Abstract
Temperature-sensitive DNA polymerase mutants (dnaE) are protected from cell death on incubation at nonpermissive temperature by mutation in the cydA gene controlling cytochrome bd oxidase. Protection is observed in complex (Luria-Bertani [LB]) medium but not on minimal medium. The cydA mutation protects a thymine-deficient strain from death in the absence of thymine on LB but not on minimal medium. Both dnaE and Δthy mutants filament under nonpermissive conditions. Filamentation per se is not the cause of cell death, because the dnaE cydA double mutant forms long filaments after 24 h of incubation in LB medium at nonpermissive temperature. These filaments have multiply dispersed nucleoids and produce colonies on return to permissive conditions. The protective effect of a deficiency of cydA at high temperature is itself suppressed by overexpression of cytochrome bo3, indicating that the phenomenon is related to energy metabolism rather than to a specific effect of the cydA protein. We propose that filamentation and cell death resulting from thymine deprivation or slowing of DNA synthesis are not sequential events but occur in response to the same or a similar signal which is modulated in complex medium by cytochrome bd oxidase. The events which follow inhibition of replication fork progression due to either polymerase inactivation, thymine deprivation, or hydroxyurea inhibition differ in detail from those following actual DNA damage.
Unbalanced growth has been invoked as a cause of death after thymine starvation (6) and as a result of restricted DNA synthesis in temperature-sensitive mutants of the Escherichia coli replicative polymerase (29). However, an exact definition in molecular terms of what constitutes unbalanced growth has not been achieved. Because the end point of the process is cell death, it should be possible to define the gene products that play a role in the process in the same manner as the gene products that play a role in apoptosis have been defined (10, 15). It has been reported that the mazEF toxin-antitoxin system is involved in thymineless death (27). However, activation of the mazF riboendonuclease (36) is likely to be only an end point in a chain of reactions leading to cell death, similar to the chain of reactions leading to apoptotic death in eukaryotic cells. An important clue to the factors leading to thymineless death must be the observation that mutation of the recQ gene makes E. coli resistant to thymineless death (23). The recQ gene product is a helicase, likely involved in the repair of stalled DNA replication forks (35). These findings imply that events at a stalled replication fork set off a chain of events which lead to cell death. We have started to investigate the steps between inhibition of replication fork movement and the death of the cell in bacteria by identifying a gene, cydA, coding a subunit of the cytochrome bd oxidase which, when in mutant form, protects temperature-sensitive dnaE mutants from death (29). In this paper, we show that this mutation also protects a thymine-requiring mutant from thymineless death. However, this protection only occurs when cells are incubated in complex (Luria-Bertani [LB]) medium; there is no protection on minimal medium. Inhibition of cell division, resulting in filamentation in E. coli, is often associated with cell death. Using the cydA mutant, we also show that filamentation due to inhibition of DNA synthesis is not itself lethal. Finally, we show that the effect of cytochrome bd oxidase mutation is related to its role in energy metabolism rather than due to a new function for this gene. We propose that filamentation and cell death are not sequential events but that both occur in response to the same or a similar signal.
MATERIALS AND METHODS
Strains.
The bacterial strains used in this study are shown in Table 1. We used an allele of the cydA gene with a C→T mutation at nucleotide 770934, resulting in a Ser→Phe change at amino acid 85 (29). The E. coli Genetic Stock Center has assigned this mutation the designation cydA85. Plasmid pBR322 was purchased from Fermentas. Plasmid pTK-1 (16), including the cydA gene, and pJRcyOhis, including the cyO operon (25), were provided by R. Gennis. The Δthy deletion was obtained from strain KL742 (λ− thyA748::Tn10 rph-1 deo77) provided by H. Engelberg-Kulka.
TABLE 1.
Strains used
| Identifying genotype of strain | Characteristics | Reference and/or source |
|---|---|---|
| BS40 (dnaE+) | metB metD Δ(proAB lac) rpsL (Smr) | 31 |
| dnaE74 (BS40dnaE74) | metB metD Δ(proAB lac)dnaE74 rpsL (Smr) | 30 |
| Δthy | λ−thyA748::Tn10 rph-1 deo77 | E. coli Genetic Stock Center (K. B. Low) via H. Engelberg-Kulka |
| BS40Δthy | metB metD Δ(proAB lac) rpsL (Smr) thyA748::Tn1 | This paper; from Δthy by P1 transduction |
| dnaE74 cydA85 Tetr | metB Δ(proAB lac) rpsL (Smr) dnaE74 cydA85 nadA57::Tn10 | 29 |
| dnaE+cydA85 Tetr | metB metD Δ(proAB lac) rpsL (Smr) cydA85 nadA57::Tn10 | 29 |
| cydA85 Tets | metB metD Δ(proAB lac) rpsL (Smr) cydA85 | This paper; by selection (19) |
| Δthy cydA85 mutant | thyA748::Tn10 metB metD Δ(proAB lac) rpsL (Smr) cydA85 | This paper; from Δthy by transduction |
| dnaE74/pBR322 | metB Δ(proAB lac) dnaE74 (Ts) rpsL/pBR322 | By CaCl2 transformation (29) |
| dnaE74/pTK1 | metB Δ(proAB lac) dnaE74 (Ts) rpsL/pTK-1 | By CaCl2 transformation with plasmid from R. Gennis (29) |
| dnaE74/pcyO | metB Δ(proAB lac) dnaE74(Ts) rpsL/pcyO | This paper; by CaCl2 transformation with plasmid from R. Gennis |
Media and general methods.
The media and general microbiological techniques used were described by Miller (22). Thymine-requiring strains do not grow on LB medium; therefore, we supplemented complex medium with 100 μg of thymidine/ml. Because cyd mutants have a high mutation frequency (34), we prepared overnight cydA85 cultures by growth on minimal medium in which the appearance of revertants appears much less frequently. Flow cytometry experiments were performed using a Becton Dickinson LSRII Laser Scanner utilizing DiVa software. Overnight cultures were diluted 1:1,000 and incubated at nonpermissive conditions. The cells were then transferred to 27°C and incubated as indicated. Cephalexin (10 μg/ml), rifampin (150 μg/ml), and chloramphenicol (25 μg/ml) were added to prevent further cell division and initiation of new rounds of DNA synthesis, and incubation was continued for an additional 3 h. The cells were then harvested, resuspended in 10 mM Tris-HCl (pH 7.5), and fixed with 70% ethanol. Fixed cells were stored overnight at 4°C. For staining, the cells were centrifuged, washed with an equal volume of 10 mM Tris-10mM MgCl2, and concentrated to 50 μl per tube. DAPI (4′,6′-diamidino-2-phenylindole) solution was prepared in 10 mM Tris-10 mM MgCl2 supplemented with 0.1 M sodium citrate and 0.1% Triton X-100 to a concentration of 3 μg/ml. All solutions used for flow cytometry were filtered through a 0.2-μm-pore-size Millipore filter. Data were analyzed using the FlowJo program. DNA sequencing was done by the University of Chicago Cancer Research Center Sequencing Facility using Applied Biosystems capillary electrophoresis. PCR products were purified using a QIAGEN Qiaquick Spin purification kit. Protein extraction for Western blotting was carried out using Sigma CelLytic B lysis buffer (Sigma B3553) and Sigma protease inhibitor cocktail (Sigma P8465).
Preparation of cells for fluorescence microscopy.
Cells were fixed directly in the medium, essentially as previously described (8). Paraformaldehyde (16%; Sigma)-60 mM dipotassium phosphate (pH 7.5) and 25% glutaraldehyde (Sigma) were added to an appropriate volume of culture (1 to 20 ml) to final concentrations of 3.5%, 13 mM, and 0.32%, respectively. Cells were then allowed to fix at room temperature for 30 min, followed by an additional 30 min on ice. Fixed cells were then washed three times in cold phosphate-buffered saline (PBS) and resuspended in 10 to 50 μl of PBS. Ten-microliter aliquots of fixed cells were spread on polylysine-coated slides and allowed to sit for 10 min. The slides were then washed three times with cold PBS and allowed to dry. The cells remaining on the slide were covered with 10 μl of 0.2 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml and incubated 15 min before being washed three times with cold PBS and mounting with 50% glycerol in PBS and sealing with a coverslip. The slides were examined on an Axiovert 200 with a 100× oil immersion objective (numeric aperture = 1.4) and a DAPI and enhanced green fluorescent protein (eGFP) filter set. Images were captured with an ORCA ER CCD camera with fluorescence exposure times of 100 to 300 ms for DAPI and 5 to 10 s for eGFP, using Improvision's Openlab software. Images were processed using Openlab and Adobe Photoshop software. Image sequences were processed and converted to MPEG format using Slidebook and the National Institutes of Health Image program.
RESULTS
Protective effect of cydA85 mutations.
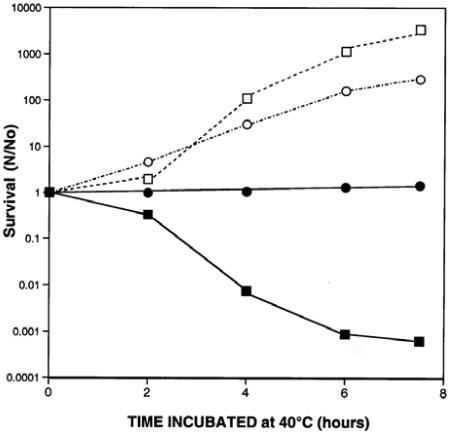
Mutation in the cydA85 gene coding for a subunit of cytochrome bd oxidase of E. coli protects cells carrying a temperature-sensitive mutation in the replicative DNA polymerase from death when incubated at nonpermissive temperature (29). However, by 24 h it is clear that cells of the dnaE74 cydA85 double mutant are protected from death only on LB medium; death of the suppressed strain occurs as rapidly as that of the dnaE74 mutant on minimal medium (Fig. 1). We have not identified the factors in LB which promote suppression (or in minimal medium which inhibit it). It has been reported that mutants of the cytochrome bd operon grow more slowly at temperatures above 37°C, although they do survive and form microcolonies (34). A dnaE+ cydA85 strain formed colonies on minimal agar plates at 40°C, and cultures incubated from frozen stocks at 27°C were able to grow in liquid minimal medium (supplemented with methionine, proline, and nicotinamide) at 40°C. CydA+ cells grow more rapidly than cydA85 cells on LB medium at 40°C but not at 27°C. After 24 h of incubation on LB plates, cydA85-carrying strains produce slightly smaller colonies than their cydA+ counterparts
FIG. 1.
The cydA85 mutation protects dnaE74 from death on LB but not on minimal medium. Cultures were grown overnight in minimal medium at 27°C, and the cells were harvested, washed with PBS, and plated on either LB plus streptomycin or on minimal medium supplemented with streptomycin, proline, l-methionine, and nicotinamide. Plates were incubated at 40°C for the indicated time and then were shifted to 27°C for an additional 24 to 48 h. The initial cell count for all samples was from 5.2 × 108 to 7 × 108 cells/ml. In a separate experiment we showed that the dnaE+ cydA85 mutant grows on minimal medium at 40°C. Open symbols, plated on minimal medium; filled symbols, plated on LB medium; squares, wild type; circles, dnaE74; triangles, dnaE74 cydA85.
Our initial hypothesis, that unbalanced growth resulted in cell death, was prompted by the similarity of the factors modulating death of the dnaE temperature-sensitive mutants to those modulating thymineless death (1). We therefore decided to test the effect of the cydA85 gene on thy mutant strains. We constructed a Δthy cydA85 strain by selecting a tetracycline-sensitive derivative of dnaE+ cydA85 (Tetr) using the fusaric acid selection technique (19). We sequenced the resulting isolated cydA85 (Tets) strain to make sure that the original ser85→phe cydA85 mutation had been retained. We then prepared a Δthy cydA85 (Tetr) strain by P1 transduction. We found that the cydA85 mutation protected from cell death during at least 7.5 h of incubation in LB medium (without added thymidine) at 40°C (Fig. 2). However, by 24 h of incubation, cell viability in the Δthy cydA85 strain had decreased to 1.1% of the original count. In agreement with the observation made with the dnaE temperature-sensitive mutant, the cydA85 mutant provided no protection from thymineless death on minimal medium (Table 2).
FIG. 2.
Protective effect of the cydA85 mutation on thymineless death on incubation at 40°C in LB medium. Overnight cultures were washed and resuspended in LB (filled symbols) or LB plus thymidine (open symbols) and incubated with shaking at 40°C for the time indicated before plating on LB plus thymidine plates. Squares, Δthy; circles, Δthy cydA85.
TABLE 2.
Protective effect of the cydA85 mutation in minimal and LB mediuma
| Mutation | Survival (N/N0) after 6 h at 37°C in
|
|
|---|---|---|
| Minimal medium | LB medium | |
| Δthy | 0.023 | 0.00049 |
| Δthy cydA85 mutant | 0.00083 | 3.6 |
Strain Δ(thy cydA85) was grown for 23 h at 27°C in 10 ml of minimal medium. The cells were harvested, washed, and resuspended in 2 ml of PBS. Ten microliters of suspension was added to 10 ml of the medium indicated and was incubated with shaking. Cultures were diluted and plated for viability count on LB plates incubated at 27°C.
Filamentation and DNA content.
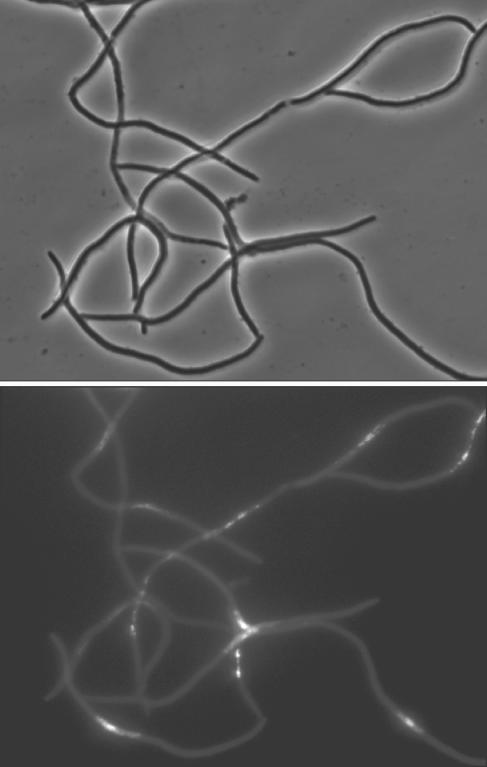
After 4 h of incubation at 40°C in LB medium, dnaE74 mutants formed long filaments, the cultures showed greatly diminished viability, and the DAPI-stained nucleoids look abnormal (Fig. 3) compared to those of cells grown under permissive conditions and to data in the literature (37). A pulsed-field electrophoresis study of the DNA extracted from dnaE74 after 4 h at 40°C in LB medium showed no sign of degradation (data not shown).
FIG. 3.
Formation of filaments by dnaE74. Cultures were grown overnight in LB medium at 27°C, harvested, diluted 1:1,000, and then incubated for 4 h (dnaE74). Cells were stained with DAPI. Top, phase microscopy; bottom, DAPI-stained fluorescence microscopy.
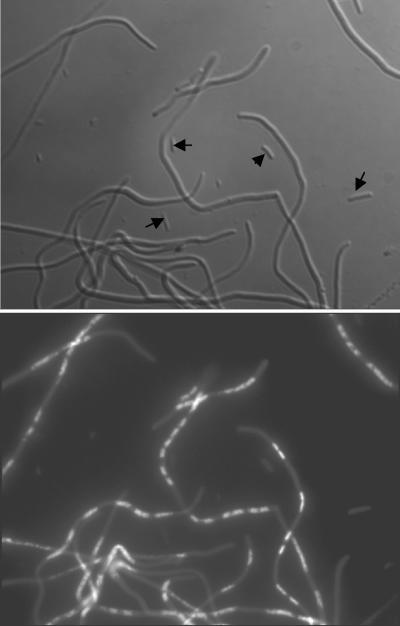
Is filamentation itself a cause of the cell death? Although dnaE74 cydA85 cells do not filament after 4 h of incubation at 40°C (29), after 22 h a culture is filled with filaments (Fig. 4). Filaments are seen after incubation in either LB or minimal medium (data not shown). Along with the filaments formed in LB medium we observe very small cells, but these do not contain DAPI-staining nucleoids. We measured the size distribution in cultures of dnaE74 incubated for 4 h at 40°C in LB medium (made up of cells which mostly do not give colonies) and of 22 h, 40°C LB cultures of dnaE74 cydA85 mutant (in which the overall viability has increased) (Fig. 5). The dnaE74 cultures include filaments of an average size of 32 μm. The 22-h dnaE74 cydA85 culture included a majority of cells of less than 2.5 μm in length, compared to an average length of 3.6 μm for BS40 wild-type cells incubated for 4 h at 40°C as well as filaments longer than the average observed in the dnaE74 culture incubated for the same time. The dnaE74 cydA85 culture incubated for 4 h at 40°C had cells of an average size of 5.0 μm, hardly larger than that of the wild type (29) (Fig. 5). In contrast to dnaE74 (Fig. 3), the larger filaments of dnaE74 cydA85 incubated for 22 h at 40°C include numerous apparently normal nucleoids (37) (Fig. 4). After 24 h, a dnaE74 culture remains highly filamented, but the filaments have a brittle appearance, which we interpret as a reflection of the nonviable nature of these bodies.
FIG. 4.
Formation of filaments by dnaE74 cydA85 cells incubated for 22 h at 40°C in LB medium. Top, phase microscopy. Arrows indicate the position of cells without DAPI staining inclusions. Bottom, DAPI-stained fluorescence microscopy.
FIG. 5.
Size distribution of cells. Open symbols, wild-type BS40 after 4 h at 40°C. Solid symbols, dnaE74 cydA85 after 22 h at 40°C. Cross-hatched symbols, dnaE74 after 4 h at 40°C.
The recovery of the suppressed strain can be observed by flow cytometry. Cultures of dnaE74 cydA85 were incubated for 24 h at 40°C and were then diluted and incubated at 27°C. At times 0, 0.5, 2, and 4 h after the switch to permissive temperature, rifampin, chloramphenicol, and cephalexin were added to stop the initiation of new rounds of DNA synthesis and to inhibit further cell division, and incubation was then continued for another 3 h at 27°C. We measured forward light scatter at different times after the shift to permissive temperature (Fig. 6). Within 2 h at 27°, the peak light scattering had greatly diminished. We interpret this alteration to mean that the culture is taken over by the smaller cells similar in size to the wild type tabulated in Fig. 5.
FIG. 6.
Forward cell scatter distributions of dnaE74 cydA85 cultures incubated 24 h at 40°C and then switched to permissive temperature (27°C) and either (A) not incubated or incubated for (B) 30 min, (C) 2 h, and (D) 4 h at 27°C before addition of rifampin, chloramphenicol, and cephalexin and further incubation for 3 h at 27°C.
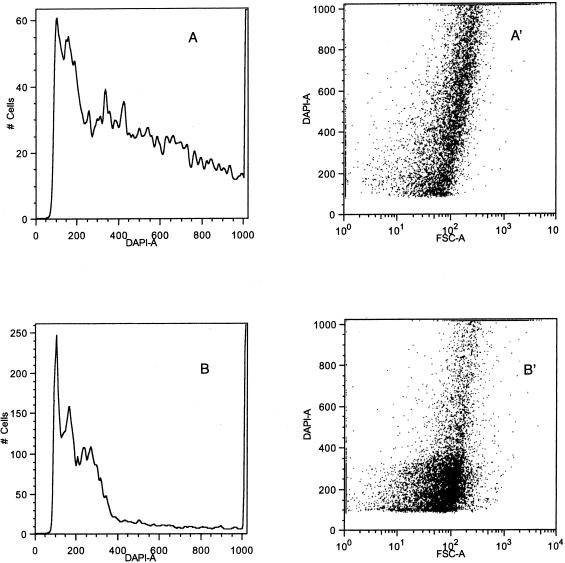
The DNA content of dnaE74 and dnaE74 cydA85 filaments was investigated by flow cytometry and DAPI staining. Overnight cultures of dnaE74 were incubated overnight at 27°C and then diluted into fresh medium to an optical density at 600 nm (OD600) of 0.12. They were then incubated for 4 h at 27°C before the addition of rifampin, chloramphenicol, and cephalexin. These cultures show cells with two peaks of DAPI staining representing cells with two and four genomes, as has been repeatedly reported (2) (Fig. 7A). Cultures of dnaE74 incubated at 40°C in LB medium showed two DAPI peaks of relatively equal size (data not shown). A dnaE74 overnight culture was diluted and incubated for 4 h at 40°C before being shifted to permissive temperature (27°C) and medium containing rifampin, cephalexin, and chloramphenicol (Fig. 7B). These cells showed a broad distribution of DNA content with filamentous cells (indicated by the higher forward cell scatter [FCS] values) containing amounts of DNA (measured as DAPI fluorescence) at least six times the peak values seen in the control. We observed that dnaE74 cydA85 cultures incubated 24 h at 40°C showed a large broad peak of DNA content along with many cells containing larger amounts of DNA similar to the picture observed with dnaE74 (compare Fig. 8A with 7B). The first peak sharpened and additional peaks became evident on incubation at permissive temperature (Fig. 8B).
FIG. 7.
(A) Flow cytometry of dnaE74 grown at permissive temperature. dnaE74 was grown for 4 h at 27°C, starting with an overnight culture and then incubated for 3 h with rifampin, chloramphenicol, and cephalexin. (B) Flow cytometry of dnaE74 after 4 h of incubation at 40°C and then incubated for 3 h at 27°C with rifampin, chloramphenicol, and cephalexin.
FIG. 8.
Flow cytometry of dnaE74 cydA85 cells. (A) Cells incubated in LB medium for 24 h at 40°C. (B) Cells depicted in panel A diluted and incubated for 4 h at 27°C before addition of rifampin, chloramphenicol, and cephalexin.
Cytochrome function.
We previously reported (29) that of 51 suppressors isolated, at least 49 mapped near cydA85 on the E. coli chromosome. At least two explanations account for this specificity: (i) the selection conditions (protection from death at high temperature) restricted the population of possible suppressor genes, or (ii) cytochrome bd oxidase has an additional and as-yet undescribed function. To distinguish between these possibilities, we utilized a plasmid designed to overproduce cytochrome bo3 (25). This plasmid (pJRHisA) includes genes for the cytochrome bo3 complex controlled by their own promoter and with a histidine tag (his6) to allow isolation. We used the his6 sequence to identify the production of protein by dnaE74 cydA85 at 40°C. Because the cytochrome bd oxidase is preferentially used at temperatures of 37°C and above (9), we thought it necessary to demonstrate that cytochrome bo3 was produced at 40°C. A Western blotting of an extract of dnaE74 cydA85 after growth at 40°C showed reactivity with anti-his antibody only when this strain carried plasmid pJRHisA but not when the strain carried either pBR322 or pTK-1, producing cytochrome bd oxidase (Fig. 9). We found that plasmid pjRHisA but not pBR322 partially reversed the protection from death at 40°C provided by a mutation in cytochrome bd oxidase (Table 4), much as has been previously reported for other cyd effects (34). This experiment implies that protection from cell death is related to the function of the cytochromes rather than to some specific effect of the cytochrome bd protein.
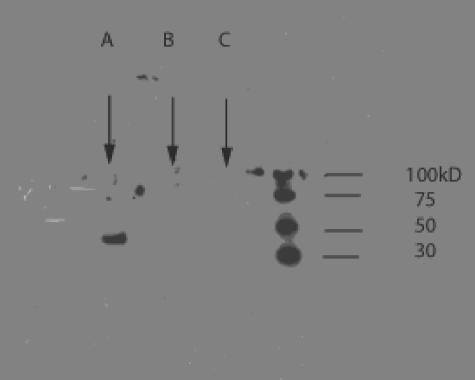
FIG. 9.
Western blot of cytochrome-overexpressing strains. Overnight cultures of dnaE74 cydA85/pJRcyOhis, dnaE74 cydA85/pTK1, and dna74 cydA85/pBR322 were diluted 1:1,000 into 5 ml of LB prewarmed to 40°C. The cultures were grown at 40°C to an OD600 of 0.5 to 1.0, and 1.5 ml of culture was harvested and frozen. The pellet was resuspended in 400 μl of CelLytic B (Sigma) working stock, treated with DNase A to a final concentration of 10 μg/ml, and centrifuged to give the soluble protein fraction. Sample (15 μl) plus loading dye was electrophoresed for 2.5 h at 125 V in a 7.5% precast polyacrylamide minigel and transferred onto nitrocellulose. The gel was blocked for 1 h in 0.2% anti-His—horseradish peroxidase conjugate blocking regent and then placed in anti-HIS—horseradish peroxidase conjugate (1:2,000 dilution; QIAGEN) at room temperature for 1 h. The gel was washed two times for 10 min with Tris-buffered saline-Tween/Triton and once for 10 min in Tris-buffered saline buffer. The chemiluminescence detection reaction was performed using the Super Signal West Pico Luminol reagent (Pierce #34080), and the blot was covered with X-ray film which was exposed for 20 s using a transilluminator. (A) dnaE74 cydA85 with pcyO; (B) dnaE74 cydA85 with pcydA85; and (C) dnaE74 cydA85 with pBR322.
TABLE 4.
Clusters of genes similarly expressed with cydAa
| Minimal medium
|
Rich medium
|
||
|---|---|---|---|
| Gene | Correlation | Gene | Correlation |
| cydA | 1 | cydA | 1 |
| daeD | 0.64 | B1491 | 0.36 |
| dnaK | 0.53 | csrA | 0.69 |
| ftsJ | 0.9 | cydB | 0.85 |
| gpmA | 0.57 | panC | 0.51 |
| hslU | 0.47 | pdxY | 0.31 |
| hslV | 0.41 | sodB | 0.5 |
| htpG | 0.53 | yafA | 0.73 |
| mopA | 0.49 | ycfM | 0.7 |
| oppB | 0.92 | ||
| oppC | 0.86 | ||
| oppD | 0.72 | ||
| oppF | 0.48 | ||
| pflB | 0.79 | ||
| rplT | 0.53 | ||
| rpoE | 0.71 | ||
| rseA | 0.84 | ||
| yhbM | 0.78 | ||
Data and analysis provided by A. Khodursky (http://gia.umn.edu/index.cgi?DynamicGeneBatchProcessing).
DISCUSSION
Unbalanced growth implies that continued synthesis of RNA and protein in the absence of concomitant DNA synthesis is in some way perceived by the cell as abnormal and leads to cell death. The concept is more difficult to apply to eukaryotes because of the separation in the cell cycle of periods of active protein and RNA synthesis in G1 from DNA synthesis, although inhibition of thymine synthesis leads to cell death, as it does in bacteria (11, 28). In prokaryotes, this separation does not occur. Transcription proceeds alongside DNA synthesis (17), and protein synthesis can start on incomplete transcripts (12). Transcription and DNA synthesis are connected, because the initiation of new rounds of DNA synthesis is dependent on new synthesis of the dnaA protein (21). What is not clear is the relationship between DNA synthesis and cell division. Intuitively, we suppose they are related. Most (but certainly not all) products of bacterial cell division must have DNA so that there is some segregation mechanism, but there is no direct linkage between DNA synthesis and cell division (3, 4, 24).
E. coli cells dying as a result of what has been called unbalanced growth form filaments, indicating a failure of the cell division (septation) process. This is observed for thymineless death and is also observed in dnaE74 mutants incubated at nonpermissive temperature. Almost any treatment which inhibits DNA synthesis leads to filamentation (18). A question to be answered is what is the precise signal that results in inhibition of septation? An additional issue is whether the inhibition of cell division is a precursor to the signal for cell death, because the signal for cell death could separately be a signal to inhibit cell division. Yet another problem is how to refine the concept of unbalanced growth to understand the events which accompany inhibition of DNA synthesis as a result of either thymine deprivation, an altered DNA polymerase, or restriction of deoxynucleotides by hydroxyurea inhibition (29). Our experiments provide some clues to the answers.
We observed that (i) the cydA85 mutation protects dnaE74 mutants incubated at nonpermissive temperature from death on complex (LB) medium but not on minimal medium; (ii) the cydA85 mutation protects a Δthy strain from death on complex (LB) medium but not on minimal medium; (iii) dnaE temperature-sensitive mutants incubated at nonpermissive temperature form filaments containing heterogeneous amounts of DNA in an amount ranging from six to eight times the normal peak amount; (iv) the dnaE74 cydA85 strain forms filaments and small anucleate cells when incubated for long periods at nonpermissive temperature, and filaments are observed on both minimal and LB medium; (v) the dnaE74 cydA85 filaments formed on LB medium contain numerous normal-appearing nucleoids; (vi) dnaE74 cydA85 cultures recover when transferred to permissive temperature; (vii) overexpression of the alternative cytochrome bo3 will substitute for wild-type cytochrome bd oxidase in making cells sensitive to nonpermissive conditions.
These observations lead us to the following conclusions. (i) Cell death is due to the production of some specific signal rather than to unbalanced growth. If unbalanced growth is defined as the ratio of the rates of RNA plus protein synthesis to those of DNA synthesis, then one might expect it to be more pronounced in LB than in minimal medium, because the rate of synthesis of macromolecules is lower in minimal medium (32). In fact, and as expected, cell death of nonsuppressed strains is observed more rapidly in LB medium. However, the protective effect of the cydA85 mutation is observed only on LB medium. On minimal medium, where one would expect the ratio of protein/DNA synthesis to be more nearly balanced, cell death occurs rapidly. (ii) The finding that overexpression of cytochrome bo3 has the same effect as normal expression of cytochrome bd oxidase in permitting cell death under nonpermissive conditions in LB medium, and that death occurs in the absence of cytochrome bd oxidase on minimal medium implies that cytochrome b03 provides an adequate energy supply in minimal medium but not in LB medium at higher temperature. (iii) Filamentation of dnaE74 cydA85 does occur in both minimal medium and LB medium at nonpermissive temperature, but cell death is only observed in minimal medium. This observation implies that the cell death response requires both an inhibition of DNA synthesis and an adequate energy supply but that the inhibition of DNA synthesis itself is sufficient to cause derangement of the septation process, as has been frequently observed (18). The formation of minicells by the dnaE74 cydA85 mutant strain at nonpermissive temperature (Fig. 4) is an example of this derangement. (iv) The reversibility of filamentation (Fig. 6 and 8B) makes it possible that inhibition of septation is not a necessary intermediate in the cell death pathway, although it may result from the same signal. The fact that filamentation can occur even with normal DNA synthesis (as in penicillin inhibition [4]) supports this interpretation. (v) The finding that the cydA85 mutation can protect Δthy strains from death supports the conclusion that cell death is not due to DNA damage but rather to some signal set off by the inhibition of DNA synthesis. This conclusion is based on our previous findings (29) that pBR322 is able to replicate in dnaE74 cells at nonpermissive temperature, that the DNA content in dnaE74 cells increases six- to eightfold on incubation at nonpermissive temperature (Fig. 7), and that we saw no obvious signs of DNA degradation in pulsed-field gels. However, because the DNA in dnaE74 cells did not resolve into sharp peaks on return to permissive conditions, incomplete chromosomes are likely to be present which could be involved in the initiation of the death signal. It would appear that although dnaE74 cells at nonpermissive temperature are able to create new DNA initiation sites (albeit slowly), they are unable to complete synthesis and segregate chromosomes. The dnaE74 cydA85 mutant appears to undergo the same sort of initiations (compare Fig. 7B and 8A), but resolution into complete molecules is possible on return to permissive conditions. Why reducing the available energy supply via introduction of the cydA85 mutation should overcome this block to completion of DNA synthesis is not clear. We speculate that the death signal in dnaE74 and Δthy strains results in degradative processes which destroy the enzymes needed for completion of DNA synthesis. This signal is either not produced or is not produced in sufficient amounts to be effective in cydA85 strains.
It is well known that the pattern of gene expression in E. coli is very different in minimal medium and in rich (LB) medium (32). An analysis of genes in the nodes surrounding cydA on rich and minimal media has been provided by A. Khodursky (http://gia.umn.edu/index.cgi?DynamicGeneBatchProcessing) (Table 4). The coexpressed genes are completely different in the two conditions. Perhaps the most interesting result of the array studies is the correlation of cydB expression with cydA on rich (LB) but not on minimal medium. The functional cytochrome bd is made of two subunits, one encoded by cydA and the other by cydB (14). We interpret this correlation as an indication that growth on rich but not on minimal medium involves functions in which cytochrome bd plays a role. This is in accord with our suggestion that the cytochrome bd oxidase is not essential on minimal medium. It is also interesting that sodB, coding for one of the superoxide dismutases, is one of the genes often coregulated with cydA85 on rich medium. One of the recurring hypotheses for the deleterious effects of cydA mutations is that the loss of cytochrome activity leads to an increase in oxygen stress due to an accumulation of the oxygen radicals of the type removed by superoxide dismutase (9, 34). cydA mutants have been observed to have high mutation frequencies. Although it is not clear that oxidative damage is a factor in the role of the cydA85 mutation in promoting survival of dnaE74 or Δthy mutants in complex medium, it is likely that the high mutation frequency is involved in understanding the behavior of cultures of dnaE74 cydA85 incubated for long periods on complex selective medium.
We suppose that inhibition of DNA synthesis by polymerase inactivation or as a result of deprivation of thymidine triggers some signal for cell death. These same conditions trigger an inhibition of septation, perhaps utilizing the same signal. Mutation in the cytochrome bd oxidase does not alter the situation in minimal medium, implying that this enzyme activity is either not required or is not active in this medium. In complex medium, the signal for cell death is not activated in the absence of cytochrome bd oxidase activity. Our finding of filaments and small cells not containing nucleoids in the cydA85 mutants at 40°C indicates that septation is not normal and is reminiscent of similar findings made years ago with cells in which DNA synthesis had been inhibited (7, 13). We suppose that the recovery of the culture on shift to permissive temperature is due to viable septation in the filaments rather than to selection and proliferation of the small (mostly anucleate) cells. It has been known for many years that filamentation can occur without setting off a cell death response (3, 5, 18, 24). Our observation of small anucleate cells accompanying filamentation in dnaE74 cydA85 mutant cells (Fig. 4 and 5) but absent from dnaE74 cydA+ cells (Fig. 3 and 5) illustrates the distinction. Our experiments imply that filamentation and cell death may actually be the result of the activation of different pathways.
The filamentation that accompanies DNA damage in E. coli is generally considered to be related to the activation of the SOS system after destruction of the lexA repressor by RecA protein. This protein is itself activated by combination with single-stranded DNA accumulating as a result of the blockage of the DNA replication fork in the face of DNA damage (33). Activation of the SOS system results in production of the SulA protein, which inhibits septation via interaction with the FtsZ ring. At some point cells are no longer viable. We suggest that the filamentation and cell death we have described results from a different but related set of reactions. First, and as we have reported previously (29), deletion of recA or sulA has no effect on the survival of dnaE74 cells incubated at nonpermissive temperature. We have been unable to demonstrate any change in the UV sensitivity of dnaE74 cydA85 cells compared to that of dnaE74 cells or of cydA85 cells compared to that of wild-type cells. We therefore suppose that the slowing down of replication fork progression as a result of either polymerase inhibition, thymine starvation, or hydroxyurea inhibition (29) does not lead to activation of the same gene products as does DNA damage, notwithstanding the likelihood that single-stranded DNA accumulates in both cases. In fact, the supposition that it is indeed single-stranded DNA which is the signal requires some special explanation because of the constant appearance of single-stranded regions in normal DNA replication. Sassanfar and Roberts (26) suppose the reason is kinetic, in that the RecA protein does not have time to combine or displace single-stranded binding protein before single-stranded DNA is covered up by replication. The situation may not be unlike that seen in eukaryotic cells in which DNA synthesis may be blocked by radiation-induced damage or by inhibitors such as hydroxyurea. In the first case the damage is mediated by the ATM kinase, and in the second case it is managed by ATR (20). The pathways are similar but not identical. Insofar as this analogy has merit, the exact components in each pathway remain to be identified.
TABLE 3.
Overproduction of cytochrome bo3 can substitute for cytochrome bd oxidasea
| Time at 40°C (h) | Surviving fraction (N/N0) with plasmid
|
||
|---|---|---|---|
| pBR322 | pcydA85 | pcyO | |
| 0 | 1.0 | 1.0 | 1.0 |
| 4 | 3.52 | 0.016 | 0.13 |
| 6 | 0.05 | ||
The dnaE74 cydA85 strain was transformed with plasmid and selected on plates of ampicillin and tetracycline. Overnight cultures were prepared on LB, diluted 1:1,000 in LB, and incubated as indicated.
Acknowledgments
This work was supported in part by a grant, CA32436, from the National Cancer Institute, NIH.
We thank R. Gennis for suggesting overexpression of cytochrome bo3 and for providing the appropriate plasmid. We thank A. Markovitz for many stimulating discussions and for his reading of the manuscript.
REFERENCES
- 1.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52:591-625. [DOI] [PubMed] [Google Scholar]
- 2.Akerlund, T., K. Nordstrom, and R. Bernander. 1995. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 177:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernander, R., and K. Nordstrom. 1990. Chromosome replication does not trigger cell division in E. coli. Cell 60:365-374. [DOI] [PubMed] [Google Scholar]
- 4.Boye, E., and K. Nordstrom. 2003. Coupling the cell cycle to cell growth. EMBO Rep. 4:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, P., and I. B. Holland. 1983. Two pathways of division inhibition in UV-irradiated. Mol. Gen. Genet. 190:309-314. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, S. S. 1971. On the nature of thymineless death. Ann. N. Y. Acad. Sci. 186:292-301. [DOI] [PubMed] [Google Scholar]
- 7.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31:725-737. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, B. S., K. K. Gabbert, and R. G. Kranz. 1996. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J. Bacteriol. 178:6348-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haupt, S., M. Berger, Z. Goldberg, and Y. Haupt. 2003. Apoptosis - the p53 network. J. Cell Sci. 116:4077-4085. [DOI] [PubMed] [Google Scholar]
- 11.Houghton, J. A., D. M. Tillman, and F. G. Harwood. 1995. Ratio of 2′-deoxyadenosine-5′-triphosphate/thymidine-5′-triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin. Cancer Res. 1:723-730. [PubMed] [Google Scholar]
- 12.Jacques, N., and M. Dreyfus. 1990. Translation initiation in Escherichia coli: old and new questions. Mol. Microbiol. 4:1063-1067. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe, A., R. D'Ari, and V. Norris. 1986. SOS-independent coupling between DNA replication and cell division in Escherichia coli. J. Bacteriol. 165:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junemann, S. 1997. Cytochrome bd terminal oxidase. Biochim. Biophys. Acta 1321:107-127. [DOI] [PubMed] [Google Scholar]
- 15.Kadenbach, B., S. Arnold, I. Lee, and M. Huttemann. 2004. The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochim. Biophys. Acta 1655:400-408. [DOI] [PubMed] [Google Scholar]
- 16.Kaysser, T. M., J. B. Ghaim, C. Georgiou, and R. B. Gennis. 1995. Methionine-393 is an axial ligand of the heme b558 component of the cytochrome bd ubiquinol oxidase from Escherichia coli. Biochemistry 34:13491-13501. [DOI] [PubMed] [Google Scholar]
- 17.Liu, B., M. L. Wong, R. L. Tinker, E. P. Geiduschek, and B. M. Alberts. 1993. The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature 366:33-39. [DOI] [PubMed] [Google Scholar]
- 18.Lutkenhaus, J., and A. Muherjee. 1996. Cell division, p. 1615-1626. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 19.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGowan, C. H., and P. Russell. 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16:629-633. [DOI] [PubMed] [Google Scholar]
- 21.Messer, W. 2002. The bacterial replication initiator DnaA, DnaA, and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 26:355-374. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Nakayama, H., K. Nakayama, R. Nakayama, N. Irino, Y. Nakayama, and P. C. Hanawalt. 1984. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. 195:474-480. [DOI] [PubMed] [Google Scholar]
- 24.Nordstrom, K., R. Bernander, and S. Dasgupta. 1991. The Escherichia coli cell cycle: one cycle or multiple independent processes that are co-ordinated? Mol. Microbiol. 5:769-774. [DOI] [PubMed] [Google Scholar]
- 25.Rumbley, J. N., E. Furlong Nickels, and R. B. Gennis. 1997. One-step purification of histidine-tagged cytochrome bo3 from Escherichia coli and demonstration that associated quinone is not required for the structural integrity of the oxidase. Biochim. Biophys. Acta 1340:131-142. [DOI] [PubMed] [Google Scholar]
- 26.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 27.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skog, S., B. Tribukait, B. Wallstrom, and S. Eriksson. 1987. Hydroxyurea-induced cell death as related to cell cycle in mouse and human T-lymphoma cells. Cancer Res. 47:6490-6493. [PubMed] [Google Scholar]
- 29.Strauss, B., K. Kelly, T. Dincman, D. Ekiert, T. Biesieda, and R. Song. 2004. Cell death in Escherichia coli dnaE(Ts) mutants incubated at a nonpermissive temperature is prevented by mutation in the cydA gene. J. Bacteriol. 186:2147-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss, B. S., R. Roberts, L. Francis, and P. Pouryazdanparast. 2000. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 182:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss, B. S., D. Sagher, and S. Acharya. 1997. Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeats in DNA. Nucleic Acids Res. 25:806-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, G. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 34.Wall, D., J. M. Delaney, O. Fayet, B. Lipinska, T. Yamamoto, and C. Georgopoulos. 1992. arc-dependent thermal regulation and extragenic suppression of the Escherichia coli cytochrome d operon. J. Bacteriol. 174:6554-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, L., and I. D. Hickson. 2002. RecQ helicases and cellular responses to DNA damage. Mutat. Res. 509:35-47. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]
- 37.Zimmerman, S. B. 2003. Underlying regularity in the shapes of nucleoids of Escherichia coli: implications for nucleoid organization and partition. J. Struct. Biol. 142:256-265. [DOI] [PubMed] [Google Scholar]