Abstract
Sequence analysis of membrane-bound glycerolipid acyltransferases revealed that proteins from the bacterial, plant, and animal kingdoms share a highly conserved domain containing invariant histidine and aspartic acid residues separated by four less conserved residues in an HX4D configuration. We investigated the role of the invariant histidine residue in acyltransferase catalysis by site-directed mutagenesis of two representative members of this family, the sn-glycerol-3-phosphate acyltransferase (PlsB) and the bifunctional 2-acyl-glycerophosphoethanolamine acyltransferase/acyl-acyl carrier protein synthetase (Aas) of Escherichia coli. Both the PlsB[H306A] and Aas[H36A] mutants lacked acyltransferase activity. However, the Aas[H36A] mutant retained significant acyl-acyl carrier protein synthetase activity, illustrating that the lack of acyltransferase activity was specifically associated with the H36A substitution. The invariant aspartic acid residue in the HX4D pattern was also important. The substitution of aspartic acid 311 with glutamic acid in PlsB resulted in an enzyme with significantly reduced catalytic activity. Substitution of an alanine at this position eliminated acyltransferase activity; however, the PlsB[D311A] mutant protein did not assemble into the membrane, indicating that aspartic acid 311 is also important for the proper folding and membrane insertion of the acyltransferases. These data are consistent with a mechanism for glycerolipid acyltransferase catalysis where the invariant histidine functions as a general base to deprotonate the hydroxyl moiety of the acyl acceptor.
Acyltransferases are central to the metabolism of fatty acids and phospholipids. The most extensively studied enzyme, sn-glycerol-3-phosphate (G3P) acyltransferase, acylates the 1 position of G3P, the first committed step in phospholipid formation (9). In Escherichia coli, G3P acyltransferase has been purified and biochemically characterized and is the membrane-bound product of the plsB gene (9). Consistent with its position at the start of the phospholipid biosynthetic pathway, the PlsB protein is thought to function as a sensor that monitors the metabolic state of the cell through its allosteric interactions with ATP and guanosine-3′,5′-tetraphosphate to coordinate the rate of G3P acylation with macromolecular biosynthesis and cell growth (20). G3P acyltransferase enzymes with a high degree of similarity to the bacterial PlsB proteins have been identified in Caenorhabditis elegans (accession no. U64847) and mammals (27, 31), and functionally equivalent, but less closely related, proteins are found in plants (13, 29). The second step in the formation of phosphatidic acid is the acylation of the 2 position of 1-acyl-G3P by 1-acyl-G3P acyltransferase, the membrane-associated product of the plsC gene in E. coli (9). Homologs of plsC have been identified in the yeast Saccharomyces cerevisiae (18), plants (5, 10, 15), and C. elegans (accession no. Z73975), and human PlsC homologs are represented in the expressed-sequence-tag database (accession no. W44905, T77083, and H44282). Although the PlsC reaction is essential for phospholipid synthesis, a regulatory role for the PlsC family of enzymes is not apparent (9). The least characterized group of acyltransferases are the enzymes that catalyze the reacylation of lysophospholipids. This class of proteins plays an important role in remodeling phospholipid fatty acid composition and in protecting the membrane bilayer from the disruptive effects of lysophospholipids. The 2-acyl-sn-glycerophosphoethanolamine (2-acyl-GPE) acyltransferase of E. coli (the aas gene product) is an example of this class of acyltransferases that has been biochemically characterized and its gene cloned (9). The primary function of Aas in bacterial metabolism is to act as a salvage pathway for the resynthesis of phosphatidylethanolamine from 2-acyl-GPE taken up from the medium or arising from either the activity of phospholipase A1 (pldA) or the transfer of 1-position acyl moieties in the acylation of membrane proteins (9). The Aas protein is unique in that it also catalyzes a synthetase reaction that transfers fatty acids to the prosthetic group sulfhydryl of a tightly bound acyl carrier protein (ACP) subunit in the presence of ATP and Mg2+.
Despite the importance and ubiquitous distribution of these enzymes, little is known about their catalytic mechanism. In general, the proteins are tightly associated with membranes, making them difficult to express at high levels and purify. The PlsB enzyme has been purified, but the kinetic properties of the purified enzyme are not the same as those of the native membrane-bound protein (24, 25). Their hydrophobic character and the need to reconstitute the isolated enzymes in either phospholipid bilayers or detergent micelles represent major impediments to obtaining crystals for high-resolution structure. Our goal was to obtain insight into the catalytic mechanism of the acyltransferase gene family by using site-directed mutagenesis to identify essential catalytic residues in the protein sequences of two representative members, PlsB and Aas.
MATERIALS AND METHODS
Materials and strains.
Sources of supplies were as follows: Promega, restriction enzymes, E. coli S30 transcription-translation extract, Taq DNA polymerase, and molecular biology supplies; DuPont-New England Nuclear, sn-[U-14C]glycerol-3-phosphate (specific activity, 148 mCi/mmol); Sigma Chemical Co., palmitoyl coenzyme A (palmitoyl-CoA) and ACP; ICN, [35S]methionine (specific activity, 1,218 Ci/mmol); ARC, [9,10-3H]palmitic acid (specific activity, 60 Ci/mmol); Analtech, preabsorbant Silica Gel G thin-layer chromatography plates; New England Biolabs, plasmid pACYC177; and Packard, Scint-A XF scintillation fluid. Protein was measured by the method of Bradford (3). 2-Acyl-GPE was prepared from phosphatidylethanolamine by using Rhizopus lipase as described elsewhere (11). Acyl-ACPs were prepared enzymatically by using the acyl-ACP synthetase method (21). All other chemicals were of reagent grade or better.
Strain SJ22 used for these experiments (plsB26 panD zad220::Tn10 glpR glpD glpK relA1 spoT1 tonA22 T2r pit-10 HfrC) (23) harbored the plsB26 mutation (2, 8).
Construction of mutants.
The H306A mutation in plsB was constructed by an overlap extension PCR protocol. Primer locations are given based on the A in the ATG initiation sequence as bp +1. Two PCRs, using plasmid pRJ22 (wild-type plsB) as the template, were performed with primer pairs plsb-A (bp 795 to 874) plus plsb-B (bp 927 to 909) (5′-CCTGACTGACCGTATTCTGG and 5′-GTGACTGCGGGCGCAAGGC) and plsb-C (bp 909 to 929) plus plsb-D (bp 1120 to 1101) (5′-GCCTTGCGCCCGCAGTCACAT and 5′-ACAGTTCGCCGAGATACTCC). These reactions produced products of 133 and 212 bp, respectively. The second round of PCR was performed by mixing equimolar amounts of first-round products and amplifying between primers plsb-A and plsb-D to produce a product of 326 bp, which was ligated into the cloning vector pCR2.1 (Invitrogen) and sequenced. A clone with the sequence for the desired H306A mutation was chosen and digested with SfiI and Bpu1102I to release a 160-bp fragment. The fragment containing the mutation was ligated into SfiI- and Bpu1102I-digested pRJ22 to create pRJ50, which expressed PlsB[H306A]. The D311A and D311E mutations were constructed in the same manner, with the internal primer pair plsbDA-A (bp 925 to 943) plus plsbDA-B (bp 934 to 916) (5′-CACATGGCCTACCTGCTGC and 5′-AGGCCATGTGACTGCGGTG) or plsbDE-A (bp 925 to 943) plus plsbDE-B (bp 934 to 916) (5′-CACATGGAGTACCTGCTGC and 5′-ACTCCATGTGACTGCGGTG). Reconstructed plasmids harboring the full-length gene with the D311A and D311E mutations were designated pRJ51 and pRJ52, respectively. Sequencing of pRJ50, pRJ51, and pRJ52 was performed by using primers plsb-A and plsb-D to confirm the presence of the desired mutations and verify that the junctions were correctly re-formed.
The H36A mutation in Aas was constructed by excising the wild-type aas gene from pLCH5 (14) by digestion with KpnI and HindIII followed by ligation into pBluescript II KS(+) to form pRJ40. PCR was performed on pRJ40 template with the primers aas-1 (bp −85 to −65) and aas-2 (bp 122 to 96) (5′-TTGGTACCCACCTAATTTACTGTCGCTCG and 5′-TCAATAAAAGAGACGGCAATTAGGCGTA) to effect the desired amino acid substitution and to create a novel KpnI site upstream of the aas promoter. Following ligation into pCR2.1 and sequencing, a KpnI-to-BsmBI cassette was removed and used to replace the wild-type sequence in pRJ40 to create pRJ41. This procedure removed 520 bp of sequence upstream of aas in pRJ40. pRJ41 was sequenced to verify that the correct mutant gene had been constructed.
G3P acyltransferase assays.
Membranes were prepared from E. coli SJ22 (plsB plsX panD2) harboring either a wild-type or mutant plsB gene or from E. coli 8 (wild-type plsB), and G3P acyltransferase assays were performed as described previously (22). Briefly, a 40-μl reaction mixture containing 0.1 M Tris-HCl (pH 8.5), 1 mg of bovine serum albumin per ml, 50 μM [14C]G3P (specific activity, 60 mCi/mmol), 25 μM 16:0-ACP (or 50 μM palmitoyl-CoA), and the indicated amount of membrane protein was incubated at 37°C for 15 min. The reaction mixtures were then deposited onto Whatman 3MM filter paper discs that were subsequently washed with 10, 5, and 1% ice-cold trichloroacetic acid (20 min, 20 ml/disc) prior to scintillation counting (22).
Acyl-ACP synthetase and 2-acyl-GPE acyltransferase assays.
Membrane preparations from strain LCH26 (aas) harboring pRJ40, pRJ41, or pBluescript KS II(+) were prepared as described previously (14). Acyl-ACP synthetase assays (14) were performed in 40 μl of 0.1 M Tris-HCl (pH 8.0) and included 5 mM ATP, 10 mM MgCl2, 0.4 M LiCl, 2% Triton X-100, 2 mM dithiothreitol, 15 μM ACP, 62.5 μM [3H]palmitic acid (specific activity, 480 mCi/mmol), and the indicated amount of membrane protein. Reactions were stopped after 10 min at 37°C by depositing the mixture onto Whatman 3MM filter discs, which were then washed twice in chloroform-methanol-acetic acid (3:6:1, vol/vol) and counted.
Reactions to measure 2-acyl-GPE acyltransferase activity (14) were performed with 0.1 M Tris-HCl (pH 8.0), 5 mM ATP, 5 mM MgCl2, 1 mM dithiothreitol, 100 μM 2-acyl-GPE, 10 μM ACP, 1% Triton X-100, 62.5 μM [3H]palmitate (specific activity, 480 mCi/mmol), and the indicated amount of membrane protein in a final volume of 40 μl. Reaction mixtures were incubated at 37°C for 10 min, reactions were then stopped by the addition of 200 μl of ethanol, and the mixtures were dried in a Speed Vac. The residue was resuspended in 100 μl of chloroform-methanol (1:1, vol/vol) and then applied to a Silica Gel G plate which was developed with chloroform-methanol-acetic acid (85:15:10, vol/vol/vol). Incorporation of radioactivity into phosphatidylethanolamine was detected with a Bioscan imaging system and measured by scraping the plate and scintillation counting.
In vitro transcription and translation.
Coupled in vitro transcription-translation assays were performed with an E. coli S30 extract as recommended by the manufacturer (Promega) except 5 μl of a 20-mg/ml concentration of phosphatidylethanolamine-phosphatidylglycerol-cardiolipin (6:1:1, wt/wt/wt) in 5 mM Tris-HCl (pH 8.5)–1 mM EDTA was added to stabilize the membrane proteins. Reactions were performed with [35S]methionine (specific activity, 1,218 Ci/mmol) and subjected to electrophoresis in sodium dodecyl sulfate–8% acrylamide gels followed by autoradiography.
Construction of flag-tag PlsB proteins and immunoblotting.
To quantitate the expression levels of the wild-type and mutant PlsB proteins, constructs containing an epitope tag at the COOH terminus of each protein were made. A novel SacI restriction site was first engineered by PCR into the wild-type plsB plasmid, pRJ22. Primers used were bnde (5′-CATATGTTGGTGCTGCCTTC) and baat (5′-ATGACGTCTTACCCTTCGAGCTCCGTCGCAC), which amplified the region of plsB from a unique internal NdeI restriction site to the end of the gene, introducing the SacI site just prior to, and a KpnI site immediately after, the stop codon. This process also altered the amino acid composition of the COOH terminus of PlsB from QGEG to ELEG and, on reinsertion into pRJ22, removed approximately 500 bp of plasmid sequences downstream of the plsB gene to create plasmid pRJ53. Oligonucleotides containing the sequence for the flag-tag epitope were constructed (5′-CGACTACAAGGACGACGATGACAAGTAAGACGT and 5′-CTTACTTGTCATCGTCGTCCTTGTAGTCGAGCT), phosphorylated, annealed, and then ligated into the SacI and KpnI sites of pRJ53 to form pRJ54. Both plasmids pRJ53 (PlsB[ELEG]) and pRJ54 (PlsB-flag) complemented the G3P auxotrophy of strain SJ22 (plsB), indicating the expression of a functional PlsB protein, and nucleotide sequencing confirmed the presence of the expected sequence.
To construct flag-tag fusions of the H306A, D311A, and D311E mutants of PlsB, plasmids pRJ50 (PlsB[H306A]), pRJ51 (PlsB[D311A]), and pRJ52 (PlsB[D311E]) were digested with Bpu1102I and NdeI and ligated into pRJ54 (PlsB-flag) to create the plasmids pRJ55 (PlsB-flag[H306A]), pRJ56 (PlsB-flag[D331A]), and pRJ57 (PlsB-flag[D311E]), respectively. Phenotype tests of these flag-tagged mutants gave the same pattern of complementation as the nontagged parents. The mutation-containing region of each plasmid was sequenced to verify correct insertion of the fragments. Membranes proteins prepared from strain SJ22 (plsB) harboring these plasmids were electrophoresed in an 8% polyacrylamide gel, which was then transferred to nitrocellulose and blotted with the anti-flag monoclonal antibody M2 (Kodak IBI, Inc.), using anti-mouse Phototope-horseradish peroxidase (New England Biolabs) detection according to the manufacturer’s instructions.
Complementation of G3P auxotrophy.
Plasmids bearing mutations in plsB were tested for complementation of the G3P requirement of strain SJ22 (plsB26). Cells were plated onto minimal medium E plates (16) containing β-alanine (1.5 μM), glucose (0.4%), Casamino Acids (0.1%), thiamine (0.001%), and ampicillin (100 μg/ml), with or without 0.04% G3P. Growth on the plates lacking G3P indicated complementation of the phenotype. Growth experiments in liquid culture used the same minimal medium.
RESULTS AND DISCUSSION
Sequence alignments and construction of the mutants.
Database searches using the BLAST algorithm (1) with the primary sequences of the PlsB and PlsC acyltransferases revealed a conserved domain in the glycerolipid acyltransferases from bacteria, plants, yeast, nematodes, and mammals (Fig. 1). The key feature of this region was the invariant His and Asp residues found in a HX4D configuration. No other areas of identity were observed among these proteins. This motif was located in different regions of the primary sequence. For example, the domain containing the HX4D arrangement was found at the amino terminus of Aas (His36 and Asp41) and near the middle of PlsB (His306 and Asp311). Mutations in plsB and aas were made by a PCR and cassette replacement protocol as outlined in Materials and Methods. Each mutation-bearing fragment was sequenced prior to reconstruction, and the re-formed junctions were sequenced to verify that the mutant plasmid was correct. To verify that the mutant constructs expressed full-length proteins, an in vitro coupled transcription-translation assay was performed with an E. coli S30 extract. The expression of the mutant genes was the same level as the wild-type genes (data not shown).
FIG. 1.
Acyltransferase domain sequence similarity. Sequences in the GenBank database from bacteria, yeast, plants, nematodes, and mammals with homology to the E. coli PlsB G3P acyltransferase (G3PAT), the E. coli PlsC lysophosphatidic acid acyltransferase (LPAAT), and 2-acyl-GPE acyltransferase (LPEAT) were aligned by using the PILEUP program in the Genetics Computer Group software package. Sequences are aligned according to overall similarity. •, invariant His and Asp; Φ, conserved hydrophobic residue. Genus names not specified in the text: A., Arabidopsis; S., Salmonella; M., Mus; H., Haemophilus; N., Neisseria; L., Limnanthes; Myc., Mycobacterium.
Analysis of the PlsB mutants.
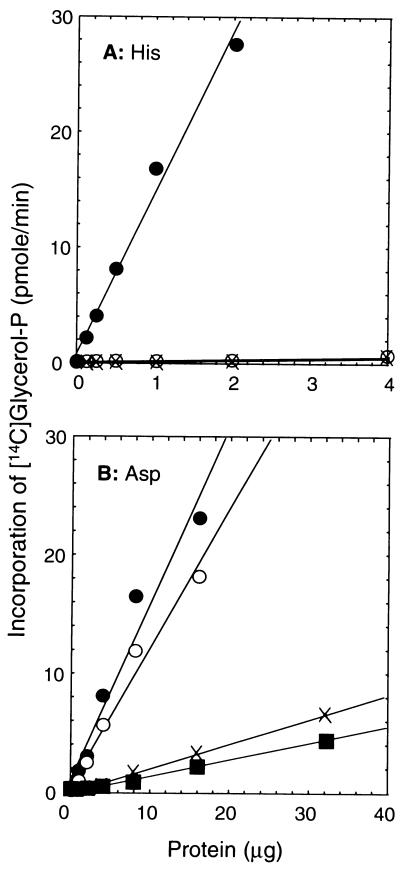
To investigate the role of the invariant His in the HX4D pattern within the conserved region of the acyltransferase gene family, we compared the activities of wild-type PlsB protein and the PlsB[H306A] mutant expressed in strain SJ22 (plsB). We also constructed a control strain harboring the empty vector, pACYC177. Membrane fractions were prepared from these strains and assayed for G3P acyltransferase activity. The wild-type gene directed the synthesis of high levels of G3P acyltransferase activity (15 nmol/min/mg of protein) with palmitoyl-ACP as the substrate (Fig. 2A), whereas membranes isolated from strain SJ22 (plsB) with the empty vector had barely detectable G3P acyltransferase activity (0.2 nmol/min/mg of protein). G3P acyltransferase specific activities in membranes isolated from strain SJ22 harboring pRJ50 (PlsB[H306A]) were indistinguishable from the background activity (0.2 nmol/min/mg of protein). The PlsB[H306A] mutant was also completely inactive when palmitoyl-CoA was used as the acyl donor, and we were unable to demonstrate any activity of the mutant at protein concentrations up to 64 μg/assay (not shown). The PlsB[H306A] protein was expressed and assembled into the membrane (see below). These data indicated that His306 was an essential residue for the G3P acyltransferase activity of PlsB.
FIG. 2.
Effects of substitutions at His306 and Asp311 on the G3P acyltransferase activity of PlsB. Membranes were prepared from strain SJ22 (plsB) harboring either a control plasmid (pACYC177) or a plasmid expressing either a wild-type or mutant plsB gene and assayed for G3P acyltransferase activity as described in Materials and Methods. (A) Effect of the PlsB[H306A] mutation, examined by assaying membranes from strain SJ22 (plsB) harboring pRJ22 (wild-type PlsB) (•), pRJ50 (PlsB[H306A]) (○) and pACYC177 (×); (B) G3P acyltransferase activity, measured in membranes prepared from strain 8 (chromosomal wild type for plsB) (•) or from strain SJ22 (plsB) harboring either pRJ52 (PlsB[D311E]) (○), pRJ51 (PlsB[D311A]) (▪), or pACYC177 (×).
The role of Asp in the conserved HX4D cluster was investigated by constructing PlsB[D311E] and PlsB[D311A] mutants and analyzing their activities. The Ala substitution was chosen to minimize unfavorable steric effects and to eliminate the acid side chain without introducing new charge interactions or hydrogen bonds. The Glu substitution was chosen to retain an acidic residue at this position, although the extra carbon atom in Glu places the carboxylate group in a slightly different orientation. Plasmids pRJ52 (PlsB[D311E]) and pRJ51 (PlsB[D311A]) were transformed into strain SJ22 (plsB), and G3P acyltransferase activity was measured in membrane preparations. Activity above the baseline established in membranes prepared from strain SJ22 (plsB) with the empty vector was not observed for the PlsB[D311A] mutant (0.14 nmol/min/mg of protein), whereas the PlsB[D311E] mutant retained significant activity (1.2 nmol/min/mg of protein) (Fig. 2B). However, the activity of the PlsB[D311E] mutant (1.2 nmol/min/mg of protein) was 10-fold less in this assay than that of overexpressed wild-type PlsB (15 nmol/min/mg of protein) (Fig. 2A). Thus, the extra methylene in the Glu residue compared to Asp significantly reduced, but did not completely eliminate, G3P acyltransferase activity.
E. coli SJ22 (plsB) requires supplementation with G3P for growth due to a defective PlsB enzyme that has a high Km for G3P (2, 8). The G3P growth phenotype was complemented by transformation with plasmid pRJ22 expressing the wild-type plsB gene. Each plasmid containing a mutant plsB gene was tested for the ability to complement this growth phenotype. Strain SJ22 (plsB) transformed with plasmids pRJ50 (PlsB[H306A]) and pRJ51 (PlsB[D311A]) failed to grow on medium lacking G3P. In contrast, strain SJ22 (plsB) transformed with pRJ52 (PlsB[D311E]) grew in the absence of G3P. Growth experiments in liquid culture showed that the growth rate of strain SJ22 (plsB) harboring the pRJ52 plasmid (PlsB[D311E]) in the absence of G3P supplementation was the same as the growth rate of strain SJ22 in the presence of a G3P supplement. The doubling times on glucose minimal medium were 57 and 58 min, respectively. These data show that the PlsB[D311E] mutant retained sufficient acyltransferase activity to support cell growth, whereas the PlsB[H306A] and PlsB[D311A] proteins did not. These in vivo data were consistent with the in vitro results.
Since the PlsB[D311E] protein complemented the growth phenotype of strain SJ22 (plsB), we also prepared membranes from E. coli 8, which is the wild-type counterpart of strain SJ22 (plsB). The specific activity of the G3P acyltransferase in membranes prepared from strain 8 was 1.6 nmol/min/mg, only slightly greater than observed for the PlsB[D311E] mutant (Fig. 2B). Therefore, despite the 10-fold reduced activity of the PlsB[D311E] protein, overexpression resulted in almost wild-type levels of total activity, thus explaining the ability of strain SJ22 (plsB) harboring plasmid pRJ52 to grow in the absence of G3P even though the PlsB[D311E] protein was catalytically impaired.
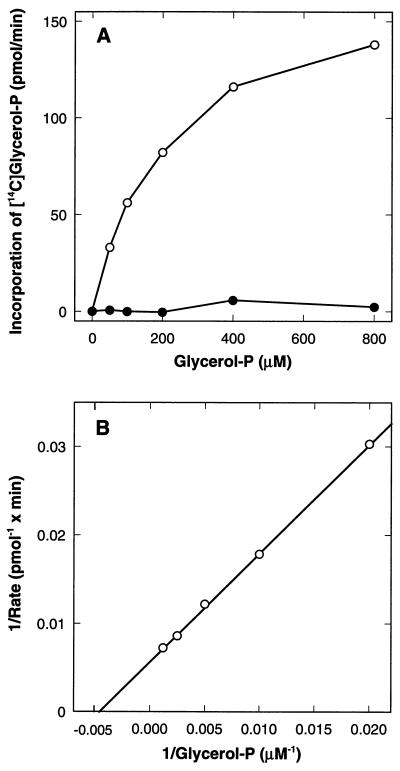
Membranes containing the PlsB[H306A] and PlsB[D311E] mutant proteins were assayed at different concentrations of G3P to determine if the mutations altered the Km for G3P (Fig. 3). To correct the data for the residual acyltransferase activity in membranes from the plsB26 mutant, the activity detected in membranes from strain SJ22 harboring the empty vector was subtracted from each point in Fig. 3A. There was no activity above background detected in membranes containing the PlsB[H306A] mutant at any G3P concentration tested (Fig. 3A). These data are consistent with the specific activity data (Fig. 2) and indicate that the PlsB[H306A] mutant was devoid of G3P acyltransferase activity. The PlsB[D311E] mutant retained acyltransferase activity, which increased as the concentration of G3P in the assay was raised (Fig. 3A). A double-reciprocal plot of these data (Fig. 3B) showed a typical Michaelis-Menten kinetic pattern, and a Km of 200 μM for G3P was calculated from these data. The G3P Km for the wild-type protein is about 100 μM (2); therefore, the compromised activity of the PlsB[D31E] mutant cannot be attributed to a large change in the affinity of the enzyme for G3P. We also examined the activity of each of the mutant enzymes as a function of increased acyl donor concentration (not shown). Again, no activity was detected with the PlsB[H306A] mutant at any acyl donor concentration tested, whereas the activity of the PlsB[D311E] as a function of the palmitoyl-CoA acyl donor concentration did not indicate a significant difference in the affinity of the enzyme for this substrate. The Km values ranged from 15 to 33 μM, comparable to the previously reported Km for palmitoyl-CoA of 50 μM (19). The PlsB[D311A] mutant was not tested in any of these experiments because it was not assembled into the membrane (see below).
FIG. 3.
G3P Kms for the PlsB[H306A] and PlsB[D311A] mutants. Membranes were prepared from strain SJ22 (plsB) harboring either a control plasmid (pACYC177) or a plasmid expressing either the PlsB[H306A] (pRJ50) or PlsB[D311E] (pRJ52) mutant and assayed for G3P acyltransferase activity as a function of G3P concentrations as described in Materials and Methods. (A) Direct plot of activity versus G3P concentration for PlsB[H306A] (•) and PlsB[D311E] (○) mutants; (B) double-reciprocal analysis of the data for the PlsB[D311E] mutant. No activity was detected in the PlsB[H306A] mutant, and the apparent Km for G3P in the PlsB[D311E] mutant was 200 μM.
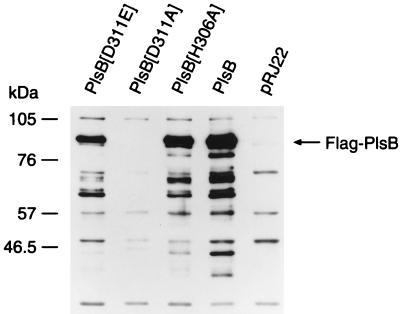
Although the mutated plsB constructs were verified by DNA sequencing and in all cases expressed proteins having the correct molecular weight in an in vitro transcription-translation assay, we verified that the PlsB mutants were expressed and properly inserted into the membrane in vivo. This was accomplished by constructing a family of plsB mutants with the flag epitope tag attached to the carboxy terminus (see Materials and Methods). These tagged PlsB proteins were detected by immunoblotting with the anti-flag antibody (Fig. 4). The wild-type PlsB-flag protein was expressed and properly inserted into the membrane. Membrane preparations from strain SJ22 transformed with the epitope-tagged construct had the same G3P acyltransferase specific activity as the construct that expressed the wild-type gene. The epitope-tagged PlsB[H306A] and PlsB[D311E] mutant proteins were also expressed and properly assembled into the membrane (Fig. 4). Biochemical analysis of the membranes from the epitope-tagged constructs confirmed the results presented in Fig. 2. The PlsB[H306A]-flag mutant lacked acyltransferase activity, whereas the PlsB[D311E]-flag enzyme had a significantly lower specific activity than the wild-type protein. In contrast, the PlsB[D311A]-flag mutant was not inserted into the membrane, and only a trace of this protein was detected in long exposures. Therefore, the lack of acyltransferase activity detected in membranes derived from cells transformed with the PlsB[D311A] construct was attributed to the absence of protein in the sample. These data show that the PlsB[H306A] and PlsB[D311E] were expressed and properly assembled into the membrane, supporting the conclusion that the deficiencies in catalytic activity noted in the biochemical assay reflected the impact of the mutations on the catalytic competence of the enzyme. The inability of the PlsB[D311A] mutant to properly assemble into the membrane precluded further analysis of this mutant.
FIG. 4.
Expression levels of wild-type and mutant epitope-tagged PlsB proteins. Flag-tagged PlsB wild-type and mutant expression vectors were transformed into strain SJ22 (plsB). Membranes were prepared, 10 μg of membrane protein was loaded into each lane, and the presence of the flag-tagged PlsB proteins was determined by immunoblotting as described in Materials and Methods. Expression of the PlsB protein was detected in membranes from cells transformed with plasmids expressing PlsB[D311E], PlsB[H306A], and PlsB. Protein expression was not detected with PlsB[D311A] and control membranes (pRJ22).
Evaluation of the Aas[H36A] mutant.
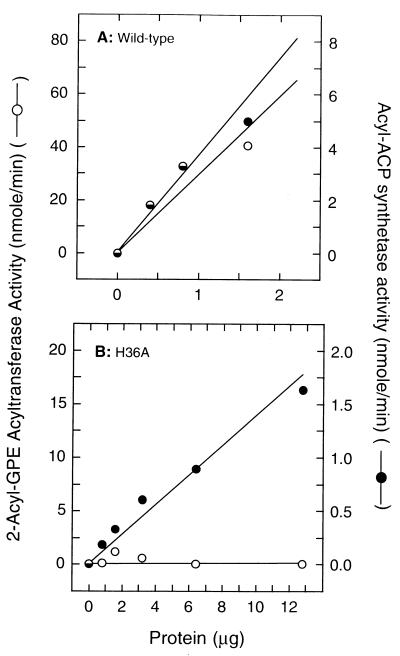
The aas gene encodes a bifunctional protein that exhibits both 2-acyl-GPE acyltransferase and acyl-ACP synthetase activities in vitro (14). Analysis of the H36A mutant of Aas thus serves as an important test of the working hypothesis. If the conserved His plays a catalytic as opposed to a structural role, we predict that the Aas[H36A] mutant should lack acyltransferase activity while retaining acyl-ACP synthetase activity. Strain LCH26 (aas) contains an inactivating mutation in the aas gene and does not express detectable levels of either 2-acyl-GPE acyltransferase or acyl-ACP synthetase activities (12). Membranes were prepared from strain LCH26 (aas) harboring either pRJ40 (wild-type Aas), pRJ41 (Aas[H36A]), or pBluescript II KS (+) and assayed for acyl-ACP synthetase activity. Membranes prepared from strain LCH26 (aas) expressing the wild-type gene had a specific activity of 2.2 μmol/min/mg of protein (Fig. 5A), whereas the synthetase activity in membranes prepared from the strain LCH26 (aas) containing the empty vector was not detectable (not shown). Membranes prepared from strain LCH26 (aas) expressing the Aas[H36A] mutant protein had an acyl-ACP synthetase specific activity of 0.13 μmol/min/mg of protein (Fig. 5B), or 6% of the activity of the wild-type protein. Thus, the Aas[H36A] mutant was expressed, assembled into the membrane, and retained acyl-ACP synthetase activity. We cannot be certain that the difference in specific activity between the wild-type protein and mutant protein are not due to differences in the level of expression. In the construction of the Aas[H36A] mutant a short region of sequence upstream of the putative aas promoter was deleted, which may have lowered the expression of the aas gene.
FIG. 5.
The Aas[H36A] mutant lacks acyltransferase activity but retains acyl-ACP synthetase activity. Wild-type Aas (A) and Aas[H36A] (B), present in membranes prepared from LCH26 (aas) containing the appropriate plasmid, were assayed for both 2-acyl-GPE acyltransferase and acyl-ACP synthetase activities as described in Materials and Methods.
The effect of the Aas[H36A] mutation on the 2-acyl-GPE acyltransferase activity of Aas was tested, using the same membrane preparations. Membranes prepared from LCH26 (aas) containing the empty vector had no detectable 2-acyl-GPE acyltransferase activity (<0.02 μmol/min/mg). In contrast, acyltransferase specific activity was 25 μmol/min/mg of protein in membranes prepared from strain LCH26 harboring pRJ40 (wild-type Aas) (Fig. 5A). The activity of the Aas[H36A] mutant (Fig. 5B) was not distinguishable from that of the empty vector control. The enzyme assay was able to detect low amounts of phosphatidylethanolamine formation. A 1% conversion of substrate yielded 2,000 cpm of [3H]phosphatidylethanolamine over the background counts. The important point was that the ratio of acyl-ACP synthetase activity to 2-acyl-GPE activity fell to zero in the Aas[H36A] mutant due to the complete lack of detectable acyltransferase activity. Thus, we concluded that the Aas[H36A] mutant lacked 2-acyl-GPE acyltransferase activity, whereas it retained acyl-ACP synthetase activity.
Conclusions.
The invariant His residue in the HX4D consensus sequence is clearly essential in the membrane- associated glycerolipid acyltransferases. Substitution of His with Ala in PlsB resulted in an inactive acyltransferase, whereas the substitution of His for Ala in Aas inactivated the acyltransferase half-reaction of the enzyme while the synthetase component was preserved. The measurements of the Km for G3P indicate that the HX4D consensus sequence is not critical for substrate binding, but it remains possible that the sequence binds a metal. However, there is no experimental evidence for a metal ion requirement for the enzyme. The utilization of His as a general base is likely to be a common feature of acyltransferases since this residue is well suited to facilitate abstraction of a proton from the hydroxyl group of the acyl acceptor. The most extensively studied acyltransferase, chloramphenicol acetyltransferase, is a soluble homotrimer, and its three-dimensional structure and catalytic mechanism are known (17). In this acetyltransferase, His195 acts as a general base to abstract a proton from the hydroxyl group of chloramphenicol and is stabilized in the correct tautomeric form by van der Waals interactions with Tyr25. By analogy, we propose that the conserved His in the glycerolipid acyltransferases also may act as a general base to abstract a proton from the hydroxyl group of the acyl acceptor to facilitate nucleophilic attack on the thioester of the acyl donor. The mechanisms of serine proteases, thioesterases, and lipases (4, 6, 7, 26, 28, 30) employ an Asp(Glu)-His-Ser catalytic triad to affect catalysis. The crystal structures of these enzymes reveal that the Asp-His charge relay system increases the nucleophilicity of the Ser, which attacks the substrate to form an acyl-Ser intermediate that is subsequently hydrolyzed. The Asp in glycerolipid acyltransferases may play a similar role in catalysis, with the hydroxyl group of the G3P acyl acceptor being analogous to the hydroxyl group of the active site Ser in the catalytic triad mechanism and the acyl-G3P product being analogous to the acyl-Ser intermediate. On the other hand, the inability of PlsB[D311A] to properly assemble into membranes in vivo suggests an important structural role for the invariant Asp. The requirement for Asp is quite specific since the PlsB[D311E] substitution has significantly reduced catalytic activity. Thus, we can conclude that the Asp that is completely conserved in all glycerolipid acyltransferases examined plays a critical role in enzyme function, but our data do not distinguish whether this role relates to catalysis, structure, or both.
ACKNOWLEDGMENTS
We thank R. Brent Calder and Jina Wang for excellent technical assistance, and we thank Irina Baburina and Suzanne Jackowski for helpful suggestions and discussions.
This work was supported by National Institutes of Health grant GM 34496, Cancer Center (CORE) support grant CA 21765, and the American and Lebanese Syrian Associated Charities.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bell R M. Mutants of Escherichia coli defective in membrane phospholipid synthesis: macromolecular synthesis in an sn-glycerol 3-phosphate acyltransferase Km mutant. J Bacteriol. 1974;117:1065–1076. doi: 10.1128/jb.117.3.1065-1076.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley S, Turkenburg J P, Christiansen L, Hughe-Jensen B, Norskov L, Thim L, Mensge U. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature (London) 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 5.Brown A P, Coleman J, Tommey A M, Watson M D, Slabas A R. Isolation and characterisation of a maize cDNA that complements a 1-acyl sn-glycerol-3-phosphate acyltransferase mutant of Escherichia coli and encodes a protein which has similarities to other acyltransferases. Plant Mol Biol. 1994;26:211–223. doi: 10.1007/BF00039533. [DOI] [PubMed] [Google Scholar]
- 6.Carter P, Wells J A. Dissecting the catalytic triad of a serine protease. Nature (London) 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 7.Craik C S, Roczniak S, Largman C, Rutter W J. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987;237:909–913. doi: 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- 8.Cronan J E, Jr, Bell R M. Mutants of Escherichia coli defective in membrane phospholipid synthesis: mapping of sn-glycerol 3-phosphate acyltransferase Km mutants. J Bacteriol. 1974;120:227–233. doi: 10.1128/jb.120.1.227-233.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtis R, Gross C A, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 10.Hanke C, Wolter F P, Coleman J, Peterek G, Frentzen M. A plant acyltransferase involved in triacylglycerol biosynthesis complements an Escherichia coli sn-1-acylglycerol-3-phosphate acyltransferase mutant. Eur J Biochem. 1995;232:806–810. [PubMed] [Google Scholar]
- 11.Homma H, Nojima S. Synthesis of various phospholipids from 2-acyl lysophospholipids by Escherichia coli extract. J Biochem (Tokyo) 1982;91:1103–1110. doi: 10.1093/oxfordjournals.jbchem.a133792. [DOI] [PubMed] [Google Scholar]
- 12.Hsu L, Jackowski S, Rock C O. Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. J Biol Chem. 1991;266:13783–13788. [PubMed] [Google Scholar]
- 13.Ishizaki O, Nishida I, Agata K, Eguchi G, Murata N. Cloning and nucleotide sequence of cDNA for the plastid glycerol-3-phosphate acyltransferase from squash. FEBS Lett. 1988;238:424–430. doi: 10.1016/0014-5793(88)80525-8. [DOI] [PubMed] [Google Scholar]
- 14.Jackowski S, Jackson P D, Rock C O. Sequence and function of the aas gene in Escherichia coli. J Biol Chem. 1994;269:2921–2928. [PubMed] [Google Scholar]
- 15.Knutzon D S, Lardizabal K D, Nelsen J S, Bleibaum J L, Davies H M, Metz J G. Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-glycerol-3-phosphate acyltransferase that accepts medium-chain-length substrates. Plant Physiol. 1995;109:999–1006. doi: 10.1104/pp.109.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Murray I A, Shaw W V. O-Acetyltransferases for chloramphenicol and other natural products. Antimicrob Agents Chemother. 1997;41:1–6. doi: 10.1128/aac.41.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagiec M M, Wells G B, Lester R L, Dickson R C. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- 19.Ray T K, Cronan J E., Jr Acylation of sn-glycerol 3-phosphate in Escherichia coli: study of the reaction with native palmitoyl-acyl carrier protein. J Biol Chem. 1975;250:8422–8427. [PubMed] [Google Scholar]
- 20.Rock C O, Cronan J E., Jr Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 21.Rock C O, Garwin J L. Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J Biol Chem. 1979;254:7123–7128. [PubMed] [Google Scholar]
- 22.Rock C O, Goelz S E, Cronan J E., Jr ATP stimulation of the sn-glycerol-3-phosphate acyltransferase of Escherichia coli. Arch Biochem Biophys. 1981;211:113–118. doi: 10.1016/0003-9861(81)90436-7. [DOI] [PubMed] [Google Scholar]
- 23.Rock C O, Jackowski S. Regulation of phospholipid synthesis in Escherichia coli. Composition of the acyl-acyl carrier protein pool in vivo. J Biol Chem. 1982;257:10759–10765. [PubMed] [Google Scholar]
- 24.Scheideler M A, Bell R M. Phospholipid dependence of homogeneous, reconstituted sn-glycerol-3-phosphate acyltransferase of Escherichia coli. J Biol Chem. 1989;264:12455–12461. [PubMed] [Google Scholar]
- 25.Scheideler M A, Bell R M. Characterization of active and latent forms of the membrane-associated sn-glycerol-3-phosphate acyltransferase of Escherichia coli. J Biol Chem. 1991;266:14321–14327. [PubMed] [Google Scholar]
- 26.Schrag J D, Li Y, Wu S, Cygler M. Ser-His-Glu triad forms the catalytic site of the lipase from Geotrichum candidum. Nature (London) 1991;351:761–764. doi: 10.1038/351761a0. [DOI] [PubMed] [Google Scholar]
- 27.Shaw-Fang Y, Lee S, Hahm Y T, Sul H S. Expression and identification of p90 as the murine mitochondrial glycerol-3-phosphate acyltransferase. Biochemistry. 1993;32:9486–9491. doi: 10.1021/bi00087a029. [DOI] [PubMed] [Google Scholar]
- 28.Upton C, Buckley J T. A new family of lipolytic enzymes? Trends Biochem Sci. 1995;20:178–179. doi: 10.1016/s0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 29.Weber S, Wolter F-P, Buck F, Frentzen M, Heinz E. Purification and cDNA sequencing of an oleate selective acyl-ACP:sn-glycerol-3-phosphate acyltransferase from pea chloroplasts. Plant Mol Biol. 1991;17:1067–1076. doi: 10.1007/BF00037145. [DOI] [PubMed] [Google Scholar]
- 30.Winkler F K, D’Arcy A, Hurst M A. Structure of human pancreatic lipase. Nature (London) 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 31.Yet S-F, Moon Y K, Sul H S. Purification and reconstitution of murine mitochondrial glycerol-3-phosphate acyltransferase. Functional expression in baculovirus-infected insect cells. Biochemistry. 1995;34:7303–7310. doi: 10.1021/bi00022a003. [DOI] [PubMed] [Google Scholar]