Abstract
Structural analysis of compounds identified as lipid I and II from Mycobacterium smegmatis demonstrated that the lipid moiety is decaprenyl phosphate; thus, M. smegmatis is the first bacterium reported to utilize a prenyl phosphate other than undecaprenyl phosphate as the lipid carrier involved in peptidoglycan synthesis. In addition, mass spectrometry showed that the muropeptides from lipid I are predominantly N-acetylmuramyl-l-alanine-d-glutamate-meso-diaminopimelic acid-d-alanyl-d-alanine, whereas those isolated from lipid II form an unexpectedly complex mixture in which the muramyl residue and the pentapeptide are modified singly and in combination. The muramyl residue is present as N-acetylmuramic acid, N-glycolylmuramic acid, and muramic acid. The carboxylic functions of the peptide side-chains of lipid II showed three types of modification, with the dominant one being amidation. The preferred site for amidation is the free carboxyl group of the meso-diaminopimelic acid residue. Diamidated species were also observed. The carboxylic function of the terminal d-alanine of some molecules is methylated, as are all three carboxylic acid functions of other molecules. This study represents the first structural analysis of mycobacterial lipid I and II and the first report of extensive modifications of these molecules. The observation that lipid I was unmodified strongly suggests that the lipid II intermediates of M. smegmatis are substrates for a variety of enzymes that introduce modifications to the sugar and amino acid residues prior to the synthesis of peptidoglycan.
Peptidoglycans are essential components of bacterial cell walls, providing both mechanical strength and shape. The basic structure of peptidoglycan is common among most of the eubacteria, consisting of glycan chains composed of alternating units of β1→4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc). The lactyl group of the MurNAc residue usually carries a short peptide (l-Ala-d-Glu-meso-diaminopimelic acid [DAP]-d-Ala-d-Ala) that forms bonds with the peptide side chains of neighboring glycan strands, thus providing mechanical strength (35). Although the basic structure of peptidoglycan remains the same among eubacterial species, there are significant variations (16, 29). The structure of the mycobacterial peptidoglycan is an example of such divergence (16, 27, 40). The glycan chains of mycobacterial peptidoglycan are composed of alternating units of β1→4-linked GlcNAc and N-glycolylmuramic acid (MurNGlyc) in which the N-acetyl function has been oxidized to an N-glycolyl function. The carbon-6 position of some of the muramic acid residues forms a phosphodiester bond with the α-l-rhamnopyranose-(1→3)-α-d-GlcNAc(1-P) linker region of the galactan chain of the arabinogalactan (24), and about a third of the peptide cross bridges occur between the carboxyl group of the l-center of one DAP residue and the amino group of the d-center of another DAP residue, forming a l,d-cross-link (40); the rest of the cross-links are between the carboxyl group of a terminal d-Ala and the amino groups of the d-center of a neighboring DAP. The overall degree of cross-linking is 70 to 80% in mycobacteria, compared to 20 to 30% for Escherichia coli (22). In addition, the free carboxylic acid groups of DAP or d-Glu of mycobacterial peptidoglycan can be amidated singly or in combination (16), and some of the d-Glu residues are also modified by the addition of a glycine residue (16).
Despite extensive structural studies on the mature peptidoglycan of Mycobacterium spp., little is known of its biosynthesis. The overall biosynthetic pathway is assumed to be the same as that of E. coli (37), in which the cytosolic steps culminate in the formation of the UDP-N-acetylmuramyl-l-alanyl-d-glutamyl-meso-diaminopimelyl-d-alanyl-d-alanine (UDP-MurNAc-pentapeptide) catalyzed by the Mur family of ligases (36). The membrane-associated steps are initiated with the formation of N-acetylmuramyl-(pentapeptide)-diphosphoryl-undecaprenol (lipid I), a reversible reaction catalyzed by MraY (38). In a subsequent step, catalyzed by MurG (25), one N-acetylglucosamine residue is added to the nonreducing end of the muramic acid forming GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-undecaprenol (lipid II). At this stage, it is believed that the disaccharide pentapeptide unit is flipped to the outer surface of the membrane, which is the site of the periplasmic steps of peptidoglycan biosynthesis that are catalyzed by penicillin binding proteins (7).
However, mycobacteria do not synthesize undecaprenyl phosphate, the activated sugar carrier utilized by E. coli for peptidoglycan synthesis. Mycobacterium tuberculosis primarily synthesizes decaprenyl phosphate, while Mycobacterium smegmatis produces a mixture of hepta-, octa-, and decaprenyl phosphate (3). Thus, it seems likely that M. tuberculosis uses decaprenyl phosphate as the carrier of activated sugars involved in peptidoglycan synthesis, but the situation for M. smegmatis is not obvious. It is clear that arabinogalactan synthesis in M. smegmatis requires decaprenyl phosphate (26, 41), but the formation of prenyl phosphoryl mannose, reported to be involved in the biosynthesis of lipomannan and lipoarabinomannan, involves all three of the hepta-, octa-, and decaprenyl moieties (1, 9, 33). In contrast, the nature of the lipid carrier used by mycobacteria in peptidoglycan synthesis is undefined.
In addition, the modifications found in the mature peptidoglycans of some bacteria occur at the lipid-linked intermediate level (6, 15, 18, 19, 23, 31, 34), but it has been reported that the oxidation of the N-acetyl function of the N-acetylmuramic acid in mycobacteria occurs at the nucleotide level (32). Therefore, the objectives of the present study were to identify which of the three candidate lipid carrier molecules are utilized by M. smegmatis in the formation of lipid I and II and to determine if the modifications in the mature mycobacterial peptidoglycan occur on lipid-linked intermediates.
MATERIALS AND METHODS
Materials.
M. smegmatis mc2155 was obtained from the American Type Culture Collection. Percoll, [1-14C]isopentenyl diphosphate ([1-14C]IPP; 55 mCi/mmol), and a Superdex peptide column were purchased from Amersham Biosciences (Piscataway, N.J.). l-[U-14C]alanine (164 mCi/mmol) was from ICN Biomedicals (Irvine, Calif.). UDP-[U-14C]N-acetylglucosamine (288 mCi/mmol) was purchased from Perkin-Elmer Life Sciences (Boston, Mass.). Geranyl diphosphate was synthesized as described previously (4). Authentic decaprenyl phosphate and undecaprenyl phosphate and prenols of various chain lengths were purchased from the Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland). Nutrient broth, aluminum-backed Kieselgel 60 F254, and Kieselgel 60 preparative thin-layer chromatography (TLC) plates were from EM Science (Gibbstown, N.J.). Baker SI-reverse phase C18 TLC plates and 2-mercaptoethanol were from J. T. Baker Inc. (Phillipsburg, N.J.). ω,E,E,E-Geranylgeranyl diphosphate, UDP-N-acetylglucosamine, and 3-[N-morpholino]propanesulfonic acid (MOPS) were purchased from Sigma (St. Louis, Mo.). PrepSep C18 columns, 1-butanol, and pyridine were from Fisher Scientific (Fair Lawn, N.J.). Ramoplanin was a gift from Biosearch Italia SpA, Geranzano, Italy. Acetonitrile, high-performance liquid chromatography (HPLC)-grade water, and methanol were from Burdick and Jackson (Muskegon, Mich.). Isobutyric acid was from Aldrich Chemical Company (Milwaukee, Wis.). SG81 chromatography paper was from Whatman (Clifton, N.J.). Trifluoroacetic acid and acetic acid were purchased from Supelco Inc. (Bellefonte, Pa.). A Hypersil C18 column was purchased from Phenomenex (Torrance, Calif.), and a C18 capillary column was purchased from Michrom BioResources, Inc. (Auburn, Calif.).
Preparation of UDP-MurNAc-pentapeptide and UDP-MurNAc-[14C]pentapeptide.
UDP-MurNAc was enzymatically synthesized from UDP-GlcNAc by use of recombinant MurA and MurB of E. coli as previously described (20).UDP-MurNAc-l-Ala-d-Glu-DAP-d-Ala-d-Ala (UDP-MurNAc-pentapeptide)was synthesized enzymatically (28, 42) by adding l-Ala, d-Glu, DAP, and d-Ala-d-Ala and the appropriate recombinant E. coli Mur enzymes to UDP-MurNAc in a single reaction mixture. UDP-MurNAc-l-[14C]Ala-d-Glu-DAP-d-Ala-d-Ala was synthesized using the same procedures, except l-Ala was replaced with l-[14C]Ala. The total yield was typically 80%.
Preparation of MurNAc-(pentapeptide)-diphosphoryl-prenol (lipid I) of M. smegmatis.
For the synthesis of radiolabeled MurNAc-(pentapeptide)-diphosphoryl-prenol (lipid I), a particulate enzyme fraction, containing cell wall, membrane, and small amounts of cytosol, from M. smegmatis was prepared as previously described (26) and preincubated with 50 μg of ramoplanin/ml for 10 min, and UDP-MurNAc-[14C]pentapeptide and ATP were added. The mixture was incubated for 45 min at 28°C. The reaction was terminated by the addition of 20 volumes of CHCl3-CH3OH (2:1) and extracted with agitation on a rotary wheel for 15 min. The supernatant was recovered after centrifugation at 3,000 × g for 10 min at room temperature and washed with deionized water. After the removal of the upper, aqueous phase, the lower, organic phase was backwashed with CHCl3-CH3OH-H2O (3:47:48), dried under a stream of N2, and redissolved in CHCl3-CH3OH-H2O-NH4OH (65:25:3.6:0.5) or CHCl3-CH3OH-NH4OH (50:25:0.025).
For large-scale, in vitro synthesis of lipid I, 2 g of protein of M. smegmatis particulate enzyme was resuspended in 400 ml of buffer A containing 50 μg of ramoplanin/ml and incubated for 10 min at 28°C followed by the addition of ATP and UDP-MurNAc-pentapeptide to final concentrations of 100 and 50 μM, respectively. The reaction mixture was incubated for another 45 min at 28°C. Lipids were extracted by the addition of 2 volumes of ice-cold 1-butanol-6 M pyridinium acetate, pH 4.0 (4:1). The resulting mixture was stirred on ice for 1 h, and the lipid containing organic phase was separated by centrifugation and transferred to a new container. The aqueous phase was extracted one more time with the same solvents. The organic extracts were pooled and washed twice with water, and 5 × 105 dpm of l-[14C]Ala-labeled lipid I was added to act as a tracer to aid in the isolation and purification of muropeptides. The mixture was dried in a rotary evaporator under a vacuum, and the muropeptides were prepared as described below.
Preparation of lipid II of M. smegmatis.
In vitro synthesis of GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-prenol (lipid II) was carried out using the particulate enzyme fraction from M. smegmatis as the source of protein and lipid. In a reaction mixture containing 2 mg of protein, 100 μM ATP, 50 μM UDP-MurNAc-pentapeptide, and 25 μM UDP-GlcNAc made up to a total volume of 300 μl in buffer A were incubated for 1 h at 28°C. The radiolabeling of lipid II at desired positions was achieved via the incorporation of specific radiolabel precursors in the reaction mixtures. In order to synthesize [U-14C]GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-prenol, UDP-GlcNAc was replaced with 1 μCi of UDP-[U-14C]GlcNAc. Similarly, UDP-MurNAc-pentapeptide was replaced with 0.25 μCi of UDP-MurNAc-[14C]pentapeptide in order to obtain GlcNAc-MurNAc-([14C]pentapeptide)-diphosphoryl-prenol. The synthesis of GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-[14C]prenol required the addition of 100 μM [1-14C]IPP, 100 μM geranyl diphosphate, and 100 μM geranylgeranyl diphosphate to the reaction mix, along with UDP-GlcNAc, UDP-MurNAc-pentapeptide, and ATP. The accumulated lipid II was extracted as described for radiolabeled lipid I.
For large-scale in vitro synthesis of lipid II, 4.8 g of cell envelope protein was suspended in 800 ml of buffer A containing 100 μM ATP, 50 μM UDP-MurNAc-pentapeptide, and 50 μM UDP-GlcNAc. The reaction mix was incubated at 28°C for 75 min, and the lipids were extracted and treated as described for the large-scale preparation of lipid I, except 5 × 105 dpm of l-[14C]Ala-labeled lipid II was added as a tracer to aid in the isolation and purification of muropeptides.
Preparation of radiolabeled lipid I and II from E. coli.
A particulate enzyme fraction was prepared from E. coli K-12 cells as described previously (38). The particulate enzyme preparation was resuspended in 40 mM Tris (pH 8.0) and 10 mM MgCl2, and UDP-MurNAc-[14C]pentapeptide and ATP were then added to final concentrations of 50 and 100 μM, respectively. The mixture was incubated at 30°C for 10 min. UDP-GlcNAc was added to achieve a final concentration of 50 μM, and the incubation was continued for another 30 min at the same temperature. To accumulate lipid I, the particulate enzyme preparation was preincubated with ramoplanin for 10 min prior to the addition of UDP-MurNAc-[14C]pentapeptide and ATP. UDP-GlcNAc was omitted in this reaction. The synthesized lipid intermediates were extracted from the reaction with CHCl3-CH3OH (2:1) as described for the radiolabeled lipid intermediates of M. smegmatis.
Purification and analysis of GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-[14C]prenol.
The CHCl3-CH3OH (2:1) extract from a 6-ml reaction mix was dried under a stream of N2 and redissolved in CHCl3-CH3OH (2:1). The dissolved material was loaded onto a DEAE-cellulose (acetate form) column equilibrated with CHCl3-CH3OH (2:1). The column was washed extensively with CHCl3-CH3OH (2:1), and bound material was eluted with CHCl3-CH3OH (2:1) containing 300 mM of CH3COONH4. The CH3COONH4 was subsequently removed from the sample by washing it twice with H2O, and the solvent was evaporated under a stream of N2. The sample was then dissolved in a small volume of CHCl3-CH3OH-H2O-NH4OH (65:25:3.6:0.5) and applied to a preparative silica gel TLC plate, which was developed with CHCl3-CH3OH-H2O-NH4OH-CH3COONH4 (5.6:4.2:0.68:0.27:0.27). The radioactive lipid II was located by autoradiography and extracted from the silica gel with CHCl3-CH3OH-H2O (10:10:3). The recovered radioactive material was applied to a second silica gel plate, which was developed with CHCl3-CH3OH-H2O-CH3COONH4 (5.6:4.2:0.68:0.27). The radiolabeled material was located and extracted from the silica gel as described above.
A portion of the radiolabeled GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-[14C]prenol was hydrolyzed in 20 mM ammonium acetate buffer (pH 4.2) at 100°C for 20 min as previously described (13). The reaction mixture was extracted twice with diethyl ether, and the extract was dried under a stream of N2. The ether-soluble material was redissolved in CHCl3-CH3OH (2:1) and analyzed by TLC on C18 silica gel reverse-phase plates developed in CH3COCH3-CH3OH (9:1) (3, 30). Authentic decaprenyl phosphate was hydrolyzed under the same conditions, and the resulting material, as well as polyprenols of different chain lengths, were used as standards for TLC analysis. Radioactivity was located by autoradiography, and the standards were detected by using an anisaldehyde-sulfuric acid spray reagent (5).
Isolation and purification of the muropeptides from lipid I and II.
Lipid extracts containing lipid I or II were suspended in aqueous 2 M trifluoroacetic acid (TFA) and incubated at 60°C for 1 h to remove the lipid from the MurNAc-peptide (lipid I) or GlcNAc-MurNAc-peptide (lipid II). The mixture was allowed to cool and was then extracted with an equal volume of chloroform. The aqueous phase was dried under a vacuum, dissolved in aqueous 0.5% TFA, and loaded onto a PrepSep C18 solid-phase extraction column preequilibrated with the same solvent. Bound material was eluted with 10% acetonitrile containing 0.5% TFA. Fractions containing radioactivity were pooled, dried under a vacuum, and applied to Whatman SG81 chromatography paper. The chromatograms were developed with isobutyric acid-1 M ammonium hydroxide (5:3), and radioactive bands corresponding to the muropeptides were visualized by autoradiography. The muropeptides were eluted from the paper with 50% methanol and dried under a vacuum.
The muropeptides were further purified by reverse-phase HPLC with a Waters model 600 controller connected to a model 600 pump, a model 2487 UV detector, and a Hypersil 5-μm C18 column (25 cm by 4.6 mm). The column was equilibrated in solvent A (0.5% TFA in water) with a flow rate of 1 ml/min at room temperature. A linear gradient of 0 to 50% solvent B (0.5% TFA in 60% acetonitrile) over a period of 25 min was applied to elute the bound muropeptides. The absorbance of the eluent was monitored at 214 nm, fractions were collected, and the radioactivity was measured by liquid scintillation spectrometry. The fractions containing radioactivity (muropeptides) were dried and used for further analysis. In some experiments, the muropeptides were reduced by use of NaBH4 (8) prior to purification by reverse-phase HPLC.
LC-MS analysis.
The HPLC-purified muropeptides were dissolved in 5% acetonitrile containing 0.5% acetic acid at a concentration of about 6 μM. An aliquot (5 μl) was applied to a 0.2- by 200-mm reverse-phase C18 capillary column that was connected to an Eldex MicroPro capillary HPLC system (Eldex Laboratories, Inc., Napa, Calif.). The muropeptides were eluted with a 5 to 50% linear gradient of acetonitrile in 0.5% acetic acid at 5 μl/min. The eluate was introduced directly into an LCQ electrospray mass spectrometer (Finnigan-Thermoquest, San Jose, Calif.), and the muropeptides were analyzed by mass spectrometry (MS) and tandem mass spectrometry (MS2). The electrospray needle was operated at 4 kV, and the capillary temperature was 200°C. MS2 was performed on the fly of the most dominant ion of the previous MS scan (N2 sheath gas flow, 20 lb/in2) or from a preselected list of [M + H]+ ions (N2 sheath gas flow, 60 lb/in2).
RESULTS
Generation of radiolabeled lipid intermediates and muropeptides of M. smegmatis and E. coli.
Lipid-linked peptidoglycan intermediates were generated from fully characterized UDP-MurNAc-pentapeptide by use of an M. smegmatis particulate enzyme fraction as the source of enzymes, as well as the native lipid carrier. The initial criterion used for the identification of lipid II was the incorporation of radiolabeled MurNAc-(pentapeptide), GlcNAc, and IPP into a single molecule. The only radiolabeled product synthesized in the presence of UDP-MurNAc-[14C]pentapeptide and cold UDP-GlcNAc was lipid II. When UDP-GlcNAc was omitted from the reaction mix, MurNAc-(pentapeptide)-diphosphoryl-prenol (lipid I) was formed.
As further support for the identification of lipid II, known inhibitors of bacterial lipid II synthesis, namely, ramoplanin and tunicamycin (2), were tested for their abilities to inhibit synthesis of the material identified as lipid II in M. smegmatis. Both drugs inhibited the incorporation of radiolabeled UDP-MurNAc-(pentapeptide) into organic-soluble material in a dose-dependent manner, confirming the identity of the lipid intermediates. TLC analysis of the organic-soluble, enzymatically synthesized products indicated that ramoplanin preferentially inhibited lipid II synthesis, as only lipid I was found in extracts from reaction mixtures containing higher levels of ramoplanin. Therefore, ramoplanin was used to obtain sufficient lipid I for subsequent analysis.
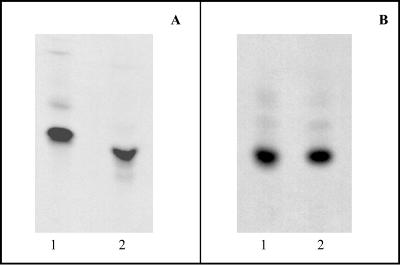
Figure 1 shows a comparison of the radiolabeled lipid intermediates from both M. smegmatis and E. coli by TLC analysis. Figure 1A shows the migration of intact lipid I from M. smegmatis and E. coli, and Fig. 1B shows the relative migration of the muropeptides obtained by acid hydrolysis of lipid I of both species. The migration of the intact lipid I from M. smegmatis is significantly slower than that of E. coli, while the migrations of the isolated muropeptides are identical, suggesting that lipid I from M. smegmatis may have a smaller lipid moiety. The migration of intact lipid II obtained from E. coli and M. smegmatis also had significant differences in Rf (data not shown). The muropeptides obtained by the hydrolysis of TLC purified lipid II from M. smegmatis separated into three distinct bands (Fig. 2), which did not cochromatograph with muropeptides isolated from lipid II of E. coli. Taken together, these results suggest that the muropeptides from mycobacteria were modified at the lipid II level. In order to characterize these modifications, detailed tandem mass spectrometry studies were undertaken on the muropeptides of the lipid intermediates isolated from M. smegmatis (see below).
FIG. 1.
TLC analysis of the lipid intermediates from E. coli and M. smegmatis. (A) Radiolabeled, intact lipid I from E. coli (lane 1) and M. smegmatis (lane 2) was loaded onto a Kieselgel 60 F254 TLC plate, which was developed in CHCl3-CH3OH-H2O-NH4OH-CH3COONH4 (5.6:4.2:0.68:0.27:0.27) and subjected to autoradiography. (B) Radiolabeled muropeptides obtained from E. coli lipid I (lane 1) or M. smegmatis lipid I (lane 2) were loaded onto a Kieselgel 60 F254 TLC plate, which was developed in isobutyric acid-1 M NH4OH (5:3) and subjected to autoradiography.
FIG. 2.
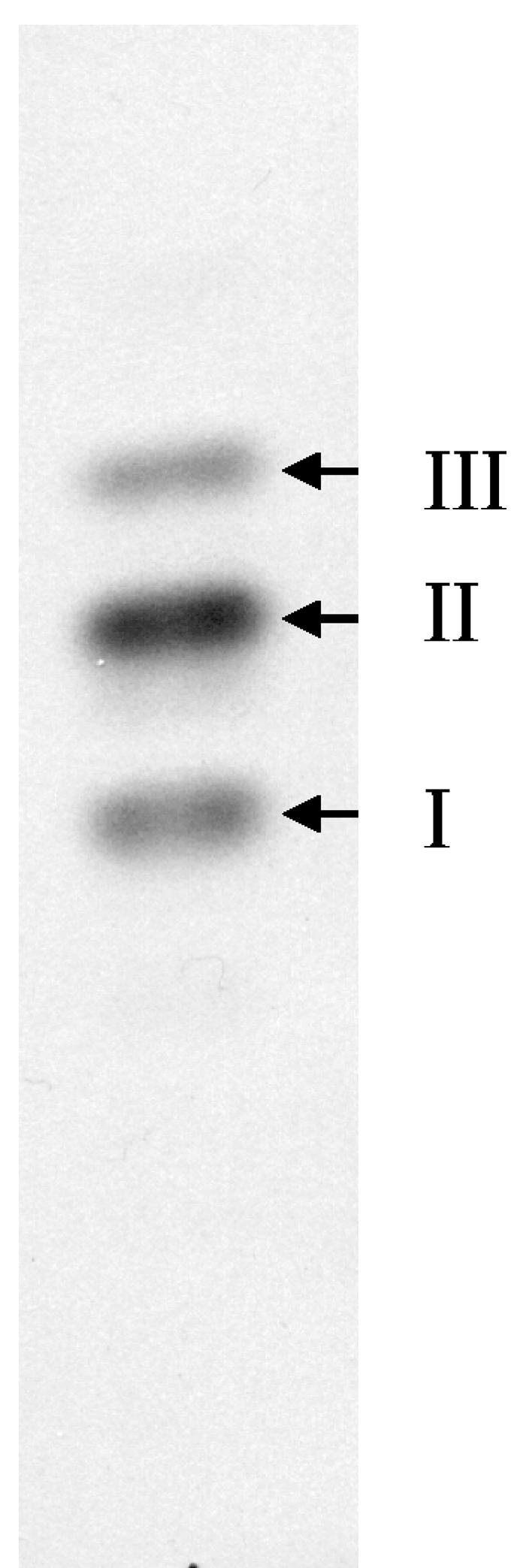
TLC analysis of muropeptides derived from M. smegmatis lipid II. Radiolabeled muropeptides obtained from M. smegmatis lipid II were loaded onto a Kieselgel 60 F254 TLC plate, which was developed in isobutyric acid-1 M NH4OH (5:3) and subjected to autoradiography. The arrows indicate bands I through III, which were subsequently isolated for further analysis.
Identification of the lipid moiety of lipid II from M. smegmatis.
In assays in which [1-14C]IPP, geranyl diphosphate, and geranylgeranyl diphosphate were added to the reaction mixtures, a number of organic solvent-soluble compounds were formed. Some of the [1-14C]IPP-labeled material comigrated with UDP-MurNAc-[14C]pentapeptide-labeled material in several solvent systems on TLC, indicating that the labeling of the lipid moiety of lipid II was successful. It was possible to resolve the GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-[14C]prenol from other 14C-labeled products formed in the reaction mixture by two simple chromatography steps, resulting in material that was pure enough for further analysis.
The GlcNAc-MurNAc-(pentapeptide)-diphosphoryl-[14C]prenol was hydrolyzed with ammonium acetate (pH 4.2) to remove the muropeptides as described in Materials and Methods, and the radioactive compounds were extracted with diethyl ether and analyzed by reverse-phase TLC (Fig. 3). The radiolabeled material was found to distribute between three major products. One of these radiolabeled products had an Rf value identical to that of decaprenol (C50). The other material migrated in a manner consistent with rearrangement products and hydrocarbons produced by acid hydrolysis of prenyl phosphates (11-13). There was no indication of an incorporation of either hepta- or octaprenyl phosphate into lipid II of M. smegmatis. Authentic decaprenyl phosphate hydrolyzed and analyzed under the same conditions produced a similar pattern when visualized with an anisaldehyde reagent (5), with the exception of the large spot near the solvent front. It is assumed that this spot is related to the presence of the sugar 1-phosphate residues found in lipid II, but no attempt was made to characterize this material.
FIG. 3.
TLC analysis of the lipid moiety of lipid II synthesized by the membrane-cell wall fraction from M. smegmatis. Autoradiogram of the radiolabeled products obtained by acid hydrolysis of lipid II from M. smegmatis (A) and generated by acid hydrolysis of decaprenyl phosphate (B). The arrows indicate the migration positions of authentic heptaprenol (C35) and decaprenol (C50). Samples were applied to a C18 silica gel reverse-phase TLC plate and developed with CH3COCH3-CH3OH (9:1).
MS analysis of the muropeptides isolated from mycobacterial lipid I.
The muropeptides isolated from the lipid I molecule were subjected to LC-MS2 analysis without borohydride reduction of the muramic acid. The positive-ion mass spectrum was dominated by a single ion (m/z 808.1) along with the corresponding monosodium (m/z 830.4) and disodium (m/z 852.4) adducts (Fig. 4). MS2 of the dominant ion (m/z 808.1) yielded a series of daughter ions, the structures of which were identical to those generated by MS2 analysis of authentic MurNAc-pentapeptide (see the supplemental material) (21). A fragment ion (m/z 605.2) representing the lactyl pentapeptide side chain and three y-type peptide fragments (according to the modified Roepstorff-Fohlman peptide fragment ion nomenclature [14]) with m/z values of 533.3, 462.2, and 333.2 were obtained. One ion (m/z 533.3) represents all five amino acids of the peptide side chain, an ion of m/z 462.2 represents d-Glu-DAP-d-Ala-d-Ala, and an ion of m/z 333.2 consists of DAP-d-Ala-d-Ala. An ion (m/z 444.2) derives from the loss of a water molecule from d-Glu-DAP-d-Ala-d-Ala (m/z 462.2). Three b-type fragments, MurNAc-l-Ala-d-Glu-DAP-d-Ala (m/z 719.3), MurNAc-l-Ala-d-Glu-DAP (m/z 648.3), and MurNAc-l-Ala-d-Glu (m/z 476.2), were observed, and two double-cleavage fragments, consisting of d-Glu-DAP-d-Ala (m/z 373.1) and d-Glu-DAP (m/z 302.1), were also identified. Thus, the dominant ion (m/z 808.1) is the molecular ion of MurNAc-l-Ala-d-Glu-DAP-d-Ala-d-Ala or MurNAc-pentapeptide.
FIG. 4.
Mass spectrum of muropeptides derived from mycobacterial lipid I. Muropeptides were isolated from mycobacterial lipid I as described in Materials and Methods and subjected to LC-MS analysis (without prior NaBH4 reduction).
Ions resulting from the neutral loss of water (m/z 790.0) were also detected, and another ion (m/z 824.0) was also present. MS2 of the ion of m/z 824.0 yielded several daughter ions identical to the daughter ions from MurNAc-pentapeptide, with the sole exception being that the b-type ions were 16 atomic mass units (amu) larger. Therefore, the additional 16 amu are associated with the muramic acid residue and not the peptide. Conversion of the N-acetyl moiety of the muramic acid to N-glycolyl would result in a net gain of 16 amu. Thus, the muropeptide of the M. smegmatis lipid I accumulated in vitro is primarily MurNAc-l-Ala-d-Glu-DAP-d-Ala-d-Ala, with smaller amounts of MurNGlyc-l-Ala-d-Glu-DAP-d-Ala-d-Ala.
Mass spectrometric analysis of the muropeptides from mycobacterial lipid II.
The muropeptides isolated from mycobacterial lipid II were analyzed by LC-mass spectrometry before and after reduction with NaBH4. Samples were first separated into the three bands seen in Fig. 2 by SG81 paper chromatography and were then processed separately in subsequent steps. The data presented are from reduced samples unless otherwise noted. The reduced and nonreduced samples produced identical spectra, except the ions containing the muramyl residue from the nonreduced samples were two mass units smaller than their reduced counterparts, as expected. It was anticipated that the sharp bands seen in Fig. 2 would be relatively homogenous in composition; however, this was not the case. The reason for the heterogeneity of compounds found in each band is not clear.
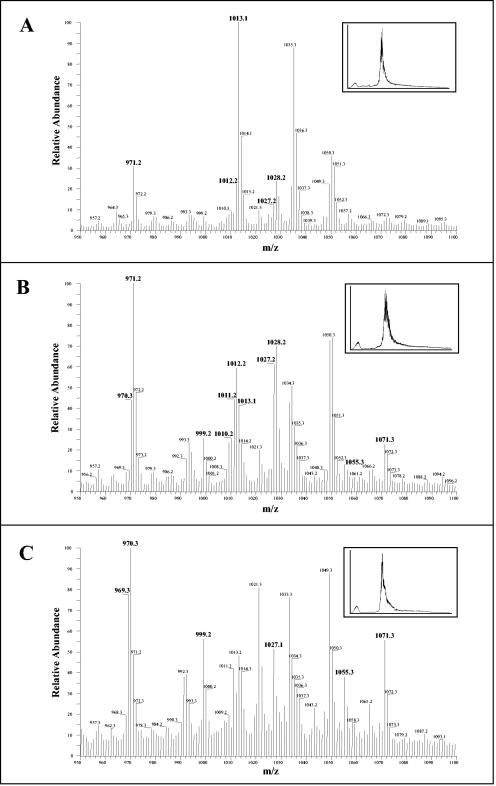
Software averaging across the total ion chromatogram provided insight into the unexpected complexity of the mixtures even after separation and purification by paper chromatography. The averaged mass spectrum of the entire total ion chromatogram generated from each of the three bands purified on SG81 paper is shown in Fig. 5. Ions with m/z values predicted for reduced muropeptides derived from lipid II (1013.2) were observed in all three.
FIG. 5.
Averaged mass spectra of entire total ion chromatograms of the reduced muropeptides of M. smegmatis lipid II. Ascending chromatography on SG81 paper was used to separate the bands shown in Fig. 2, which were then extracted as described in Materials and Methods. The reduced and purified muropeptides were subjected to LC-MS analysis, and software averaging was used across the entire total ion chromatogram (inset in each panel) to approximate a direct liquid injection. The averaged mass spectra of band I (A), band II (B), and band III (C) are shown. Ions from the spectra (labeled in bold) were selected for further characterization by MS2 (see Fig. 6 for an example).
The major ions observed in Fig. 5 were subjected to tandem mass spectrometry. In order to obtain unambiguous MS2 results for the various samples, time-resolved separation of molecules by LC was important and helped distinguish between [M + H]+ and fragment ions. Thus, spectra obtained from material in band I contained [M + H]+ ions with m/z values of 971.2 to 972.2, 1,012.2 to 1,013.2, and 1,027.2 to 1,028.2 and the corresponding [M + Na]+ ions at 22 amu higher (Fig. 5). Spectra obtained from material in band II contained [M + H]+ ions with m/z values of 970.3 to 972.2, 999.2 to 1,000.2, 1,010.2 to 1,013.2, 1,027.2 to 1,028.2, and 1,071.3 to 1,072.3. Except for the last cluster, the corresponding [M + Na]+ ions were also seen. Spectra generated from band III yielded [M + H]+ ions with m/z values of 969.2 to 970.2, 1,011.2, 1,027.2, 1,055.3, and 1,071.3, along with the corresponding [M + Na]+ ions for all but m/z 1,071.3.
A comparison of the daughter ions of a given molecular ion with the daughter ions of the MurNAc-pentapeptide standard positively identified the molecule as a muropeptide and also served to identify and localize the modifications (see the supplementary material). The relative abundance of the daughter ions varied from scan to scan; therefore, comparisons of several scans were necessary to identify the major fragment ions.
Table 1 shows the molecular ions observed from the muropeptides of M. smegmatis lipid II and their inferred structures based on analysis of MS2 spectra. A number of modifications were observed on the muramyl residue and the peptide. Interestingly, the GlcNAc moiety was not observed to be modified.
TABLE 1.
Inferred structures of muropeptides isolated from M. smegmatis lipid IIa
| Source | m/z values for [M + H]+ ions | Inferred structures of amino acids and sugars found in muropeptidesb
|
||||||
|---|---|---|---|---|---|---|---|---|
| GlcNAc | MurNAc* | l-Ala | d-Glu* | DAP* | d-Ala | d-Ala* | ||
| Standard | 808.2 | NP | + | + | + | + | + | + |
| Band I | 971.2 | + | MurNH2 | + | + | + | + | + |
| 1,012.2 | + | + | + | + | DAP(NH2) | + | + | |
| 1,013.2 | + | + | + | + | + | + | + | |
| 1,027.2 | + | MurNGlyc | + | d-Gln | DAP(NH2) | + | + | |
| 1,028.2 | + | MurNGlyc | + | + | DAP(NH2) | + | + | |
| Band II | 970.3 | + | MurNH2 | + | d-Gln | + | + | + |
| 971.2 | + | MurNH2 | + | + | + | + | + | |
| 999.2 | + | + | + | + | + | + | d-Alaninol | |
| 1,010.2 | + | + | + | d-Gln | DAP(NH2) | + | d-Ala(NH2) | |
| 1,011.2 | + | + | + | d-Gln | DAP(NH2) | + | + | |
| 1,011.2 | + | + | + | + | DAP(NH2) | + | d-Ala(NH2) | |
| 1,012.0 | + | + | + | + | DAP(NH2) | + | + | |
| 1,013.2 | + | + | + | + | + | + | + | |
| 1,027.2 | + | MurNGlyc | + | d-Gln | DAP(NH2) | + | + | |
| 1,028.2 | + | MurNGlyc | + | + | DAP(NH2) | + | + | |
| 1,029.0 | + | MurNGlyc | + | + | + | + | + | |
| 1,055.3 | + | + | + | d-Glu(CH3) | DAP(CH3) | + | d-Ala(CH3) | |
| 1,071.3 | + | MurNGlyc | + | d-Glu(CH3) | DAP(CH3) | + | d-Ala(CH3) | |
| Band III | 969.3 | + | MurNH2 | + | d-Gln | DAP(NH2) | + | + |
| 970.3 | + | MurNH2 | + | d-Gln | DAP | + | + | |
| 999.2 | + | + | + | + | + | + | d-Alaninol | |
| 1,027.2 | + | + | + | + | + | + | d-Ala(CH3) | |
| 1,055.3 | + | + | + | d-Glu(CH3) | DAP(CH3) | + | d-Ala(CH3) | |
| 1,071.3 | + | MurNGlyc | + | d-Glu(CH3) | DAP(CH3) | + | d-Ala(CH3) | |
Muropeptides were prepared from lipid II as described in Materials and Methods, separated into bands (Fig. 2) by ascending chromatography on Whatman SG81 paper and analyzed by LC-MS2. Structures were determined by comparing MS2 spectra of molecular ions to those of known standards (see the supplemental material).
*, residues that were always modified at the positions designated R1, R2, R3, or R4 in Fig. 7. NP, not present. +, unmodified.
The muramic acid residues were observed in three different forms: MurNAc, MurNGlyc, and MurNH2. The carboxylic acid residues of the peptide side chains were also found to have three different modifications, singly or in combination (Table 1). The dominant form of peptide modification was the amidation of DAP, in some cases in combination with the amidation of d-Glu and terminal d-Ala. The formation of methyl ester of the terminal d-Ala alone or together with DAP and d-Glu was also observed. Interestingly, there were no dimethylated species found (Table 1). The third modification observed was the addition of a Gly residue. The low abundance of these molecules prevented the detailed MS2 analysis required for localization of the Gly attachment site. All modifications of the peptide side chains occurred in combination with the modifications of the muramic acid residue resulting in at least 14 different species of muropeptide (Table 1).
DISCUSSION
The radiolabeled lipid moiety obtained from M. smegmatis was identified as decaprenyl phosphate. The mild hydrolysis conditions used for the isolation of the lipid also generated a series of other products, as has been previously reported (11-13). These products are believed to be the result of cleavage and rearrangement during hydrolysis. Hydrolysis and analysis of authentic decaprenyl phosphate under the same conditions produced a similar pattern on the TLC plate. Thus, M. smegmatis appears to be the first bacterium reported to utilize a polyprenyl phosphate other than undecaprenyl phosphate as the lipid moiety of lipid I and lipid II. Interestingly, even though the M. smegmatis particulate fraction also synthesizes heptaprenyl phosphate and octaprenyl phosphate under the conditions reported here (data not shown) and similar conditions (3), these lipids were not found in mycobacterial lipid I or II. These results indicate that prenyl phosphates of various chain lengths are compartmentalized in M. smegmatis, likely to reduce competition between various biosynthetic pathways. The mechanism of compartmentalization is not known but may be due to the substrate specificity of MraY or represent an example of biosynthetic channeling.
Although the muropeptides isolated from mycobacterial lipid I migrated as a single band when analyzed by TLC, the muropeptides isolated from M. smegmatis lipid II showed significantly different mobilities on TLC plates, suggesting the presence of structural differences. Tandem mass spectrometry showed that the muropeptides obtained from mycobacterial lipid I are predominantly MurNAc-pentapeptide with a small amount of MurNGlyc-pentapeptide, possibly derived from the endogenous UDP-linked precursors. No modifications in the peptide side chains were seen. However, the muropeptides isolated from mycobacterial lipid II form a complex mixture. The muramyl residues alone are present in three different forms: MurNAc, MurNGlyc, and MurNH2. Although MurNAc and MurNGlyc residues are found in both nucleotide-linked peptidoglycan precursors and peptidoglycan isolated from mycobacteria (21), the dominant form observed in the muropeptides isolated from mycobacterial lipid II was MurNAc, which was the form presented in the substrate (UDP-MurNAc-pentapeptide) used for the accumulation of lipid II in the experiments presented in this report. Thus, the relative abundance of MurNAc residues seen in this study may be somewhat higher than is the case in vivo.
Significant amounts of MurNH2 containing muropeptides were also identified in all preparations of mycobacterial lipid II. The loss of the acetyl residue could have been due to ionization in the mass spectrometer. However, the retention times of muropeptides containing MurNH2 were different from those of the muropeptides containing MurNAc or MurNGlyc with identical peptide side chains on the HPLC column, strongly suggesting that the deacylation is not an ionization artifact. Although the presence of MurNH2 in the mature peptidoglycan has been reported for Streptococcus pneumoniae (39), M. smegmatis peptidoglycan has only been reported to contain MurNGlyc (17). Early estimations of the extent of N-glycolylation relied on colorimetric estimations of glycolic acid after hydrolysis and comparison with DAP content. It is possible that these relatively crude and indirect assays may have missed the presence of a population of MurNH2 residues.
The carboxylic acid functions of the peptide side chains of the mycobacterial lipid II also underwent three types of modification. The dominant modification is amidation. The preferred site for amidation in the pentapeptide appears to be the free carboxyl group of the DAP residue. Almost all of the monoamidated muropeptides observed were amidated at this site. Diamidated species in which the free carboxylic functions of d-Glu or the terminal d-Ala were also amidated and triamidated muropeptides were observed. The mature peptidoglycan of M. tuberculosis is known to have amidated DAP and d-Glu (16), but the physiological significance of these modifications is unknown.
Some of the carboxylic functions of the terminal d-Ala of mycobacterial lipid II were methylated. A species of lipid II in which all three of the carboxylic acid functions were methylated was also observed. Interestingly, these molecules appeared to give weak ions for the fragments labeled F through K in Fig. 6. However, sufficient fragments were identified to be able to assign the methyl group on one ion (m/z 1,027.2) to the terminal d-Ala residue (Table 1). As further evidence of methylation, an ion (m/z 999.2) was observed in bands 2 and 3 (Fig. 5 and Table 1), which, when subjected to MS2, generated daughter ions indicating that the terminal d-Ala had lost 14 amu. Since an ion having an m/z of 999.2 was not seen in samples that were not reduced, this outcome suggests that the ion is the result of the chemical reduction of a methyl ester bond by the NaBH4, strengthening the evidence that the terminal d-Ala residue was methylated. The presence of methyl esters in mycobacterial or other bacterial peptidoglycan has not been reported; thus, it is possible that the esterified forms of lipid II represent intermediates which are not present in mature peptidoglycan.
FIG. 6.
Tandem mass spectroscopic analysis of an ion with an m/z value of 1,013.1. (A) The MS2 spectrum of an ion (m/z 1,013.1) from a reduced sample (Fig. 5) is shown. (B) The deduced structure and fragmentation pattern of the [M + H]+ ion is shown. Structure I shows the origin of fragments A through E and I. Fragments F through H, J, and K are the result of double cleavage events; the origins of these fragments are shown in structures II and III. Fragment B is not found in the scan shown in panel A but was present in others.
The occurrence of a glycine residue on the d-Glu residue of peptidoglycan of several organisms, including M. tuberculosis, has been reported (29). Molecular ions and some daughter ions were observed in the course of these experiments which had m/z values consistent with the addition of a glycine residue to lipid II. However, it was not possible to assign a position to the modification from the ions observed.
All of the observed modifications of the muropeptides appeared to occur independently, forming a complex mixture of at least 14 species with modifications of the muramyl residue and the peptide side chain and various combinations (Fig. 7). Some of the observed modifications have previously been reported for mature peptidoglycan, and the search for analogous structures in mature mycobacterial peptidoglycan is currently under way. The observation that lipid I was largely unmodified in these studies strongly suggests that the lipid II intermediates of M. smegmatis are substrates for a variety of enzymes. Although amidation of the peptide side chains of peptidoglycan has been reported from a variety of microorganisms and previous work has pointed to lipid II as the substrate (31), the enzymes involved have yet to be identified.
FIG. 7.
Observed variations in the structure of mycobacterial lipid II. The various substituents found at the R positions (indicated on the figure) can occur in essentially any combination. However, R3 appears to be preferentially amidated. Dimethylated species were not observed, although mono- and trimethylated species were both present. The stereochemistry of the prenyl moiety was not determined and is drawn to conform to earlier structural analysis of mycobacterial decaprenyl phosphate; similarly, the stereochemistry of the peptide chain was not determined.
While it is known that modifying the sugar residues of peptidoglycan can result in resistance to muramidases (10), it is not obvious what the physiological relevance of the modifications of the pentapeptide are. However, one can speculate that modification of the carboxylic acid residue of the d-center of DAP could be involved in regulation of cross-linking in the mature peptidoglycan, even though it is not directly involved in the formation of the resulting peptide bond.
Supplementary Material
Acknowledgments
This work was supported by grants (AI18357, AI33706, and AI49151) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Besra, G. S., C. B. Morehouse, C. M. Rittner, C. J. Waechter, and P. J. Brennan. 1997. Biosynthesis of mycobacterial lipoarabinomannan. J. Biol. Chem. 272:18460-18466. [DOI] [PubMed] [Google Scholar]
- 2.Billot-Klein, D., D. Shlaes, D. Bryant, D. Bell, R. Legrand, L. Gutmann, and J. van Heijenoort. 1997. Presence of UDP-N-acetylmuramyl-hexapeptides and -heptapeptides in enterococci and staphylococci after treatment with ramoplanin, tunicamycin, or vancomycin. J. Bacteriol. 179:4684-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick, D. C., M. C. Schulbach, E. E. Zink, M. Macchia, S. Barontini, G. S. Besra, and P. J. Brennan. 2000. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 182:5771-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davisson, V. J., A. B. Woodside, and C. D. Poulter. 1985. Synthesis of allylic and homoallylic isoprenoid pyrophosphates. Methods Enzymol. 110:130-144. [DOI] [PubMed] [Google Scholar]
- 5.Dunphy, P. J., J. D. Kerr, J. F. Pennock, K. J. Whittle, and J. Feeney. 1967. The plurality of long chain isoprenoid alcohols (polyprenols) from natural sources. Biochim. Biophys. Acta 136:136-147. [DOI] [PubMed] [Google Scholar]
- 6.Filipe, S. R., M. G. Pinho, and A. Tomasz. 2000. Characterization of the murMN operon involved in the synthesis of branched peptidoglycan peptides in Streptococcus pneumoniae. J. Biol. Chem. 275:27768-27774. [DOI] [PubMed] [Google Scholar]
- 7.Ghuysen, J. M. 1997. Penicillin-binding proteins. Wall peptidoglycan assembly and resistance to penicillin: facts, doubts and hopes. Int. J. Antimicrob. Agents 8:45-60. [DOI] [PubMed] [Google Scholar]
- 8.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid-chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 9.Gurcha, S. S., A. R. Baulard, L. Kremer, C. Locht, D. B. Moody, W. Muhlecker, C. E. Costello, D. C. Crick, P. J. Brennan, and G. S. Besra. 2002. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem. J. 365:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi, H., Y. Araki, and E. Ito. 1973. Occurrence of glucosamine residues with free amino groups in cell-wall peptidoglycan from bacilli as a factor responsible for resistance to lysozyme. J. Bacteriol. 113:592-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemming, F. W. 1974. Lipids in glycan synthesis, p. 39-97. In T. W. Goodwin (ed.), Biochemistry, series one, vol. 4. Biochemistry of lipids. Butterworths, London, United Kingdom. [Google Scholar]
- 12.Higashi, Y., J. L. Strominger, and C. C. Sweeley. 1967. Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of a C55 isoprenoid alcohol. Proc. Natl. Acad. Sci. USA 57:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higashi, Y., J. L. Strominger, and C. C. Sweeley. 1970. Biosynthesis of the peptidoglycan of bacterial cell walls. XXI. Isolation of free C55-isoprenoid alcohol and of lipid intermediates in peptidoglycan synthesis from Staphylococcus aureus. J. Biol. Chem. 245:3697-3702. [PubMed] [Google Scholar]
- 14.Johnson, R. S., S. A. Martin, K. Biemann, J. T. Stults, and J. T. Watson. 1987. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass-spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 59:2621-2625. [DOI] [PubMed] [Google Scholar]
- 15.Katz, W., M. Matsuhashi, C. P. Dietrich, and J. L. Strominger. 1967. Biosynthesis of the peptidoglycan of bacterial cell walls. IV. Incorporation of glycine in Micrococcus lysodeikticus. J. Biol. Chem. 242:3207-3217. [PubMed] [Google Scholar]
- 16.Kotani, S., I. Yanagida, K. Kato, and T. Matsuda. 1970. Studies on peptides, glycopeptides and antigenic polysaccharide-glycopeptide complexes isolated from an L-11 enzyme lysate of the cell walls of Mycobacterium tuberculosis strain H37Rv. Biken J. 13:249-275. [PubMed] [Google Scholar]
- 17.Lederer, E. 1971. The mycobacterial cell wall. Pure Appl. Chem. 25:135-165. [DOI] [PubMed] [Google Scholar]
- 18.Linnett, P. E., R. J. Roberts, and J. L. Strominger. 1974. Biosynthesis and cross-linking of the gamma-glutamylglycine-containing peptidoglycan of vegetative cells of Sporosarcina ureae. J. Biol. Chem. 249:2497-2506. [PubMed] [Google Scholar]
- 19.Linnett, P. E., and J. L. Strominger. 1974. Amidation and cross-linking of the enzymatically synthesized peptidoglycan of Bacillus stearothermophilus. J. Biol. Chem. 249:2489-2496. [PubMed] [Google Scholar]
- 20.Mahapatra, S., D. C. Crick, and P. J. Brennan. 2000. Comparison of the UDP-N-acetylmuramate:l-alanine ligase enzymes from Mycobacterium tuberculosis and Mycobacterium leprae. J. Bacteriol. 182:6827-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahapatra, S., H. Scherman, P. J. Brennan, and D. C. Crick. 2005. N glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J. Bacteriol. 187:2341-2347. [DOI] [PMC free article] [PubMed]
- 22.Matsuhashi, M. 1994. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation., p. 55-72. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 23.Matsuhashi, M., C. P. Dietrich, and J. L. Strominger. 1965. Incorporation of glycine into the cell wall glycopeptide in Staphylococcus aureus: role of sRNA and lipid intermediates. Proc. Natl. Acad. Sci. USA 54:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil, M., M. Daffe, and P. J. Brennan. 1990. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 265:18200-18206. [PubMed] [Google Scholar]
- 25.Mengin-Lecreulx, D., L. Texier, M. Rousseau, and J. van Heijenoort. 1991. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 173:4625-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 27.Petit, J. F., A. Adam, J. Wietzerbin-Falszpan, E. Lederer, and J. M. Ghuysen. 1969. Chemical structure of the cell wall of Mycobacterium smegmatis. I. Isolation and partial characterization of the peptidoglycan. Biochem. Biophys. Res. Commun. 35:478-485. [DOI] [PubMed] [Google Scholar]
- 28.Reddy, S. G., S. T. Waddell, D. W. Kuo, K. K. Wong, and D. L. Pompliano. 1999. Preparative enzymatic synthesis and characterization of the cytoplasmic intermediates of murein biosynthesis. J. Am. Chem. Soc. 121:1175-1178. [Google Scholar]
- 29.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulbach, M. C., S. Mahapatra, M. Macchia, S. Barontini, C. Papi, F. Minutolo, S. Bertini, P. J. Brennan, and D. C. Crick. 2001. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J. Biol. Chem. 276:11624-11630. [DOI] [PubMed] [Google Scholar]
- 31.Sewert, G., and J. L. Strominger. 1968. Biosynthesis of the peptidoglycan of bacterial cell walls. XI. Formation of the isoglutamine amide group in the cell walls of Staphylococcus aureus. J. Biol. Chem. 243:783-790. [PubMed] [Google Scholar]
- 32.Takayama, K., H. L. David, L. Wang, and D. S. Goldman. 1970. Isolation and characterization of uridine diphosphate-N-glycolylmuramyl-l-alanyl-gamma-d-glutamyl-meso-alpha,alpha′-diaminopimelic acid from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 39:7-12. [DOI] [PubMed] [Google Scholar]
- 33.Takayama, K., and D. S. Goldman. 1970. Enzymatic synthesis of mannosyl-1-phosphoryl-decaprenol by a cell-free system of Mycobacterium tuberculosis. J. Biol. Chem. 245:6251-6257. [PubMed] [Google Scholar]
- 34.Tipper, D. J., W. Katz, J. L. Strominger, and J. M. Ghuysen. 1967. Substituents on the alpha-carboxyl group of d-glutamic acid in the peptidoglycan of several bacterial cell walls. Biochemistry 6:921-929. [DOI] [PubMed] [Google Scholar]
- 35.van Heijenoort, J. 1994. Biosynthesis of bacterial peptidoglycan unit, p. 39-54. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 36.van Heijenoort, J. 1998. Assembly of the monomer unit of bacterial peptidoglycan. Cell. Mol. Life Sci. 54:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 38.van Heijenoort, Y., M. Gomez, M. Derrien, J. Ayala, and J. van Heijenoort. 1992. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J. Bacteriol. 174:3549-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 40.Wietzerbin, J., B. C. Das, J. F. Petit, E. Lederer, M. Leyh-Bouille, and J. M. Ghuysen. 1974. Occurrence of d-alanyl-(d)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of mycobacteria. Biochemistry 13:3471-3476. [DOI] [PubMed] [Google Scholar]
- 41.Wolucka, B. A., M. R. McNeil, E. de Hoffmann, T. Chojnacki, and P. J. Brennan. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269:23328-23335. [PubMed] [Google Scholar]
- 42.Yagi, T., S. Mahapatra, K. Mikusova, D. C. Crick, and P. J. Brennan. 2003. Polymerization of mycobacterial arabinogalactan and ligation to peptidoglycan. J. Biol. Chem. 278:26497-26504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.