Abstract
The type II secretion system (main terminal branch of the general secretion pathway) is used by diverse gram-negative bacteria to secrete extracellular proteins. Proteins secreted by this pathway are synthesized with an N-terminal signal peptide which is removed upon translocation across the inner membrane, but the signals which target the mature proteins for secretion across the outer membrane are unknown. The plant pathogens Erwinia chrysanthemi and Erwinia carotovora secrete several isozymes of pectate lyase (Pel) by the out-encoded type II pathway. However, these two bacteria cannot secrete Pels encoded by heterologously expressed pel genes from the other species, suggesting the existence of species-specific secretion signals within these proteins. The functional cluster of E. chrysanthemi out genes carried on cosmid pCPP2006 enables Escherichia coli to secrete E. chrysanthemi, but not E. carotovora, Pels. We exploited the high sequence similarity between E. chrysanthemi PelC and E. carotovora Pel1 to construct 15 hybrid proteins in which different regions of PelC were replaced with homologous sequences from Pel1. The differential secretion of these hybrid proteins by E. coli(pCPP2006) revealed M118 to D175 and V215 to C329 as regions required for species-specific secretion of PelC. We propose that the primary targeting signal is contained within the external loops formed by G274 to C329 but is dependent on residues in M118 to D170 and V215 to G274 for proper positioning.
Secretion of extracellular proteins is essential for virulence in many bacterial pathogens. Of the three major secretion pathways present in gram-negative bacteria, the type II pathway, or main terminal branch of the general secretion pathway, is used for the largest and most diverse group of proteins (41). The first type II pathway genes to be identified were those in the pul cluster, required for secretion of the starch-degrading enzyme pullulanase by Klebsiella oxytoca. Pugsley and coworkers demonstrated that pulS and a 13-gene operon, consisting of pulC to pulO, are necessary and sufficient for secretion of pullulanase across the outer membrane (40). Subsequent research has demonstrated that components of this secretion pathway are conserved among diverse gram-negative bacteria (39), including many pathogens of plants and animals.
Erwinia chrysanthemi and Erwinia carotovora are agents causing soft rot diseases in a variety of plant hosts. Characteristic symptoms of tissue maceration and cell death are caused by extracellular pectate lyase (Pel) and polygalacturonase, which are secreted along with other cell wall-degrading enzymes, including cellulases and pectin methyl esterase (4). Secretion of these enzymes by the type II pathway is essential for virulence, as out mutants, which are blocked in the pathway, do not elicit symptoms on host plants (26). A cosmid clone, pCPP2006, which complemented E. chrysanthemi out mutants was identified from a library of strain EC16 DNA, partially sequenced, and found to contain 12 genes, outC to outM and outO, highly similar to the pul cluster (14, 28). out genes have also been cloned from E. carotovora, but only the cosmid from E. chrysanthemi enables Escherichia coli to secrete cloned Erwinia enzymes (33, 45). Genes homologous to the pul and out clusters have been cloned from other pathogens, including xps of Xanthomonas campestris (10, 18) and eep of Pseudomonas solanacearum (21), involved in secretion of plant cell wall-degrading enzymes, xcp of Pseudomonas aeruginosa for secretion of elastase, exotoxin A, lipase, and alkaline phosphatase (2, 3, 11), tcp of Vibrio cholerae for secretion of cholera toxin (34), and exe in Aeromonas hydrophila for secretion of aerolysin (17, 20).
Exoproteins employing the type II pathway appear to cross the bacterial envelope in two stages. Synthesized with an N-terminal signal peptide, they are directed first to the Sec machinery for export across the cytoplasmic membrane (15, 42). Translocation of the mature protein across the outer membrane follows rapidly, but the mechanism of transport and the specific targeting features which distinguish exoproteins from those proteins remaining in the periplasm are poorly understood. Comparisons of the sequences of the diverse proteins which use the type II pathway have revealed no obvious regions of similarity (41). Deletion analyses, linker insertions, point mutations, and construction of hybrids with β-lactamase or alkaline phosphatase have all been used in the search for discrete targeting regions in the primary sequence (5, 9, 12, 15, 19, 24, 25, 35, 53). Studies on K. oxytoca pullulanase and P. aeruginosa exotoxin A have led to identification of relatively limited domains sufficient for targeting β-lactamase fusion proteins across the outer membrane (30, 31, 48). However, the observations that (i) two noncontiguous regions are required for targeting of pullulanase (48) and (ii) additional regions of exotoxin A either enhance or independently promote secretion (31) support the hypothesis that targeting features may be dependent on higher-order structure for their formation and/or proper positioning. For other exoproteins studied, including two from E. chrysanthemi, the vast majority of deletions and fusion constructs resulted in loss of all secretion capability, possibly because the structural context of the targeting signal had been disrupted (15, 43).
The Erwinia out-encoded type II pathway provides an attractive system for identification of targeting signals on exoproteins. Multiple enzymes employ the Out type II pathway, providing the opportunity for development of a general targeting model using proteins with various structures and sequences. Furthermore, three-dimensional structures have been determined for two E. chrysanthemi Pels, enabling structural analyses of any targeting regions that are identified (27, 54). Finally, species-specific secretion of E. chrysanthemi and E. carotovora Pels provides a novel avenue for identification of targeting regions in these proteins. E. chrysanthemi and E. carotovora secrete similar arsenals of plant cell wall-degrading enzymes, but the exoproteins of one species cannot be secreted by the other (14, 44). Furthermore, the cluster of E. chrysanthemi out genes carried on cosmid pCPP2006 retains this specificity for E. chrysanthemi Pels when functioning in E. coli (14).
Here, we have constructed hybrid proteins in which one region of a secreted Pel is substituted with the same region from a nonsecreted homolog, to systematically test each region for involvement in species-specific targeting while minimizing changes to their overall structural context. By mapping the regions required for species-specific secretion on the known structure of PelC, the locations of these regions within the protein structure were determined. Using this approach, we have demonstrated that elements within the regions V215 to C329 near the C terminus and M118 to D175 in the central part of the PelC protein are required for targeting PelC to the Out machinery. It is proposed that the primary signal determining species-specific secretion is contained within external loops near the C terminus whereas the required regions in the β-helix core are necessary for their proper positioning.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli DH5α (13) was used as the standard strain for propagating recombinant plasmids and assaying for protein secretion. DH5α was grown in Terrific broth (47) at 37°C for isolation of plasmids and in King’s B medium (22) at 30°C for protein secretion assays. The following concentrations of antibiotics were used where appropriate: ampicillin, 100 μg/ml; spectinomycin, 50 μg/ml; and kanamycin, 50 μg/ml.
Recombinant DNA techniques.
General procedures for isolation, analysis, and manipulation of DNA fragments were as described by Sambrook et al. (47). Subcloning was routinely performed by digesting vector and insert DNA with appropriate restriction enzymes, separating fragments by electrophoresis through 0.7% agarose gels, purifying the DNA with the Prep-a-Gene kit (Bio-Rad Laboratories, Richmond, Calif.), and ligating the vector and insert according to standard procedures. Before ligation, incompatible restriction sites were cloned by blunting the incompatible ends with T4 DNA polymerase (New England Biolabs, Beverly, Mass.) as described in reference 1.
Plasmids and subclones used in construction of PelC-Pel1 chimeras.
Plasmids carrying E. carotovora pel1 and E. chrysanthemi pelC, used for construction of the PelC-Pel1 chimeras, were made as follows. The NspI site in pBluescript II SK(−) and pBluescript KS(−) (Stratagene, La Jolla, Calif.) was deleted by digesting each with NspI, removing the 3′ overhangs with T4 DNA polymerase, and religating. The KpnI site was subsequently deleted from each plasmid by the same procedure. The EcoRV site was deleted from pBluescript KS(−) NspI− KpnI− by digesting with EcoRV and HindIII, filling in the HindIII 5′ overhang, and religating. The EcoRI fragment containing the pel1 open reading frame from pAKC617 (7) was cloned into the EcoRI site of both pBluescript SK NspI− KpnI− and pBluescript KS NspI− KpnI− EcoRV− to create pAKC692 and pAKC693, respectively. For both pAKC692 and pAKC693, pel1 is transcribed in the orientation opposite the lac promoter. pCPP2192, used for construction of pCPP2180, pCPP2183, pCPP2185, and pCPP2186, was made by cloning the XbaI-SphI fragment from pPEL405 (52) into the XbaI-PstI site of pBluescript SK. pCPP2193 was created by cloning the NciI-XhoI fragment from pPEL403 (52) into the StyI-XhoI site of pAKC617.
Site-directed mutagenesis of residues in the C-terminal branch.
In preparation for site-directed mutagenesis, a 1.4-kb XbaI-HindIII fragment was cloned into the same sites in pRSET5A (49). Mutagenesis of divergent residues in the pelC region encoding the C-terminal branch to the corresponding sequences in pel1 was performed by PCR using the overlapping extension method (32), with modifications described by Kita et al. (23). Following selection of desired pelC mutants, the genes were sequenced in entirety to confirm fidelity, using a Sequenase version 2.0 kit from United States Biochemical.
Assays for Pel activity and secretion.
Pel secretion was assayed by fractionating 1 ml of culture at late logarithmic phase into cell and supernatant fractions by centrifugation. The cell pellets were washed once in cold fresh medium and sonicated in 1 ml of cold medium for 4 min with a model W-225R sonicator (Heat Systems Ultrasonics Inc., Plainview, N.Y.) at a duty cycle of 40% and an output of 4. Culture supernatants and sonicated cell pellets were assayed by an A235 assay and expressed in micromoles of unsaturated product liberated per minute per milligram of protein (8). Protein concentrations were determined by the Bradford assay (6). To account for nonspecific leakage, β-lactamase activity was determined for both cell and supernatant fractions, using the chromogenic cephalosporin compound nitrocefin (Glaxo, Greenford, Middlesex, England) and monitored at 540 nm. The percentage of Pel activity specifically secreted was determined by subtracting the percentage of total β-lactamase activity in the supernatant from the percentage of total Pel activity in the supernatant. In control experiments, with cells containing pel genes but no out genes, the percentage of total β-lactamase in the supernatant was 10 to 20% higher than the total percentage of Pel activity in the supernatant, indicating that use of β-lactamase as a periplasmic marker gives a conservative estimate of secretion efficiency.
Isoelectric focusing, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and activity staining of the PelC-Pel1 chimeric proteins.
Three-milliliter cultures of E. coli DH5α cells carrying pCPP2193, pAKC617, pCPP2194, pCPP2195, and each of the 14 chimeric constructs were centrifuged, washed with 5 ml of H2O, centrifuged again, and sonicated for 5 min to break open the cells. Cell debris was removed by centrifugation, samples were concentrated as needed in Centricon microconcentrators (Amicon, Beverly, Mass.) and then washed with two starting volumes of H2O to remove salts. Isoelectric focusing was performed on a PhastSystem (Pharmacia, Uppsala, Sweden), using gels rehydrated with 60% 3-10 and 40% 9-11 ampholytes (Sigma). pCPP2194 and pCPP2195 were run on premade sodium dodecyl sulfate-polyacrylamide gels, using the PhastSystem. Activity staining was performed as previously described (46), with the addition of 1% Triton X-100 to the wash buffer.
Computer analyses of protein sequences and structures.
The PelC and Pel1 sequences were compared by using the FASTA program (36). The PelC structure was analyzed by using RasMol v2.5 (Roger Sayle, Biomolecular Structures Group, Glaxo Research & Development). Atomic coordinates for PelC and PelE were kindly provided by Frances Jurnak. Molecular modeling of Pel1 using the Pel1 sequence and PelC structure predicts a very high degree of similarity in overall folding (20a).
RESULTS
E. carotovora Pel1 is highly similar to E. chrysanthemi PelC but is not secreted by the E. chrysanthemi type II pathway.
The amino acid sequences of the E. chrysanthemi PelC and E. carotovora Pel1 mature proteins are 71% identical (Fig. 1). The sequence lengths differ by 1 residue, with mature PelC being 353 and Pel1 352 residues in length. When PelC is expressed in Out+ E. coli DH5α(pCPP2006), 40 to 60% of the protein is specifically secreted to the supernatant, with efficiency increasing with reduced levels of expression. When Pel1 is expressed in DH5α(pCPP2006), no activity is observed in the supernatant.
FIG. 1.
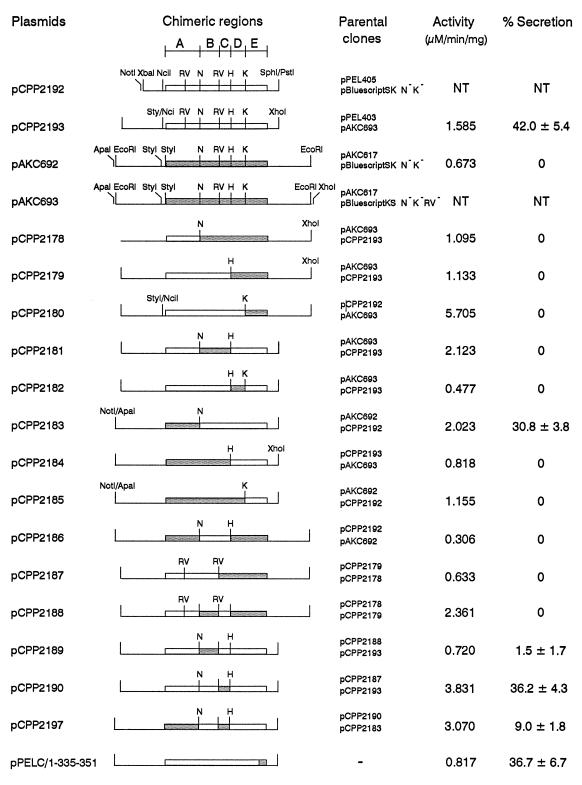
Alignment of amino acid sequences for E. chrysanthemi PelC and E. carotovora Pel1. The N-terminal signal peptide is shaded, and the residues composing the C-terminal branch are underlined. Locations of four conserved restriction sites relative to the amino acid sequences are shaded and labeled. These sites have been used to divide the two sequences into five separate regions, labeled A, B, C, D, and E above the restriction sites.
Fourteen chimeric proteins constructed from E. chrysanthemi PelC and E. carotovora Pel1 retain Pel activity and have intermediate isoelectric points.
Alignment of the coding regions for PelC and Pel1 revealed four conserved restriction sites in the two sequences. Their locations relative to the PelC sequence are as follows: NspI centered over the codon for M118, EcoRV centered over D170, HpaI centered over V215, and KpnI centered over G274 (Fig. 1). Using these sites, the sequences have been divided into five regions, designated A, B, C, D, and E (Fig. 1 and 2). Construction of hybrid genes from pelC and pel1 required a set of parent clones for which the four conserved restriction sites were absent from the vector, the inserts were flanked by other restriction sites useful for subcloning, and the genes were expressed at appropriate levels. Four such clones were made as follows. (i) The NspI and KpnI sites were deleted from pBluescript SK, and the NspI, KpnI, and EcoRV sites were deleted from pBluescript KS. (ii) The EcoRI fragment from pAKC617 carrying the pel1 gene driven by the tet promoter from pBR322 was cloned into the EcoRI site of pBluescript SK NspI− KpnI− and pBluescript KS NspI− KpnI− EcoRV− in orientations opposite the lac promoter to create pAKC692 and pAKC693, respectively. (iii) The NciI-XhoI fragment carrying pelC from pPEL403 was cloned into the StyI-XhoI sites of pAKC693 to create pCPP2193 such that pelC is driven by the tet promoter in pBluescript KS NspI− KpnI− EcoRV−. (iv) pCPP2192 was created by cloning the XbaI-SphI fragment from pPEL405 into the XbaI-PstI sites of pBluescript SK NspI− KpnI− EcoRV−. Using the four conserved sites described above and other sites flanking the coding regions, we constructed 14 hybrid genes in which different parts of pelC were replaced with homologous sequences from pel1 (Fig. 2). For example, pCPP2178, in which regions B to E of pelC are replaced with pel1 sequences, was constructed by cloning the NspI-XhoI fragment from pAKC693 into the NspI-XhoI sites of pCPP2193. All exchanges were verified by using diagnostic restriction digests.
FIG. 2.
Diagram of 2 pelC clones, 2 pel1 clones, and 15 pel1-pelC hybrid constructs with parental clones, with the activity and percent secretion by Out+ E. coli shown for each. Regions derived from pelC are indicated as open bars, and regions derived from pel1 are shaded. Regions are designated A, B, C, D, and E according to the key at the top. Abbreviations for the four conserved sites used in construction of each hybrid: RV, EcoRV; N, NspI; H, HpaI; K, KpnI (all other sites are indicated by full names). Parental clones for each construct are shown with the source of the insert listed first and the vector-containing part of the construct listed below. For example, the hybrid clone pCPP2178 was constructed by inserting the NspI-XhoI fragment from pAKC693 into the NspI-XhoI sites of pCPP2193. For pPELC/1-335-351, the region corresponding to the pel1 sequence was introduced by site-directed mutagenesis. Activity is shown as micromoles of unsaturated product liberated per minute per milligram of protein. Percent total extracellular Pel activity was obtained by determining the percentage of total Pel activity in the supernatant minus the percentage of total β-lactamase activity in the supernatant. Values indicated represent the averages of four separate trials for each clone. NT, not tested.
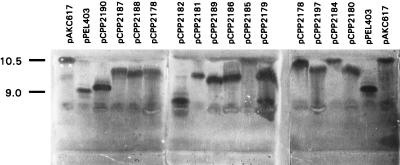
E. coli DH5α cells carrying each of the 14 hybrids were grown to stationary phase and lysed, and total protein was concentrated 2- to 10-fold. Samples were run on an isoelectric focusing gel and activity stained. As seen in Fig. 3, all of the chimeric proteins from Fig. 2 retained Pel activity. The band for pCPP2185, although not as clear as the others, can be distinguished at the top of the gel, in close agreement with its predicted isoelectric point of 9.98. Hybrid proteins showed isoelectric points differing from PelC and Pel1, further confirming their status as novel proteins.
FIG. 3.
Activity-stained isoelectric focusing profiles for PelC, Pel1, and 14 PelC-Pel1 hybrids. Names of each are indicated at the top, and the isoelectric points for PelC (pPEL403) and Pel1 (pAKC617) are indicated on the left. Faint artifactual bands are visible in each lane at the site of sample application (below 9.0), with intensities proportional to the amount of protein loaded.
Three of the 14 PelC-Pel1 chimeric proteins retain secretion ability.
Each of the 14 chimeric proteins made from PelC and Pel1 was expressed in E. coli DH5α carrying pCPP2006 and tested for its ability to be secreted. As shown in Fig. 2, proteins encoded by pCPP2183, where region A has been replaced, and pCPP2190, where region C has been replaced, were secreted at only slightly lower levels than wild-type PelC from pCPP2193. pCPP2197, in which both regions A and C of PelC are replaced with Pel1 sequences, was secreted at approximately one-quarter of wild-type levels. These results suggest that residues 1 to 118 at the N terminus of PelC and 170 to 215 in the central core of the protein are not absolutely required for species-specific targeting to the E. chrysanthemi Out system. Substitution of region B with the homologous sequences from Pel1 resulted in slightly higher levels of secretion than for wild-type Pel1, but when this region was replaced together with region C or both regions A and C, secretion ability was completely lost, indicating that it is in some way required for species-specific targeting. Replacement of PelC regions D and E, alone or in combination, resulted in complete loss of secretion.
Species-specific targeting of PelC is controlled by external loops at the C terminus as well as several noncontiguous turns of the β-helix core.
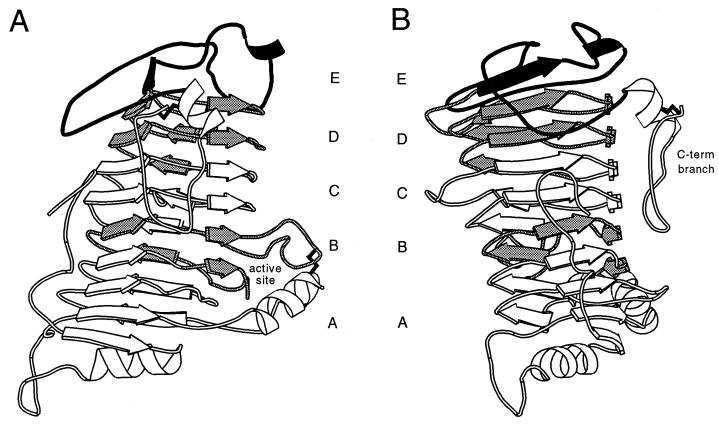
As shown in Fig. 4, the PelC protein is composed of a parallel β-helix core covered at the N terminus by a short α-helix and at the C terminus by three loops (54). The results presented in Fig. 2 indicate that all or part of the three-loop cap at the C terminus (region E) is required for species-specific targeting as are elements within the 2.5 turns at the C-terminal end of the β-helix (region D) and 1.5 turns (region B) in the central part of the helix. The three-loop cap contained in region E is stabilized primarily by region D, although the third loop, or C-terminal branch (16), defined by C329 to C352, extends down the external face of the helix, passing over regions C and B in the β-helix core (Fig. 4). Structural analysis of PelC suggests that region B may influence the conformation of the three-loop cap by virtue of its direct interaction with the C-terminal branch.
FIG. 4.
Two views of a ribbon diagram of PelC with regions required for species-specific secretion shaded. Regions A and C and the C-terminal (C-term) branch of region E, none of which are required for species-specific secretion, are indicated in white. Regions B and D, required for targeting, are lightly shaded. Region E, excluding the C-terminal branch, is shaded darkly because the external loops in this region are considered the best candidates for a primary targeting signal. Disulfide bonds linking C72 to C155 and C329 to 352, where visible, are indicated in black.
The C-terminal branch defined by C329 to C352 is not a primary determinant of species-specific targeting.
To better define the parts of the C-terminal region required for species-specific targeting, the third loop (C terminal branch) was chosen as a target of mutagenesis. Divergent residues 335 and 336, 346, and 349 to 351 in PelC were replaced with the corresponding sequences from Pel1 by site-directed mutagenesis. The resulting construct, PelC/1-335-351, was efficiently secreted by E. coli(pCPP2006), suggesting that the C-terminal branch does not play a direct role in species-specific targeting (Fig. 2).
DISCUSSION
The type II secretion system is present in diverse gram-negative bacteria and is required for secretion of virulence proteins by many pathogens. E. chrysanthemi and E. carotovora secrete similar Pels, using type II secretion pathways whose individual components share a high degree of sequence conservation; however, the two species cannot reciprocally secrete their Pel proteins (14). This observation reveals that species-specific secretion signals are embedded within otherwise similar proteins. Thus, the E. chrysanthemi Out system fails to secrete Pel1 from E. carotovora EC71 even though Pel1 is 71% identical to E. chrysanthemi PelC. By constructing 15 hybrid proteins in which different regions of PelC were substituted with homologous sequences from Pel1, the C-terminal cap, excluding the C-terminal branch, and several turns in the β-helix core were identified as regions required for species-specific secretion of PelC.
Most attempts to define targeting signals through the use of deletions or gene fusions with alkaline phosphatase or β-lactamase have failed to define discrete regions required for targeting (9, 19, 24, 25). For example, the two functional domains from E. chrysanthemi endoglucanase Z were stable when expressed independently, but neither retained secretion ability (43). Although specific regions have been implicated in the secretion of P. aeruginosa exotoxin A and K. oxytoca pullulanase, their actual role has been difficult to establish. In P. aeruginosa exotoxin A, residues 60 to 120 were found to be sufficient for targeting a β-lactamase fusion protein across the outer membrane. However, a deletion construct of exotoxin A containing the N-terminal 30 amino acids attached to the C-terminal 370 residues is also secreted (31), suggesting that targeting of the native protein may involve more than a single region within the primary sequence. Studies with K. oxytoca pullulanase–β-lactamase fusions revealed that two noncontiguous regions composed of residues 1 to 78 and 735 to 814 are sufficient and jointly necessary to promote secretion of β-lactamase across the outer membrane (48). However, deletion of either of these regions from native pullulanase only partially reduces secretion.
The elusiveness of a discrete element in the primary sequence involved in targeting has led to the proposal that the secretion signal could be dependent on higher-order structure for its formation. For example, targeting signals could be determined by a patch signal located in one region of the final structure but composed of amino acids from diverse parts of the linear sequence. Alternatively, the signal could be defined by a region of primary sequence but highly dependent on the overall structure of the protein for proper presentation to the secretion machinery (39, 48). Disruption of overall structure, the likely reason that other attempts to identify targeting regions have been unsuccessful, is minimized by the PelC-Pel1 hybrid approach described here.
Using the PelC-Pel1 hybrid approach, elements within the following three regions of PelC were identified as being required for species-specific targeting: (i) region E, composed of the loops at the C-terminal end of the β-helical core of the protein; (ii) region D, containing the four helical turns partially covered by the C-terminal loops; and (iii), region B, containing 1.5 helical turns in the central portion of the β-helical core. Further mutagenesis of region E revealed that the C-terminal branch is not directly involved in species-specific targeting. The PelC-Pel1 hybrids could not be used to reciprocally identify regions in Pel1 required for species-specific targeting because the E. carotovora out gene cluster is not functional in E. coli (29) and an appropriate Pel-deficient E. carotovora strain is not available.
Several lines of evidence indicate that proteins secreted by the type II pathway are secreted in a largely folded conformation. It has been shown that disulfide bonds are made prior to secretion, and cholera toxin, a multimeric protein secreted by the V. cholerae type II pathway, is assembled in the periplasm prior to secretion (5, 37, 38, 50, 55). Assuming that the Pel proteins are folded prior to secretion, we suggest that external loops of the protein are more likely to be involved in targeting than regions of the β-helical core. Examination of structural elements in regions B, D, and E reveals two places where loops extend away from the body of the protein. One is composed of the three loops at the C terminus in region E, and the other consists of a loop in region B which forms part of the active site. Neither of these loop regions is completely conserved at the sequence level between PelC and Pel1, but without structural information on Pel1 it is difficult to predict how they structurally differ. However, comparison of PelC with the known structure for E. chrysanthemi PelE can be used to identify which of these regions are most similar between the cosecreted Pels. Although PelC and PelE differ substantially in sequence, the comparison done by Lietzke et al. (27) reveals that the β-helical core and several elements in the C-terminal loops are structurally conserved between the two proteins. In contrast, the loop in the active site is highly divergent in both size and folding, making it an unlikely candidate for a conserved targeting signal among the E. chrysanthemi Pels. We therefore propose that the primary signal for species-specific targeting is contained within the C-terminal loops in region E whereas regions D and B play a secondary role in the structural positioning of these residues.
In an attempt to better define the putative targeting signal contained within the three C-terminal loops, all divergent residues in the third loop, or C-terminal branch, of PelC were mutagenized to the corresponding sequence in Pel1. It was hypothesized that if the C-terminal branch plays a key role in species-specific targeting, changing the divergent sequences should disrupt secretion by the E. chrysanthemi type II pathway. However, the resulting protein, encoded by pPELC/1-335-351, was secreted as efficiently as wild-type PelC, indicating that the signal for species-specific targeting is most likely located in the other loops of the C-terminal cap.
Mutagenesis of the these other loops is a major undertaking, as the number of divergent residues is substantial and the structural implications of changes difficult to predict without knowledge of the Pel1 structure. Structural determination of additional Pels from both E. chrysanthemi and E. carotovora is anticipated to yield further clues regarding structurally conserved regions. Comparison of the location of putative targeting regions on PelC with the targeting regions identified in pullulanase and exotoxin A indicates that the location of these signals among proteins using the type II pathway is not conserved at the level of primary sequence. However, the pullulanase and exotoxin A fusions to β-lactamase were made with the goal of identifying general targeting signals that could confer secretion on a normally periplasmic enzyme, while the PelC-Pel1 hybrid strategy was directed at identification of those signals involved in species-specific recognition by the Out pathway. Given their overall structural similarity, it is highly possible that additional regions in the Pels play a role in targeting but, being conserved between the two proteins, are not revealed by this approach.
As a corollary to our experiments involving hybrid E. chrysanthemi-E. carotovora Pel proteins, we have used pCPP2006 as the basis for constructing hybrid E. chrysanthemi-E. carotovora type II secretion systems. This has revealed OutD, an outer membrane protein, as a candidate gatekeeper for the species-specific secretion of E. chrysanthemi Pels (29). Furthermore, the E. chrysanthemi OutD protein interacts with E. chrysanthemi, but not E. carotovora, Pel proteins (51). The species-specific interactions of cognate gatekeepers and structurally defined, secreted proteins could provide a new avenue for determining the structural features that control the secretion of proteins through the type II pathway.
ACKNOWLEDGMENTS
We thank Arun K. Chatterjee for providing the Pel1 sequence before publication and for helpful discussion throughout this work, Cathy Zumoff for assistance with the Pel secretion assays, and Frances Jurnak for helpful discussions on Pel structural features and atomic coordinates for PelC and PelE.
This work was supported by NSF grant DCB-9106431 (A.C.) and NSF grant MCB 9408999 (N.T.K.). M.L. was supported by the NSF/DOE/USDA Plant Science Center and the Cornell Biotechnology Program and NIH training grant 5T32GM08384.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 2.Bally M, Ball G, Badere A, Lazdunski A. Protein secretion in Pseudomonas aeruginosa: the xcpA gene encodes an integral inner membrane protein homologous to Klebsiella pneumoniae secretion function protein PulO. J Bacteriol. 1991;173:479–486. doi: 10.1128/jb.173.2.479-486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepillin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 4.Barras F, Van Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 5.Bortoli-German I, Brun E, Py B, Chippaux M, Barras F. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol Microbiol. 1994;11:545–553. doi: 10.1111/j.1365-2958.1994.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;92:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee A, Liu Y, Chatterjee A K. Nucleotide sequence of a pectate lyase structural gene, pel1 of Erwinia carotovora subsp. carotovora strain 71 and structural relationship of pel1 with other pel genes of Erwinia species. Mol Plant-Microbe Interact. 1995;8:92–95. doi: 10.1094/mpmi-8-0092. [DOI] [PubMed] [Google Scholar]
- 8.Collmer A, Ried J L, Mount M S. Assay methods for pectic enzymes. Methods Enzymol. 1988;161:329–335. [Google Scholar]
- 9.d’Enfert C, Pugsley A P. A gene fusion approach to the study of pullulanase export and secretion in Escherichia coli. Mol Microbiol. 1987;1:159–168. doi: 10.1111/j.1365-2958.1987.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 10.Dums F, Dow J M, Daniels M J. Structural characterization of protein secretion genes of the bacterial phytopathogen Xanthomonas campestris pathovar campestris: relatedness to secretion systems of other gram-negative bacteria. Mol Gen Genet. 1991;229:357–364. doi: 10.1007/BF00267456. [DOI] [PubMed] [Google Scholar]
- 11.Filloux A, Bally M, Ball G, Akrim M, Tommassen J, Lazdunski A. Protein secretion in gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 1990;9:4323–4329. doi: 10.1002/j.1460-2075.1990.tb07881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamood A N, Olson J C, Vincent T S, Iglewski B H. Regions of toxin A involved in toxin A excretion in Pseudomonas aeruginosa. J Bacteriol. 1989;171:1817–1824. doi: 10.1128/jb.171.4.1817-1824.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He S Y, Schoedel C, Chatterjee A K, Collmer A. Extracellular secretion of pectate lyase by the Erwinia chrysanthemi Out pathway is dependent upon Sec-mediated export across the inner membrane. J Bacteriol. 1991;173:4310–4317. doi: 10.1128/jb.173.14.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat B, Heffron S E, Yoder M D, Lietzke S, Jurnak F. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu N-T, Hung M-N, Chiou S-J, Tang F, Chiang D-C, Huang H-Y, Wu C-Y. Cloning and characterization of a gene required for the secretion of extracellular enzymes across the outer membrane by Xanthomonas campestris pv. campestris. J Bacteriol. 1992;174:2679–2687. doi: 10.1128/jb.174.8.2679-2687.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Sukordhaman M, Schell M A. Excretion of the egl gene product of Pseudomonas solanacearum. J Bacteriol. 1989;171:3767–3774. doi: 10.1128/jb.171.7.3767-3774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang B, Howard S P. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol Microbiol. 1992;6:1351–1361. doi: 10.1111/j.1365-2958.1992.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 20a.Jurnak, F. Personal communication.
- 21.Kang Y, Huang J, Guozhang M, He L-Y, Schell M A. Dramatically reduced virulence of mutants of Pseudomonas solanacearum defective in export of extracellular proteins across the outer membrane. Mol Plant-Microbe Interact. 1994;7:370–377. [Google Scholar]
- 22.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Med. 1954;22:301–307. [PubMed] [Google Scholar]
- 23.Kita N, Boyd C M, Garrett M R, Jurnak F, Keen N T. Differential effect of site-directed mutations in pelC on pectate lyase activity, plant tissue maceration, and elicitor activity. J Biol Chem. 1996;271:26529–26535. doi: 10.1074/jbc.271.43.26529. [DOI] [PubMed] [Google Scholar]
- 24.Kornacker M G, Pugsley A P. Molecular characterization of pulA and its product, pullulanase, a secreted enzyme of Klebsiella pneumoniae UNF5023. Mol Microbiol. 1989;4:73–85. doi: 10.1111/j.1365-2958.1990.tb02016.x. [DOI] [PubMed] [Google Scholar]
- 25.Kornacker M G, Pugsley A P. The normally periplasmic enzyme β-lactamase is specifically and efficiently translocated through the Escherichia coli outer membrane when it is fused to the cell-surface enzyme pullulanase. Mol Microbiol. 1990;4:1101–1109. doi: 10.1111/j.1365-2958.1990.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 26.Kotoujansky A. Molecular genetics of pathogenesis by soft-rot erwinias. Annu Rev Phytopathol. 1987;25:405–430. [Google Scholar]
- 27.Lietzke S E, Yoder M D, Keen N T, Jurnak F. The three-dimensional structure of pectate lyase E, a plant virulence factor from Erwinia chrysanthemi. Plant Physiol. 1994;106:849–862. doi: 10.1104/pp.106.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindeberg M, Collmer A. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J Bacteriol. 1992;174:7385–7397. doi: 10.1128/jb.174.22.7385-7397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindeberg M, Salmond G P C, Collmer A. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologs reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 30.Lu H-M, Lory S. A specific targeting domain in mature exotoxin A is required for its extracellular secretion from Pseudomonas aeruginosa. EMBO J. 1996;15:429–436. [PMC free article] [PubMed] [Google Scholar]
- 31.McVay C S, Hamood A N. Toxin A secretion in Pseudomonas aeruginosa: the role of the first 30 amino acids of the mature toxin. Mol Gen Genet. 1995;249:515–525. doi: 10.1007/BF00290577. [DOI] [PubMed] [Google Scholar]
- 32.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murata H, Fons M, Chatterjee A, Collmer A, Chatterjee A K. Characterization of transposon insertion Out− mutants of Erwinia carotovora subsp. carotovora defective in enzyme export and of a DNA segment that complements out mutations in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Erwinia chrysanthemi. J Bacteriol. 1990;172:2970–2978. doi: 10.1128/jb.172.6.2970-2978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overbye L J, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- 35.Palomaki T, Saarilathi H T. The extreme C-terminus is required for secretion of both the native polygalacturonase (PehA) and PehA-Bla hybrid proteins in Erwinia carotovora subsp. carotovora. Mol Microbiol. 1995;17:449–459. doi: 10.1111/j.1365-2958.1995.mmi_17030449.x. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peek J A, Taylor R K. Characterization of periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc Natl Acad Sci USA. 1992;89:6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugsley A P. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugsley A P. The complete general protein secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugsley A P, d’Enfert C, Reyss I, Kornacker M G. Genetics of extracellular protein secretion by gram-negative bacteria. Annu Rev Genet. 1990;24:67–90. doi: 10.1146/annurev.ge.24.120190.000435. [DOI] [PubMed] [Google Scholar]
- 41.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 42.Pugsley A P, Poquet I, Kornacker M G. Two distinct steps in pullulanase secretion by Escherichia coli K12. Mol Microbiol. 1991;5:865–873. doi: 10.1111/j.1365-2958.1991.tb00760.x. [DOI] [PubMed] [Google Scholar]
- 43.Py B, Chippaux M, Barras F. Mutagenesis of cellulase EGZ for studying the general protein secretory pathway. Mol Microbiol. 1993;7:785–793. doi: 10.1111/j.1365-2958.1993.tb01169.x. [DOI] [PubMed] [Google Scholar]
- 44.Py B, Salmond G P C, Chippaux M, Barras F. Secretion of cellulases in Erwinia chrysanthemi and E. carotovora is species-specific. FEMS Microbiol Lett. 1991;79:315–322. [Google Scholar]
- 45.Reeves P J, Whitcombe D, Wharam S, Gibson M, Allison G, Bunce N, Barallon R, Douglas P, Mulholland V, Stevens S, Walker D, Salmond G P C. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 46.Ried J L, Collmer A. An activity stain for the rapid characterization of pectic enzymes in isoelectric focusing and sodium dodecyl sulfate-polyacrylamide gels. Appl Environ Microbiol. 1985;50:615–622. doi: 10.1128/aem.50.3.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sauvonnet N, Pugsley A P. Identification of two regions of Klebsiella oxytoca pullulanase that together are capable of promoting beta-lactamase secretion by the general secretory pathway. Mol Microbiol. 1996;22:1–7. doi: 10.1111/j.1365-2958.1996.tb02650.x. [DOI] [PubMed] [Google Scholar]
- 49.Schoepfer R. The pRSET family of T7 promoter expression vectors for Escherichia coli. Gene. 1993;124:83–85. doi: 10.1016/0378-1119(93)90764-t. [DOI] [PubMed] [Google Scholar]
- 50.Shevchik V E, Condemine G, Robert-Baudouy J. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 1994;13:1007–2012. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shevchik V E, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamaki S J, Gold S, Robeson M, Manulis S, Keen N T. Structure and organization of the pel genes from Erwinia chrysanthemi EC16. J Bacteriol. 1988;170:3468–3478. doi: 10.1128/jb.170.8.3468-3478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong K R, Buckley J T. Site-directed mutagenesis of a single tryptophan near the middle of the channel-forming toxin aerolysin inhibits its transfer across the outer membrane of Aeromonas salmonicida. J Biol Chem. 1991;266:14451–14456. [PubMed] [Google Scholar]
- 54.Yoder M D, Keen N T, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 55.Yu J, Webb H, Hirst T R. A homolog of the Escherichia coli DsbA protein involved in disulphide bond formation is required for exotoxin biogenesis in Vibrio cholerae. Mol Microbiol. 1992;6:1949–1958. doi: 10.1111/j.1365-2958.1992.tb01368.x. [DOI] [PubMed] [Google Scholar]