of both temporal and spatial regulation of AC activity. For example, we expect that specific cyclases are activated to produce cAMP in response to different physiological conditions, such as hypoxia and/or the intramacrophage environment, as suggested by this study. Selective availability of cAMP-binding effector molecules under each condition would allow for different outcomes depending on the environmental condition. M. tuberculosis encodes 10 putative cNMP-binding proteins with diverse functions that could fill this role (35). Preliminary studies indicate that one of these proteins is a CRP-like DNA-binding protein that is likely to play an important role in cAMP-mediated gene regulation in M. tuberculosis (G. Bai, L. A. McCue, and K. A. McDonough, unpublished data). Subcellular localization of ACs within the bacterium could provide another level of regulation, particularly if specific cAMP-binding effector molecules colocalize with specific cyclases.
cAMP signaling may represent a third global regulatory system that should be considered in conjunction with sigma factors and two-component systems in M. tuberculosis. As the regulatory methods used by mycobacteria in response to environmental stimuli are determined, we will be better able to decipher the biology of this deadly organism and its response to the host environment during infection.
Acknowledgments
We thank Matt Florczyk and Guangchun Bai for helpful discussions and review of the manuscript and Damen Schaak for expert technical assistance. We also thank the Mass Spectrometry cores of the NYSDOH Wadsworth Center and University at Albany Center for Functional Genomics, along with the Molecular Genetics Core of the Wadsworth Center. We are grateful to the TB Resource Materials and Vaccine Testing Contract, NIH NIAID NO1 AI75320, at Colorado State University for providing antibodies.
This work was supported in part by National Institutes of Health grant AI4565801 (K.A.M.). M.A.G. also received support from a David Axelrod Fellowship from University at Albany and NIH training grant AI055429.
REFERENCES
- 1.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bijlsma, J. J., and E. A. Groisman. 2003. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 11:359-366. [DOI] [PubMed] [Google Scholar]
- 4.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, M. J., and G. Delogu. 2002. The PE multigene family: a “molecular mantra” for mycobacteria. Trends Microbiol. 10:246-249. [DOI] [PubMed] [Google Scholar]
- 6.Cann, M. J., A. Hammer, J. Zhou, and T. Kanacher. 2003. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J. Biol. Chem. 278:35033-35038. [DOI] [PubMed] [Google Scholar]
- 7.Cohn, D. L., F. Bustreo, M. C. Raviglione, et al. 1997. Drug-resistant tuberculosis: review of the worldwide situation and the W.H.O./IUATLD Global Surveillance Project. Clin. Infect. Dis. 24:1-30. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. R. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 10.DeMaio, J., Y. Zhang, C. Ko, D. B. Young, and W. R. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed] [Google Scholar]
- 13.Fernie, A. R., F. Carrari, and L. J. Sweetlove. 2004. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7:254-261. [DOI] [PubMed] [Google Scholar]
- 14.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florczyk, M. A., L. A. McCue, R. F. Stack, C. R. Hauer, and K. A. McDonough. 2001. Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect. Immun. 69:5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukamauchi, F., N. Mataga, Y. J. Wang, and D. M. Chuang. 1996. Differential effects of butyrate and dibutyryl cAMP on mRNA levels of muscarinic acetylcholine receptor subtypes expressed in neurohybrid cell lines. Neurosci. Lett. 212:49-52. [DOI] [PubMed] [Google Scholar]
- 17.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, A., M. Bouaboula, P. Casellas, J. P. Liautard, and J. Dornand. 2003. Subversion and utilization of the host cell cyclic adenosine 5′-monophosphate/protein kinase A pathway by Brucella during macrophage infection. J. Immunol. 170:5607-5614. [DOI] [PubMed] [Google Scholar]
- 19.Guo, Y. L., T. Seebacher, U. Kurz, J. U. Linder, and J. E. Schultz. 2001. Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. EMBO J. 20:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guyton, A. C. 1991. Textbook of medical physiology, 8th ed. W.B. Saunders Company, Philadelphia, Pa.
- 21.Hobson, R. J., A. J. McBride, K. E. Kempsell, and J. W. Dale. 2002. Use of an arrayed promoter-probe library for the identification of macrophage-regulated genes in Mycobacterium tuberculosis. Microbiology 148:1571-1579. [DOI] [PubMed] [Google Scholar]
- 22.Kaur, H., and G. Khuller. 1995. Role of cyclic adenosine monophosphate in phospholipid synthesis in Mycobacterium smegmatis ATCC 607. Lipids 30:345-349. [DOI] [PubMed] [Google Scholar]
- 23.Kendall, S. L., F. Movahedzadeh, S. C. Rison, L. Wernisch, T. Parish, K. Duncan, J. C. Betts, and N. G. Stoker. 2004. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinburgh) 84:247-255. [DOI] [PubMed] [Google Scholar]
- 24.Li, M. S., I. M. Monahan, S. J. Waddell, J. A. Mangan, S. L. Martin, M. J. Everett, and P. D. Butcher. 2001. cDNA-RNA subtractive hybridization reveals increased expression of mycocerosic acid synthase in intracellular Mycobacterium bovis BCG. Microbiology 147:2293-2305. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y. J., M. Petrofsky, and L. E. Bermudez. 2002. Mycobacterium tuberculosis uptake by recipient host macrophages is influenced by environmental conditions in the granuloma of the infectious individual and is associated with impaired production of interleukin-12 and tumor necrosis factor alpha. Infect. Immun. 70:6223-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebmann, B., S. Gattung, B. Jahn, and A. A. Brakhage. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420-435. [DOI] [PubMed] [Google Scholar]
- 27.Linder, J. U., A. Hammer, and J. E. Schults. 2004. The effect of HAMP domains on class IIIb adenylyl cyclases from Mycobacterium tuberculosis. Eur. J. Biochem. 271:2446-2451. [DOI] [PubMed] [Google Scholar]
- 28.Linder, J. U., A. Schultz, and J. E. Schultz. 2002. Adenylyl cyclase Rv1264 from Mycobacterium tuberculosis has an autoinhibitory N-terminal domain. J. Biol. Chem. 277:15271-15276. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Grp1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowrie, D. B., V. R. Aber, and P. S. Jackett. 1979. Phagosome-lysosome fusion and cyclic adenosine 3′:5′-monophosphate in macrophages infected with Mycobacterium microti, Mycobacterium bovis BCG or Mycobacterium lepraemurium. J. Gen. Microbiol. 110:431-441. [DOI] [PubMed] [Google Scholar]
- 31.Malhotra, V., D. Sharma, V. D. Ramanathan, H. Shakila, D. K. Saini, S. Chakravorty, T. K. Das, Q. Li, R. F. Silver, P. R. Narayanan, and J. S. Tyagi. 2004. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231:237-245. [DOI] [PubMed] [Google Scholar]
- 32.Mangan, J. A., K. M. Sole, D. A. Mitchison, and P. D. Butcher. 1997. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 25:675-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extracytoplasmic function sigma factor σE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCue, L. A., K. A. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10:204-219. [DOI] [PubMed] [Google Scholar]
- 36.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. J. Jacobs, and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 37.Mehrotra, J., and W. R. Bishai. 2001. Regulation of virulence genes in Mycobacterium tuberculosis. Int. J. Med. Microbiol. 291:171-182. [DOI] [PubMed] [Google Scholar]
- 38.Merril, C. R., D. Goldman, and M. L. Van Keuren. 1984. Gel protein stains: silver stain. Methods Enzymol. 104:441-447. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. V., D. A. Estell, and R. A. Lazarus. 1987. Purification and characterization of 2,5-diketo-d-gluconate reductase from Corynebacterium sp. J. Biol. Chem. 262:9016-9020. [PubMed] [Google Scholar]
- 40.Mollenkopf, H. J., P. R. Jungblut, B. Raupach, J. Mattow, S. Lamer, U. Zimny-Arndt, U. E. Schaible, and S. H. Kaufmann. 1999. A dynamic two-dimensional polyacrylamide gel electrophoresis database: the mycobacterial proteome via Internet. Electrophoresis 20:2172-2180. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, H., P. Rafiee, P. J. Fisher, N. A. Johnson, M. F. Otterson, and D. G. Binion. 2003. Butyrate modulates gene and protein expression in human intestinal endothelial cells. Biochem. Biophys. Res. Commun. 309:512-519. [DOI] [PubMed] [Google Scholar]
- 42.Padh, H., and T. A. Venkitasubramanian. 1977. Adenosine 3′,5′-monophosphate in mycobacteria. Life Sci. 20:1273-1280. [DOI] [PubMed] [Google Scholar]
- 43.Padh, H., and T. A. Venkitasubramanian. 1976. Adenosine 3′,5′ monophosphate in Mycobacterium phlei and Mycobacterium tuberculosis H37Ra. Microbios 16:183-189. [PubMed] [Google Scholar]
- 44.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423-1435. [DOI] [PubMed] [Google Scholar]
- 45.Peterkofsky, A., A. Reizer, J. Reizer, N. Gollop, P. P. Zhu, and N. Amin. 1993. Bacterial adenylyl cyclases. Prog. Nucleic Acid Res. Mol. Biol. 44:31-65. [DOI] [PubMed] [Google Scholar]
- 46.Petersen, S., and G. M. Young. 2002. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70:3665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purkayastha, A., L. A. McCue, and K. A. McDonough. 2002. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr gene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 70:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 49.Raviglione, M. C. 2003. The TB epidemic from 1992 to 2002. Tuberculosis (Edinburgh) 83:4-14. [DOI] [PubMed] [Google Scholar]
- 50.Reddy, S. K., M. Kamireddi, K. Dhanireddy, L. Young, A. Davis, and P. T. Reddy. 2001. Eukaryotic-like adenylyl cyclases in Mycobacterium tuberculosis H37Rv: cloning and characterization. J. Biol. Chem. 276:35141-35149. [DOI] [PubMed] [Google Scholar]
- 51.Rickman, L., J. W. Saldanha, D. M. Hunt, D. N. Hoar, M. J. Colston, J. B. Millar, and R. S. Buxton. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem. Biophys. Res. Commun. 214:259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenkrands, I., K. Weldingh, S. Jacobsen, C. V. Hansen, W. Florio, I. Gianetri, and P. Andersen. 2000. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis 21:935-948. [DOI] [PubMed] [Google Scholar]
- 53.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 54.Shenoy, A. R., N. Srinivasan, M. Subramaniam, and S. S. Visweswariah. 2003. Mutational analysis of the Mycobacterium tuberculosis Rv1625c adenylyl cyclase: residues that confer nucleotide specificity contribute to dimerization. FEBS Lett. 545:253-259. [DOI] [PubMed] [Google Scholar]
- 55.Smith, R. S., M. C. Wolfgang, and S. Lory. 2004. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Starck, J., G. Kallenius, B. Marklund, D. I. Anderrsson, and T. Akerlund. 2004. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150:3821-3829. [DOI] [PubMed] [Google Scholar]
- 57.Wiechmann, A. F., and M. A. Burden. 1999. Regulation of AA-NAT and HIOMT gene expression by butyrate and cyclic AMP in Y79 human retinoblastoma cells. J. Pineal Res. 27:116-121. [DOI] [PubMed] [Google Scholar]
- 58.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 59.Zahrt, T. C., C. Wozniak, D. Jones, and A. Trevett. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71:6962-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]