Abstract
Listeria monocytogenes is a gram-positive bacterial pathogen that multiplies in the cytosol of host cells and spreads directly from cell to cell. During cell-to-cell spread, bacteria become temporarily confined to secondary vacuoles. The broad-range phospholipase C (PC-PLC) of L. monocytogenes contributes to bacterial escape from secondary vacuoles. PC-PLC requires cleavage of an N-terminal propeptide for activation, and Mpl, a metalloprotease of Listeria, is involved in the proteolytic activation of PC-PLC. Previously, we showed that cell wall translocation of PC-PLC is inefficient, resulting in accumulation of PC-PLC at the membrane-cell wall interface. In infected cells, rapid cell wall translocation of PC-PLC is triggered by a decrease in pH and correlates with cleavage of the propeptide in an Mpl-dependent manner. To address the role of the propeptide and of Mpl in cell wall translocation of PC-PLC, we generated a cleavage site mutant and a propeptide deletion mutant. The intracellular behavior of these mutants was assessed in pulse-chase experiments. We observed efficient translocation of the proform of the PC-PLC cleavage site mutant in a manner that was pH sensitive and Mpl dependent. However, the propeptide deletion mutant was efficiently translocated into host cells independent of Mpl and pH. Overall, these results suggest that Mpl regulates PC-PLC translocation across the bacterial cell wall in a manner that is dependent on the presence of the propeptide but independent of propeptide cleavage. In addition, similarly to Mpl-mediated cleavage of PC-PLC propeptide, Mpl-mediated translocation of PC-PLC across the bacterial cell wall is pH sensitive.
Listeria monocytogenes is an opportunistic bacterial pathogen that has the ability to multiply in the cytosol of eukaryotic cells (9, 32) and spread to neighboring cells without entering the extracellular milieu. Successful infection by L. monocytogenes is dependent on the bacterium's ability to escape membrane vacuoles formed upon initial entry into a host cell and upon direct cell-to-cell spread. Among the factors involved in bacterial escape from these vacuoles is a secreted broad-range phospholipase C (PC-PLC) (Fig. 1a) (28, 34), whose activity requires proteolytic cleavage of a 24-amino-acid N-terminal propeptide (24, 25). A metalloprotease of L. monocytogenes (Mpl) is associated with the proteolytic activation of PC-PLC (19, 25). During intracellular infection, PC-PLC activation is dependent on cell-to-cell spread and vacuolar acidification (19), indicating that cleavage of the propeptide occurs specifically in acidified vacuoles. We are interested in defining the mechanism regulating PC-PLC activity during intracellular infection.
FIG. 1.
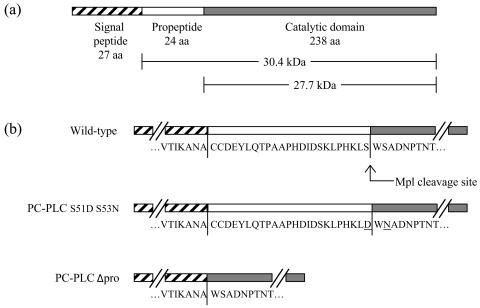
Schematic diagrams of L. monocytogenes PC-PLC. (a) PC-PLC is translated as a preproprotein. The proform of PC-PLC encompasses the propeptide and the catalytic domain. The pro and active forms of PC-PLC have calculated molecular masses of 30.4 and 27.7 kDa but run as 33- and 28-kDa species on SDS-PAGE. (b) The amino acid sequence of the propeptide is flanked by partial amino acid sequence of the signal peptide and catalytic domain. The Mpl cleavage site is indicated for the wild-type strain. For the PC-PLC S51D S53N mutant, two amino acid residues are mutated, decreasing the efficiency of Mpl cleavage. The PC-PLCΔpro mutant was generated by deletion of the entire propeptide.
A number of prokaryotic and eukaryotic proteins are synthesized as preproproteins, in which the signal peptide is followed by a propeptide whose proteolytic cleavage is required to generate the mature protein (5, 27). There are two classes of propeptides, class I and II. Class I propeptides generally function as intramolecular chaperones, facilitating folding of the mature protein. In some instances the propeptide can function in trans, but native conformation cannot be reached without the propeptide (21, 36). Class I propeptides are primarily associated with proteases. Class II propeptides exert their biological function by a variety of mechanisms, including oligomerization, intermolecular interactions, maturation, sorting, folding, and secretion (27). Among others are the propeptide of Bacillus amyloliquefaciens small RNase barnase, which decreases the rate of protein folding in the presence of GroEL (11), the propeptides of Staphylococcus aureus nuclease and of Rhizopus oryzae lipase, which increase the rate of protein folding and secretion (3, 17), and that of human myeloperoxidase, which is required for the maturation process and sorting of the protein to azurophil granules of neutrophils (1).
Studies have shown that processing of the PC-PLC propeptide is a prerequisite to enzymatic activity (25), but the mechanism by which the propeptide prevents activity is unknown. The propeptide could interact with the phospholipase active site or interfere with folding of the catalytic domain. In a recent study, we showed that the proform of PC-PLC does not translocate very efficiently across the bacterial cell wall and that processing of the PC-PLC propeptide correlates with a rapid increase in the rate of PC-PLC translocation (20, 30). This observation is consistent with a role of the propeptide in protein folding, as there are examples of proteins whose rate of folding correlates with their rate of translocation across the cell wall (14, 33, 35). Alternatively, intermolecular interactions involving the prodomain could prevent translocation of PC-PLC across the cell wall.
Mpl is predicted to be a 55-kDa secreted protein composed of a 20-kDa propeptide and a 35-kDa catalytic domain (22). Like PC-PLC, Mpl is bacterium associated and secreted (30). In infected mammalian cells, rapid translocation of bacterium-associated PC-PLC is triggered by a decrease in pH in an Mpl-dependent manner. Therefore, Mpl-mediated processing of the PC-PLC propeptide might control translocation of PC-PLC across the bacterial cell wall. Alternatively, Mpl may target secondary molecules whose processing or degradation is the prerequisite to the efficient translocation of PC-PLC across the cell wall. Moreover, Mpl may carry a dual function as a chaperone and a protease contributing to PC-PLC folding and proteolytic activation. DegP, also known as HtrA, is a good example of a bacterial extracytoplasmic protein carrying such a dual function (2, 15, 31).
The present study investigates the role of the PC-PLC propeptide and of Mpl in the mechanism regulating translocation of PC-PLC across the bacterial cell wall. To address this point, we created a PC-PLC cleavage site mutant (PC-PLC S51D S53N) and a prodomain deletion mutant (PC-PLCΔpro). The intracellular behaviors of the mutated PC-PLC proteins as a function of pH and of Mpl were investigated. Our results indicate that Mpl contributes to the translocation of PC-PLC independently of cleavage of the propeptide and that, in absence of the propeptide, PC-PLC translocation across the bacterial cell wall is no longer regulated by Mpl and pH.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and relevant genotypes are listed in Table 1. For the in vitro experiments, expression of the PrfA-regulated genes was induced by growing bacteria in Luria-Bertani (LB) broth with 50 mM morpholinepropanesulfonic acid (MOPS) adjusted to pH 7.3, 25 mM glucose-1-phosphate (G1P), and 0.2% activated charcoal (LB-MOPS-G1P) (26, 30). For J774 infections, bacteria were grown overnight at 30°C in brain heart infusion broth.
TABLE 1.
L. monocytogenes strains and genotypes
| Strain | Genotypea | Source of strain or reference |
|---|---|---|
| 10403S | Wild type | 4 |
| DP-L1935 | ΔplcB | 28 |
| DP-L2296 | Δmpl | 18 |
| DP-L2787 | ΔplcB Δmpl | 19 |
| DP-L3525 | φ(plcB′-plc+)b | 38 |
| HEL-333 | plcBΔpro Δmpl | This study |
| HEL-335 | plcBΔpro | This study |
| HEL-337 | plcB S51D S53N | This study |
| HEL-338 | plcB S51D S53N Δmpl | This study |
All mutants are derivatives of 10403S.
Gene fusion of L. monocytogenes prepro-PC-PLC plcB sequence and B. cereus catalytic domain of plc sequence.
Construction of plcBΔpro.
An internal in-frame deletion was created in the plcB gene to delete the propeptide coding sequence (Fig. 1b). A 672-bp fragment containing the 5′ end of the plcB gene (coding for the signal sequence) and upstream region was amplified by PCR with primers Marq-46 and Marq-123 (Table 2), using 10403S genomic DNA as a template. A second fragment of 1,300 bp containing sequence coding for the catalytic domain of PC-PLC and downstream sequence was amplified by PCR with primers Marq-124 and Marq-107 (Table 2). PCR fragments were purified, digested with AvaIII, and ligated. Ligation product was used as template for PCR amplification of plcBΔpro using flanking primers Marq-46 and Marq-107. The resulting product (1,938 bp) was digested with HindIII and XbaI and cloned into the shuttle vector pKSV7 (29), creating pHEL-332. pHEL-332 was electroporated into strains 10403S and the Δmpl mutant (DP-L2296), and the mutation was introduced into the chromosome of these strains by allelic exchange as previously described (6), creating HEL-335 and HEL-333 (Table 1).
TABLE 2.
Oligonucleotide primers used in this study
| Marq no. | Nucleotide sequence (5′-3′) with underlined restriction sites | Characteristics |
|---|---|---|
| 46 | AATAAGCGCACCAAGCTTGATAAGTGACATAACTA | 5′ plcB HindIII fwd |
| 101 | CCACATAAGCTTRRYTGGRRYGCGGATAAC | 5′ plcB cleavage HindIII fwd |
| 102 | GCGGACCAACTAAGCTTATGGGGTAATTTGCTG | 3′ plcB propeptide HindIII rev |
| 105 | GGTGAATGTCATGAAGTTCAAAAAGGTGGTT | 5′ plcB signal peptide BspHI fwd |
| 106 | CTAGCTGCAGCTTTTCCATGGGTTTCACTCTCC | 3′ hly P NcoI rev |
| 107 | TTACATCTAGAACTGCGGTACCCTTTT | 3′ plcB XbaI and KpnI rev |
| 122 | TTTAAAGAATTCGGATCCTGGTAAATGTTGAG | 5′ hly P BamHI fwd |
| 123 | GTAAGTATGGATCCCAACATGCATTTGCTTTTA | plcB Δpro AvaIII rev |
| 124 | CCACATAAACATGCATGGTCAGCGGATAAC | plcB Δpro AvaIII fwd |
| 151 | CACATAAACTTGATTGGAACGCGGATAAC | plcB S51D S53N fwd |
| 152 | GTTATCCGCGTTCCAATCAAGTTTATGTG | plcB S51D S53N rev |
Construction of plcB cleavage site mutant.
The plcB cleavage site mutant was generated by random mutagenesis. As described in more detail below, a degenerate primer with the sequence RRYTGGRRY was used, which would put one of four amino acids (D, N, S, and G) at S51 and S53 of the plcB open reading frame. D, N, and G are not generally favorable for protease activity of the Mpl family (23).
To construct PC-PLC cleavage site mutants, three separate fragments were amplified by PCR using purified 10403S genomic DNA as a template. The hly promoter region (621 bp) was amplified with primers Marq-122 and Marq-106 (Table 2); the 5′ end of the plcB gene, containing the signal sequence and propeptide (170 bp) was amplified with primers Marq-105 and Marq-102 (Table 2); and the 3′ end of plcB (1,300 bp) was amplified with primer Marq-101, which contains degenerate codons for the cleavage site mutants, and primer Marq-107 (Table 2). The hly promoter fragment was digested with BamHI and NcoI, the plcB 5′ fragment was digested with BspHI and Hind III, and the plcB 3′ fragment was digested with HindIII and KpnI. All three fragments were sequentially ligated together and cloned into pPL2. pPL2 is an L. monocytogenes phage-specific integration vector (16). Resulting constructs were conjugated into DP-L1935, a ΔplcB derivative of 10403S, and integrants were screened for chloramphenicol resistance and PC-PLC activity on LB-G1P-EY plates. Chloramphenicol-resistant clones showing no or decreased levels of PC-PLC activity were selected. The plcB cleavage site region was sequenced from the selected clones, and Western immunoblot assays were performed for detection of PC-PLC from bacterial supernatants. One mutant was selected for its defect in PC-PLC processing: plcB S51D S53N (Fig. 1b).
The plcB cleavage site mutant was recreated by allelic exchange in wild-type strain 10403S and DP-L2296. For this purpose, the construct was generated by SOEing PCR and cloned into the shuttle vector pKSV7 as described above for the plcBΔpro mutants. Primers used for plcB S51D S53N were Marq-46 and Marq-152 for the 5′ DNA fragment (738 bp) and Marq-151 and Marq-107 for the 3′ DNA fragment (1,299 bp) (Table 2). The construct was sequenced and integrated into the chromosome of L. monocytogenes. Mutant clones were identified by PCR amplification of the plcB gene using primers Marq 46 and Marq 107 and restriction digest of the PCR product with BsaJ1. Acquisition of the mutation correlated with the loss of a BsaJ1 site.
Detection of phospholipase activity.
LB agar was supplemented with 0.2% activated charcoal granules before autoclaving. The medium was cooled down to 55°C before adding glucose-1-phosphate (25 mM), egg yolk (1.25%), and streptomycin (200 μg/ml). LB-G1P-EY plates were poured without the charcoal. Inoculated plates were incubated at 37°C, and PC-PLC activity was detected by the formation of a zone of opacity in the medium.
The egg yolk overlay assay was performed as previously described (19). Briefly, trichloroacetic acid-concentrated supernatants from LB-MOPS-G1P bacterial cultures were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gel was washed successively with isopropanol and phosphate-buffered saline, overlaid with egg yolk soft agar, and incubated at 37°C. PC-PLC activity was detected by the formation of a zone of opacity in the overlay.
Proteolytic digests.
To investigate the folding of secreted PC-PLC, bacterial culture supernatants were treated with different endopeptidases prior to Western immunoblotting. Briefly, L. monocytogenes wild-type and mutant strains were grown in LB-MOPS-G1P at 37°C without shaking to an optical density at 600 nm of 1.2 to 1.4. Bacterial cells were pelleted by centrifugation at 8,820 × g for 10 min at 4°C. Supernatants were collected and treated with 200 μg of trypsin/ml for 15 min at 25°C, 50 μg of chymotrypsin/ml for 15 min at 25°C, 10 μg of thermolysin/ml for 18 h at 37°C, or 2 U of glutamyl endopeptidase (V8)/ml for 18 h at 37°C. Endopeptidase concentrations and incubation times used in these reactions were optimized experimentally. Digested products were precipitated on ice by adding 10% trichloroacetic acid. Precipitates were pelleted by centrifugation at 20,000 × g at 4°C for 10 min and washed with acetone. Precipitates were solubilized in 2× sample buffer (4% SDS, 20% glycerol, 125 mM Tris-HCl [pH 6.8], 10% β-mercaptoethanol, bromophenol blue) and boiled for 5 min. Proteins from an equivalent of 250 μl of bacterial supernatant were resolved by electrophoresis on an SDS-11% polyacrylamide gel and electrotransferred to a polyvinylidene difluoride membrane by using a semidry transfer apparatus (Bio-Rad Laboratories, Hercules, Calif.). PC-PLC was detected by Western immunoblotting.
Western immunoblotting.
Western immunoblot assays were performed as described previously (30). Primary antibodies used were affinity-purified rabbit anti-PC-PLC (1/5,000) and affinity-purified rat anti-PC-PLC propeptide (1/1,000). Secondary antibodies used were alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) and anti-rat IgG (1/25,000).
Generation of anti-propeptide antibody and affinity purification.
An 18-mer peptide (CDEYLQTPAAPHDIDSKL), encompassing three-fourths of the PC-PLC propeptide (Fig. 1b), was synthesized (Protein Chemistry Laboratory of the Medical School of the University of Pennsylvania) and conjugated to keyhole limpet hemocyanin. Generation of the rat immune serum was contracted by Cocalico Biologicals, Inc. For affinity purification of antibodies, the propeptide was conjugated to SulfoLink coupling gel (Pierce Biotechnology Inc.), and specific antibodies were purified according to the manufacturer's instructions.
Metabolic labeling and immunoprecipitation experiments.
For metabolic labeling of intracellularly grown bacteria, the mouse macrophage-like cell line J774 was used and the experiment was performed as described previously (19, 30) with minor modifications. Briefly, at 3 h postinfection, bacterial cell-to-cell spread was blocked with cytochalasin D, an inhibitor of actin polymerization, to test specifically for Mpl-mediated processing of PC-PLC and prevent PC-PLC activation by a host vacuolar cysteine protease (19). At 4 to 5 h postinfection, cells were pulse-labeled for 5 min with [35S]Met and chased for 5 min. When indicated, cells were chased for an additional 5 min in nigericin-containing buffer at pH 7.3 or 6.5. The pulse-chase was performed in the presence of proteasome and host protein synthesis inhibitors. Chloramphenicol was also present during the chase. Labeled samples were lysed, and bacteria were separated from host cell lysates by centrifugation. Bacterial cells were lysed, and PC-PLC or PLCBC was immunoprecipitated from host and bacterial cell lysates using affinity-purified antibodies. PLCBC is a chimera composed of the PC-PLC propeptide and the catalytic domain of the Bacillus cereus PC-PLC ortholog (38). Bacterial counts were determined in parallel dishes and ranged from 1.5 × 107 to 6 × 107 per sample. Equivalent numbers of bacteria were loaded per lane of an SDS-11% polyacrylamide gel. Immunoprecipitates were resolved by SDS-PAGE and detected by autoradiography with a BioMax Transcreen LE and by phosphorimaging using a STORM 860 scanner. Results were analyzed with ImageQuant software (Molecular Dynamics).
RESULTS
Active PC-PLC is generated independently of the propeptide.
Protein propeptides may function as intramolecular chaperones and in some instances are required for a protein to reach its native conformation. To test whether the propeptide of PC-PLC is required for protein folding, we created a mutant in which the prodomain was deleted (plcBΔpro) (Fig. 1b). The plcBΔpro strain was tested for phospholipase activity on egg yolk plates and showed a strong zone of opacity, which is indicative of phospholipid hydrolysis (Fig. 2). Therefore, the catalytic domain of PC-PLC can reach native conformation independently of the propeptide. In addition, as expected, the plcBΔpro strain generated active PC-PLC independently of Mpl (Fig. 2).
FIG. 2.
Detection of PC-PLC activity. L. monocytogenes wild-type strain 10403S and isogenic mutants were grown on LB-G1P-EY plates for 36 h. PC-PLC activity was detectable by the formation of a zone of opacity in the medium.
The zone of PC-PLC activity generated by the plcBΔpro strain was easily detectable after an 18-h incubation at 37°C, whereas that of the wild-type strain appeared after 36 to 48 h of incubation. In the wild-type strain, proteolytic activation of PC-PLC requires Mpl and a decrease in pH (20). The observation that the plcBΔpro strain showed activity early during growth was not surprising, as it bypasses the requirements for activation. In addition, this result substantiates that PC-PLC activity per se is not dependent on acidic pH (10).
It is also worth noting that the plcBΔpro strain did not show any growth defect in broth (data not shown) or on plates (Fig. 2), indicating that constitutive PC-PLC activity is not toxic for the bacterial cell. Since PC-PLC is capable of hydrolyzing phosphatidylglycerol and cardiolipin, two phospholipids present in the cytoplasmic membrane of L. monocytogenes (8, 10), these two phospholipids may not be predominant in the outer layer of the bacterial cytoplasmic membrane.
The propeptide of PC-PLC does not prevent native folding of the PC-PLC catalytic domain.
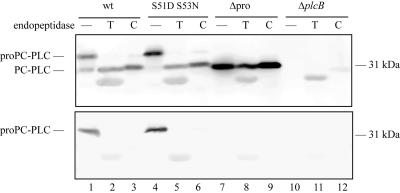
Translocation across the cell wall of gram-positive bacteria is the rate-limiting step for many secreted proteins. Protein structure is one of the factors influencing protein translocation, as correct protein folding is a prerequisite to efficient translocation across the bacterial cell wall (13, 14, 35). For PC-PLC, efficient translocation correlates with proteolytic activation, and the propeptide may interfere with folding of the catalytic domain, hampering translocation of PC-PLC across the cell wall. We performed proteolytic digests of the secreted pro and active forms of PC-PLC to investigate their structure. Trypsin, chymotrypsin, and glutamyl endopeptidase (V8), which are serine peptidases, and the zinc-dependent peptidase thermolysin were used for proteolytic digests. There are over 30 putative sites for each one of these peptidases in the pro and active forms of PC-PLC (37). Partial proteolysis was observed for the proform of PC-PLC (Fig. 3, upper panel, and data not shown). Trypsin, chymotrypsin, and thermolysin digests generated one major product that reacted with the affinity-purified PC-PLC antibody and comigrated with the active form of PC-PLC. It was further determined that the N terminus of the proform of PC-PLC was being cleaved by these three peptidases, as the PC-PLC product generated was not recognized by a propeptide-specific antibody (Fig. 3, bottom panel, and data not shown). The proform of PC-PLC was also partially sensitive to proteolysis by glutamyl endopeptidase. The product generated comigrated with the active form of PC-PLC and did not react with the propeptide-specific antibody (data not shown). Interestingly, cleavage of PC-PLC propeptide by these peptidases did not generate active PC-PLC, as tested by the egg yolk overlay assay (data not shown). According to structural studies on phospholipase C from B. cereus, a PC-PLC ortholog, the first amino acid residue of the PC-PLC catalytic domain (W52) would be an important zinc-coordinating residue (12). Perhaps specific cleavage between S51 and W52 is essential to generate an active enzyme.
FIG. 3.
PC-PLC proteolytic digests. Bacterial supernatants from strains expressing PC-PLC, PC-PLC S51D S53N, PC-PLCΔpro, or no PC-PLC were subjected to the proteolytic activity of trypsin (T) and chymotrypsin (C). Proteolytic digests were analyzed by Western immunoblotting with affinity-purified rabbit anti-PC-PLC (upper panel) or rat anti-propeptide (bottom panel) antibodies. Samples incubated without proteases were loaded in lanes 1, 4, 7, and 10. The extra band migrating below the processed form of PC-PLC in all the trypsin digests is due to a nonspecific reaction with trypsin. Chymotrypsin, which migrates slightly below the processed form of PC-PLC, can also be seen as a very faint band in lane 12, upper panel.
The active form of PC-PLC (PC-PLCΔpro) was completely resistant to proteolysis by all four peptidases and the protein maintained its full activity, indicating that none of the putative cleavage sites are accessible on active PC-PLC (Fig. 3, upper panel, and data not shown). Overall, these results suggest that the catalytic domain of PC-PLC reaches native conformation before cleavage of the propeptide, except for a small region encompassing amino acid residues flanking the propeptide cleavage site, which was accessible to proteolysis.
Efficient translocation of PC-PLC across the bacterial cell wall is independent of propeptide cleavage but dependent of Mpl.
Rapid translocation of PC-PLC across the bacterial cell wall occurs in response to a decrease in pH and correlates with processing of the PC-PLC propeptide in an Mpl-dependent manner (20, 30). To investigate the specific role of the propeptide in PC-PLC translocation across the cell wall, we created a PC-PLC cleavage site mutant. The cleavage site between the pro and catalytic domains of PC-PLC is K49-L50-S51//W52-S53-A54 (Fig. 1b). The catalytic domain of the B. cereus ortholog also starts with WSA, and the crystal structure indicates that the tryptophan residue is one of the nine zinc-coordinating residues (12). Site-directed mutagenesis was used to mutate the last amino acid residue of the propeptide (S51) and the second amino acid residue of the catalytic domain (S53). The resulting mutant, PC-PLC S51D S53N (Fig. 1b), was negative for PC-PLC activity when grown on egg yolk semisolid medium (Fig. 2), although a small amount of processed PC-PLC was detectable from infected cells (see below). PC-PLC S51D S53N behaved similarly to PC-PLC when subjected to trypsin, chymotrypsin, thermolysin, and glutamyl endopeptidase activities, suggesting that the double mutation did not affect PC-PLC native conformation (Fig. 3 and data not shown).
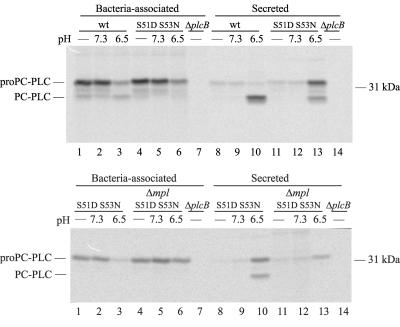
Pulse-chase experiments were performed to follow translocation of PC-PLC S51D S53N across the bacterial cell wall as a function of pH. Infected cells were pulse-labeled with [35S]Met and then chased in a potassium-based buffer at either pH 7.3 or 6.5 in the presence of nigericin to equilibrate host intracellular pH (20). As usual, the labeling experiment was performed in the presence of a proteasome inhibitor to stabilize PC-PLC, as PC-PLC secreted in the cytosol of infected cells is rapidly degraded by the proteasome (19). In addition, an inhibitor of bacterial cell-to-cell spread was used to prevent PC-PLC activation by a host vacuolar cysteine protease (19). Similar to wild-type PC-PLC, PC-PLC S51D S53N remained largely bacteria-associated at pH 7.3 (9.4% translocated) (Table 3 and Fig. 4). At pH 6.5, approximately 29.3% of secreted PC-PLC S51D S53N was processed, as opposed to 90.2% for PC-PLC (Fig. 4, upper panel, lanes 13 and 10). Nevertheless, under these conditions, PC-PLC S51D S53N was translocated as efficiently as PC-PLC (79.2% translocation for PC-PLC S51D S53N versus 73.8% for PC-PLC). This result indicates that a decrease in pH is sufficient for efficient translocation of the proform of PC-PLC S51D S53N across the bacterial cell wall.
TABLE 3.
Quantification of PC-PLC translocation across the bacterial cell wall
| Strain genotype | Translocation at pHa:
|
|
|---|---|---|
| 7.3 | 6.5 | |
| Wild type | 7.9 ± 0.5b | 73.8 ± 2.9*c |
| plcB S51D S53N | 9.4 (7.2, 11.6) | 79.2 (72.7, 85.8) |
| Δmpl plcB S51D S53N | 11.0 (8.0, 14.0) | 18.6 (14.0, 23.3) |
| plcBΔpro | 64.4 ± 9.3 | 82.8 ± 7.7 |
| Δmpl | 6.5 ± 4.5 | 15.1 ± 3.6 |
| Δmpl plcBΔpro | 55.9 ± 22.0 | 73.5 ± 12.6 |
pH at which the pulse-labeled infected cells were chased in the presence of nigericin as described in Materials and Methods.
Mean percent ± standard deviation of PC-PLC immunoprecipitated from lysates of infected cells, determined by quantitative analysis of phosphorscans from three independent experiments, except for the cleavage site mutants for which two independent experiments were performed. Results from each individual experiment performed with the cleavage site mutants are shown in parentheses.
P values (*, P < 0.0001) for comparison of PC-PLC translocation at either pH 7.3 or 6.5 by two-sample t test. Statistical analyses were not performed on samples with less than three replicates.
FIG. 4.
Cell wall translocation of PC-PLC and PC-PLC S51D S53N during intracellular infection. Bacteria grown in J774 cells were pulse-labeled for 5 min and chased for 5 min at 37°C as described in Materials and Methods. When indicated, samples were treated for an additional 5 min with nigericin in buffer of pH 7.3 or 6.5. PC-PLC was immunoprecipitated from bacterial lysates (bacteria-associated) and host cell lysates (secreted), and samples were fractionated by SDS-PAGE. The gel was scanned with a phosphorimager. Proform and processed forms of PC-PLC migrated as indicated on the left side of the figure.
Next, to investigate the specific role of Mpl in PC-PLC translocation across the cell wall, we transferred the plcB S51D S53N mutation into a Δmpl strain and evaluated the pH-dependent behavior of this mutant in the absence of Mpl. In the absence of Mpl, PC-PLC S51D S53N remained entirely in its proform and largely bacterium-associated at either pH: 11.0 and 18.6% translocation at pH 7.3 and 6.5, respectively (Table 3 and Fig. 4, lower panel). Overall, these results reveal that Mpl controls PC-PLC translocation across the bacterial cell wall by a mechanism that is pH sensitive but independent of its role in the proteolytic activation of PC-PLC.
PC-PLCΔpro is efficiently translocated across the bacterial cell wall at physiological pH.
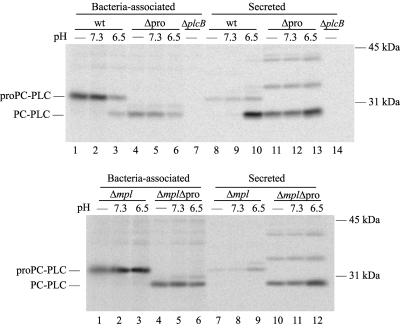
To investigate the specific role of the PC-PLC propeptide in protein translocation, the intracellular behavior of PC-PLCΔpro was evaluated as a function of pH and Mpl. At pH 7.3, PC-PLC remained largely bacterium-associated (7.9% translocated), whereas 64.4% of PC-PLCΔpro was immunoprecipitated from the host cell lysate (Table 3 and Fig. 5, upper panel). At pH 6.5, PC-PLC was activated and translocation increased by 9.3-fold (73.8% translocated), whereas PC-PLCΔpro translocation only increased by 1.3-fold (82.8% translocated) (Table 3 and Fig. 5, upper panel). These results indicate that in the absence of a propeptide, translocation of PC-PLC across the bacterial cell wall is relatively insensitive to pH.
FIG. 5.
Cell wall translocation of PC-PLC and PC-PLCΔpro during intracellular infection. The experiment was performed as described in the legend of Fig. 4.
Next, the requirement for Mpl for translocation of PC-PLCΔpro across the bacterial cell wall was evaluated. At pH 7.3 and in the absence of Mpl, PC-PLC remained largely bacterium-associated (6.5% translocated), whereas 55.9% of PC-PLCΔpro was immunoprecipitated from the host cell lysate (Table 3 and Fig. 5, lower panel). At pH 6.5 and in the absence of Mpl, PC-PLC remained largely bacteria-associated (15.1% translocated), whereas 73.5% of PC-PLCΔpro was immunoprecipitated from the host cell lysate (Table 3 and Fig. 5, lower panel). Although the small changes in protein translocation at pH 6.5 were reproducible, these changes were not statistically significant. These results indicate that, in the absence of a propeptide, translocation of PC-PLC across the bacterial cell wall is no longer regulated by Mpl.
Immunoprecipitates from cells infected with the plcBΔpro strain revealed the presence of secreted protein species larger than monomeric PC-PLC (Fig. 5, upper panel, lanes 11 to 13, and lower panel, lanes 10 to 12). These protein species were not detected by Western immunoblotting of the supernatant of bacterial cultures. The identity of these large protein species is unknown.
The propeptide of PC-PLC does not regulate translocation of a chimera across the bacterial cell wall.
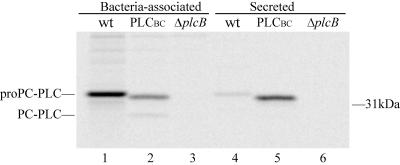
Results from the previous experiment suggest that the PC-PLC propeptide contributes to the formation of a bacterium-associated pool of PC-PLC. To determine if the catalytic domain influences propeptide function in protein translocation across the bacterial cell wall, the behavior of a chimera was evaluated. For these experiments, we used an isogenic 10403S derivative expressing a chimera composed of the PC-PLC propeptide and the catalytic domain of the B. cereus PC-PLC ortholog, PLCBC (38). The catalytic domains of PC-PLC and PLCBC are nearly 40% identical at the amino acid level. Like PC-PLC, the chimera is processed by Mpl in a pH-dependent manner, generating an active enzyme (reference 38 and data not shown). In a previous study, this chimera was shown to be secreted at a higher concentration than PC-PLC in infected J774 cells (38). However, it was not established whether the difference in protein concentration was due to an increase in the total amount of protein or to an increase in the rate of protein translocation across the bacterial cell wall. In the present study, we evaluated the behavior of PLCBC in infected J774 cells maintained at physiological pH. Affinity-purified antibodies specific to PC-PLC and PLCBC were used to immunoprecipitate these proteins from bacterial and host cell lysates, respectively. We observed that PC-PLC and PLCBC were present in similar amounts in the combined cell wall-associated and secreted fractions (Fig. 6). However, at physiological pH, the proform of the chimera was more efficiently translocated across the bacterial cell wall of L. monocytogenes than that of PC-PLC (62% translocation for PLCBC and 5% for PC-PLC) (Fig. 6). These results indicate that the PC-PLC propeptide does not have an intrinsic ability to regulate protein translocation across the bacterial cell wall, but rather the ability of the PC-PLC propeptide to regulate protein translocation is influenced by the nature of the protein to which it is bound.
FIG. 6.
Cell wall translocation of PC-PLC and the chimera PLCBC during intracellular infection. The experiment was performed as described in the legend of Fig. 4, except that the samples were not chased with nigericin in buffer pH 7.3 or 6.5.
DISCUSSION
L. monocytogenes has the remarkable ability to maintain a pool of PC-PLC at its membrane-cell wall interface and to promptly release a bolus of active PC-PLC within vacuoles formed in the process of cell-to-cell spread (19, 20, 30). Efficient translocation of PC-PLC across the bacterial cell wall occurs upon a decrease in pH and correlates with the proteolytic cleavage of an N-terminal propeptide in an Mpl-dependent manner. In the absence of Mpl, bacteria-associated PC-PLC remains in its proform and translocation across the bacterial cell wall is very inefficient independent of pH. In this study, we investigated the role of the propeptide and of Mpl in the mechanism regulating translocation of PC-PLC across the bacterial cell wall. Our results revealed that Mpl regulates PC-PLC translocation across the bacterial cell wall in a manner that is dependent on the presence of the propeptide, but independent of propeptide cleavage. In addition, similar to Mpl-mediated cleavage of the PC-PLC propeptide, Mpl-mediated translocation of PC-PLC across the bacterial cell wall is regulated by pH. Therefore, both Mpl functions are pH sensitive and dependent on the PC-PLC propeptide.
A cleavage site mutant (PC-PLC S51D S53N) was created to determine whether Mpl possesses a biological function that is independent of cleavage of the PC-PLC propeptide. PC-PLC S51D S53N was processed upon a decrease in intracellular pH, although to a much lesser extend than wild-type PC-PLC. However, in absence of Mpl, processing of PC-PLC S51D S53N was undetectable. These results suggest that Mpl is responsible for the proteolytic activation of PC-PLC S51D S53N, even though D and N are not generally favorable for protease activity of the Mpl family (23).
Proteolytic digests of PC-PLC and PC-PLC S51D S53N suggested that the proteins are structurally identical. Similarly to PC-PLC, PC-PLC S51D S53N remained largely bacteria-associated at physiological pH. At acid pH, PC-PLC S51D S53N was as efficiently translocated across the bacterial cell wall as PC-PLC, even though 70% of the translocated mutant protein was in its proform. These results indicate that Mpl carries a pH-sensitive function that is independent of cleavage of the PC-PLC propeptide and that contributes to the regulation of PC-PLC translocation across the bacterial cell wall.
A prodomain deletion mutant was created to address the role of the propeptide in translocation of PC-PLC across the bacterial cell wall. This mutant makes constitutively active PC-PLC, indicating that the PC-PLC propeptide is not required for folding of the mature protein, suggesting that it is a class II propeptide (27). Class II propeptides are associated with a variety of biological functions, including intermolecular interactions, sorting, and secretion. Remarkably, PC-PLCΔpro was very efficiently translocated across the bacterial cell wall at physiological pH independent of Mpl, a behavior that is contrary to that of PC-PLC. This result suggests that the PC-PLC propeptide is a regulatory element, as the propeptide enables the formation of a pool of PC-PLC at the membrane-cell wall interface and prevents premature PC-PLC activity. However, the propeptide does not regulate translocation of the PLCBC chimera, even though it regulates its activity (38). It seems that the sorting function of the propeptide is influenced by the nature of the protein to which it is bound.
Efficient translocation of PC-PLC across the bacterial cell wall and cleavage of the PC-PLC propeptide are both mediated by Mpl upon a decrease in pH. Mpl is active at pH values ranging from 5 to 9 (7), suggesting that Mpl proteolytic activity per se is unlikely to be the pH-limiting factor. However, similar to PC-PLC, Mpl is made as a proenzyme whose activation requires cleavage of an N-terminal propeptide, so proteolytic activation of Mpl could be the pH-sensitive step. It is possible that Mpl functions as a chaperone in its proform, preventing PC-PLC to translocate across the bacterial cell wall and perhaps contributing to PC-PLC folding. DegP, also known as HtrA, is an example of a protein carrying a dual function as a chaperone and a protease (2, 15, 31). If Mpl contributes to PC-PLC folding, PC-PLC translocation across the bacterial cell wall would be impaired in the absence of Mpl as a result of PC-PLC misfolding. Alternatively, Mpl may target molecules other than PC-PLC whose processing or degradation is the prerequisite for the efficient translocation of PC-PLC across the cell wall. Future studies will assess the requirement for Mpl proteolytic activity in the mechanism regulating PC-PLC translocation across the bacterial cell wall.
In conclusion, results from this study and previous studies indicate that Mpl contributes to the regulation of PC-PLC activity by two independent mechanisms, both of which are pH sensitive. First, Mpl regulates cell wall translocation of PC-PLC. Second, Mpl regulates the proteolytic activation of PC-PLC. Importantly, both functions require the presence of the PC-PLC propeptide, because if the propeptide is absent, then PC-PLC translocation and activity are no longer regulated by Mpl and pH. These studies emphasize the complexity of protein translocation across the cell wall of gram-positive bacteria and the ability of a bacterial pathogen to regulate in a spatial and temporal manner the activity of an important virulence factor.
Acknowledgments
We thank D. A. Portnoy for the anti-propeptide rabbit immune serum. We are grateful to Darren Higgins for valuable advice.
This work was supported by U.S. Public Health Service grant AI52154 to H.M.
REFERENCES
- 1.Andersson, E., L. Hellman, U. Gullberg, and I. Olsson. 1998. The role of the propeptide for processing and sorting of human myeloperoxidase. J. Biol. Chem. 273:4747-4753. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 3.Beer, H. D., J. E. McCarthy, U. T. Bornscheuer, and R. D. Schmid. 1998. Cloning, expression, characterization and role of the leader sequence of a lipase from Rhizopus oryzae. Biochim. Biophys. Acta 1399:173-180. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 5.Braun, P., and J. Tommassen. 1998. Function of bacterial propeptides. Trends Microbiol. 6:6-8. [DOI] [PubMed] [Google Scholar]
- 6.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffey, A., B. van den Burg, R. Veltman, and T. Abee. 2000. Characteristics of the biologically active 35-kDa metalloprotease virulence factor from Listeria monocytogenes. J. Appl. Microbiol. 88:132-141. [DOI] [PubMed] [Google Scholar]
- 8.Fischer, W., and K. Leopold. 1999. Polar lipids of four Listeria species containing l-lysylcardiolipin, a novel lipid structure, and other unique phospholipids. Int. J. Syst. Bacteriol. 49:653-662. [DOI] [PubMed] [Google Scholar]
- 9.Gaillard, J.-L., P. Berche, J. Mounier, S. Richard, and P. Sansonetti. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldfine, H., N. C. Johnston, and C. Knob. 1993. Nonspecific phospholipase C of Listeria monocytogenes: activity on phospholipids in Triton X-100-mixed micelles and in biological membranes. J. Bacteriol. 175:4298-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, T. E., J. Eder, M. Bycroft, A. G. Day, and A. R. Fersht. 1993. Refolding of barnase mutants and pro-barnase in the presence and absence of GroEL. EMBO J. 12:4145-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hough, E., L. K. Hansen, B. Birknes, K. Jynge, S. Hansen, A. Hordvik, C. Little, E. Dodson, and Z. Derewenda. 1989. High-resolution (1.5 Å) crystal structure of phospholipase C from Bacillus cereus. Nature 338:357-360. [DOI] [PubMed] [Google Scholar]
- 13.Kontinen, V. P., P. Saris, and M. Sarvas. 1991. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol. Microbiol. 5:1273-1283. [DOI] [PubMed] [Google Scholar]
- 14.Kontinen, V. P., and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8:727-737. [DOI] [PubMed] [Google Scholar]
- 15.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 16.Lauer, P., M. Y. N. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Loir, Y., S. Nouaille, J. Commissaire, L. Bretigny, A. Gruss, and P. Langella. 2001. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marquis, H., V. Doshi, and D. A. Portnoy. 1995. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63:4531-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquis, H., H. Goldfine, and D. A. Portnoy. 1997. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J. Cell Biol. 137:1381-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquis, H., and E. J. Hager. 2000. pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 35:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIver, K. S., E. Kessler, J. C. Olson, and D. E. Ohman. 1995. The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol. Microbiol. 18:877-889. [DOI] [PubMed] [Google Scholar]
- 22.Mengaud, J., C. Geoffroy, and P. Cossart. 1991. Identification of a new operon involved in Listeria monocytogenes virulence: its first gene encodes a protein homologous to bacterial metalloproteases. Infect. Immun. 59:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami, Y., K. Chiba, T. Oda, and A. Hirata. 2001. Novel kinetic analysis of enzymatic dipeptide synthesis: effect of pH and substrates on thermolysin catalysis. Biotechnol. Bioeng. 74:406-415. [DOI] [PubMed] [Google Scholar]
- 24.Niebuhr, K., T. Chakraborty, P. Köllner, and J. Wehland. 1993. Production of monoclonal antibodies to the phosphatidylcholine-specific phospholipase C of Listeria monocytogenes, a virulence factor for this species. Med. Microbiol. Lett. 2:9-16. [Google Scholar]
- 25.Raveneau, J., C. Geoffroy, J.-L. Beretti, J.-L. Gaillard, J. E. Alouf, and P. Berche. 1992. Reduced virulence of a Listeria monocytogenes phospholipase-deficient mutant obtained by transposon insertion into the zinc metalloprotease gene. Infect. Immun. 60:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ripio, M. T., K. Brehm, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179:7174-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinde, U., and M. Inouye. 2000. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11:35-44. [DOI] [PubMed] [Google Scholar]
- 28.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 30.Snyder, A., and H. Marquis. 2003. Restricted translocation across the cell wall regulates secretion of the broad-range phospholipase C of Listeria monocytogenes. J. Bacteriol. 185:5953-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 32.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Wely, K. H., J. Swaving, R. Freudl, and A. J. Driessen. 2001. Translocation of proteins across the cell envelope of gram-positive bacteria. FEMS Microbiol. Rev. 25:437-454. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Boland, J.-A., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitikainen, M., T. Pummi, U. Airaksinen, E. Wahlstrom, H. Wu, M. Sarvas, and V. P. Kontinen. 2001. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of alpha-amylase in Bacillus subtilis. J. Bacteriol. 183:1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wetmore, D. R., S. L. Wong, and R. S. Roche. 1992. The role of the pro-sequence in the processing and secretion of the thermolysin-like neutral protease from Bacillus cereus. Mol. Microbiol. 6:1593-1604. [DOI] [PubMed] [Google Scholar]
- 37.Wilkins, M. R., I. Lindskog, E. Gasteiger, A. Bairoch, J. C. Sanchez, D. F. Hochstrasser, and R. D. Appel. 1997. Detailed peptide characterization using PEPTIDEMASS—a World-Wide-Web-accessible tool. Electrophoresis 18:403-408. [DOI] [PubMed] [Google Scholar]
- 38.Zuckert, W. R., H. Marquis, and H. Goldfine. 1998. Modulation of enzymatic activity and biological function of Listeria monocytogenes broad-range phospholipase C by amino acid substitutions and by replacement with the Bacillus cereus ortholog. Infect. Immun. 66:4823-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]