Abstract
Quorum sensing is a process by which bacteria communicate by using secreted chemical signaling molecules called autoinducers. Many bacterial species modulate the expression of a wide variety of physiological functions in response to changes in population density by this mechanism. In this study, the opportunistic pathogen Klebsiella pneumoniae was observed to secrete type 2 signaling molecules. A homologue of luxS, the gene required for AI-2 synthesis in Vibrio harveyi, was isolated from the K. pneumoniae genome. A V. harveyi bioassay showed the luxS functionality in K. pneumoniae and its ability to complement the luxS-negative phenotype of Escherichia coli DH5α. Autoinducer activity was detected in the supernatant, and maximum expression of specific messengers detected by quantitative reverse transcription-PCR analysis occurred during the late exponential phase. The highest levels of AI-2 were observed in minimal medium supplemented with glycerol. To determine the potential role of luxS in colonization processes, a K. pneumoniae luxS isogenic mutant was constructed and tested for its capacity to form biofilms in vitro on an abiotic surface and to colonize the intestinal tract in a murine model. No difference was observed in the level of intestinal colonization between the wild-type strain and the luxS mutant. Microscopic analysis of biofilm structures revealed that the luxS mutant was able to form a mature biofilm but with reduced capacities in the development of microcolonies, mostly in the early steps of biofilm formation. These data suggest that a LuxS-dependent signal plays a role in the early stages of biofilm formation by K. pneumoniae.
Many species of bacteria regulate gene expression through an intercellular communication mechanism called quorum sensing (QS) (17). QS is a mechanism of cell density-dependent regulation of bacterial gene expression and was first described in the marine luminous bacteria Vibrio fischeri and Vibrio harveyi, which use them to control the expression of bioluminescence (28). Bacteria synthesize, release, and detect specific small signaling molecules, referred to as autoinducers (AI), which accumulate in the external environment in conjunction with the population growth. When a critical threshold concentration of autoinducer is reached within a population, a signal transduction cascade is triggered, which forms the basis for change in the expression of specific target genes and thereby modifies the bacterial phenotypes.
At present, there are two major independent types of recognized quorum-sensing systems in bacteria. Type I quorum sensing is a highly specific system and is used for intraspecies communication. In gram-negative bacteria, the autoinducers of signal system I (AI-1) have been identified as derivates of an acyl homoserine lactone (acyl-HSL) backbone with species-specific substitutions. These molecules diffuse freely in and out of cells. Two genes are crucially involved in quorum sensing through acyl-HSL: the synthesis of acyl-HSL is dependent on luxI, while luxR encodes a transcriptional activator protein that is responsible for the detection of the cognate acyl-HSL and induction of the appropriate output (1, 16).
Type II QS, most thoroughly characterized for V. harveyi, responds to an autoinducer signal (AI-2) that is produced by LuxS, the luxS product. LuxS converts S-ribosylhomocysteine to 4,5-dihydroxyl-2,3-pentanedione, catalyzing AI-2 formation. The AI-2 molecule produced by V. harveyi is a furanosyl borate diester (7). In V. harveyi, AI-2 binds to a specific receptor/sensor complex (LuxP/Q), and subsequent phosphorelay via either LuxU or a LuxU homologue leads to modification of the transcriptional activator LuxO (15). Many gram-positive and gram-negative bacteria contain highly conserved luxS homologues and produce autoinducer molecules that are functionally similar to V. harveyi AI-2 (41). The LuxS-dependent quorum-sensing system has been referred to as an interspecies communication system and may operate as a universal quorum system for many bacteria possessing the characteristic luxS gene. Several reports have shown the involvement of this type II QS in the regulation of expression of virulence-related factors, motility, secretion systems, regulatory proteins, and polypeptides involved in the acquisition of hemin (for a review, see reference 41). It was shown that bacterial communication mediated by LuxS is also involved in biofilm formation by Streptococcus gordonii (4), Streptococcus mutans (26), and Salmonella enterica serovar Typhimurium (32).
In their natural environment, bacteria do not exist as isolated cells but grow and survive in organized communities and persist attached to solid surfaces, where they are frequently encased in an exopolysaccharide matrix. These microbial communities, composed of single or multiple bacterial species, are termed biofilms and have become accepted as being the predominant natural life for most bacteria (30). These groupings of bacteria are associated with a higher level of resistance to antimicrobial agents than planktonic cells (24). It is now clear that the formation of these sessile communities and their inherent resistance have an enormous impact on medicine. Biofilms are at the root of several persistent and chronic bacterial infections and form on many medical implants. Moreover, many nosocomial infections are considered to be the consequence of surface contamination of indwelling medical devices (24) or are associated with intestinal colonization (11, 25).
Klebsiella pneumoniae is an important gram-negative opportunistic pathogen frequently associated with nosocomially acquired infections. This ubiquitous enterobacterium is involved in urinary tract infections, pneumonia, infections of surgical wounds, bacteremia, and septicemia. Whatever the infection site, the first stage of nosocomial infections due to K. pneumoniae consists of colonization of the patient's gastrointestinal tract (11, 25). K. pneumoniae is characterized by its ability to produce several distinct types of adherence factors and copious amounts of an acid polysaccharide capsule (39), which allow it to adhere to epithelial cells and form biofilms on abiotic surfaces. Previous studies on gram-negative bacilli established that some quorum-sensing mechanisms are involved in the regulation of diverse factors involved in virulence and biofilm formation, so we looked for comparable regulation in K. pneumoniae. Our study showed that K. pneumoniae possesses a luxS ortholog and synthesizes functional AI-2 molecules. By creating an isogenic luxS-deficient mutant of K. pneumoniae, we were able to determine the role of type II QS in biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmid, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. V. harveyi BB120 strain and its derivates V. harveyi BB886 and V. harveyi BB170 were kindly provided by B. Bassler (Princeton University). All bacterial strains were stored at −20°C in appropriate media containing 15% glycerol. Luria-Bertani (LB) broth, 2% glycerol AB medium (3), brain heart infusion broth (BHI), and Dulbecco's modified Eagle's medium (DMEM) were used for the experiments. Biofilm assays were performed in 0.4% glucose M63B1 minimal medium at 37°C. Antibiotics were added to the media for the appropriate bacterial strains at the following concentrations: ampicillin, 50 μg · ml−1; kanamycin, 50 μg · ml−1; gentamicin, 50 μg · ml−1; chloramphenicol, 50 μg · ml−1; streptomycin, 50 μg · ml−1; tetracycline, 20 μg · ml−1.
TABLE 1.
Strains and plasmids useda
| Strain or plasmid | Description | Antibiotic resistance(s) | Growth medium/temp (°C) | Source or reference |
|---|---|---|---|---|
| Strains | ||||
| K. pneumoniae | ||||
| LM21 | Clinical isolate, serogroup O25 | Apr | LB/37 | 14 |
| LM21-ΔluxS | LM21 isogenic mutant with deletion of luxS gene | Kmr, Apr | LB/37 | This study |
| LM21 gfp | LM21 strain SHV-1::gfpmut3-cat | Cmr | LB/37 | This study |
| LM21-ΔluxS gfp | LM21-ΔluxS strain SHV-1::gfpmut3-cat | Kmr, Cmr | LB/37 | This study |
| E. coli K-12 | ||||
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14-(McrA-) Δ(lac-proAB) thi gyrA96(Nalr) endAI hsdRI7 (rK−mK+) relA1 supE44 recA1 | LB/37 | ||
| DH5α | φ80 ΔlacZ ΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK+) supE44 thi− d− | LB/37 | ||
| S17 λpir | Tpr SmrrecA thi pro hsdR M+ RP4::2-Tc::Mu::Km Tn7 λpir lysogen | LB/37 | 29 | |
| CH158 | E. coli strain harboring the kanamycin resistance gene | Kmr | LB/37 | 6 |
| CH295 | E. coli TG1 strain λ-att gfpmut3-cat | Cmr | LB/37 | |
| V. harveyi | ||||
| BB120 | Wild type (AI-1+, AI-2+), positive control, synthesizes AI-1 and AI-2 | AB/30 | 2 | |
| BB170 | luxN::Tn5 (S-1−, S-2+), sensor 1−, sensor 2+ | AB/30 | 2 | |
| P. aeruginosa | ||||
| PAO-I | Wild type | 0.4% glucose A/37 | 18 | |
| PAO-RI | PAO-I derivative mutant lasR−lasI− | 0.4% glucose A/37 | 18 | |
| A. tumefaciens NTL4 | pZLR4 (traG::lacZ fusion and traR) | Gmr, Cbr | 0.4% mannitol AB/28 | 5 |
| Plasmids | ||||
| pBR322 | E. coli cloning vector, oriColE1 | Tetr | Biolabs | |
| pKNG101 | Suicide conjugative vector with an R6K origin and sacBR genes | Str | 22 | |
| pCR2.1 | High-copy-number TA cloning vector | Apr, Kmr | Invitrogen | |
| pZLR4 | Contained a traG::lacZ fusion | Gmr, Cbr | 33 | |
| pKOBEG199 | pBR322 derivative containing the KpnI-HindIII fragment of pKOBEG | Tetr | This study | |
| pCR-luxΔKm | pCR2.1 derivative containing Km cassette flanking by 5′ and 3′ region of luxS | Apr, Kmr | This study | |
| pDB-luxΔKm | pKNG101 derivative Km cassette flanking by 5′ and 3′ region of luxS | Str, Kmr | This study | |
| pBDluxS | pBR322 derivative containing KpLM21 luxS and its own promoter | Tetr | This study |
Abbreviations: Ap, ampicillin; Km, kanamycin; Gm, gentamicin; Cb, carbenicillin; St, streptomycin; Tet, tetracycline; Cm, chloramphenicol.
Conjugative plasmid pKNG101 contains the sacB gene of Bacillus subtilis as a positive selection marker. Plasmid pKOBEG199 was obtained by blunt subcloning the KpnI-HindIII fragment containing the λ Red region and araC gene of pKOBEG (6) at the ScaI site of pBR322.
Determination of AI-1 activity; TLC.
Reverse-phase chromatography was used to examine K. pneumoniae acyl-HSL production. Extracts were prepared from a 5-ml culture of K. pneumoniae LM21 and Pseudomonas aeruginosa PAO-I and PAO-RI. K. pneumoniae LM21 and the P. aeruginosa strains were grown overnight in AB medium, 0.4% glucose A medium, and 0.4% mannitol AB medium, respectively. Extraction of culture supernatants and analytical thin-layer chromatography (TLC) were realized as described by Shaw et al. (33).
Determination of AI-2 activity; V. harveyi bioluminescence assay.
Relative levels of AI-2 in cell-free culture fluids of K. pneumoniae LM21, V. harveyi, and Escherichia coli DH5α strains were measured by using a V. harveyi bioluminescence assay, as described previously (2, 3, 35). Briefly, cell-free supernatants were prepared as follows. V. harveyi strain BB120 was grown in AB medium overnight at 30°C with aeration. K. pneumoniae LM21 and E. coli DH5α strains were cultured in AB medium at 30°C with aeration, and growth was monitored by readings of optical density at 620 nm (OD620). After the appropriate incubation period, bacterial cells were removed by centrifugation and the resulting supernatant was subsequently filter sterilized through a 0.22-mm-pore-size filter (Millipore). The cell-free supernatants were either used immediately or stored frozen at −20°C.
The luminescent reporter strain V. harveyi BB170 (sensor 1−, sensor 2+) was grown for 18 h at 30°C with aeration in AB medium and diluted 1:5,000 into fresh AB medium. The cell-free supernatants (20 μl) were added to the diluted BB170 cells (180 μl) at a 10% (vol/vol) final concentration in hemolysis tubes, which were then shaken at 30°C for 3 h. To determine levels of background luminescence, a sample containing 180 μl of V. harveyi BB170 (dilution, 1:5,000) and 20 μl of sterile AB medium was also included. Positive and negative control samples were obtained by adding cell-free supernatant from V. harveyi BB120 and E. coli DH5α, respectively, to a final concentration of 10%.
After the incubation period, the resulting light production of each sample was measured by a luminometer (Sirius; Berthold). AI-2 activity was expressed as arbitrary luminescence units or as a percentage of bioluminescence induction of the V. harveyi BB170 reporter strain compared to the positive control (V. harveyi BB120). Induction values were obtained by dividing the relative light unit values obtained from the experimental samples by the background values (sterile culture medium).
DNA manipulation and PCRs.
Restriction enzymes, Taq DNA polymerase, and TaKaRa LA Taq were purchased from Roche Molecular Biochemicals (Rotkreuz, Switzerland), QBiogene (Illkirch, France), and Cambrex Compagny (Emerainville, France), respectively, and used according to the manufacturers' recommendations. PCRs were performed by using a Perkin-Elmer GeneAmp PCR system 2400 thermal cycler as reported in Table 2.
TABLE 2.
Oligonucleotides used and PCR product sizes
| Primer | Oligonucleotide sequence (5′-3′)a | PCR product size (bp) | Use |
|---|---|---|---|
| luxS-1 | GCCGTTGTTAGATAGTTTCACAG | 477 | LM21-ΔluxS isogenic mutant construction |
| luxS-2 | CAGTTCGTCGTTGCTGTTGATG | ||
| luxS-A1-5′ | AGGGCGTAAGCCGGCTTAAC | 611 | LM21-ΔluxS isogenic mutant construction |
| luxS-A2 GBL-3′ | GATTTTGAGACACAACGTGGCTTTTGAAACTATCTAACAACGGC | ||
| luxS-B1-3′ | TGCTGAAAATGGCGGTTTAG | 697 | LM21-ΔluxS isogenic mutant construction |
| luxS-B2 GBLnp-5′ | TGCTCGATGAGTTTTTCTAATCAACAGCAACGACGAACT | ||
| A2 GBL-3′ | AGCCACGTTGTGTCTCAAAATC | 977 | Kanamycin resistance cassette amplification |
| B2 GBLnp-5′ | TTAGAAAAACTCATCGAGCA | ||
| luxS-5′ | TGAGTATATCGAAGTGCGTTCG | 1,464 | luxS cloning |
| luxS-3′ | TACGTACCGGCGTCGCGACC | ||
| SHV-1gfp-5′ | ACTTTCTTATAGTTCATCACGGCCTTGAGTCAAAAAATAGCGTGCTTAGGCAGGGCTAGAATGGCGCAATGCCATCTGGT | 2,411 | gfp-tagged strain construction |
| SHV-1gfp-3′ | AAGCGACAAATAAGAATAACCCGGCGTTTTGCTGATTCACAATTCCTCTTTTTTCCTTCACAGGTTGCTCCGGGCTATG | ||
| RT-luxS-5′ | GGAACGCGGTATCCACAC | 226 | RT-PCR |
| RT-luxS-3′ | TGAGCTCCGGGATCTGGT | ||
| RNA 16S 1 | ATGACCAGCCACACTGGAAC | 157 | RT-PCR |
| RNA 16S 2 | CTTCCTCCCCGCTGAAAGTA | ||
| luxS-int | CGAACAGATGCTCCAGGGTG | Sequencing |
Boldface characters: luxS-B2 GBLnp-5′ and luxS-A2 GBL-3′, homologous, respectively, to the 3′ and 5′ end regions of the kanamycin cassette; SHV-1 gfp-5′ and SHV-1gfp-3′, homologous, respectively, to the 5′ and 3′ end regions of the cat-gfpmut3 cassette.
RNA manipulations, real-time RT-PCR.
Total RNA was extracted from bacteria as described by Gosink et al. (19) and treated twice with DNase I (Roche Diagnostics) to remove any contaminating genomic DNA. Total RNA was reverse transcribed and amplified by using primers specific to luxS mRNA (RT-luxS-5′-RT-luxS-3′) and 16S rRNA (RNA 16S 1-RNA 16S 2) (Table 2). Amplification of a single expected product was confirmed by melting curve analysis and electrophoresis on 2% agarose gels. Real-time reverse transcription-PCR (RT-PCR) was performed by using a LightCycler (Roche Diagnostics). Quantification of luxS mRNA levels or 16S rRNA (endogenous control) was done by using RNA master SYBR Green I (Roche Diagnostics) with 0.5 μg of total RNA.
Cloning and analyses of the K. pneumoniae LM21 luxS gene.
Primer sequences (luxS-5′, luxS-3′, and luxS-int) were designed on the basis of the nucleic K. pneumoniae MGH 78578 sequence information available on the National Center for Biotechnology Information (NCBI) database. A 1,464-bp region was amplified from the genomic DNA of K. pneumoniae LM21 with the primers luxS-5′ and luxS-3′ (Table 2). The resulting fragment was sequenced with primers luxS-3′ and luxS-int. The reconstituted 1,235-nucleotide (nt) sequence containing a putative K. pneumoniae LM21 luxS open reading frame (ORF) was investigated for homologies with the NCBI BLAST, version 2.0, algorithm.
Construction and analysis of a K. pneumoniae LM21 ΔluxS mutant.
For functional analysis, a luxS deletion mutant strain was created by allelic exchange. The basic strategy was to replace a chromosomal sequence with a selectable antibiotic resistance gene (kanamycin) generated by a two-step PCR procedure. A 977-bp kanamycin cassette PCR product was generated with primers A2 GBL-3′ and B2 GBLnp-5′ (Table 2) and the template E. coli CH158 strain harboring the kanamycin resistance gene. The K. pneumoniae LM21 DNA was used as a template to amplify the 611- and 697-bp regions flanking, respectively, the 5′ upstream and the 3′ end of luxS with primer pairs luxS-A1-5′-luxS-A2 GBL-3′ and luxS-B1-3′-luxS-B2 GBLnp-5′ (Table 2). At their 5′ ends, primers luxS-A2 GBL-3′ and luxS-B2 gblnp-5′ contained, respectively, a 24- and a 21-bp region homologous to the extremities of the kanamycin cassette. After purification with the QIAquick PCR purification kit (QIAGEN) and quantification, the three resulting amplified fragments were then used for the second step of amplification with primers luxS-A1-5′ and luxS-B1-3′. One hundred nanograms of each fragment was mixed and used as a template to generate a 2,240-bp DNA luxS::Km fragment. The amplicon luxS::Km fragment was purified, ligated into the TOPO TA cloning vector (Invitrogen), and electrotransformed in E. coli JM109. This recombinant plasmid, pCR-luxΔKm, was then XbaI and BamHI restricted, and the kanamycin cassette-containing fragment was subcloned into the corresponding sites in the nonreplicative vector pKNG101 to yield pDB-luxΔKm. The plasmid construct was then transformed into the donor strain E. coli S17 for further conjugal transfer into K. pneumoniae LM21. After overnight incubation on LB plates, mating mixtures were plated on LB agar containing kanamycin. Kmr colonies were then replicated on LB agar containing kanamycin and 5% sucrose to identify isolates that had lost the sacB-associated vector. The replacement of luxS by the kanamycin resistance cassette in the LM21 ΔluxS isogenic mutant was confirmed by PCR with primer couples luxS-1-luxS-2 (internal to luxS), luxS-5′-luxS-3′ (hybridizing 715 bp upstream and 189 bp downstream of luxS, respectively), and A2 GBL-3′-B2 GBLnp-5′ (for kanamycin cassette amplification).
Transcomplementation of LM21 ΔluxS mutant and E. coli DH5α.
A 1,464-bp fragment containing the entire luxS gene under the control of its own putative promoter, as detected by sequence analysis, was amplified from K. pneumoniae LM21 genomic DNA by using luxS-5′ and luxS-3′ primers (Table 2). The resulting fragment was cloned into the TOPO TA cloning vector (Invitrogen) and then transformed into E. coli JM109. Purified recombinant plasmid (QIAGEN plasmid mini kit) was EcoRI digested, and the fragment containing luxS was cloned into the pBR322 vector to yield the pBDluxS plasmid. This construct was used to transform the LM21 ΔluxS isogenic mutant and E. coli DH5α.
Construction of green fluorescent protein (GFP)-tagged strains.
The strains LM21 gfp and LM21 ΔluxS gfp were constructed by integrating inside the SHV-1 β-lactamase-encoding gene (chromosomal ampicillin resistance) a gfpmut3-cat cassette amplified from E. coli strain CH295 (Table 1) by using a procedure previously described by Chaveroche et al. (6) and Derbise et al. (13). Strain CH295 was a gift from J. M. Ghigo, in which the gfpmut3 gene (10) is inserted at the λ att site and controlled by the lambda right promoter.
Briefly, a PCR fragment formed of the cat-gfpmut3 cassette flanked by short portions (i.e., ∼60 bp) of the regions surrounding the SHV-1 gene target sequence was generated using primers SHV-1gfp-5′ and SHV-1gfp-3′ (Table 2) and a DNA template from E. coli CH295. The resulting PCR product was transformed by electroporation in the pKOBEG199-containing K. pneumoniae strains (Table 1). Mutants LM21 gfp and LM21 ΔluxS gfp were selected on LB agar containing chloramphenicol.
Biofilm assay and flow chamber biofilms.
We used a slightly modified version of the microtiter plate assay developed by O'Toole and Kolter (31) to quantify early phase biofilm formation. Briefly, 4 μl of overnight culture was inoculated into 150 μl of DMEM in a 96-well culture-treated polystyrene microtiter plate (Nunc). Wells filled with growth medium alone were included as negative controls. After 4 h of incubation at 37°C, surface-adherent biofilm formation was measured by staining bound cells for 15 min with a 0.5% (wt/vol) aqueous solution of crystal violet. After rinsing with distilled water, the bound dye was released from stained cells by using 95% ethanol and absorbance at 570 nm was determined.
For flow chamber biofilm cultivation, modified FAB medium (20) supplemented with 0.01% glycerol was used. Biofilms were grown at 30°C in flow chambers with individual channel dimensions of 1 by 4 by 40 mm. The flow system was assembled and prepared as described previously (8). The flow chambers were inoculated by injecting 250 μl of overnight culture diluted to an OD600 of 0.001 into each flow channel with a small syringe. After inoculation, flow channels were left without flow for 1 h, after which medium flow was started by using a Watson Marlow 205S peristaltic pump.
All microscopic observations were done with an LSM510 confocal scanning laser microscope (CSLM; Carl Zeiss, Jena, Germany) equipped with an argon laser and detector and filter sets for monitoring GFP (excitation, 488 nm; emission, 517 nm). Images were obtained by using a 40×/1.3 Plan-Neofluar oil objective. Multichannel simulated fluorescence projection (a shadow projection) images and vertical cross sections were generated using the IMARIS software package (Bitplane AG, Zürich, Switzerland).
For the quantification of the K. pneumoniae wild type and ΔluxS mutant, three independent flow channels were quantified by acquiring six image stacks randomly down through to each flow channel.
Images were further treated by using COMSTAT (21).
RESULTS
Production of autoinducer by K. pneumoniae LM21.
To investigate whether K. pneumoniae possesses a quorum-sensing system, we looked for autoinducer molecule production. Two assays allowing detection of AI-1 and/or AI-2 were used: a TLC assay and a V. harveyi bioluminescence assay.
To detect acyl-HSL signal molecules (AI-1), supernatant extracts from K. pneumoniae LM21 cultures were tested for their ability to induce β-galactosidase activity in the Agrobacterium tumefaciens traG::lacZ/traR reporter plasmid pZLR4 (33). Figure 1 shows the chromatogram obtained with extracts from strain LM21 and the control strains, P. aeruginosa PAO-1 and P. aeruginosa PAO-RI. As expected, different structures of AI-1 were visible in the P. aeruginosa PAO-1 positive control sample (Fig. 1, lane 2) and no spot was detected in the lane corresponding to the negative control P. aeruginosa PAO-RI extract (Fig. 1, lane 1). We were unable to detect the presence of any AI-1-like activity in the strain LM21 extract (Fig. 1, lane 3), which suggests that K. pneumoniae does not produce any AI-1 detectable with the traG::lacZ/traR reporter system, nor were we able to detect any AI-1 activity in the strain LM21 supernatant with the bioluminescent V. harveyi BB886 reporter strain (data not shown). Moreover, the screening of the partial K. pneumoniae MGH 78578 genome sequence available on the NCBI microbial genome database revealed no protein showing homology with members of the LuxI family of autoinducer synthase. Taken together, these data provide supporting evidence for the lack of AI-1 activity production by K. pneumoniae.
FIG. 1.
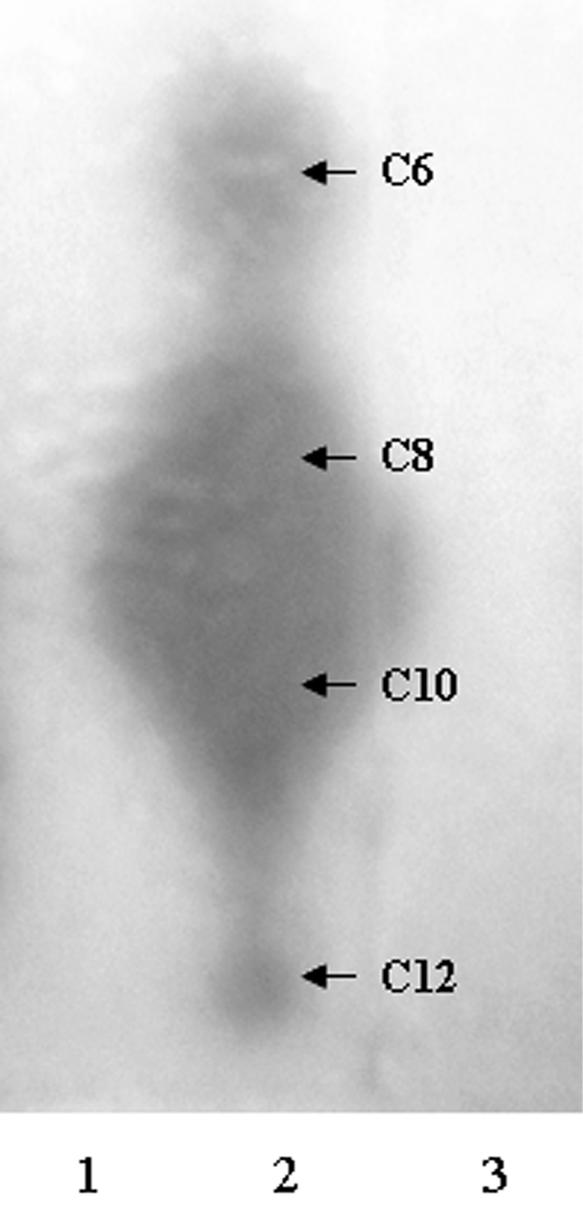
K. pneumoniae LM21 does not produce any acyl-HSL detectable by TLC. Samples were chromatographed on C18 reversed-phase thin-layer plates and developed with methanol-water (60:40, vol/vol), and the spots were visualized with the A. tumefaciens NLT4 reporter strain. Each blue spot visible on the TLC plate corresponded to a β-galactosidase activity induction, resulting from a TraR-mediated transcriptional activation of the traG::lacZ fusion. Samples are from culture extracts of the following: 1, P. aeruginosa PAO-R1; 2, P. aeruginosa PAO-I; 3, K. pneumoniae LM21. The arrows show the position at which P. aeruginosa PAO-I secreted acyl-HSL migrate, and the acyl chain lengths are indicated for each compound. P. aeruginosa PAO-I served as a positive control, whereas P. aeruginosa PAO-R1 served as a negative control. No acyl-HSL was detected in the K. pneumoniae LM21 culture extract with the A. tumefaciens NLT4 reporter strain.
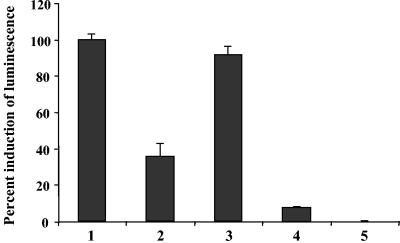
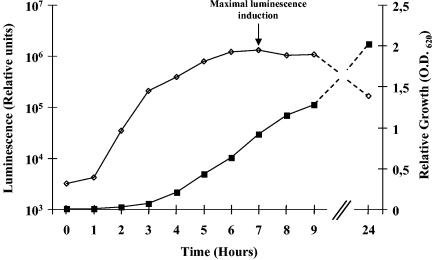
To determine whether strain LM21 had endogenous AI-2 activity, we used the deficient AI-1 sensor V. harveyi BB170 reporter strain (luxN::Tn5, AI-1 sensor− AI-2 sensor+) (35). These BB170 cells were exposed to exogenous cell-free supernatants, and the following induction of bioluminescence was quantified. Since the highest stimulation of light production occurred 3 h after the addition of the cell-free culture supernatants (data not shown), measurements of bioluminescence were all performed after 3 h of incubation at 30°C. It has been reported that AI-2 activity in E. coli and S. enterica serovar Typhimurium is influenced by the presence of certain preferred carbon sources, medium pH, or osmotic pressure (35, 36). Thus, we tested cell-free supernatants from strain LM21 grown in four different growth media (AB, LB, BHI, and DMEM) (OD620, ∼1). Samples from AB medium culture induced the greatest luminescence by the reporter strain (more than 90% of that produced by the positive control, the V. harveyi wild-type strain BB120), whereas supernatant from the DMEM culture induced none (Fig. 2). When the K. pneumoniae strain was grown in LB and BHI media, it produced the signaling factor at a reduced level (35 and 8% of induction over the background level, respectively) compared to culture in AB medium (Fig. 2). This result showed that strain LM21 produces AI-2-like molecules, maximally in AB medium. Therefore, the following experiments were done by using this medium. To determine the AI-2 production kinetics during growth, cell-free supernatants of strain LM21 collected at different time points on the growth curve were tested in the V. harveyi reporter system. Whatever the extract, induction of the V. harveyi BB170 luciferase operon expression was observed (Fig. 3), indicating that K. pneumoniae synthesizes AI-2 or AI-2-like molecules. There was an accumulation of signaling molecules during the exponential growth phase, leading to a maximal induction during the early stationary phase (OD620 of ∼1).
FIG. 2.
Detection of K. pneumoniae LM21 AI-2 production by measuring the level of bioluminescence induced in the V. harveyi BB170 reporter strain (see Materials and Methods). V. harveyi BB120 grown in AB medium was used as a positive control, and the AI-2 activity over the background level of this strain was normalized to 100%. Luminescence induction of experimental samples was expressed as the percent relative to the activity obtained with V. harveyi BB120. Data represent the means and standard errors of the means of the results from four independent experiments. Cell-free supernatants from K. pneumoniae LM21 grown at 30°C in LB, AB, BHI, and DMEM to an OD of ∼1 were tested for AI-2 activity. Bars: 1, V. harveyi BB120 cell-free supernatant; 2, LB medium; 3, AB medium; 4, BHI medium; 5, DMEM medium. K. pneumoniae AI-2 production occurred in AB and LB medium (induction of ∼92 and ∼35%, respectively), whereas no significant production (induction of <10%) was detectable in BHI and DMEM medium. AB, autoinducer bioassay.
FIG. 3.
K. pneumoniae LM21 exhibits maximum AI-2 production in late-exponential and early stationary growth phases (OD620 of ∼1). The expression of AI-2 activity by K. pneumoniae LM21 (open diamonds) is depicted in relation to the growth of the bacteria in AB medium (filled squares). The cell density was determined by measurement of the OD620, and AI-2 activity in bacterial cell-free supernatants was determined by using the V. harveyi reporter strain BB170 as described in Materials and Methods.
Identification of a luxS ortholog in K. pneumoniae LM21.
The synthesis of type II quorum-sensing molecules is dependent on the product of the luxS gene, which is highly conserved among diverse bacteria. To determine whether K. pneumoniae LM21 harbors a luxS-like gene, the unfinished genomic sequence of K. pneumoniae MGH 78578 was BLAST searched by using the E. coli K-12 MG1655 ygaG (ortholog of luxS gene) sequence as a probe. Inside the 2,445-bp K. pneumoniae MGH 78578 contig 1032, a 516-bp candidate ORF exhibited 83% sequence identities with the E. coli ygaG sequence. Using the K. pneumoniae sequence data identified above, primers luxS-5′ and luxS-3′ were designed to amplify a fragment from strain LM21 genomic DNA containing the luxS-like ORF. The resulting fragment was sequenced, and the 1,235-nt obtained sequence and its translated amino acid sequence were submitted for BLASTN and BLASTP programs, respectively. The nucleotide sequence yielded a high level of identity (99%) with the K. pneumoniae MGH 78578 putative luxS gene, and the translated amino acid sequence showed strong homologies and identities with known LuxS sequences, particularly with LuxS from other gram-negative species.
To determine whether the K. pneumoniae luxS ortholog was the ORF responsible for synthesis of AI-2-like molecules, the gene's function was examined by complementing the AI-2 production defect in E. coli DH5α. We took advantage of the inability of E. coli DH5α to synthesize a functional AI-2 molecule, since this laboratory strain fails to produce AI-2 due to a frameshift mutation in the 3′ portion of the luxS ORF that renders it inactive (37). To accomplish this, the recombinant plasmid pDBluxS containing the luxS gene (Materials and Methods) was introduced into E. coli DH5α, and cell-free supernatant prepared from this recombinant strain was tested for its ability to induce luminescence in V. harveyi BB170. As expected, no bioluminescence induction was observed with the cell-free supernatant from DH5α alone, whereas the presence of the luxS-containing plasmid in this strain was sufficient to induce luminescence approximately 70% of that produced by the V. harveyi positive control strain. The data provided evidence that luxS is a critical gene for AI-2 synthesis in strain LM21.
Regulation of luxS expression according to growth phase and medium used.
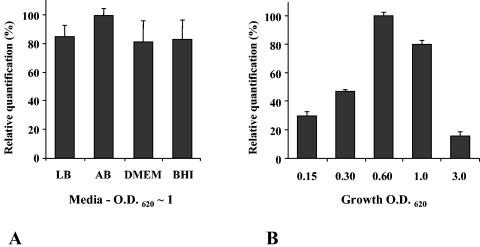
We used RT-PCR to investigate whether the variations of AI-2 activity observed between different media and during the growth was caused by a variation in luxS expression. K. pneumoniae LM21 total RNA was extracted from cultures in different media (LB, AB, DMEM, and BHI medium) at an OD of ∼1 and from cultures at different points of growth in AB medium. Total RNA was reverse transcribed with specific primers, and the levels of luxS and 16S rRNA (endogenous control) expression were analyzed. Results of RT-PCR experiments are shown in Fig. 4. Values indicate a relative quantification of luxS mRNA under the different tested conditions relating to the condition with a maximal level, which was normalized at 100%. As shown in Fig. 4A, the mRNA levels of luxS were similar in experiments made with total RNA from different media. This suggests that the difference in medium-dependent AI-2 activity was not due to transcriptional regulation of luxS. In contrast, the copy number of luxS transcripts seems to vary during the growth of strain LM21 (Fig. 4B). The greatest expression of luxS was observed at an OD of 0.6, which corresponds to the late-exponential growth phase. The transcript levels of luxS were significantly decreased during latency (OD of 0.15) and stationary (OD of 3.0) phases, respectively, 3.4- and 6.4-fold decreased in regard to the condition at an OD of 0.6. This profile was similar to that observed with the AI-2 activity profile.
FIG. 4.
K. pneumoniae luxS expression is growth phase dependent. RT-PCR was carried out by using 0.5 μg of K. pneumoniae LM21 DNA-free total RNA. Primers RT-luxS-5′ and RT-luxS-3′, specific to the luxS ORF, were used. The maximum luxS mRNA level was normalized to 100%, and values are indicated as relative quantifications. (A) Results of RT-PCRs of total RNA isolated from cultures of K. pneumoniae LM21 in LB, AB, BHI, and DMEM at an OD620 of ∼1. (B) Results of RT-PCRs of total RNA isolated at different time points of K. pneumoniae LM21 growth. The bacterial growth was monitored by measurement of OD620. The endogenous control, which consists of amplification of 16S rRNA with primers TM1 and TM2, was not represented. The figure represents the means and standard errors of the means of the results from three independent experiments.
Construction and phenotypic analysis of a K. pneumoniae LM21 ΔluxS mutant.
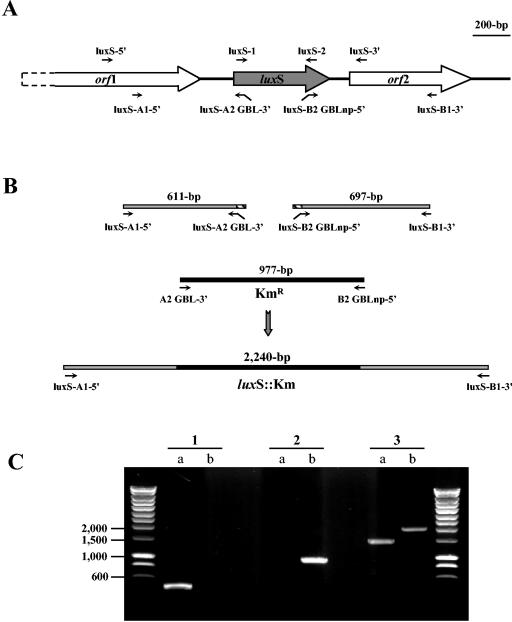
To further analyze the role of luxS-dependent signaling in K. pneumoniae LM21, we created an isogenic luxS deletion mutant by allelic replacement with a kanamycin resistance cassette. A 416-nt internal fragment of luxS was deleted. The allelic exchange was confirmed by various PCRs as shown in Fig. 5C.
FIG. 5.
Construction and PCR verifications of a luxS null mutant. Short arrows indicate approximate locations and directions of primers used. (A) Map of the K. pneumoniae contig 1032, containing two complete and one partial ORF. The arrows indicate locations and directions of transcription. (B) Fragments containing the upstream region of luxS or the 3′ end of the luxS ORF and the downstream region (gray boxes), which have, respectively, 24 and 21 bp of homology to the 5′ and 3′ regions of the kanamycin cassette (hatched boxes), were PCR generated with primer couples luxS-A1-5′-luxS-A2 GBL-3′ and luxS-B2 GBLnp-5′-luxS-B1-3′, respectively. The kanamycin cassette (black box) was amplified from the E. coli CH158 strain with primer couple A2 GBL-3′-B2 GBLnp-5′. The three PCR products were submitted to a second amplification with primers luxS-A1-5′ and luxS-B1-3′. This fragment was cloned into the suicide plasmid pKNG101, resulting in pDB-luxΔKm. (C) PCR amplification product analysis. Amplification products were generated by using primers specific for the intragenic region of the luxS gene (luxS-1-luxS-2) (lanes 1), kanamycin resistance cassette sequence (A2 GBL-3′-B2 GBLnp 5′) (lanes 2), and to amplify the extragenic region of the luxS gene (luxS-5′-luxS-3′) (lanes 3). DNA to be amplified was obtained by lysing strains LM21 (lanes a) and LM21-ΔluxS (lanes b).
The resulting luxS mutant derived from wild-type strain LM21 was designated LM21 ΔluxS. Its growth rate was similar to that of the wild type (data not shown). To determine the effect of the mutation on AI-2 synthesis, LM21 ΔluxS was then compared with the wild-type strain for AI-2 production by using the V. harveyi BB170 bioassay. The cell-free supernatant from the LM21 ΔluxS mutant did not induce any luminescence in the reporter strain. To restore the AI-2 production, the luxS null mutant was transformed by pDBluxS. The defect in AI-2 activity production was partially restored by the wild-type allele (up to 60% of the wild-type level). In contrast, no induction was observed when the plasmid vector alone was introduced into LM21 ΔluxS.
Biofilm growth.
Our next investigation of the mutant phenotype was to determine if there were any alterations of biofilm development as a result of loss of AI-2 production. The ability of K. pneumoniae to adhere and to form a young biofilm was assessed in a microtiter plate experimental model. The biofilm formed by the luxS defective mutant and the LM21 wild-type strains showed no major noticeable differences between both strains. Optical densities after staining of 4-h-old biofilms were similar (OD of 2.209 ± 0.227 versus OD of 2.220 ± 0.267).
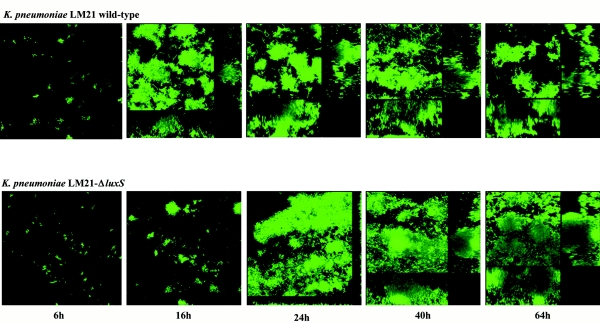
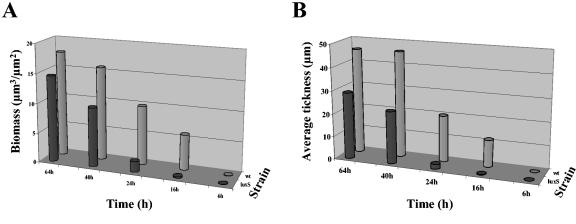
We then determined the three-dimensional structures of the LM21 wild-type and LM21 ΔluxS biofilms by CSLM with GFP-tagged strains. The wild type and the LM21 ΔluxS mutant were grown as biofilms in flow chambers with glycerol minimal media, and development was investigated by acquiring CSLM micrographs at 6, 16, 24, 40, and 64 h after inoculation (Fig. 6). After 6 h, there was little or no difference between biofilms of the wild type and the ΔluxS mutant. Small clusters of cells were initiated at this stage. After 16 and 24 h, the wild-type strain differentiated into a mature (but relatively loose) biofilm exhibiting huge microcolony structures separated by medium-filled channels. At this stage, the ΔluxS mutant was distributed as a flatter biofilm with single cells spreading out on the substratum mainly from initiated microcolony structures, however, not developed to the same degree as the wild-type biofilm. Streamers from the initiated microcolonies in the ΔluxS mutant indicated a loose binding between the cells at this stage. After 40 h, there was still a difference in biomass and biofilm structure between the wild type and the ΔluxS mutant, but they began to resemble each other. After 64 h and during further biofilm growth, the difference between the wild type and the mutant was not pronounced concerning biomass, but there is still a difference in average thickness between the two strains after 64 h. Biomass distribution and average thickness for the mutant and the wild-type strain were compared by quantification with COMSTAT and underlined the description above (Fig. 7).
FIG. 6.
luxS is a critical gene for successfully promoting the first steps of K. pneumoniae biofilm development. Spatial distribution of biofilm formation for the K. pneumoniae LM21 wild type and the luxS-deficient mutant expressing GFP. Biofilms were grown in flow chambers, and development was monitored by SCLM at 6, 16, 24, 40, and 64 h after inoculation. Micrographs represent simulated three-dimensional images. Shown in the right and lower frames are vertical sections though the biofilms.
FIG. 7.
COMSTAT analysis of biofilm structures. Diagrams of biomass (A) and average thickness (B) of biofilms of the luxS-deficient mutant (luxS) and the wild-type strain (wt) of K. pneumoniae LM21 were determined by the COMSTAT program at five different time points (6, 16, 24, 40, and 64 h). Values are means of data from 18 image stacks (6 image stacks from three independent channels). Biomass is measured in cubic micrometers per micrometer squared, and thickness is measured in micrometers.
DISCUSSION
Quorum-sensing mechanisms are used by numerous species of bacteria to regulate virulence factor expression or phenotypic changes. Bacteria control gene expression on a community-wide scale by producing, secreting, detecting, and responding to extracellular signaling molecules called autoinducers. They accumulate in the environment in proportion to cell density, and two major classes of quorum-sensing molecules have been described for gram-negative bacteria. By far the most intensively investigated family of signal molecules are the derivates of acyl-HSL, the so-called AI-1. In this study, we looked for production of type 1 signal molecules by K. pneumoniae LM21 by using two distinct bioassays with A. tumefaciens and V. harveyi reporter strains, respectively. Both of them failed to detect any AI-1-like molecule in K. pneumoniae LM21 culture supernatant. However, this result should be interpreted with caution for several reasons. First, detection is limited to those acyl-HSL to which the reporter strain will respond. Second, the signal must be present at levels detectable by the reporter. Finally, the medium and culture conditions used to grow K. pneumoniae LM21 in this study may not have been optimal for AI-1 production. Likewise, Escherichia and Salmonella species failed to produce detectable levels of acyl-HSL under laboratory growth conditions (27, 38).
The second type of signaling molecule, called AI-2, was first described for V. harveyi and mediates type 2 quorum sensing. A remarkably wide variety of gram-negative and gram-positive bacteria produce AI-2, and many reports have led to the proposal that type 2 quorum sensing is an interspecies communication system and controls an assortment of apparently niche-specific genes (41). In the present study, we showed that culture supernatants from K. pneumoniae LM21 elicit bioluminescence in the V. harveyi AI-2 reporter system, and sequence analysis allowed us to identify a monocistronic luxS homologue in the K. pneumoniae LM21 genome. The putative LuxS exhibited high degrees of similarity to other LuxS proteins in the NCBI database. The laboratory strain E. coli DH5α possesses a luxS gene containing a frameshift mutation in the coding sequence and hence does not produce any AI-2 signaling molecule, and expression of recombinant luxS from various bacterial species restores AI-2 production (37). The putative K. pneumoniae luxS under the control of its own promoter was able to transcomplement the E. coli DH5α LuxS defect. We constructed a K. pneumoniae LM21 luxS isogenic deletion mutant. Abolition of its AI-2 production and previous results confirmed the involvement of luxS in quorum-sensing molecule production by K. pneumoniae LM21.
K. pneumoniae LM21 AI-2 production was evaluated in a variety of growth media. It is known that carbohydrate source, osmolarity, and pH are factors that influence signal production and degradation in S. enterica serovar Typhimurium LT2. Signal production is enhanced in nutrient-rich, high-osmolarity, and low-pH conditions, whereas the conditions favoring its degradation are nutrient poor and low osmolarity (36). Bassler et al. (2) considered an autoinducer-like activity as negative if it is less than 10% of the V. harveyi BB120 luminescence stimulation. On this basis, no significant activity was detected with K. pneumoniae LM21 cultivated in DMEM containing 0.45% glucose or in BHI medium. Only a relatively low level of activity was measured from LB culture. The optimal medium for AI-2 production by K. pneumoniae LM21 was the AB minimum medium supplemented with 2% glycerol. These results were particularly puzzling because E. coli and S. enterica serovar Typhimurium do not produce any significant signaling activity in nutrient-poor media and when glycerol is used as a carbon source (35). Nevertheless, it is known that the regulation of AI-2 production differs between pathogenic and nonpathogenic strains. E. coli O157:H7 strains produce AI-2 at 30 and 37°C with or without glucose, whereas E. coli K-12 strains do not produce AI-2 in the absence of the preferred carbon source (37). There also exists the possibility that AI-2 is still produced in each medium but that the reporter strain is inhibited by some factors secreted by K. pneumoniae grown under these conditions. The great differences observed in AI-2 activity were not due to a differential transcription rate of luxS, as demonstrated by RT-PCR experiments with total RNA from K. pneumoniae LM21 grown in the different media. Controlled steps in AI-2 biosynthesis or degradation could be involved. Moreover, as in S. enterica serovar Typhimurium LT2, AI-2 may also be subjected to protein-dependent degradation in K. pneumoniae. Hence, environmental conditions may affect the level of these degradation proteins and consequently bring about AI-2 degradation.
The kinetics of AI-2 production by strain LM21 indicated that autoinducer production occurred during exponential growth. Using RT-PCR analysis, we were able to detect a maximal luxS transcription during the exponential growth phase, whereas the amounts of luxS transcripts during latency and stationary phase were 3.4- to 6.4-fold lower, respectively. The correlation between luxS expression and AI-2 activity indicated that the production of AI-2 by K. pneumoniae LM21 is probably partially regulated at the transcriptional level of the luxS gene. Accumulation of autoinducer molecules led to a maximal activity in late-exponential to early stationary phase. This activity in culture fluid was severely reduced by the stationary phase. Surette and Bassler (35) showed that AI-2 production by E. coli K-12 MG1655 and S. enterica serovar Typhimurium LT2 strains is influenced by the growth phase of bacteria with activity production maximal during mid- to late exponential phase, when the cells are actively utilizing glucose in rich media. Subsequently, protein-dependent degradation of the signal occurs by the time the cells reach stationary phase or when the carbohydrate is depleted from the medium (35). The highest AI-2 activity produced by K. pneumoniae occured at a relatively high bacterial density. In pathogenic microorganisms, quorum sensing allows bacteria to reach a high cell density before virulence determinants are expressed. By doing so, the bacteria are able to make a concerted attack to produce ample virulence factors and to be present in sufficient numbers to overwhelm the host defenses (12).
Quorum sensing plays an important role in adherence and biofilm formation in several bacterial species. Most of the well-characterized examples involved intraspecies quorum sensing. A few studies have reported a connection between LuxS-dependent AI-2 signals and biofilm formation in S. gordonii, S. mutans, S. enterica serovar Typhimurium, and Helicobacter pylori species (4, 9, 26, 32). Like many bacteria, K. pneumoniae forms characteristic biofilms on abiotic surfaces (unpublished data). To assess the role played by luxS in K. pneumoniae biofilm formation, we tested wild-type and luxS-deficient mutant strains in a microtiter plate bioassay, with a static culture model. The LuxS-deficient strain did not show any major impairment in biofilm formation. This bioassay is very useful for selecting mutants impaired in biofilm formation but is not able to identify mutants with an altered biofilm structure. In the same way, Wen and Burne have used microtiter plate bioassay and concluded that luxS was not involved in biofilm formation by S. mutans (40). However, microscopic analyses showed that the S. mutans luxS mutant had several altered biofilm phenotypes (26). In our study, CSLM analyses of biofilms grown in flow chambers indicated a difference in biofilm development between the K. pneumoniae wild type and luxS mutant. The progression of the biofilm formation by strain LM21 mimics other biofilms described in the literature, beginning with individual bacteria adhering to the surface, expansion into microcolonies, and formation of a three-dimensional structures. In the early steps of biofilm development, the luxS-deficient mutant did not mature with the same speed as the wild type. Cells were mainly distributed on the substratum, and microcolonies were not initiated and developing in size to the same degree as the wild-type strain. The attachment and binding ability of the ΔluxS mutant were reduced compared to the wild type.
Our data suggest that luxS could have a regulatory role for one or more genes related to the early steps of biofilm formation in K. pneumoniae. Several structures (capsule, fimbriae, and lipopolysaccharide) have been implicated in other species (23). Reverse transcription-PCR experiments showed that the capsular biosynthesis operon and the lipopolysaccharide biosynthesis operon were not regulated by LuxS-mediated quorum sensing (data not shown). Other genes are probably regulated by LuxS signaling during the process of biofilm formation, but they remain to be identified.
It was recently shown that quorum sensing may affect intestinal colonization by Vibrio cholerae through a mechanism that involves a phenotype expressed in biofilm (42). K. pneumoniae is an opportunist pathogen responsible for nosocomial infections, and gastrointestinal colonization is an integral part of its infection process. However, the ability of the LM21 ΔluxS to colonize the intestines of mice was the same as that of its parent strain (data not shown), indicating that type 2 quorum sensing was probably not required for these processes. Another hypothesis would be that the ΔluxS LM21 is able to respond to exogenous signaling molecules released from other commensal flora bacterial species. Stroeher et al. (34) recently showed that after intranasal mouse administration of the Streptococcus pneumoniae wild type and its luxS mutant, both strains were able to establish mucosal colonization. However, the luxS mutant was less able to spread from the nasopharynx to the lungs or the blood. Given that the K. pneumoniae wild-type and luxS mutant strains colonized the intestine at similar densities, the quorum-sensing molecule could also be critical for K. pneumoniae to spread out from the intestine to colonize different sites.
In summary, K. pneumoniae expresses luxS and secretes a signal related to AI-2 of V. harveyi. LuxS-dependent signaling was shown to be greatest in minimal media when bacterial densities were highest. As an opportunist pathogen, K. pneumoniae evolves under various conditions. In an external hostile environment, the bacteria may use their type 2 quorum-sensing mechanism to rapidly produce a highly structured biofilm on abiotic surfaces. We speculate that the ability of K. pneumoniae to produce and detect AI-2 may be an important component of the process by which bacterial resistance and spreading occur in the environment.
Acknowledgments
We are grateful to B. Bassler for kindly providing V. harveyi strains prior to publication. We thank C. Lartigue for assistance with chromatography experiments and N. Charbonnel for technical help. We thank J. M. Ghigo for providing GFP tools and for helpful discussions.
This work was supported by the “Contrat Quadriennal de Recherche, Equipe d'Accueil 3844” and by the program “PRFMMIP, Réseau Infections Nosocomiales.”
REFERENCES
- 1.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 4.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 6.Chaveroche, M. K., J. M. Ghigo, and C. d′Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 11.De Champs, C., M. P. Sauvant, C. Chanal, D. Sirot, N. Gazuy, R. Malhuret, J. C. Baguet, and J. Sirot. 1989. Prospective survey of colonization and infection caused by expanded-spectrum-β-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J. Clin. Microbiol. 27:2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113-116. [DOI] [PubMed] [Google Scholar]
- 14.Favre-Bonte, S., B. Joly, and C. Forestier. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 17.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosink, M. M., N. M. Franklin, and G. P. Roberts. 1990. The product of the Klebsiella pneumoniae nifX gene is a negative regulator of the nitrogen fixation (nif) regulon. J. Bacteriol. 172:1441-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 21.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 22.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 23.Lejeune, P. 2003. Contamination of abiotic surfaces: what a colonizing bacterium sees and how to blur it. Trends Microbiol. 11:179-184. [DOI] [PubMed] [Google Scholar]
- 24.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 25.Markowitz, S. M., J. M. Veazey, Jr., F. L. Macrina, C. G. Mayhall, and V. A. Lamb. 1980. Sequential outbreaks of infection due to Klebsiella pneumoniae in a neonatal intensive care unit: implication of a conjugative R plasmid. J. Infect. Dis. 142:106-112. [DOI] [PubMed] [Google Scholar]
- 26.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael, B., J. N. Smith, S. Swift, F. Heffron, and B. M. Ahmer. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O′Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 32.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stroeher, U. H., A. W. Paton, A. D. Ogunniyi, and J. C. Paton. 2003. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 37.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swift, S., M. J. Lynch, L. Fish, D. F. Kirke, J. M. Tomas, G. S. Stewart, and P. Williams. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarkkanen, A. M., B. L. Allen, P. H. Williams, M. Kauppi, K. Haahtela, A. Siitonen, I. Orskov, F. Orskov, S. Clegg, and T. K. Korhonen. 1992. Fimbriation, capsulation, and iron-scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect. Immun. 60:1187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]