Abstract
Eosinophils (Eos) reside in multiple organs during homeostasis and respond rapidly to an inflammatory challenge. Although Eos share chemical staining properties, they also demonstrate phenotypic and functional plasticity that is not fully understood. Here, we used a murine model of allergic lung inflammation to characterize Eos subsets and determine their spatiotemporal and functional regulation during inflammation and its resolution in response to resolvin D2 (RvD2), a potent specialized proresolving mediator. Two Eos subsets were identified by CD101 expression with distinct anatomic localization and transcriptional signatures at baseline and during inflammation. CD101low Eos were predominantly located in a lung vascular niche and responded to allergen challenge by moving into the lung interstitium. CD101high Eos were predominantly located in bronchoalveolar lavage (BAL) and extravascular lung, only present during inflammation, and had transcriptional evidence for cell activation. RvD2 reduced total Eos numbers and changed their phenotype and activation by at least two distinct mechanisms: decreasing interleukin 5-dependent recruitment of CD101low Eos and decreasing conversion of CD101low Eos to CD101high Eos. Collectively, these findings indicate that Eos are a heterogeneous pool of cells with distinct activation states and spatiotemporal regulation during resolution of inflammation and that RvD2 is a potent proresolving mediator for Eos recruitment and activation.
Keywords: specialized proresolving mediators, resolvin D2, eosinophils, allergic inflammation
Eosinophils (Eos) play important roles in health and disease. In addition to their homeostatic roles, including in the early stages of lung development (1), Eos participate in the host response to parasitic infection (2) and the pathogenesis of asthma and other allergic diseases (3, 4). On the basis of their earliest identification using chemical stains (5), Eos were considered to be a homogeneous population of cells with proinflammatory actions. Recently, several reports have identified distinct subsets of Eos with varying activation states and roles in disease models (6). In mouse lung, CD101 expression has been used to identify tissue-resident Eos (CD101low Eos), which may regulate type 2 immune responses (7, 8), and to distinguish them from inflammatory Eos (CD101high Eos), which are present in exudates and have an activated phenotype (6, 7). In addition, functions in tissue homeostasis for type 2 inflammation and Eos have recently been assigned (9). With this growing recognition that Eos are a heterogeneous population of immune effectors, a more integrated understanding of the spatiotemporal and functional regulation of Eos subsets is urgently needed.
Eos differentiation, proliferation, migration, and survival have been shown to be centrally regulated by IL-5 (4). This important role for IL-5 in eosinophilia has been the basis for the development of several anti–IL-5 targeted therapies for conditions characterized by excess Eos numbers and activation, such as severe asthma (10). Of interest, anti–IL-5 therapies have only been effective for some patients with asthma, and despite reducing Eos numbers, evidence for changing Eos phenotype or function has been challenging to demonstrate (11, 12).
Resolution of inflammation, including type 2 inflammation, is an active process governed by specific proresolving mediators and cellular effectors (13, 14). A family of specialized proresolving mediators (SPMs) have been identified as products of essential fatty acid metabolism (13). Recently, genetic variation for DRV2, a high-affinity receptor for the SPM resolvin D2 (7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-docosahexaenoic acid; RvD2), was linked to asthma risk (15). RvD2 is produced by enzymatic conversion of docosahexaenoic acid (DHA) via the sequential actions of 15-lipoxygenase and 5-lipoxygenase with additional enzymatic steps (14). Eos are the major source of human 15-lipoxygenase (16). The mouse ortholog of the human enzyme is 12/15-lipoxygenase, and its role in mouse Eos is amplified during the resolution phase of inflammatory peritonitis (17). Of interest, the production of 15-lipoxygenase–derived SPMs is diminished in uncontrolled or severe asthma (18–20).
Here, using a mouse model of self-limited allergic lung inflammation, we determined the spatiotemporal regulation of Eos subsets and uncovered pivotal proresolving roles for RvD2 in the regulation of IL-5–induced Eos subset numbers, phenotype, and function.
Methods
Mice
We used 6–8-week-old female wild-type BALB/c mice from The Jackson Laboratory, and all animal experiments were performed in accordance with NIH Guidelines for the Care and Use of Laboratory Animals and as prescribed by the institutional animal care and use committee at Brigham and Women’s Hospital (2016N000357) (AAALAC 1729).
Allergic Airway Inflammation Model
Mice were anesthetized and exposed to house dust mite (HDM) (Dermatophagoides pteronyssinus, B70 source material; Greer Laboratories) (100 μg) on protocol Days 0, 7, and 14. Some mice were exposed to RvD2 or O-1918 (Cayman Chemical) (1 mg/kg in 100 μl). Tissues and fluids were harvested at the indicated time points (see Materials and Methods in the data supplement).
Flow Cytometry
Cells from bronchoalveolar lavage (BAL) were obtained with three aliquots of PBS (0.5 ml each). Subsequently, lung tissue was dissociated to obtain single-cell suspensions. Cells were fixed and permeabilized to stain for surface and intracellular markers using a FoxP3 staining kit (Thermo Fisher Scientific). A list of antibodies used for flow cytometry can be found in the data supplement. All flow cytometry data were acquired on a BD FACS Fortessa device, then analyzed using FlowJo version 10 software.
Targeted Metabololipidomics
CD101low and CD101high Eos were flow sorted at Day 17 after HDM challenge and incubated in complete media with DHA (1 μM, 37°C, pH 7.45). Data were acquired on a 6500+ triple quadrupole QTRAP mass spectrometer in low mass negative mode (SCIEX) equipped with an ExionLC system (Shimadzu) using Analyst version 1.7.1 (SCIEX). Data were analyzed and presented as screen captures using SCIEX OS-Q version 1.7.0.36606 (SCIEX) (see extended Materials and Methods in the data supplement).
Gene Expression
Gene expression analysis was performed using NanoString nCounter technology (NanoString Technologies Inc.) to determine the transcriptional profile of flow-sorted CD101low and CD101high Eos from naive and HDM-challenged mice at Days 17 and 21 of the HDM protocol (see extended Materials and Methods in the data supplement). Expression of mouse 5-lipoxygenase (Alox5), 12/15-lipoxygenase (Alox15), and 18S ribosomal RNA (18S rRNA) (primer sequences are Qiagen proprietary material) was performed by real-time quantitative PCR (RT-qPCR) (Qiagen) (see extended Materials and Methods in the data supplement).
Eos Peroxidase Assay
Flow-sorted CD101low and CD101high Eos from HDM-challenged mice on Day 17 were incubated with vehicle (Veh), RvD2 (10 nM), prostaglandin D2 (PGD2; 10 nM), and RvD2 and PGD2 (10 nM of RvD2 and PGD2), and Eos peroxidase (EPO) release was measured in cell supernatants.
Bone Marrow–derived Eos
Bone marrow–derived Eos (BMEos) were generated as previously described (21) (see Materials and Methods in the data supplement).
Statistics
Results are expressed as mean ± SEM. Statistical differences between two groups were calculated by two-tailed unpaired t test. Statistical differences between three or more groups were calculated by one-way ANOVA and Tukey test for multiple comparisons. P < 0.05 was defined as statistically significant.
Results
Eos Are Spatiotemporally Regulated in a Self-Limited Model of Allergic Lung Inflammation
To investigate Eos phenotypes and functions over the time course of allergic lung inflammation, mice were airway sensitized and challenged with the common household allergen HDM (Figure 1A). The number of total lung Eos peaked on protocol Day 15, which corresponds to 24 hours after the last HDM challenge, and gradually decreased thereafter on protocol Days 17, 21, and 28 (Figure 1B). Different anatomical compartments of the lungs were assessed by sampling proximal and distal airways by BAL for comparison with lung tissue after BAL (lung). Eos numbers peaked at Day 15 in lung and at Day 17 in BAL (Figure 1C). Notably, BAL Eos decreased more rapidly to lower numbers than lung Eos. Concentrations of the Eos chemoattractant CCL11 (eotaxin-1) were determined in BAL and lung homogenates by ELISA. At Day 17 of the HDM protocol, lung CCL11 concentrations were significantly increased compared with baseline (Day 0) (Figure E1). Of note, BAL CCL11 concentrations were not significantly increased at this time point (Figure E1), suggesting a role for CCL11 in recruiting Eos from the vasculature to the lung tissue. Together, these data suggest distinct regulatory mechanisms for lung and BAL Eos during allergic lung inflammation and its resolution.
Figure 1.
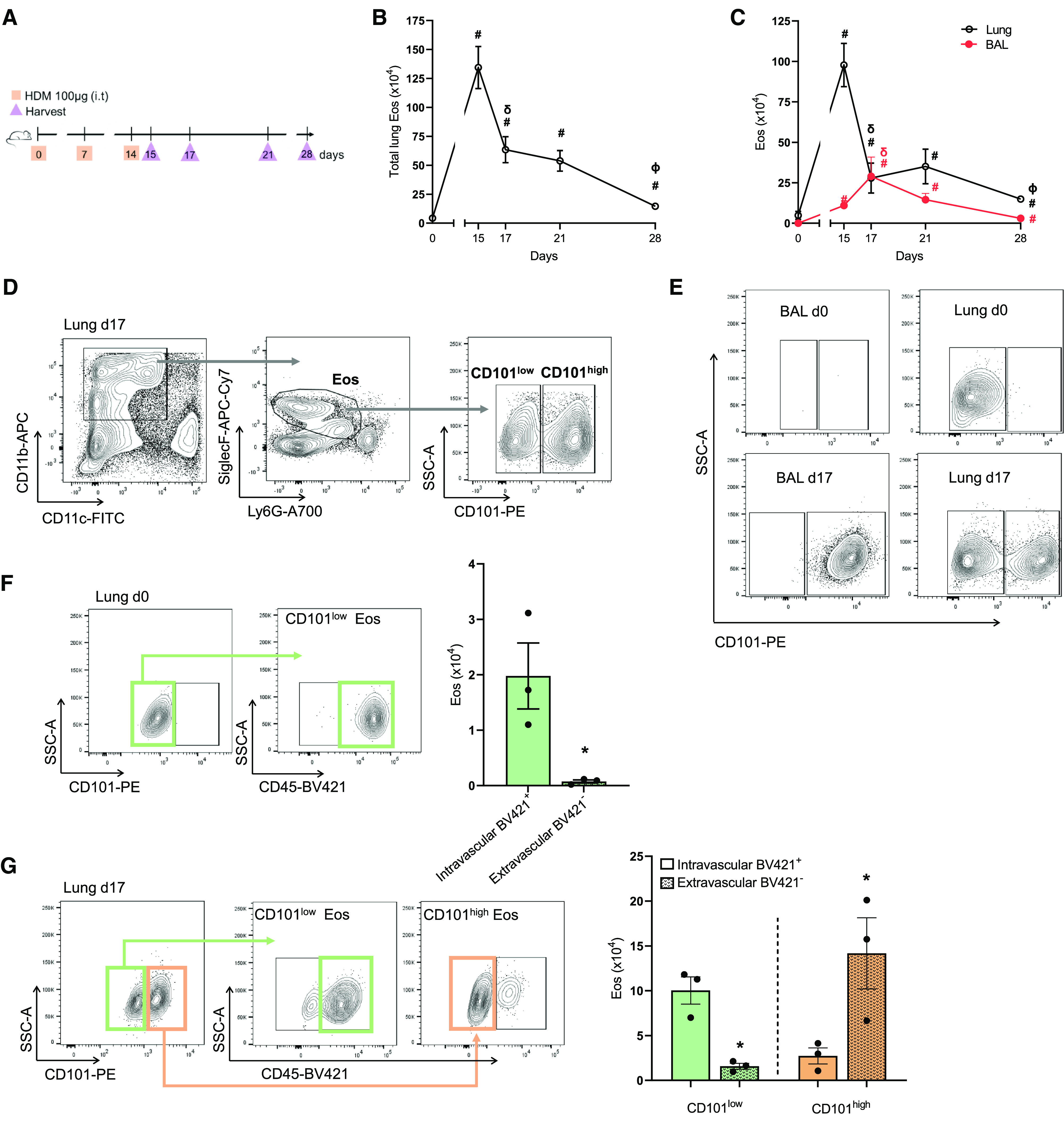
Eosinophils (Eos) are spatiotemporally regulated in a self-limited model of allergic lung inflammation. (A) Allergic lung inflammation model (see Methods). (B) Time course of Eos numbers in whole lung. (C) Time course of Eos numbers in BAL (red) and lung tissue after BAL (black). (D) Representative flow cytometry plots of CD101low and CD101high Eos from lung at Day 17. Full gating strategy in Figure E2A. (E) Representative flow cytometry plots of CD101low and CD101high Eos from BAL and lung at Days 0 and 17. Full gating strategy in Figures E2A and E2B. (F) Representative flow cytometry plot of whole-lung CD101low Eos at Day 0 after CD45-BV421 staining (see Methods), with quantification of CD45-BV421 positive or negative Eos on the right. (G) Representative flow cytometry plot of whole-lung CD101low and CD101high Eos at Day 17 after CD45-BV421, with quantification of CD45-BV421 positive or negative CD101low and CD101high Eos on the right. #P < 0.05 compared with Day 0. δP < 0.05 compared with Day 15. ΦP < 0.05 compared with Day 21. *P < 0.05 compared with BV421+ within same cell type (CD101low and CD101high). Comparisons between two groups were performed by unpaired t test. Bars represent mean ± SEM. HDM = house dust mite.
On the basis of prior reports that defined different subsets of Eos by distinct expression of CD11c, CD11b, Siglec F, and CD101 (7, 22, 23), we next identified these Eos subsets in lung and BAL, as well as their time course during allergic lung inflammation. Eos numbers in BAL and lung tissue were similar at Day 17 (Figure 1C), so this time point was chosen for further investigation of the Eos subsets because sufficient cell numbers were present in both anatomic compartments. CD101 differentially marked two main Eos subsets, referred to as CD101low Eos and CD101high Eos (Figure 1D; complete flow cytometry gating strategy is shown in Figure E2A). In the absence of inflammation (baseline, protocol Day 0), Eos were not present in BAL (BAL Day 0), (Figure 1E, top left; complete flow cytometry gating strategy is shown in Figure E2B); however, in the lung tissue (lung Day 0), CD101low Eos were identified (Figure 1E, top right). During inflammation (protocol Day 17), BAL Eos were nearly all CD101high Eos (BAL Day 17) (Figure 1E, bottom left). In contrast, lung Eos were a mixed population of CD101low Eos and CD101high Eos (lung Day 17) (Figure 1E, bottom right). To identify intravascular leukocytes and distinguish them from interstitial leukocytes that had transmigrated into the lung, intravascular CD45-BV421–conjugated antibody was injected intravenously 5 to 10 minutes before euthanasia and harvest to stain cells in the intravascular space (see Methods). The lung circulation was not cleared with saline before harvesting so that lung vascular and interstitial compartments could be distinguished. In naive mice at Day 0, more than 95% of lung CD101low Eos stained positive for CD45 (CD45-BV421+) (Figure 1F), suggesting that this Eos subset was principally intravascular at baseline. During lung inflammation on Day 17 (lung Day 17), ∼85% of the CD101low Eos were CD45-BV421+ with ∼14% of CD101low Eos CD45-BV421− (Figure 1G). In contrast, CD101high Eos, which were only present in the lung during inflammation (Day 17), were ∼15% CD45-BV421+ and ∼84% CD45-BV421− (Figure 1G). Together, these data indicate that lung and BAL Eos are spatiotemporally regulated during allergic lung inflammation and that Eos subsets exist with specific anatomic preferential location: CD101low Eos intravascular and CD101high Eos extravascular.
CD101low and CD101high Eos Have Distinct Transcriptional Signatures
To further characterize the different Eos subsets, CD101low and CD101high Eos were first sorted by flow cytometry (Figure 2A). The morphology of CD101low Eos was similar at baseline (Day 0) and during inflammation (Day 17), having a circular nucleus and increased granule density, whereas CD101high Eos, present only during inflammation (Day 17), had a multisegmented nucleus and appeared to be less granulocytic (Figure 2A). Although the staining morphology of CD101low Eos was similar at Days 0 and 17, there were several transcriptional differences evident by NanoString assay of inflammation gene expression (Figure 2B). Of the 254 genes in the array, 33 genes were differentially expressed in CD101low Eos at Day 0 compared with CD101low Eos at Day 17. There was selective upregulation of 29 genes in the CD101low Eos at Day 17, including the asthma-related genes Chi3l3, Gpr44, Ccr3, and Ltb4r1 and the bioactive lipid mediator biosynthetic enzymes Alox5 and Alox15 (Figure 2B). Pathway analysis revealed upregulation of several inflammatory pathways, including leukocyte migration, chemotaxis, and cell–cell adhesion on CD101low Eos on Day 17 compared with Day 0 (Figure E3A), and upregulation of cell activation signaling, including extracellular signal-regulated kinase 1 (ERK1) and ERK2 on CD101high Eos relative to CD101low Eos on Day 17 (Figure E3B). Comparing CD101low Eos with CD101high Eos at Day 17, there were 29 differentially expressed genes (Figure 2C). Three noteworthy genes that gave relatively increased expression in CD101high Eos, namely Il5, C3ar1, and Ccr1, are known to be linked to Eos recruitment and activation. Also of interest, expression of the Eos recruitment receptor Ccr3 was downregulated in CD101high Eos relative to CD101low Eos at Day 17 (Figure 2C). The bioactive lipid mediator biosynthetic enzyme Alox15 was expressed at even higher levels in CD101high Eos compared with CD101low Eos at Day 17 (Figure 2C).
Figure 2.
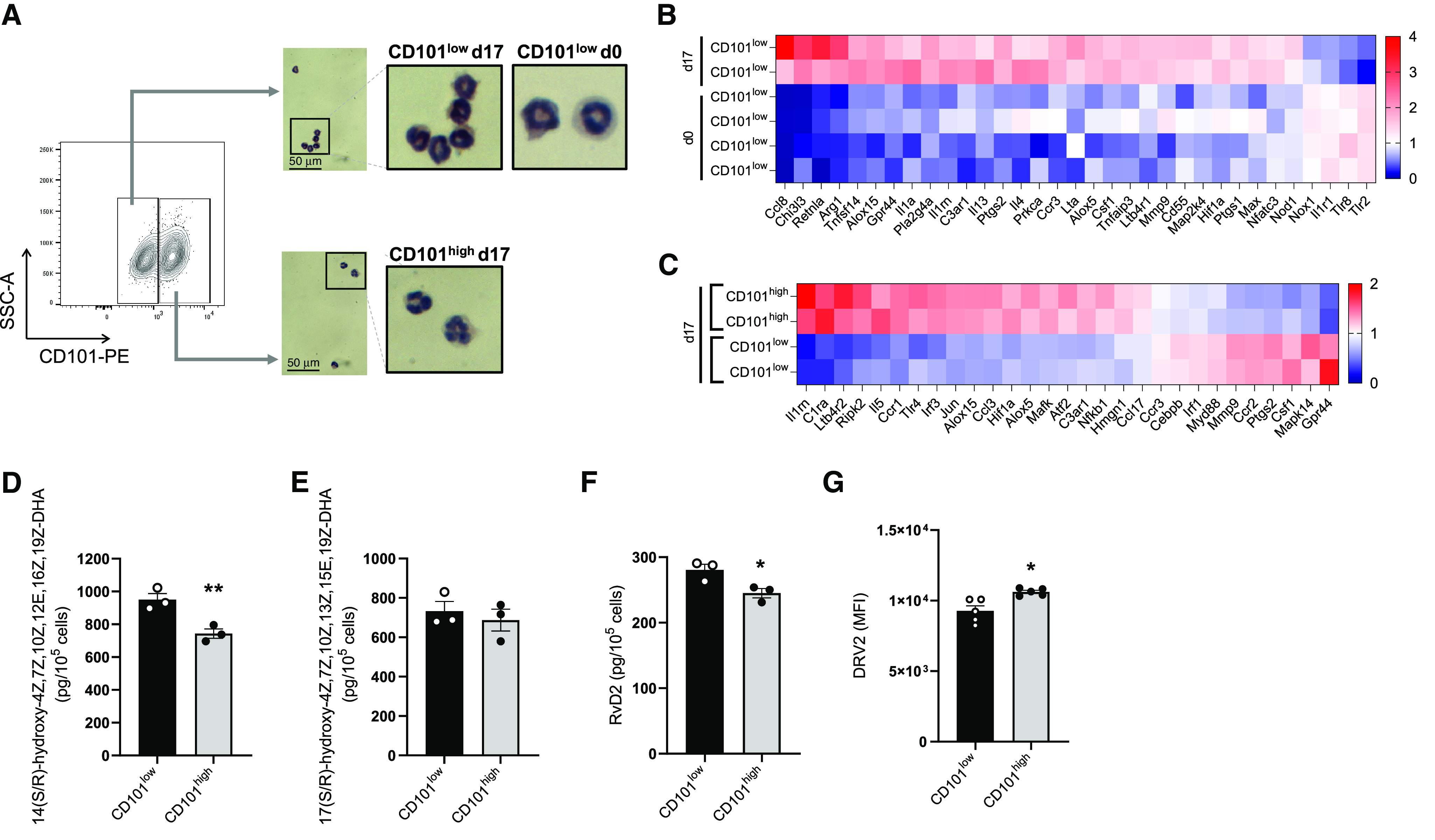
CD101low and CD101high Eos have distinct transcriptional signatures. (A) Representative images of cytospin performed on flow-sorted CD101low Eos from Days 0 and 17 and CD101high Eos from Day 17 of house dust mite (HDM) protocol. Scale bars, 50 μm. (B and C) mRNA analysis of flow-sorted CD101low and CD101high Eos using NanoString (see Methods). Data are represented as a heatmap of differentially expressed genes (P < 0.05 by unpaired t test) comparing CD101low Eos from Day 0 with Day 17 of HDM protocol (B) and comparing CD101low Eos with CD101high Eos at Day 17 of HDM protocol (C). (D–F) Concentrations of 14-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14-HDHA), 17-hydroxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid (17-HDHA), and resolvin D2 (RvD2) from flow-sorted CD101low and CD101high Eos stimulated with docosahexaenoic acid (DHA) by liquid chromatography–tandem mass spectrometry. (G) Level of DRV2 expression on CD101low and CD101high Eos at Day 17 of HDM protocol, as mean fluorescence intensity measured by flow cytometry. *P < 0.05 and **P < 0.01 by unpaired t test. Bars represent mean ± SEM.
Because Alox5 and Alox15 were differentially expressed in CD101low Eos and CD101high Eos, we next investigated the metabolic functions of these enzymes in the Eos subsets. CD101low Eos and CD101high Eos were sorted by flow cytometry from inflamed lungs at Day 17 and incubated with DHA in vitro (see Methods). Targeted lipid mediator metabolomic analyses demonstrated that CD101high Eos had significantly lower concentrations of 14-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14-HDHA) (Figures 2D and E4A), but there was no significant difference in the concentrations of 17-hydroxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid (17-HDHA) between CD101low Eos and CD101high Eos (Figures 2E and E4B). Each was identified by matching liquid chromatography retention times and reported characteristic ions (17-HDHA mass-to-charge ratio [m/z], 343 > 245; 14-HDHA m/z 343 > 205) present in the respective tandem mass spectrometry spectra. In addition, CD101high Eos produced less RvD2 (m/z 375 > 141) compared with CD101low Eos (Figures 2F and E4C). In contrast, expression of the RvD2 receptor DRV2 (24) was increased by CD101high Eos (Figure 2G). Together, these data indicate that CD101low and CD101high Eos subsets were distinct morphologically and at the transcriptional level. CD101high Eos had a lower capacity to produce RvD2 and 14-HDHA, which was in contrast to the increased expression of the RvD2 receptor DRV2 on CD101high Eos.
RvD2 Regulates CD101low and CD101high Eos Numbers during Inflammation
Because RvD2 production differed among these Eos subsets, we next used the HDM model (Figure 1A) to determine the impact of RvD2 on allergen-evoked eosinophilic lung inflammation. After allergic lung inflammation was established, mice were exposed to either RvD2 (100 ng/mouse, once daily, intranasally [i.n.]) or its Veh on protocol Days 15 and 16, then lung inflammation was analyzed on protocol Day 17 (Figure 3A). In addition to Day 17, later assessments were performed 1 week (Day 21) and 2 weeks (Day 28) after the last challenge with HDM. For Day 21, mice were also exposed to RvD2 or Veh on protocol Days 18 and 20; and for Day 28, mice were additionally exposed to RvD2 on protocol Days 22, 24, and 26 (Figure 3A). Authentication of the identity and physical properties of RvD2 used in these experiments was ascertained by liquid chromatography–tandem mass spectrometry (Figures 3B and 3C). RvD2 gave a single peak on liquid chromatography and a fragmentation pattern on tandem mass spectrometry with a spectral library fit score of 95.8% based on matching authentic RvD2 (Figures 3B and 3C). Next, inflammation was quantitated by specific resolution indices (25). Total lung (lung tissue and BAL) Eos numbers were maximal on Day 15 (Tmax) in Veh- and RvD2-exposed cohorts (Ψmax = 134.3 ± 53.3 × 104 cells) (Figure 3D). RvD2 accelerated resolution of total lung Eos, reaching 50% of maximum (Ψ50) at 16.1 days (T50), which was slightly faster than the Veh cohort with a T50 of 16.8 days (Figure 3D; blue vs. red dotted line, respectively). This gave a resolution interval (Ri; the difference between Tmax and T50,) of 1.8 days with Veh and 1.1 days with RvD2 (Figure 3D). The difference in the Ri became even more evident when calculated as the time to reach 70% reduction of maximum (T70). RvD2 gave an Ri to reach T70 of 1.6 days in comparison with Veh that had a significantly longer Ri to reach T70 of 8.1 days (Figure 3D). This difference was related in part to transient further increases in BAL Eos with Veh that were not observed with RvD2 (Figure 3E). When analyzing BAL Eos and Lung Eos separately, the Ri for BAL Eos was significantly faster (∼50%) with RvD2 (4.7 d) than Veh (10.2 d) (Figure 3E; blue vs. red dotted line, respectively). Although RvD2 decreased lung Eos at Day 17, no significant changes in Ri were observed between RvD2 and Veh cohorts (Figure 3F).
Figure 3.
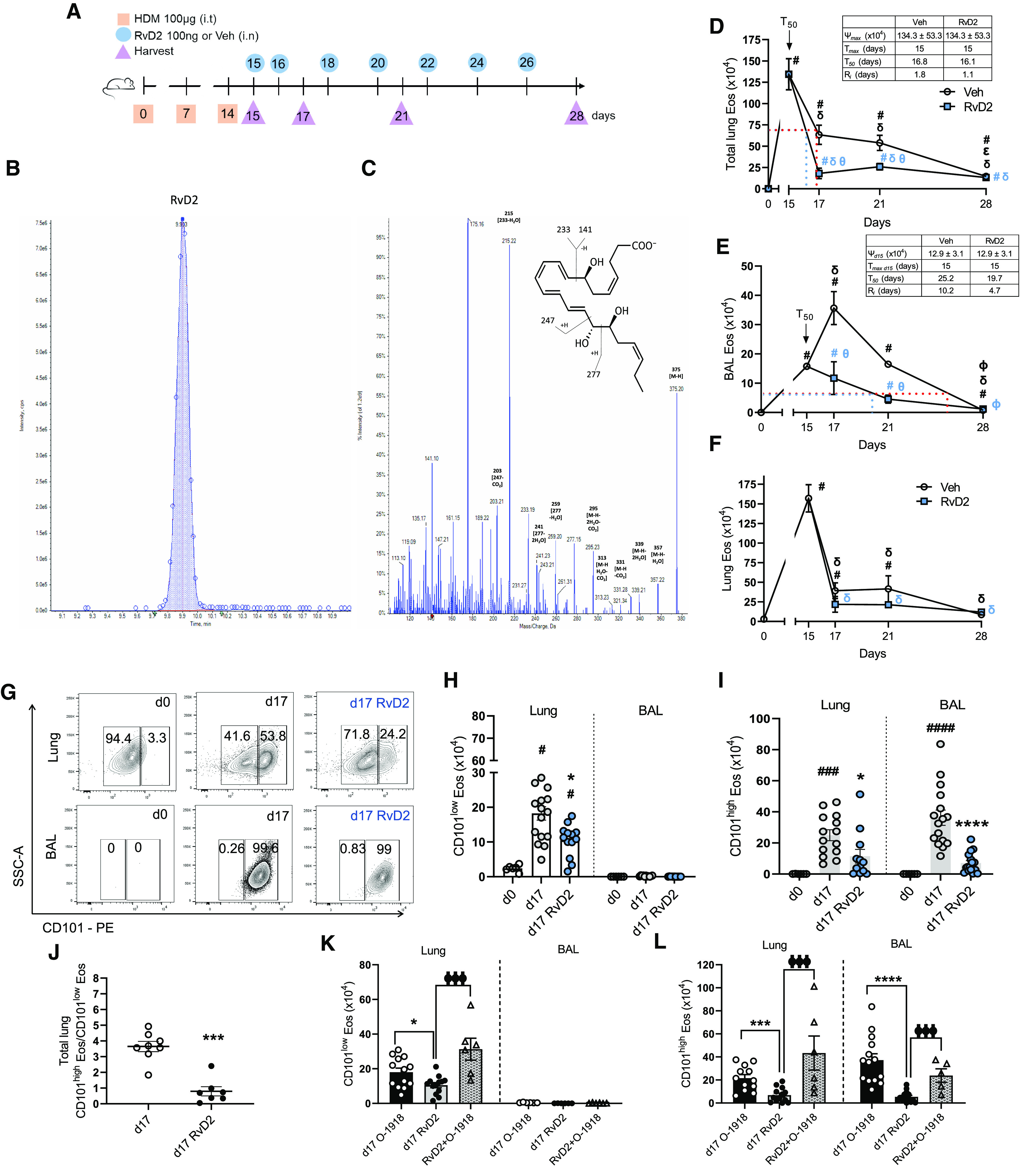
RvD2 regulates CD101low and CD101high Eos numbers during inflammation. (A) Animal model (see Methods). (B and C) Authentication of RvD2 (see Methods). (D–F) Eos numbers in whole lung (D), BAL (E), and lung after BAL (F). (G) Flow cytometry plots of CD101low and CD101high Eos from lung (top) and BAL (bottom) at Days 0 and 17. (H and I) Number of CD101low (H) and CD101high (I) Eos in lung and BAL at Days 0 and 17. (J) CD101high/CD101low Eos ratio in whole lungs at Day 17. (K and L) Number of CD101low (K) and CD101high (L) Eos in lung and BAL at Day 17. #P < 0.05, ###P < 0.001, ####P < 0.0001 compared with Day 0. δP < 0.05 compared with Day 15 in D–F. ΦP < 0.05 compared with Day 21 in D and E. ɵP < 0.05 comparing vehicle (Veh) with RvD2 within the same day in D and E. *P < 0.05, ***P < 0.001, ****P < 0.0001 compared with Day 17 within the same compartment lung or BAL in H–L. ΦΦΦP < 0.001 compared with Day 17 RvD2 in K and L. Comparisons between more than two groups were performed by one-way ANOVA and Tukey test for multiple comparisons (D–F and H, I, K, and L). Comparison between two groups was performed by unpaired t test (J). Bars represent mean ± SEM.
RvD2 exposure decreased both CD101low Eos and CD101high Eos in lung and markedly decreased CD101high Eos in BAL compared with Veh at Day 17 (Figures 3G–3I). The total lung ratio of CD101high Eos/CD101low Eos was significantly decreased in the RvD2 group (Figure 3J). At Day 21, the difference in CD101low Eos in the lung between Veh and RvD2 groups was no longer apparent; however, there was a sustained decrease of CD101high Eos in lung and BAL (Figures E5A–E5C). At Day 28 of the HDM protocol, Eos numbers were still higher than at Day 0, but with similar numbers of CD101low and CD101high Eos and no significant changes after RvD2 exposure (Figures E5D–E5F). Replotted as a time course for CD101low Eos and CD101high Eos in the presence of Veh or RvD2 over 28 days demonstrated a marked and sustained proresolving action for RvD2, especially on CD101high Eos (Figures E5G and E5H).
To determine whether RvD2 actions were receptor dependent, select mice in this HDM protocol were exposed to the DRV2 receptor antagonist O-1918. On Days 15 and 16 of the HDM protocol, animal cohorts included Veh with O-1918 (1 mg/kg in 100 μl intraperitoneally [i.p.]) (Day 17 O-1918), RvD2 (100 ng i.n.) with saline 0.9% (100 μl i.p.) (Day 17 RvD2), or RvD2 with O-1918 (1 mg/kg in 100 μl i.p.) (RvD2 + O-1918) (Figure E5I). At Day 17, exposure to O-1918 blocked the proresolving actions of RvD2 on CD101low Eos in lung (Figure 3K) and on CD101high Eos in lung and BAL (Figure 3L). Together, these findings indicated that the RvD2-DRV2 signaling axis was pivotal to RvD2’s potent regulation of lung allergic eosinophilic inflammation, specifically for the CD101high Eos phenotype.
RvD2 Regulates CD101low and CD101high Eos Gene Signature and Function during Inflammation
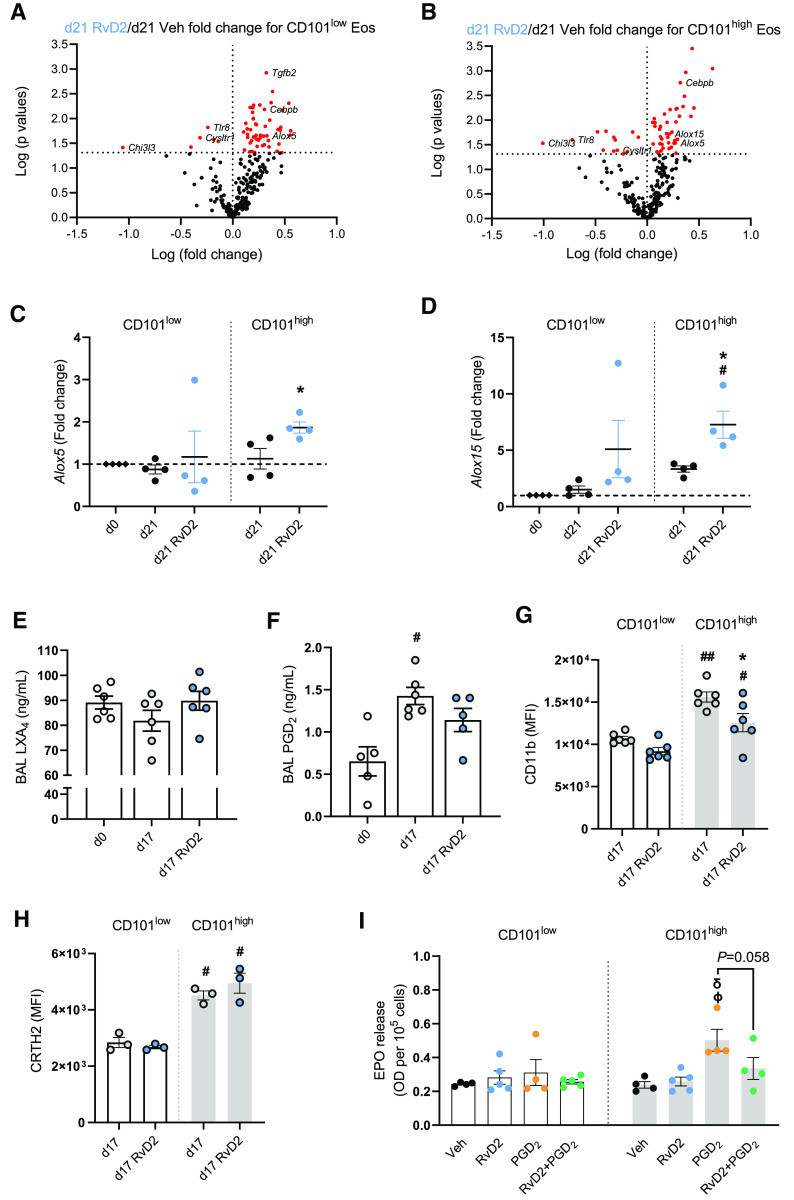
For further mechanistic insight into the proresolving actions of RvD2 on Eos, we flow sorted CD101low and CD101high Eos collected from lungs during the resolution phase (Day 21) after in vivo exposure to RvD2 or Veh on Days 15 and 16, and we performed mRNA analyses using the NanoString inflammation panel (Figure E6A). RvD2 markedly downregulated the expression of proinflammatory genes, including Chi3l3, Tlr8, and Cysltr1, in both CD101low and CD101high Eos (Figures 4A and 4B). RvD2 also increased the expression of the lipid mediator biosynthetic genes Alox5 and Alox15 in the CD101high Eos (Figures 4A–4D).
Figure 4.
RvD2 regulates CD101low and CD101high Eos gene signature and function during inflammation. (A and B) Volcano plot of RNA analysis using NanoString in CD101low (A) and CD101high (B) Eos. Fold change of differentially expressed genes (P < 0.05 by unpaired t test; red dots) comparing CD101low (A) and CD101high (B) Eos at Day 21 of RvD2 versus Veh. (C and D) Gene expression of Alox5 (C) and Alox15 (D) by RT-PCR. (E) Lipoxin A4 (LXA4) concentrations in BAL fluid at Day 17. (F) Prostaglandin D2 (PGD2) concentrations in BAL fluid at Day 17. (G) CD11b expression on CD101low and CD101high Eos. (H) CRTH2 expression on CD101low and CD101high Eos. (I) Eos peroxidase (EPO) release from flow-sorted CD101low and CD101high Eos (see Methods). #P < 0.05 compared with Day 0 in D and F and CD101low Day 17 in G and H. ##P < 0.01 compared with CD101low Day 17 in G. *P < 0.05 compared with Day 21 within CD101high Eos in C and D and with CD101high Day 17 in G. ϕP < 0.05 compared with Veh within CD101high and δP < 0.05 compared with RvD2 within CD101high in I. Comparisons were performed by one-way ANOVA and Tukey test for multiple comparisons. Bars represent mean ± SEM.
In HDM-challenged mice, BAL concentrations of lipoxin A4 (LXA4), a product of ALOX5 and ALOX15 activity (26), trended toward a decrease on Day 17 compared with baseline (Day 0), and in vivo RvD2 exposure returned LXA4 to baseline levels (Figure 4E). In contrast, BAL concentrations of the prophlogistic lipid mediator PGD2 significantly increased at Day 17, and in vivo RvD2 exposure showed a trend toward decreased PGD2 (Figure 4F). In addition to these effects of RvD2 on the concentrations of LXA4 and PGD2 in BAL fluid, RvD2 had a significant and selective effect on Eos activation (Figure 4G). Flow cytometric analyses showed higher expression of the activation marker CD11b on CD101high Eos than CD101low Eos at Day 17, and in vivo RvD2 exposure significantly decreased CD11b expression on CD101high Eos (Figure 4G). CD101high Eos also had significant increases in the expression of the cell activation marker CD69 at Day 17; however, RvD2 exposure had no effect on CD69 expression (Figure E6B). The type 2 inflammation–associated proteins chitinase-like 3 and 4 (YM1/2) had higher expression levels in CD101high Eos than in CD101low Eos at Day 17 (Figure E6C). RvD2 gave trends for decreased concentrations of chitinase-like 3 (Chi3l3) in BAL at this time point (Figures E6C and E6D).
Given the changes in PGD2 concentrations in BAL fluid in the HDM-challenged mice, we next investigated the expression of the PGD2 receptor (CRTH2) on Eos. This activating receptor was present on lung Eos with higher concentrations on the CD101high Eos than on CD101low Eos without significant changes with in vivo RvD2 exposure (Figure 4H). To test the impact of RvD2 on the functional response of the CD101low and CD101high Eos, we determined PGD2-evoked degranulation ex vivo by monitoring EPO release (see Methods). Flow-sorted cells were incubated with either Veh, RvD2 (10 nM), PGD2 (10 nM), or PGD2 and RvD2. CD101high Eos selectively responded to PGD2 with increased EPO release (Figure 4I), and RvD2 exposure lowered PGD2-stimulated EPO release (P = 0.058) (Figure 4I). Together, these results indicate that RvD2 has direct actions on CD101low Eos and CD101high Eos with changes in gene expression and altered cell activation and functional responses to local environmental proinflammatory stimuli.
RvD2 Regulates IL-5-induced Eos Responses
Given the potent proresolving actions of RvD2 and the pivotal role of IL-5 in eosinophilic lung inflammation, we next determined the impact of RvD2 on IL-5 concentrations in the BAL fluid of HDM-challenged mice. After HDM sensitization and challenge (Figure 3A), IL-5 concentrations in BAL fluid were significantly higher at Day 17 than at baseline (Day 0), and RvD2 exposure significantly decreased IL-5 concentrations at Day 17 (Figure 5A). Flow cytometric analyses revealed that RvD2 decreased numbers of type 2 innate lymphoid cells (ILC2) and IL-5–producing CD4+ T cells (Figures E7A–E7D). Peripheral blood CD101low Eos, which were elevated at Day 17, were also markedly decreased by RvD2 (Figure 5B). In addition to regulating IL-5, RvD2 also significantly decreased the expression of integrin-α4 on lung CD101low Eos at Day 17 (Figure E7E), which is pivotal to lung Eos recruitment (27). Of interest, mouse CD101low and CD101high lung Eos expressed IL-5 receptor-α (IL-5Rα, also known as CD125) at Day 17, with slightly higher concentrations in CD101high Eos (Figure 5C). In addition, in vivo RvD2 exposure decreased IL-5Rα expression on CD101high Eos (Figure 5C).
Figure 5.
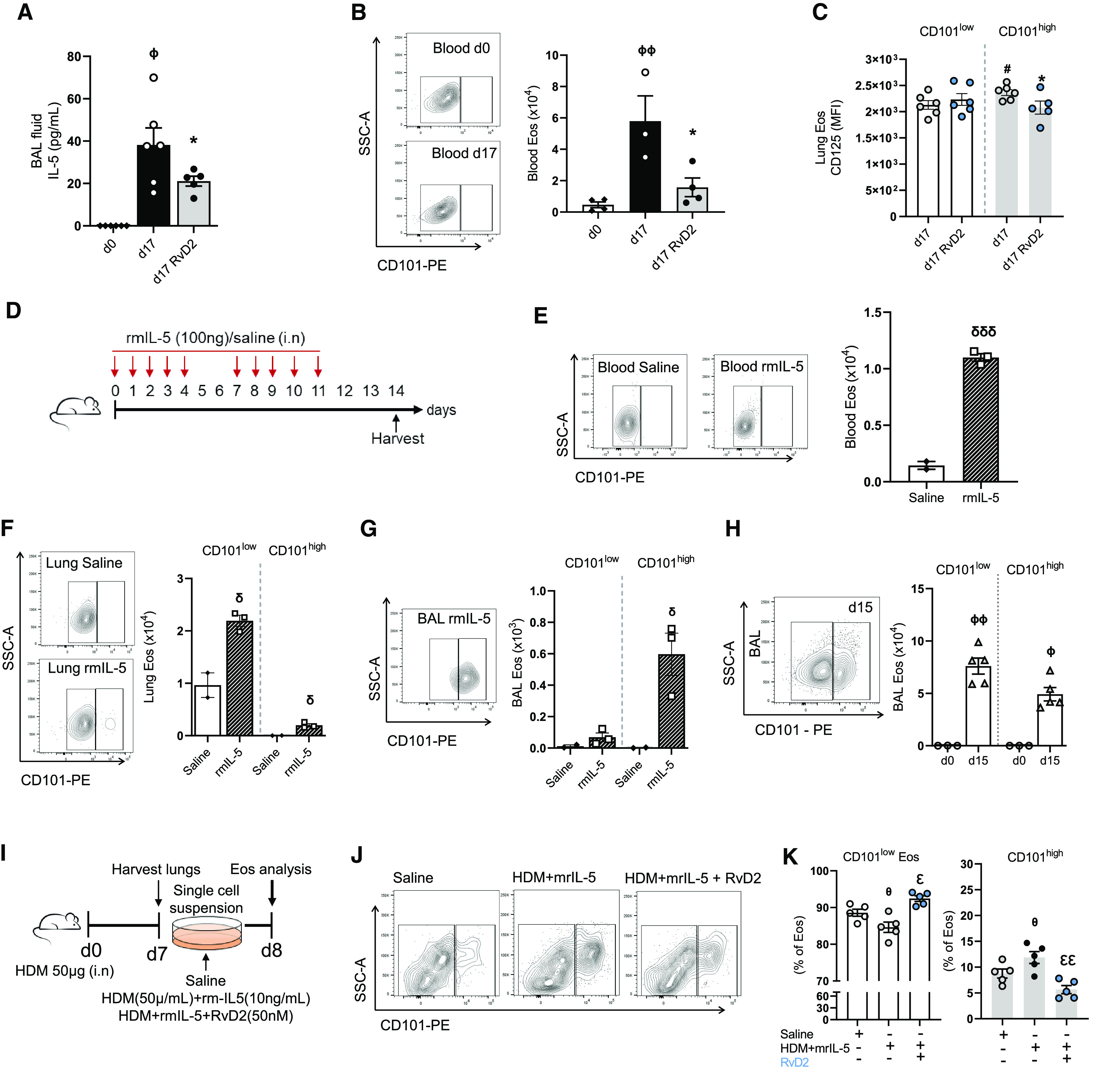
RvD2 regulates IL-5–induced Eos responses. (A) IL-5 concentrations in BAL fluid by cytometric bead array. (B) Blood Eos numbers at Days 0 and 17. (C) CD125 expression. (D) Timeline of recombinant mouse IL-5 (rmIL-5) exposure; quantification of Eos in E–G. (E) Blood Eos numbers at Day 14 in D. (F and G) Lung and BAL CD101low and CD101high Eos, respectively. (H) Number of Eos in BAL at Day 15 of the HDM protocol (Figure 1A). (I) Timeline of lung cells ex vivo stimulation (see Methods). (J) Flow cytometry plots of CD101low Eos after ex vivo stimulation. (K) Percentage of CD101low and CD101high Eos after ex vivo stimulation. ϕP < 0.05 and ϕϕP < 0.01 compared with Day 0 in A, B, and H. *P < 0.05 compared with Day 17 in A–C (within CD101high). #P < 0.05 compared with CD101low d17 in C. δP < 0.05 and δδδP < 0.001 compared with saline within the same cell type. θP < 0.05 compared with saline and εP < 0.05 and εεP < 0.01 compared with HDM and rmIL-5 in K. Comparisons between two groups were performed by unpaired t test (C and E–H). Comparisons between more than two groups were performed by one-way ANOVA and Tukey test for multiple comparisons (A, B, and K). Bars represent mean ± SEM.
To further investigate the role of IL-5 on Eos, mice were exposed i.n. to recombinant mouse IL-5 (rmIL-5) (Figure 5D). Repetitive airway exposure to rmIL-5 (100 ng i.n.) over 2 weeks on protocol Days 0–4 and 7–11 led to a significant increase in blood Eos on protocol Day 14 relative to saline-exposed mice (Figure 5E). CD101high Eos were not detected in peripheral blood, despite the substantial exogenous IL-5 exposure to the mouse airways (Figure 5E). CD101low Eos and CD101high Eos were present in the lungs with significantly higher numbers with rmIL-5 exposure than with the saline control (Figure 5F). Lung CD101high Eos were not detected in mice exposed to saline control (Figure 5F). In addition, rmIL-5 significantly increased CD101high Eos in BAL compared with saline control mice (Figure 5G). CD101low Eos were present only in very low numbers in BAL but were increased by rmIL-5 (Figure 5G). Together, these findings point to IL-5 as a pivotal mediator for Eos recruitment and maturation from lung CD101low Eos to CD101high Eos.
Because airway administration of rmIL-5 recruited CD101low Eos to BAL (Figure 5G), we next investigated the possibility that CD101low Eos were present in BAL at earlier time points than Day 17 in the HDM model, when essentially all Eos in BAL were CD101high (Figure 1E). When BAL cells were collected at Day 15, 1 day after the last HDM challenge, both CD101low Eos and CD101high Eos were present (Figure 5H). At this time point, only CD101low Eos were detected in blood, and their numbers were significantly increased compared with Day 0 (Figure E7F). In lungs, CD101low Eos predominated, and low numbers of CD101high Eos had become detectable at Day 15 (Figure E7G). Together, the data from Day 15 and Day 17 in the HDM sensitization and challenge model indicate that CD101low Eos were recruited to the lungs and transitioned to CD101high Eos in the lungs, driven in part by IL-5, during the allergic inflammatory response.
We next addressed the role of IL-5 in the maturation of Eos immunophenotype from CD101low Eos to CD101high Eos. Single-cell suspensions were prepared from lungs after HDM sensitization at protocol Day 7 (Figure 5I). At this point, only CD101low Eos were present in the lungs (Figure E7H). Total lung cells were plated in complete media with either saline (control); HDM and rmIL-5 (challenge/stimulation); or HDM, rmIL-5, and RvD2 (Figure 5I; see Methods). Cells were incubated for 24 hours, and then CD101 expression on Eos was assessed by flow cytometry. Sample preparation and saline exposure during the incubation resulted in ∼10% presence of CD101high Eos (Figures 5J and 5K). Challenge with HDM and rmIL-5 led to a significant increase in CD101high Eos relative to control incubations. With RvD2 addition to the challenge, the percentage of CD101high Eos significantly decreased to approximate the control results (Figures 5J and 5K). To confirm that IL-5 played a critical role in CD101 expression and Eos maturation, BMEos were obtained (see Methods). BMEos were cultured in the presence of rmIL-5 (10 ng/ml) starting at Day 4. From Day 6 to Day 12, ∼14–30% of immature Eos expressed CD101 (Figure E7I). Eos were fully mature at Day 14, and, at this time interval, most of the cells (∼70%) had high expression of CD101 (Figure E7I). Together, these findings indicate that IL-5 is pivotal to CD101low Eos recruitment and maturation to CD101high Eos and that RvD2 is a potent regulator of IL-5 production and its downstream actions on Eos maturation.
Discussion
Eos are found in multiple organs at baseline, and their numbers significantly expand in the lungs during type 2 inflammation, such as in asthma (reviewed in [2, 28]). The significant decrease of lung Eos from Day 15 to 17 indicates that there are endogenous regulatory mechanisms triggered early after allergen challenge. On the basis of a recent report linking the RvD2 receptor to asthma risk (15), this SPM was administered after allergen sensitization and challenge to ascertain proresolving Eos responses. The administration of exogenous RvD2 propelled the endogenous resolving mechanisms with further decreases in BAL Eos. Different subsets or activation states of Eos have recently been identified that can carry select functions, depending on the inflammatory environment (2). Here, we used a mouse model of self-limited allergic lung inflammation to investigate the spatiotemporal and functional regulation of two Eos subsets during the onset and resolution of type 2 inflammation. CD101 expression distinguished the two mouse Eos subsets. CD101low Eos were located in a lung vascular niche at baseline, infiltrated the lung in response to allergen challenge, and then converted to CD101high Eos. These Eos subsets had distinct transcriptional signatures, with CD101high Eos demonstrating an activation phenotype and a proinflammatory response to PGD2. RvD2 selectively regulated lung Eos subsets, specifically decreasing CD101low Eos recruitment and their activation to CD101high Eos. In addition, RvD2 decreased IL-5 concentrations in BAL, which reduced the number of CD101high Eos and controlled their phenotype to that of a less activated state.
CD101 is a regulatory monocyte and lymphocyte marker (29); however, its function in Eos is uncertain. Expression of CD101 differentiates lung Eos into phenotypically and functionally distinct subsets, with low expression levels in tissue-resident Eos and high expression levels in infiltrating Eos during lung inflammation (7). Here, after HDM sensitization and challenge, CD101low Eos in the lung vasculature expanded and upregulated the expression of leukocyte migration, chemotaxis, and cell–cell adhesion-related genes in response to allergen. In the lung tissue and airways, CD101low Eos transitioned to an activated state with high expression of CD101. The CD101high Eos were predominantly located in BAL, colocalizing with HDM in the airways. In addition to increased CD101 expression, the CD101high Eos displayed some distinguishing morphologic features in the shape of their nucleus and apparent granule contents, suggesting that CD101 expression marked an activation phenotype for mouse Eos. In addition, CD101high Eos had upregulated gene expression of cell activation signaling, including intracellular ERK1 and ERK2 transduction pathways, suggesting that these proinflammatory pathways for CD101high Eos govern their cell activation state. Although useful in identifying distinct subsets of mouse Eos, further experiments will be needed to provide further clarity on CD101’s function in mouse Eos.
Eos have inflammatory roles in different organs and disease models (2). There is a growing appreciation that during inflammation, Eos can also exert proresolving roles by producing SPMs, including RvD2, via lipoxygenase-mediated enzymatic conversion of DHA (reviewed in [9, 13]). Peripheral blood Eos isolated from healthy human subjects produce, upon DHA exposure, 14-HDHA, 17-HDHA, and SPMs, including RvD2 (20). Here, the mouse Eos subsets produced differing concentrations of 14-HDHA and RvD2 when incubated with DHA. There was lower RvD2 production by CD101high Eos than by CD101low Eos. In contrast, expression of the RvD2 receptor, DRV2, was higher on the CD101high Eos, suggesting potential direct roles for RvD2-DRV2 signaling in regulating mouse Eos transitions to a more activated state (from CD101low to CD101high expression).
Allergen challenge induced expression of the mouse ortholog of 15-lipoxygenase (i.e., 12/15-lipoxygenase) in CD101low Eos, and it was even further upregulated in CD101high Eos. Of interest, the CD101high Eos had relatively lower concentrations of 14-HDHA, despite the increased 12/15-lipoxygenase gene expression, suggesting a discordance with decreased mouse 12/15-lipoxygenase activity in this Eos subset. Of note, 12/15-lipoxygenase can be inhibited by direct interaction with Ym1/2 (chitinase-like proteins), and this inhibition has been linked to increased type 2 inflammatory cytokines in a related preclinical mouse model (30). Ym1/2 expression was higher in CD101high Eos and is one potential explanation for the discrepancy between 12/15-lipoxygenase gene expression and enzymatic activity. Ym1 has been closely related to lung type 2 immune responses (30–32). In the present experiments, in vivo RvD2 exposure significantly decreased chi3l3 (Ym1 gene) expression in both CD101low and CD101high Eos, demonstrating a new counterregulatory mechanism for SPMs.
There are several mediators, including cytokines and chemokines derived from structural cells (epithelial cells and endothelial cells) and immune cells (innate lymphoid cells and CD4+ T cells), that are involved in Eos recruitment, retention, and function during inflammation (2, 6). Here, consistent with its role in Eos recruitment from the bloodstream into lung tissue, CCL11 (eotaxin-1) concentrations were increased in the lungs after repeated allergen challenge. Among the cytokines, IL-5 plays a critical role in the differentiation, maturation, activation, and survival of Eos during the type 2 immune response in asthma (33). The use of anti–IL-5-based therapies in some patients with severe asthma can reduce peripheral blood Eos numbers in conjunction with clinical efficacy (34). Here, BAL IL-5 concentrations and Eos numbers were increased during allergic lung inflammation. Direct IL-5 administration into the airways increased Eos numbers and induced the transition from CD101low Eos to CD101high Eos. Local administration of RvD2 into the airways decreased the allergen-induced BAL IL-5 concentrations more rapidly than vehicle, in part by decreasing IL-5–producing CD4+ T-helper cell type 2 and type 2 innate lymphoid cell numbers. RvD2 potently regulated IL-5–mediated direct actions on lung Eos, including the CD101low Eos transition to the further activated state in CD101high Eos. Collectively, these findings have uncovered direct and indirect mechanisms for RvD2-mediated regulation of the clinically relevant target IL-5 that promoted the resolution of mouse lung eosinophilia and Eos activation.
In summary, phenotypically and functionally distinct Eos subsets, namely CD101low and CD101high Eos, were spatiotemporally regulated in a mouse model of self-limited airway type 2 inflammation. RvD2 signaling mediated proresolving actions that accelerated resolution by decreasing allergen-induced lung Eos numbers in a DRV2 receptor-dependent manner. RvD2 regulated Eos phenotype and function by at least two distinct mechanisms: decreasing IL-5–dependent recruitment of CD101low Eos into the airways and decreasing the transition of CD101low Eos to CD101high Eos. These findings support the notion that Eos subsets contribute to the resolution of inflammation via the biosynthesis and release of proresolving mediators. Our findings indicate that mouse Eos are a heterogeneous pool of cells with distinct activation states and spatiotemporal regulation during the resolution of type 2 inflammation and that RvD2 is a potent proresolving mediator for inflammatory Eos recruitment and activation.
Acknowledgments
Acknowledgment
The authors thank Guangli Zhu for technical assistance.
Footnotes
Supported by funds from National Institutes of Health grants R01HL122531 (B.D.L.), P01GM0955467 (B.D.L., A.E.S., and C.N.S.), and R35GM139430 (C.N.S.).
Author Contributions: T.R.B., H.Y.P., L.P.T., A.E.S., R.M.R., T.B.L., and C.N.S. designed and performed experiments, analyzed data, performed statistical analyses, and reviewed and edited the manuscript. T.R.B. and J.N. wrote the initial manuscript draft. B.D.L. conceived the study, designed the experiments, analyzed the data, and reviewed and edited the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2023-0121OC on August 8, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Loffredo LF, Coden ME, Jeong BM, Walker MT, Anekalla KR, Doan TC, et al. Eosinophil accumulation in postnatal lung is specific to the primary septation phase of development. Sci Rep . 2020;10:4425. doi: 10.1038/s41598-020-61420-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wechsler ME, Munitz A, Ackerman SJ, Drake MG, Jackson DJ, Wardlaw AJ, et al. Eosinophils in health and disease: a state-of-the-art review. Mayo Clin Proc . 2021;96:2694–2707. doi: 10.1016/j.mayocp.2021.04.025. [DOI] [PubMed] [Google Scholar]
- 3. Fanta CH. Asthma. N Engl J Med . 2009;360:1002–1014. doi: 10.1056/NEJMra0804579. [DOI] [PubMed] [Google Scholar]
- 4. Jacobsen EA, Jackson DJ, Heffler E, Mathur SK, Bredenoord AJ, Pavord ID, et al. Eosinophil knockout humans: uncovering the role of eosinophils through eosinophil-directed biological therapies. Annu Rev Immunol . 2021;39:719–757. doi: 10.1146/annurev-immunol-093019-125918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kay AB. Paul Ehrlich and the early history of granulocytes. Microbiol Spectr . 2016;4 doi: 10.1128/microbiolspec.MCHD-0032-2016. [DOI] [PubMed] [Google Scholar]
- 6. Abdala-Valencia H, Coden ME, Chiarella SE, Jacobsen EA, Bochner BS, Lee JJ, et al. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J Leukoc Biol . 2018;104:95–108. doi: 10.1002/JLB.1MR1117-442RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest . 2016;126:3279–3295. doi: 10.1172/JCI85664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yi S, Zhai J, Niu R, Zhu G, Wang M, Liu J, et al. Eosinophil recruitment is dynamically regulated by interplay among lung dendritic cell subsets after allergen challenge. Nat Commun . 2018;9:3879. doi: 10.1038/s41467-018-06316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Isobe Y, Kato T, Arita M. Emerging roles of eosinophils and eosinophil-derived lipid mediators in the resolution of inflammation. Front Immunol . 2012;3:270. doi: 10.3389/fimmu.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farne HA, Wilson A, Milan S, Banchoff E, Yang F, Powell CV. Anti-IL-5 therapies for asthma. Cochrane Database Syst Rev . 2022;7:CD010834. doi: 10.1002/14651858.CD010834.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corren J. Inhibition of interleukin-5 for the treatment of eosinophilic diseases. Discov Med . 2012;13:305–312. [PubMed] [Google Scholar]
- 12. Kelly EA, Esnault S, Liu LY, Evans MD, Johansson MW, Mathur S, et al. Mepolizumab attenuates airway eosinophil numbers, but not their functional phenotype, in asthma. Am J Respir Crit Care Med . 2017;196:1385–1395. doi: 10.1164/rccm.201611-2234OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krishnamoorthy N, Abdulnour RE, Walker KH, Engstrom BD, Levy BD. Specialized proresolving mediators in innate and adaptive immune responses in airway diseases. Physiol Rev . 2018;98:1335–1370. doi: 10.1152/physrev.00026.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, et al. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med . 2002;196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valette K, Li Z, Bon-Baret V, Chignon A, Bérubé JC, Eslami A, et al. Prioritization of candidate causal genes for asthma in susceptibility loci derived from UK Biobank. Commun Biol . 2021;4:700. doi: 10.1038/s42003-021-02227-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Serhan CN, Hirsch U, Palmblad J, Samuelsson B. Formation of lipoxin A by granulocytes from eosinophilic donors. FEBS Lett . 1987;217:242–246. doi: 10.1016/0014-5793(87)80671-3. [DOI] [PubMed] [Google Scholar]
- 17. Arita M. Eosinophil polyunsaturated fatty acid metabolism and its potential control of inflammation and allergy. Allergol Int . 2016;65:S2–S5. doi: 10.1016/j.alit.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 18. Levy BD, Bonnans C, Silverman ES, Palmer LJ, Marigowda G, Israel E, Severe Asthma Research Program, National Heart, Lung, and Blood Institute Diminished lipoxin biosynthesis in severe asthma. Am J Respir Crit Care Med . 2005;172:824–830. doi: 10.1164/rccm.200410-1413OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levy BD, Kohli P, Gotlinger K, Haworth O, Hong S, Kazani S, et al. Protectin D1 is generated in asthma and dampens airway inflammation and hyperresponsiveness. J Immunol . 2007;178:496–502. doi: 10.4049/jimmunol.178.1.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyata J, Fukunaga K, Iwamoto R, Isobe Y, Niimi K, Takamiya R, et al. Dysregulated synthesis of protectin D1 in eosinophils from patients with severe asthma. J Allergy Clin Immunol . 2013;131:353–360.e1-2. doi: 10.1016/j.jaci.2012.07.048. [DOI] [PubMed] [Google Scholar]
- 21. Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol . 2008;181:4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdala Valencia H, Loffredo LF, Misharin AV, Berdnikovs S. Phenotypic plasticity and targeting of Siglec-Fhigh CD11clow eosinophils to the airway in a murine model of asthma. Allergy . 2016;71:267–271. doi: 10.1111/all.12776. [DOI] [PubMed] [Google Scholar]
- 23. Zhu C, Weng QY, Zhou LR, Cao C, Li F, Wu YF, et al. Homeostatic and early-recruited CD101- eosinophils suppress endotoxin-induced acute lung injury. Eur Respir J . 2020;56:1902354. doi: 10.1183/13993003.02354-2019. [DOI] [PubMed] [Google Scholar]
- 24. Chiang N, Dalli J, Colas RA, Serhan CN. Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J Exp Med . 2015;212:1203–1217. doi: 10.1084/jem.20150225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bannenberg GL, Chiang N, Ariel A, Arita M, Tjonahen E, Gotlinger KH, et al. Molecular circuits of resolution: formation and actions of resolvins and protectins. J Immunol . 2005;174:4345–4355. doi: 10.4049/jimmunol.174.7.4345. [DOI] [PubMed] [Google Scholar]
- 26. Levy BD, Clish CB, Schmidt B, Gronert K, Serhan CN. Lipid mediator class switching during acute inflammation: signals in resolution. Nat Immunol . 2001;2:612–619. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 27. Barthel SR, Johansson MW, McNamee DM, Mosher DF. Roles of integrin activation in eosinophil function and the eosinophilic inflammation of asthma. J Leukoc Biol . 2008;83:1–12. doi: 10.1189/jlb.0607344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holgate ST, Wenzel S, Postma DS, Weiss ST, Renz H, Sly PD. Asthma. Nat Rev Dis Primers . 2015;1:15025. doi: 10.1038/nrdp.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schey R, Dornhoff H, Baier JL, Purtak M, Opoka R, Koller AK, et al. CD101 inhibits the expansion of colitogenic T cells. Mucosal Immunol . 2016;9:1205–1217. doi: 10.1038/mi.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai Y, Kumar RK, Zhou J, Foster PS, Webb DC. Ym1/2 promotes Th2 cytokine expression by inhibiting 12/15(S)-lipoxygenase: identification of a novel pathway for regulating allergic inflammation. J Immunol . 2009;182:5393–5399. doi: 10.4049/jimmunol.0803874. [DOI] [PubMed] [Google Scholar]
- 31. Draijer C, Robbe P, Boorsma CE, Hylkema MN, Melgert BN. Dual role of YM1+ M2 macrophages in allergic lung inflammation. Sci Rep . 2018;8:5105. doi: 10.1038/s41598-018-23269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass CK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem . 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 33. Greenfeder S, Umland SP, Cuss FM, Chapman RW, Egan RW. Th2 cytokines and asthma. The role of interleukin-5 in allergic eosinophilic disease. Respir Res . 2001;2:71–79. doi: 10.1186/rr41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leckie MJ. Anti-interleukin-5 monoclonal antibodies: preclinical and clinical evidence in asthma models. Am J Respir Med. 2003;2:245–259. doi: 10.1007/BF03256653. [DOI] [PubMed] [Google Scholar]