Abstract
Speciation in enterobacteria involved horizontal gene transfer. Therefore, analysis of genes acquired by horizontal transfer that are present in one species but not its close relatives is expected to give insights into how new bacterial species were formed. In this study we characterize iroN, a gene located downstream of the iroBC operon in the iroA locus of Salmonella enterica serotype Typhi. Like iroBC, the iroN gene is present in all phylogenetic lineages of S. enterica but is absent from closely related species such as Salmonella bongori or Escherichia coli. Comparison of the deduced amino acid sequence of iroN with other proteins suggested that this gene encodes an outer membrane siderophore receptor protein. Mutational analysis in S. enterica and expression in E. coli identified a 78-kDa outer membrane protein as the iroN gene product. When introduced into an E. coli fepA cir fiu aroB mutant on a cosmid, iroN mediated utilization of structurally related catecholate siderophores, including N-(2,3-dihydroxybenzoyl)-l-serine, myxochelin A, benzaldehyde-2,3-dihydroxybenzhydrazone, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine amide, and enterochelin. These results suggest that the iroA locus functions in iron acquisition in S. enterica.
The genera Salmonella and Escherichia diverged from a common ancestor some 100 million to 160 million years ago (26). The lineage of the genus Salmonella subsequently split into two species, S. enterica and S. bongori (22, 28). During formation of these species a large amount of DNA was acquired by plasmid- or phage-mediated horizontal gene transfer (3). As a result of horizontal transfer, more than 10% of the S. enterica serotype Typhimurium genome consists of genetic material that is not present in Escherichia coli (25, 30). To fully comprehend the biological characteristics that distinguish S. enterica from closely related species such as E. coli and S. bongori, it is necessary to study the functions of proteins that are encoded by genetic material that was acquired by way of horizontal transfer.
Genetic material that is absent from closely related bacteria but is present in all phylogenetic lineages of S. enterica was likely received during horizontal transfer events that contributed to the formation of this species. Two genetic regions on the S. enterica chromosome, Salmonella pathogenicity island 2 and the iroBC operon, indeed show this phylogenetic distribution (4, 19, 24). In a study on the distribution of iroB among 197 bacterial isolates, this gene was found to be present in all S. enterica serotypes tested but absent from S. bongori serotypes and from 15 other bacterial species tested (4). The phylogenetic distribution thus suggests that the gene products encoded in the iroA locus confer properties that set S. enterica apart from other bacterial species. What are the characteristics that were obtained by S. enterica during acquisition of the iroA locus?
The iroA locus was first described in S. enterica serotype Typhimurium based on its iron-regulated expression (12). An insertion that created a transcriptional fusion between lacZ and the iroA locus was mapped close to the tct locus (13), an area of the S. enterica serotype Typhimurium chromosome that is not present in E. coli (31). The first genes of the iroA locus, designated iroBC, were identified in S. enterica serotype Typhi during a genetic screen for genes that are regulated by the iron response regulator Fur (5). Regulation by Fur results in expression of iroBC under iron-limited growth conditions. In contrast, during growth under iron sufficiency expression is prevented by binding of the Fur-Fe2+ repressor complex to a Fur DNA binding site in the iroB promoter region. In addition to iroBC, the Fur repressor protein controls expression of some 28 genes in S. enterica, including those that function in iron acquisition and some genes involved in defense against oxidative stress (38–40). However, the iroBC gene products show homology to proteins that have so far not been associated with iron uptake or defense against oxidative stress in other bacteria. IroB shows homology with bacterial glycosyltransferases, and IroC is a member of the ATP binding cassette (ABC) family of transport proteins (5). Interestingly, IroC has little homology to bacterial ABC transport proteins involved in the import of siderophores but rather shows strong homology to ABC export proteins of the eukaryotic multidrug resistance family. To obtain further clues about the function of genes encoded in the iroA locus, we analyzed iroN, an open reading frame located downstream of the iroBC operon.
MATERIALS AND METHODS
Bacterial strains, media, growth conditions, and outer membrane preparations.
A collection of Salmonella serotypes representing S. bongori and six subspecies of S. enterica has been described by Reeves et al. (28). All other bacterial strains used in this study are listed in Table 1. All bacteria were routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB plates. Antibiotics, when required, were included in the culture medium or plates at the following concentrations: kanamycin, 100 mg/liter; chloramphenicol, 30 mg/liter; and carbenicillin, 100 mg/liter. To create iron-limiting or iron-sufficient growth conditions, 0.2 mM 2,2′-dipyridyl or 0.04 mM FeSO4, respectively, was added. Desferal (desferrioxamine B) was purchased from Ciba Geigy (Basel, Switzerland). N-(2,3-Dihydroxybenzoyl)-l-serine (DBS) and benzaldehyde-2,3-dihydroxybenzhydrazone were synthesized and kindly provided by L. Heinisch, Hans-Knölle Institut, Jena, Germany. The myxochelin derivatives 2-N,6-N-bis(2,3-dihydroxybenzoyl) lysine (9), 2-N,6-N-bis(2,3-dihydroxybenzoyl) lysine amide (37), myxochelin A (21), myxochelin B, and myxochelin C (36) were synthesized in the l or d configuration and kindly provided by W. Trowitzsch-Kienast and H. D. Ambrosi, Technische Fachhochschule, Berlin, Germany. Cross-feeding with bacterial supernatants and utilization of siderophores was detected by an agar diffusion assay (29). The strain to be tested was poured in 3 ml of 2% Noble agar onto a nutrient broth-dipyridyl (NBD) agar plate. Filter paper disks impregnated with ferrioxamine B (5 μl of a 1-mg/ml solution of Desferal in 0.1 M FeCl3), DBS (5 μl of a 1-mg/ml solution), benzaldehyde-2,3-dihydroxybenzhydrazone (5 μl of a 1-mg/ml solution), or myxochelin derivatives (5 μl of a 1-mg/ml solution) were laid onto the top agar, and after incubation overnight at 37°C, growth stimulation around the filter disk was recorded. Growth promotion in broth culture was determined in NBD supplemented with a 500× siderophore stock solution (1 mg/ml). Bacterial outer membranes were isolated as previously described (17). Outer membrane proteins were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were visualized by Coomassie blue staining.
TABLE 1.
Bacterial strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| E. coli K-12 | ||
| DH5α | endA1hsdR17 (rK−mK−) supE44thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80 dlac Δ(lacZM15)] | 15 |
| S17-1 λpir | prp thi recA hsdR; chromosomal RP4-2 (Tn1::ISR1 tet::Mu Km::Tn7); λpir | 33 |
| AB2847 | aroB malT tsx thi | 17 |
| H5058 | aroB malT tsx thi cir fiu fepA | K. Hantke |
| S. enterica serotype Typhimurium | ||
| 14028 | Wild type (isolated from bovine septicemia) | American Type Culture Collection |
| CL1509 | 14028 aroA::Tn10 | 8 |
| AR1258 | 14028 entB::MudJ | 38 |
| IR715 | 14028 Nalr | 35 |
| AJB20 | 14028 NalriroBC::Kmr | 5 |
| AIR49 | 14028 NalriroBC::KmraroA::Tn10 | This study |
| AJB52 | 14028 NalriroN::pGP704 | This study |
| AJB64 | 14028 NalriroN::pGP704 aroA::Tn10 | This study |
| S. enterica serotype Typhi | ||
| AJB70 | Wild type (clinical isolate, San Diego, Calif.) | 5 |
| TY21a | galE viaB rpoS | 14 |
| AJB22 | TY21a Nalr | 5 |
| AJB54 | TY21a NalriroN::pGP704 | This study |
Recombinant DNA techniques.
Plasmid DNA was isolated by using ion-exchange columns from Qiagen. Standard methods were used for restriction endonuclease analyses, ligation, and transformation of plasmid DNA (2). Sequencing was performed with an ALF automated sequencer (Pharmacia).
Suicide vector constructs and isolation of mutants.
For mutational analysis of iroN, a 0.4-kb XbaI-KpnI fragment of pTY961 containing an internal part of the iroN open reading frame was introduced into the suicide vector pGP704 (20) to give rise to pTY966. Plasmid pTY966 was conjugated into S. enterica serotype Typhimurium IR715 and S. enterica serotype Typhi AJB22. Exconjugants were designated AJB52 and AJB54, respectively. Strains AJB64 and AIR49 were generated by P22 transduction of aroA::Tn10 from S. enterica serotype Typhimurium CL1509 into strains AJB52 and AJB20, respectively. E. coli S17-1 λpir was used for propagation of all suicide vector constructs and as a donor for introduction of these constructs into IR715 or AJB22 by conjugation. Chromosomal DNA of mutants was routinely tested by Southern hybridization with suitable DNA probes to confirm mutational inactivation of the gene of interest.
Southern hybridization.
The inserts of plasmids pTY961 and pTY911 (5) were used to generate DNA probes specific for iroN and iroCDE, respectively. Chromosomal DNA was isolated as recently described (2). Chromosomal DNA of strains shown in Fig. 4 was restricted with EcoRI, and the fragments were separated on a 0.5% agarose gel. Southern transfer of DNA onto a nylon membrane was performed as previously described (2). Hybridization was performed at 65°C in solutions without formamide. Two 15-min washes were performed under nonstringent conditions at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS. Hybrids were detected by using a labeling and detection kit (nonradioactive) from Boehringer Mannheim.
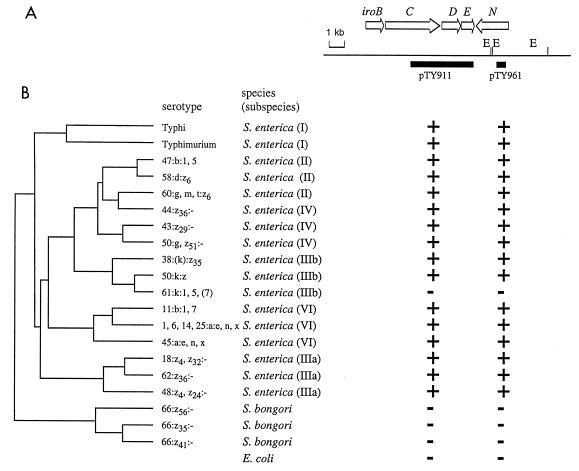
FIG. 4.
Phylogenetic distribution of genes of the iroA locus. The phylogenetic tree on the left was established by Reeves and coworkers (28). (A) Restriction map of the region from S. enterica serotype Typhi AJB70 (E, EcoRI). Positions of genes (arrows) identified in the iroA locus and of DNA fragments used as probes (black bars) are indicated. (B) Results of hybridization with these DNA probes. +, hybridization signal; −, no hybridization signal.
Computer analysis.
The nucleotide sequences were compared to SWISS-PROT, PIR, and GenPept at the National Center for Biotechnology Information by using the program blastX and to GenBank and EMBL by using the program blastN (1). Multiple alignments were performed with the program CLUSTAL, which is part of the program package PCGENE.
Nucleotide sequence accession number.
The sequence reported (Fig. 1) has been deposited at GenBank under accession no. U97227.
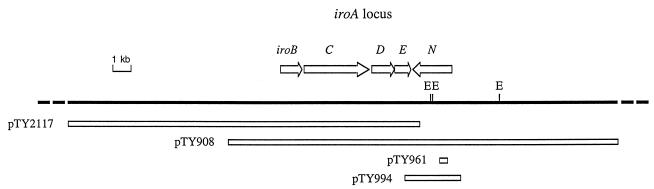
FIG. 1.
Restriction map of a DNA region located at about four centisomes on the S. enterica serotype Typhi chromosome. Positions and sizes of inserts carried in cosmids (pTY908 and pTY2117) or plasmids depicted were determined previously (5). Arrows above the map indicate positions and orientations of open reading frames identified by sequence analysis. E, EcoRI.
RESULTS
Sequence analysis of a DNA region located downstream of iroBC.
The nucleotide sequence of a 4,837-bp DNA region of S. enterica serotype Typhi AJB70 that is located downstream of iroBC was determined. Directly downstream of iroC were two open reading frames transcribed in the same orientation. These open reading frames were designated iroDE (Fig. 1). The close proximity of the open reading frames and the lack of transcriptional terminators suggest that the iroBCDE cluster forms an operon. The deduced amino acid sequence of iroD showed 28% sequence identity with Fes, the E. coli enterochelin esterase (27). The amino-terminal 169 amino acids of IroE displayed 38% identity with the deduced amino acid sequence of an open reading frame located downstream of pfeA, the enterochelin receptor gene of Pseudomonas aeruginosa (data not shown) (10). Downstream of iroDE was a third open reading frame transcribed in the opposite orientation. This open reading frame, termed iroN, encoded a polypeptide of 727 amino acids with a calculated molecular mass of 79.5 kDa. Cleavage of a predicted N-terminal signal sequence of 25 amino acids would yield a mature protein with a calculated molecular mass of 76.8 kDa. The region between iroE and iroN contained a putative transcriptional terminator (stem bp 2261 to 2270; loop bp 2271 to 2273; stem bp 2274 to 2283 of the GenBank sequence). A putative Fur-DNA binding site (4576-GATAATTATTATCATTAGC-4558) that matches the E. coli consensus sequence (34) in 16 of 19 bases was located 86 bp upstream of the iroN start codon. The G+C content of the entire 10,837-bp DNA region containing the iroBCDE and iroN genes was 55%, which is slightly higher than the 52% average G+C content characteristic of S. enterica.
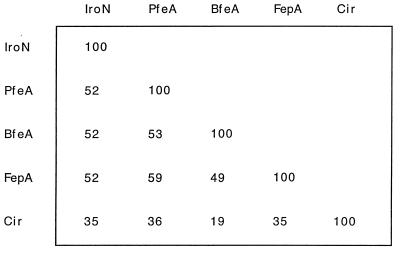
Sequence homology identified IroN as a member of the family of TonB-dependent outer membrane receptor proteins. The highest degree of sequence identity was found with outer membrane receptor proteins that mediate uptake of the siderophore enterochelin (Fig. 2). Multiple alignment between IroN and enterochelin receptors from E. coli (FepA) (23), Bordetalla pertussis (BfeA) (6), and P. aeruginosa (PfeA) (10) showed that 36% of the amino acids were identical in all four receptors (Fig. 3). The conservation of amino acid sequences was strongest between amino acids 77 and 171 of IroN, where all four receptors had 85% identical amino acids.
FIG. 2.
Percentage sequence identity determined by pairwise alignment of amino acid sequences from IroN, PfeA, BfeA, FepA, and Cir with the program CLUSTAL.
FIG. 3.
Multiple sequence alignment of IroN, PfeA, FepA, and BfeA with the program CLUSTAL. Dashes represent gaps introduced by the program to improve the alignment; identical amino acids are indicated by asterisks; dots indicate amino acids with similar properties.
Phylogenetic distribution of iroN.
To obtain information on the distribution of iroN, we used 20 strains representing all phylogenetic lineages of the genus Salmonella, including S. bongori and S. enterica subspecies I, II, IIIa, IIIb, IV, and VI. The phylogenetic relationship among these strains has previously been established by multilocus enzyme electrophoresis (28). Analysis of the distribution of iroN among these strains can therefore identify the branch of the phylogenetic tree in which this gene was acquired. To compare the distribution of iroN with that of other genes of the iroA locus DNA, probes specific for iroCDE (pTY911), and iroN (pTY961) were used for Southern hybridization (Fig. 4).
The distribution of iroB among 197 bacterial isolates collected in Germany revealed the presence of this gene in all S. enterica isolates tested. However, iroB was absent from 26 bacterial isolates representing 16 different species, including the closely related organisms S. bongori and E. coli (4). Like iroB, the genes iroCDE and iroN were present in all lineages of S. enterica but absent from S. bongori and E. coli, as shown by Southern blot analysis with probes pTY911 and pTY961, respectively (Fig. 4). Only one strain of S. enterica subspecies IIIb did not contain the genes iroCDE and iroN. The phylogenetic distribution of the iroA locus is most likely the result of acquisition of iroBCDE and iroN by a single horizontal transfer event in a lineage ancestral to S. enterica. Subsequent loss of the iroA locus by deletion is infrequent and was detected only in S. enterica subspecies IIIb serotype 61:k:1,5,(7). A possible mechanism for acquisition by way of horizontal transfer is suggested by the presence of a phage attachment site (atdA) located close to iroA in S. enterica (32). However, alternate scenarios that could explain the phylogenetic distribution of iroA (e.g. deletion of the iroA locus from S. bongori and E. coli) cannot at this point be ruled out.
Identification of the iroN gene product.
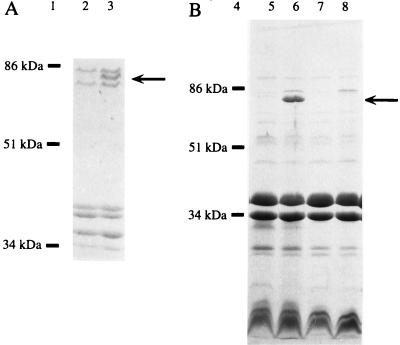
Homologies to siderophore receptors from other bacteria suggested that the iroN gene product is localized in the outer membrane. To detect IroN in outer membrane preparations, we constructed mutants of S. enterica serotype Typhimurium (AJB52 and AJB64) and S. enterica serotype Typhi (AJB54) in which the iroN open reading frame was disrupted by integration of suicide vector pTY966 via homologous recombination. S. enterica serotype Typhi and S. enterica serotype Typhimurium have been shown to contain three major iron-regulated outer membrane proteins which are 69, 78, and 83 kDa in size, respectively (7, 11). The 69- and 83-kDa proteins likely represent the S. enterica FhuA and FepA receptor proteins, respectively. Inactivation of iroN in AJB52, AJB54 (data not shown), and AJB64 (Fig. 5) resulted in loss of the 78-kDa outer membrane protein. These data therefore identify the 78-kDa outer membrane protein as the iroN gene product. The size predicted for the mature IroN protein (76.8) is in good agreement with the apparent molecular weight determined by SDS-PAGE. Furthermore, a 78-kDa protein could be detected in outer membrane preparations of strain H5058 upon introduction of a cosmid (pTY908) carrying the iroN gene of S. enterica serotype Typhi (Fig. 5), indicating that IroN also localizes to the outer membrane when expressed in E. coli. Outer membrane preparations of E. coli strains that were lacking the iroN gene [H5058 or H5058(pTY2117)] did not contain this 78-kDa protein. Sequence analysis of the insert of cosmid pTY908 revealed no open reading frames other than iroN that could encode this 78-kDa outer membrane protein (3a).
FIG. 5.
Outer membrane profiles of bacterial strains which carry the iroN gene (lanes 3, 5, and 6) or in which iroN is lacking (lanes 7 and 8) or inactivated (lane 2). The position of IroN is indicated by an arrow. Positions and sizes of bands from standard proteins are indicated (lanes 1 and 4). (A) SDS-PAGE of outer membrane preparations of S. enterica serotype Typhimurium AJB64 (lane 2) and IR715 (lane 3) grown under iron limitation. (B) Outer membrane profiles of E. coli H5058 (lanes 7 and 8) and H5058(pTY908) (lanes 5 and 6) grown in LB supplemented with 0.2 mM 2,2′-dipyridyl (lanes 6 and 8) or 0.04 mM FeSO4 (lanes 5 and 7).
The presence of a Fur DNA binding site in the iroN promoter region suggested that expression of this gene is iron regulated. To investigate iron responsiveness of IroN expression, outer membrane profiles of strain H5058(pTY908) were compared after growth in LB supplemented with either 0.04 mM FeSO4 (iron sufficiency) or 0.2 mM 2,2′-dipyridyl (iron deficiency). This analysis revealed that expression of IroN is repressed in iron-rich medium and strongly induced during growth under iron deficiency (Fig. 5). Thus, IroN is a typical iron-regulated outer membrane protein, as shown by its molecular weight, sequence homology, and iron-regulated expression.
IroN serves as a receptor for catecholate siderophores.
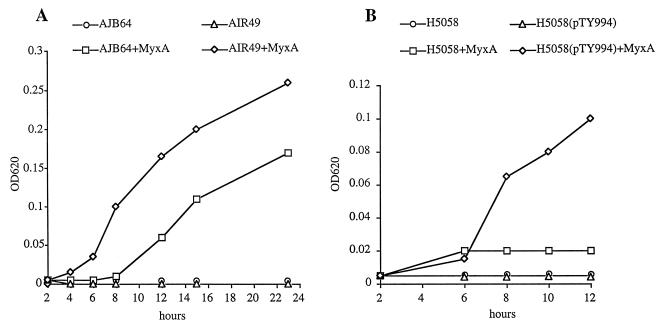
The effect of mutations in iroN on siderophore utilization was tested in S. enterica serotype Typhimurium strains carrying a mutation in aroA. S. enterica aroA mutants are unable to produce the siderophore enterochelin and therefore exhibit strongly reduced growth under iron deficiency (NBD plates). Levels of growth stimulation of strains CL1509 (aroA) and AJB64 (aroA iroN) and AIR49 (aroA iroBC) by different siderophores were compared on NBD agar plates (Table 2). In E. coli, the catecholate-type siderophore enterochelin, composed of a circular trimer of DBS, is transported across the outer membrane via the receptor protein FepA. During its transport into the cytosol, enterochelin is hydrolyzed by Fes esterase to N,N′,N"-tri-(2,3,-dihydroxybenzoyl)-dipeptide (DBS3), dimers (DBS2), and monomers of DBS. These breakdown products of enterochelin can be used as siderophores, and each is translocated across the outer membrane by any of three different E. coli outer membrane receptor proteins: FepA, Cir, and Fiu (16). Since IroN showed the highest degree of homology to enterochelin receptors of E. coli, B. pertussis, and P. aeruginosa, we investigated the ability of an S. enterica serotype Typhimurium iroN mutant to utilize catecholate-type siderophores, including enterochelin, DBS, benzaldehyde-2,3-dihydroxybenzhydrazone, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine amide, myxochelin A, myxochelin B, and myxochelin C (Fig. 6). As a control, we studied uptake of ferrioxamine B, a hydroxamate siderophore. All siderophores stimulated growth of all S. enterica strains, although the growth stimulation by benzaldehyde-2,3-dihydroxybenzhydrazone and DBS was less in the iroN aroA mutant (AJB64) than in the aroA mutant (CL1509) and the iroBC mutant (AIR49) (Table 2). When utilization of myxochelin A was tested in broth culture, addition of the siderophore promoted growth of an S. enterica serotype Typhimurium iroBC mutant (AIR49) better than of an iroN mutant (AJB64) (Fig. 7). These data indicated that IroN contributes to but is not the sole receptor involved in myxochelin A uptake in S. enterica.
TABLE 2.
Utilization of siderophores by S. enterica and E. coli strains
| Substance | Growth zone around filter disk placed on NBD agar (mm)
|
||||||
|---|---|---|---|---|---|---|---|
|
S. enterica
|
E. coli
|
||||||
| CL1509 | AJB64 | AIR49 | H5058(pTY2117) | H5058(pTY908) | AB2847 | H5058(pTY994) | |
| Ferrioxamine B | 30 | 30 | 30 | 4 | 4 | 4 | 4 |
| Benzaldehyde-2,3-dihydroxybenzhydrazone | 16 | 8 | 16 | 0 | 22 | ND | 14 |
| Enterochelin | 28 | 28 | 28 | 0 | 15 | ND | 16 |
| DBS | 22 | 12 | 22 | 0 | 21 | 15 | 21 |
| 2-N,6-N-Bis(2,3-dihydroxybenzoyl)-l-lysine | 40 | 40 | 40 | 0 | 22 | 22 | 22 |
| 2-N,6-N-Bis(2,3-dihydroxybenzoyl)-l-lysine amide | 38 | 38 | NDa | 0 | 14 | 24 | 15 |
| Myxochelin A | 38 | 38 | ND | 0 | 10 | 22 | 12 |
| Myxochelin B | 30 | 28 | ND | 0 | 0 | 21 | 0 |
| Myxochelin C | 14 | 18 | ND | 0 | 0 | 10 | 0 |
ND, not determined
FIG. 6.
Structures of catecholate siderophores used in this study.
FIG. 7.
Growth curves of S. enterica serotype Typhimurium (A) and E. coli (B) strains in NBD broth culture without supplements or supplemented with myxochelin A (MyxA). Growth was measured as optical density at 620 nm (OD620).
To study siderophore transport via IroN in the absence of other S. enterica outer membrane receptors that are likely to have overlapping substrate specificities (e.g., FepA or Cir) (18, 38), we used E. coli H5058. Due to a mutation in aroB, H5058 is unable to produce the siderophore enterochelin. In addition, H5058 carries mutations in the genes fepA, cir, and fiu and is therefore deficient for enterochelin and DBS uptake. The iroN gene was introduced into E. coli via either a cosmid (pTY908) or a 2,691-bp SalI-PstI fragment cloned into vector pBluescript SK (pTY994) (Fig. 1). Expression of the cloned iroN gene of S. enterica serotype Typhi (pTY908 and pTY994) in E. coli H5058 conferred the ability to utilize several catecholate siderophores during growth under iron deficiency (NBD), including DBS, enterochelin, benzaldehyde-2,3-dihydroxybenzhydrazone, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine amide, and myxochelin A (Table 2). Growth of H5058(pTY994) but not of H5058 was promoted by myxochelin A in broth culture (Fig. 7). These data show that IroN can serve as an outer membrane siderophore receptor which can in part complement mutations in the E. coli fepA, cir, and fiu receptor genes.
All siderophores transported by E. coli H5058(pTY908) and H5058(pTY994) possessed a N-linked (2,3-dihydroxybenzoyl) moiety, suggesting that substrate specificity of IroN is restricted to substances containing this group. No differences were observed between the utilization of d and l configurations of myxochelin derivatives (data not shown). However, myxochelin B and C, two siderophores that are closely related to myxochelin A, were not transported by IroN, an indication that additional structural features are required for the interaction between IroN and its substrate. In this context, it should be mentioned that for myxochelin derivatives, substrate specificity was strongly influenced by which group was linked to the C-1 atom of lysine. For instance, presence of a carboxy, hydroxy (myxochelin A), or amide group on C-1 allowed utilization of the respective siderophore via IroN, whereas derivatives substituted at C-1 by an amino (myxochelin B) or a N-(2,3-dihydroxybenzoyl) moiety (myxochelin C) were not utilized (Fig. 6).
DISCUSSION
Several lines of evidence suggest that the genes iroBCDE and iroN, part of the S. enterica iroA locus, form a functional unit. Expression of iroBCDE and that of iroN are both iron regulated, indicating that these genes may be functionally linked. The iron response regulator Fur is likely involved in iron-responsive expression of genes in the iroA locus, as typical Fur DNA binding sites are present in the iroB and iroN promoter regions (5). Both the iroBCDE operon and the iroN gene are present in S. enterica but are absent from closely related bacteria (4) (Fig. 4). This phylogenetic distribution can best be explained by acquisition of the entire iroA locus during a single horizontal transfer event in a lineage ancestral to the species S. enterica. A function of the iroA locus in iron acquisition is suggested by homologies of iroDE and iroN to genes associated with siderophore utilization. The iroN gene encodes an outer membrane siderophore receptor with high homology to FepA, BfeA, and PfeA, the enterochelin receptors of E. coli, B. pertussis, and P. aeruginosa, respectively (Fig. 2 and 3) (6, 10, 23). The iroD gene product shows homology to Fes, an E. coli enzyme involved in enterochelin utilization (27). Finally, iroE has homology to an open reading frame located downstream of pfeA, the enterochelin receptor gene of P. aeruginosa (10). Although these data suggest that the iroA locus functions in iron acquisition, there is at present no evidence implicating the iroBC genes in siderophore biosynthesis or uptake.
Acquisition of the iroN gene by S. enterica introduced two characteristics which now set this species apart from related bacteria. First, localization of IroN in the outer membrane exposes this protein to the host immune system. Fur-regulated siderophore receptors of S. enterica serotype Typhimurium are highly expressed during infection, as shown by in vivo studies (18). Furthermore, antibodies against iron-regulated outer membrane proteins have been shown to be present in sera from patients who recovered from typhoid fever, an infection caused by S. enterica serotype Typhi (11). Thus, acquisition of iroN introduced a new antigen that is characteristic of S. enterica and likely presents a target for the host immune system. Second, acquisition of iroN provided S. enterica with a new protein involved in iron uptake. Some of the substrates that are utilized by the S. enterica IroN receptor protein are excreted by soil bacteria. For instance, the siderophores 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine and 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine amide are produced by Azotobacter vinelandii (9), and myxochelin A is a product of the myxobacterium Angiococcus disciformans (21). It could therefore be speculated that IroN facilitates growth of S. enterica in soil, a step frequently encountered during the fecal oral transmission of this ubiquitous pathogen. Although it is possible that some soil bacteria produce siderophores which are transported exclusively by IroN in S. enterica, this was not the case for the catecholates used in this study (Table 2). However, the presence of IroN resulted in an increased growth rate of S. enterica in broth culture containing myxochelin A (Fig. 7), suggesting that this receptor may confer a selective advantage in the environment under certain growth conditions.
In E. coli, IroN mediated uptake of a variety of catecholate siderophores, including DBS, enterochelin, benzaldehyde-2,3-dihydroxybenzhydrazone, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine, 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine amide, and myxochelin A. However, transport of these siderophores was not abolished in a S. enterica iroN mutant. A possible explanation for the utilization of catecholate siderophores by an S. enterica iroN mutant is that the substrate specificity of IroN overlaps with that of other siderophore receptor proteins present in serotype Typhimurium, such as orthologs of FepA and Cir (18, 38). Overlapping substrate specificities of catecholate receptors were first identified in E. coli, where DBS is transported by the outer membrane receptors FepA, Cir, and Fiu (16). Furthermore, all catecholates transported by E. coli fepA cir fiu mutants expressing IroN [H5058(pTY908) and H5058(pTY994)] were also utilized by the isogenic E. coli parent (AB2847), indicating that these siderophores are substrates of the E. coli FepA, Cir, and/or Fiu receptor proteins (Table 2). In analogy, uptake of catecholate siderophores in an S. enterica iroN mutant may thus be mediated by the orthologs of the FepA and/or Cir receptor proteins present in this organism (18, 38).
ACKNOWLEDGMENTS
We thank K. Hantke for experimental advice and for providing bacterial strains.
This work was supported by Public Health Service grant AI 22933 to F.H. from the National Institutes of Health. Work in A.J.B.’s laboratory is supported by grant AI40124 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley & Sons; 1994. [Google Scholar]
- 3.Bäumler A J. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 3a.Bäumler, A. J., and F. Heffron. Unpublished data.
- 4.Bäumler A J, Heffron F, Reissbrodt R. Rapid detection of Salmonella enterica with primers specific for iroB. J Clin Microbiol. 1997;35:1224–1230. doi: 10.1128/jcm.35.5.1224-1230.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler A J, Tsolis R M, van der Velden A W M, Stojiljkovic I, Anic S, Heffron F. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;193:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 6.Beall B, Sanden G N. A Bordetella pertussis FepA homologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 7.Bennet L, Rothfield L I. Genetic and physiological regulation of intrinsic proteins of the outer membrane of Salmonella typhimurium. J Bacteriol. 1976;127:498–504. doi: 10.1128/jb.127.1.498-504.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchmeier N A, Lipps C J, So M Y, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 9.Corbin J L, Bulen W A. The isolation and identification of 2,3-dihydroxy-benzoic acid and 2-N,6-N-bis(2,3-dihydroxybenzoyl)-l-lysine formed by iron deficient Azotobacter vinelandii. Biochemistry. 1969;8:757. doi: 10.1021/bi00831a002. [DOI] [PubMed] [Google Scholar]
- 10.Dean C R, Poole K. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J Bacteriol. 1993;175:317–324. doi: 10.1128/jb.175.2.317-324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Beros M E, Gonzalez C, McIntosh M, Cabello F C. Immune response to the iron-deprivation-induced proteins of Salmonella typhi in typhoid fever. Infect Immun. 1989;57:1271–1275. doi: 10.1128/iai.57.4.1271-1275.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster J W, Hall H K. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J Bacteriol. 1992;174:4317–4323. doi: 10.1128/jb.174.13.4317-4323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster J W, Park Y K, Bang I S, Karem K, Betts H, Hall H K, Shaw E. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology. 1994;140:341–352. doi: 10.1099/13500872-140-2-341. [DOI] [PubMed] [Google Scholar]
- 14.Germanier R, Fürer E. Isolation and characterization of galE mutant Ty21a of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975;131:553–558. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- 15.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hantke K. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol Lett. 1990;67:5–8. doi: 10.1016/0378-1097(90)90158-m. [DOI] [PubMed] [Google Scholar]
- 17.Hantke K. Regulation of the ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 18.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel M, Shea J E, Bäumler A J, Gleeson C, Blattner F, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R-M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 21.Kunze B, Bedorf N, Kohl W, Höfle G, Reichenbach H. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales) J Antibiot. 1989;42:14–17. doi: 10.7164/antibiotics.42.14. [DOI] [PubMed] [Google Scholar]
- 22.Le Minor L, Popoff M Y. Designation of Salmonella enterica sp. nov., nom. rev., as the type and only species of the genus Salmonella. Int J Syst Bacteriol. 1987;37:465–468. [Google Scholar]
- 23.Lundrigan M D, Kadner R J. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 24.Ochman H, Groisman E A. Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996;64:5410–5412. doi: 10.1128/iai.64.12.5410-5412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H, Lawrence J G. Phylogenetics and the amelioration of bacterial genomes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2627–2637. [Google Scholar]
- 26.Ochman H, Wilson A C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 27.Pettis G S, Brickman T J, McIntosh M A. Transcriptional mapping and nucleotide sequence of the Escherichia coli fepA-fes enterobactin region. Identification of a unique iron-regulated bidirectional promoter. J Biol Chem. 1988;263:18857–18863. [PubMed] [Google Scholar]
- 28.Reeves M W, Evins G M, Heiba A A, Plikaytis B D, Farmer J J., III Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reissbrodt R, Heinisch L, Möllmann U, Rabsch W, Ulbricht H. Growth promotion of synthetic catecholate derivatives on gram-negative bacteria. Biol Metals. 1993;6:155–162. [Google Scholar]
- 30.Riley M, Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1976;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- 31.Riley M, Krawiec S. Genome organization. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 967–981. [Google Scholar]
- 32.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 34.Stojiljkovic I, Bäumler A J, Hantke K. Fur regulon in Gram-negative bacteria: characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 35.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trowitzsch-Kienast, W., V. Hartmann, R. Reissbrodt, and H. D. Ambrosi. 1994. Anm. 22.12.1994, Offenlegung 27.6.1996. German patent. Offenlegungsschrift DE 44 47 374 A1.
- 37.Trowitzsch-Kienast W, Kunze B, Irschik H, Wray V, Reichenbach H, Höfle G. Presented at the DECHEMA-Jahrestagung der Biotechnologen, Berlin, Germany. 1991. [Google Scholar]
- 38.Tsolis R, Bäumler A J, Stoljilkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsolis R M, Bäumler A J, Heffron F. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun. 1995;63:1739–1744. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsolis R M, Bäumler A J, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]