Abstract
Amylin is a pancreatic β-cell hormone that acts as a satiating signal to inhibit food intake by binding to amylin receptors (AMY) and activating a specific neuronal population in the area postrema (AP). AMY are heterodimers that include a calcitonin receptor (CTR) subunit (CTRa or CTRb) and a member of the receptor activity modifying proteins (RAMPs). Here, we used single cell qPCR to assess co-expression of AMY subunits in AP neurons from rats that were injected with amylin or vehicle. Because amylin interacts synergistically with the adipokine leptin to reduce body weight, we also assessed the co-expression of AMY and the leptin receptor isoform LepRb in amylin-activated AP neurons. Single cells were collected from Wistar rats and from transgenic Fos-GFP rats that express green-fluorescent protein (GFP) under the control of the Fos promoter. We found that the mRNAs of CTRa, RAMP1, RAMP2 and RAMP3 were all co-expressed in single AP-neurons. Amylin down-regulated RAMP1 and RAMP3 but not CTR mRNAs in AMY positive neurons, suggesting a possible negative feedback mechanism of amylin at its own primary receptors. Interestingly, amylin up-regulated RAMP2 mRNA. Moreover, RAMP2 appears to be co-expressed in CTRa+ cells with either one or both RAMP1 and RAMP3. We also found that a high percentage of single cells that co-expressed all components of a functional AMY expressed LepRb mRNA. Thus, single AP-cells express both AMY and LepRb that form a population of first-order neurons that presumably can be directly activated by amylin and at least in part also by leptin.
Keywords: CTR, RAMP1, RAMP2, RAMP3, laser capture microdissection
Graphical Abstract
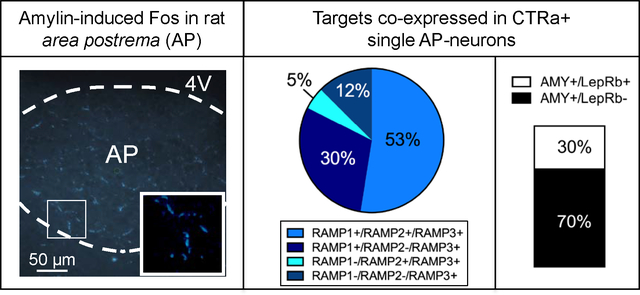
Single amylin-activated AP neurons co-express all necessary components of functional amylin receptors, i.e. CTRa and RAMP1, RAMP2, RAMP3, or a combination of RAMPs. The graph shows the co-expression in CTR-positive cells of amylin injected rats. A high percentage of these cells also co-express the leptin receptor (LepRb). Thus, single AP-cells form a population of first-order neurons that presumably can be directly activated by amylin and at least in part also by leptin.
Introduction
Amylin, also known as islet amyloid polypeptide, is co-secreted with insulin by pancreatic β-cells in response to nutrient stimuli (Lutz, 2010). Amylin reduces food intake and body weight (Lutz et al., 2001; Roth et al., 2012) and may also act as adiposity signal to control energy expenditure (Wielinga et al., 2007; Zhang et al., 2011). Circulating amylin acts centrally to control energy balance by primarily activating neurons of the area postrema (AP), a circumventicular organ located in the hindbrain (Riediger et al., 2001; Riediger et al., 2004; Lutz, 2009; Potes & Lutz, 2010; Potes et al., 2012).
A functional amylin receptor (AMY) results from a heterodimer of the calcitonin receptor (CTR) with one member of the receptor activity modifying proteins (RAMPs) (Christopoulos et al., 1999). The rat CTR exists in two different isoforms, CTRa and CTRb; but the exact functional relevance of action mediated by either isoform is not yet fully understood. In situ hybridization studies that mapped the localization of CTRa/b and RAMPs suggested that only CTRa is present in the rat AP (Ueda et al., 2001; Barth et al., 2004). Three members of the RAMP family have been identified (McLatchie et al., 1998; Sexton et al., 2001): RAMP1, RAMP2 and RAMP3. They are associated in the endoplasmic reticulum and are co-trafficked to the cell surface in order to form stable complexes that act as chaperons to form different receptors with selective ligand specificity. The dimerization of RAMP1, RAMP2 and RAMP3 with CTRa generates AMY1, AMY2 and AMY3, respectively (Alexander et al., 2013).
The presence of CTR and RAMPs has been shown in different brain areas (Sexton et al., 1994; Christopoulos et al., 1995; Skofitsch et al., 1995; Becskei et al., 2004; Mietlicki-Baase et al., 2013). However, none of these studies tested the co-localization of the AMY components at the single cell level; the latter is necessary to study the physiological relevance CTR and RAMPs exert in vivo.
Amylin is also known to synergistically interact with leptin to decrease body weight and food intake (Roth et al., 2008; Trevaskis et al., 2010b; Gautron & Elmquist, 2011). Leptin exerts its effects by binding to the b isoform of the leptin receptor (LepRb), and the ventromedial nucleus of the hypothalamus (VMH) may be the site mediating the amylin-leptin interaction (Trevaskis et al., 2010b; Turek et al., 2010; Le Foll et al., 2015). Other brain areas, such as the AP (Roth et al., 2008; Trevaskis et al., 2010b) or the VTA may also play a role (Mietlicki-Baase et al., 2015); however a population of first-order neurons receptive to both amylin and leptin has yet to be reported (Turek et al., 2010). Evidence of single neurons expressing both hormone receptors is lacking.
To establish that all components of the AMY are present in single AP cells and to investigate whether AP neurons co-express AMY and leptin receptors, we examined the expression of CTRa and RAMPs in single AP cells, and of the LepRb in AMY-positive neurons.
Materials and methods
Animals and tissue collection
Male Wistar rats (Janvier, Le Genest Saint Isle, France; Experiment 1 – 2; 200–225 g) and male and female Fos-GFP rats (NIDA/NIH, Baltimore, USA; Experiments 3–5; 250–320 g) were single-housed in a temperature controlled environment (21±1°C) on an artificial 12h/12h light/dark cycle. Rats had ad libitum access to water and standard chow, except during fasting periods as described below. All procedures involving animals and their care were approved by the Veterinary Office of the Canton Zurich, Switzerland, and in accordance with the EU Directive 2010/63/EU on the protection of animals used for scientific purposes.
Drugs
Amylin (Bachem AG, Bubendorf, Switzerland; catalogue number: H-9475.1000) was reconstituted with sterile 0.9% NaCl.
Experimental design
Rat injections and sample preparation: For all experiments, rats were fasted for twelve hours and injected i.p. with vehicle or amylin (20 μg/kg for Wistar and 50 μg/kg for Fos-GFP rats, respectively) at dark onset. Ninety minutes after drug administration, rats were anesthetized with isoflurane and decapitated.
Experiment 1:
Brains from Wistar rats (n=4) were promptly processed under a light microscope. The AP was surgically removed from the brainstem and then the subfornical organ (Takahashi et al., 1997) and the hypothalamus were dissected. All the tissues were placed immediately in TRI Reagent® (Sigma-Aldrich) and processed for RT-PCR, as described below.
Experiment 2–5:
Brains were rapidly isolated, snap-frozen in cold-isopentane and embedded in Tissue-Tek® OCT™ (Sakura Finetek) compound just prior to sectioning. Coronal sections were cut at 10-μm thickness on a cryostat (Leica, Germany) through the entire AP and affixed on pre-cleaned, Superfrost glass slides (Thermo Scientific, Waltham, MA USA). The sections were immediately placed in a slide box on dry ice until completion of sectioning. The slide box was rinsed with RNase Away (Molecular BioProduct, Mexico) prior to usage to prevent RNA degradation. Samples were then stored at −80 °C until laser capture microdissection (LCM) procedure.
In Experiment 2, Wistar rats (n=4 per group) were treated with amylin (20 μg/kg; i.p.) and single neurons (n=8–10 cells per rat) were identified with the cresyl violet staining as previously described (Kadar et al., 2009). CTRa, RAMP1 and RAMP3 have been extensively described to be present in the rat AP, thus generating AMY1 and AMY3 as the primary amylin receptors in this brain nucleus. RAMP2 mainly contributes to the generation of AMY2 by coupling with CTRb. Because CTRb is not expressed in the rat AP, we mainly focused on the analysis of RAMP1 and RAMP3 in combination with CTRa.
In Experiment 3–5, Fos-GFP transgenic males and females rats (n=4 rats/treatment/gender) were treated with vehicle or amylin (50 μg/kg; i.p.). Fos-GFP rats express the green-fluorescence-protein (GFP) under the Fos promoter in strongly activated neurons (Cifani et al., 2012), therefore no additional immunohistochemical procedure was required to visualize Fos-GFP-activated neurons. Amylin-stimulated cells (n=9–10 cells/per rat) were readily detectable under the eGFP filter (490–560 nm).
RT-PCR
RNA was extracted according to the manufacturer’s instructions (TRI Reagent®; Sigma-Aldrich) and then purified following the cleanup protocol of RNeasy® Mini kit (Quiagen, Germany), including the DNase step. The concentration and the integrity of RNA were measured using a nanodrop system (NanoDrop 1000 Spectrophotometer, Thermo Scientific). cDNA was generated from the extracted RNA using the Terto cDNA synthesis kit (Bioline, Switzerland).
The primer pairs for rat CTRa/b were previously reported (Mori et al., 2006) and primers for rat GAPDH were manually designed using Primer 3 web version 3.0.0. The primer sequences for rat CTRa/b were, forward: 5’-TGGTTGAGGTTGTGCCCAATGGA-3’, and reverse: 5’-TCCATGGGTTTGCCTCATCTTGGTC-3’. This primer pair was able to differentiate the two isoform of CTR, giving a 392 bp product for CTRa and a 503 bp product for CTRb (Accession L14617 and L13040). For rat GAPDH, forward: 5’-GCCAGCCTCGTCTCATAGACA-3’ and reverse: 5’-GTGCGATACGGCCCAATC-3’ (Accession NM_017008.4). RT-PCR conditions were the following: initial denaturation of 3 min at 95°C, followed by 35 cycles alternation of 15 s at 95 °C, 15 s at 60 °C and a final extension step of 20 s at 72°C. RT-PCR amplification products were separated by 2% agarose gel electrophoresis (BioRad 3000 Xi).
Laser capture microdissection (LCM)
Slides containing AP sections, collected from Wistar rats and from vehicle-treated Fos-GFP animals, were Nissl-stained to allow cell identification. Slides were removed from −80 °C and thawed at 4°C for 2 minutes; sections where stained in 0.1% cresyl violet acetate (Sigma-Aldrich) solution dissolved in 70% ethanol for 5 minutes on ice. The excess stain was drained on filter paper. Sections were then dehydrated once in 96% EtOH and once in 100% EtOH, 30 seconds each, on ice, and finally dried in a fume hood for 2 minutes at room temperature before being processed under the LCM microscope (Arcturus X MDS, Life Technologies; Carlsbad, California). As described above, no additional immunohistochemical procedure was required to identify Fos-GFP-positive cells in Fos-GFP transgenic rats. Brains slices were observed under the microscope and the AP was localized based on morphology (Bregma −13.68 to −14.28 mm; Paxinos & Watson, 2007).
In Wistar rats and in vehicle-treated Fos-GFP rats, single cells were visualized with the Nissl staining and they were randomly selected and individually collected in caps (CapSure® HS LCM caps, Arcturus Bioscience). In amylin-treated transgenic rats, Fos-GFP positive cells were identified under the eGFP filter and collected in individual caps. Total RNA was extracted and isolated, including a DNase step, using the vendor’s protocol (PicoPure™ RNA isolation kit, Arcturus) and subsequently stored at −80 °C.
Quantitative PCR (qPCR)
RNA extracted from each individual cell was amplified using Ribo-SPIA technology (Ovation One-Direct System, PART NO. 3500, NuGEN; San Carlos, California); cDNA was subsequently purified using Agencourt RNAClean XP beads. A post-SPIA modification process was run to complete the process and to generate sense-target cDNA, according to the manufacturer’s instructions. Single-cell qPCR was performed using the 7500 Fast system (Applied Biosystem/Life Technologies) with QuantiTect® SYBR® green PCR kit (QUIAGEN). Rat GAPDH was used as housekeeping gene. To avoid the possibility of amplifying contaminating DNA, intron-spanning primer pairs for rat CTRa, RAMP1 and RAMP3 were designed with Primer 3 web version 4.0.0. The primer sequences were the following; for rat GAPDH, forward: 5’-AGACAGCCGCATCTTCTTGT-3’ and reverse: 5’-CTTGCCGTGCGTAGAGTCAT -3’ (Accession: NM_017008.4), for rat CTRa, forward: 5’-TTTCCAGGGATTCTTTGTCG-3’ and reverse: 5’-TTCGTTGCTGACTGGAG-3’(Accession: NM_001034015.1), for rat RAMP1, forward: 5’-GGCAAACAAGATTGGCTGTT-3’ and reverse: 5’-AATGGGGAGCACAATGGAAG-3’ (Accession: NM_031645.1); for rat RAMP2, primer pairs were previously reported (Qi et al., 2003), forward: 5’-TGAGGACAGCCTTCTGTCAA-3’ and reverse: 5’-CATCGCCGTCTTTACTCCTC-3’ (Accession: NM_031646.1). For rat RAMP3, forward: 5’-AGGTCATCTGGAAGGTGTGG-3’ and reverse: 5’-AATGGGGAGCACAATGGAAG-3’ (Accession: NM_02100.2). The intron-spanning primer pairs for rat LepRb were designed with IDT (Integrated DNA Technologies); forward: 5’-GGTTGGATGGACTAGGGTATTG-3’ and reverse: 5’-CAGAATTCAGGCCCTCTCATAG-3’ (Accession: NM_012596.1).
200 ng of cDNA were subjected to an initial heat activation at 95°C for 15 minutes, followed by 40 cycles of alternating between 94°C for 15 s then 60°C for 30 s, and final extension at 72°C for 30 s. The relative transcriptional levels of CTRa, RAMP1 and RAMP3 mRNA were calculated using the comparative ΔΔCt method that generates relative CTRa, RAMP1 and RAMP3 mRNA levels adjusted for the GAPDH endogenous control mRNA. Each sample was run in duplicate.
Statistical Analysis
CTRa-positive cells versus CTRa-negative cells (n=4 rats/8–10 cells per rat) and the related presence or absence of RAMPs, respectively, were analyzed with Chi-square test (Experiment 2). Relative mRNA expression levels were assessed by unpaired Student’s t-test (Experiment 4; n=4 rats/9–10 cells per rat; Experiment 5; n=4 rats/8–10 cells per rat) by using GraphPad Software version 5.0 (San Diego, CA, USA).
Results
Experiment 1: Only the CTRa isoform is expressed in native rat AP tissue
Confirming previous studies (Hilton et al., 1995; Barth et al., 2004), we observed that only the CTRa isoform is expressed in native rat AP tissue (Figure 1). The subfornical organ and hypothalamus were used as positive controls, with both brain regions expressing CTRa and CTRb isoforms (Becskei et al., 2004; Mori et al., 2006).
Figure 1.
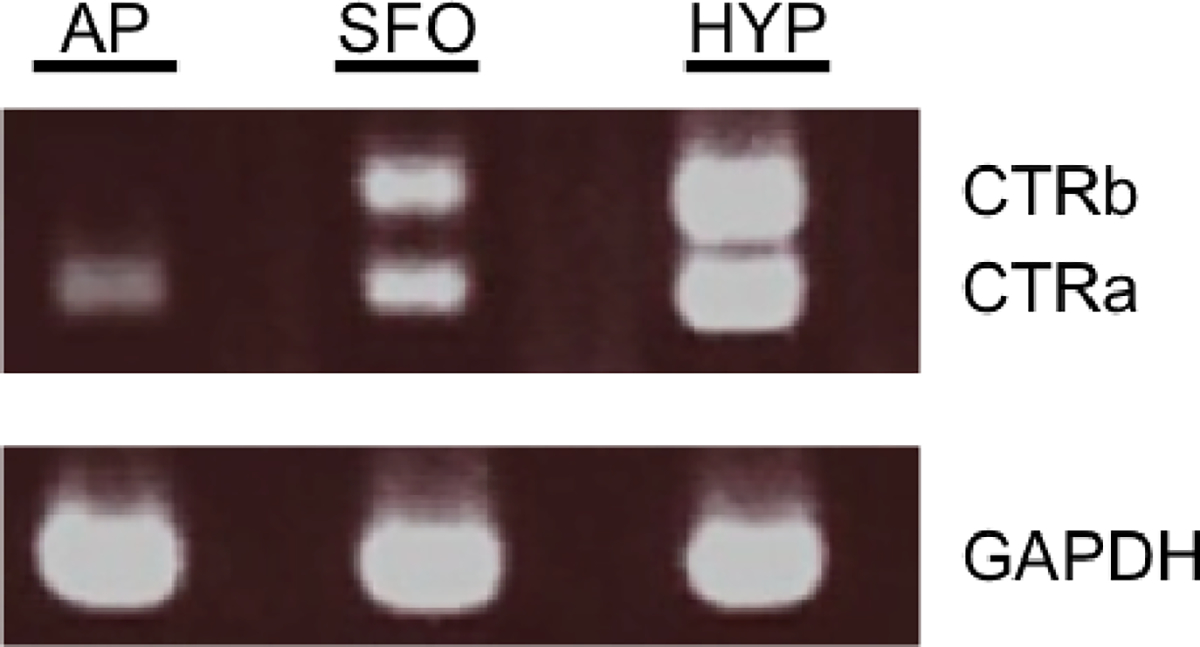
RT-PCR describing the distribution of CTR isoforms in different brain nuclei. Agarose gel electrophoresis of CTRa (392bp), CTRb (503 bp) PCR products from rat area postrema (AP), subfornical organ (SFO) and hypothalamus (HYP). GADPH (207 bp) was used as the reference gene and run in different lanes. Only the CTRa is detected in the rat AP.
Experiment 2: Randomly collected cells from the AP of amylin treated rats present a complex neuronal landscape. Most of CTRa+ cells are RAMP1+ and RAMP3+; whereas most of CTRa− cells are also RAMP1− and RAMP3−.
The rapid Nissl staining procedure before LCM enabled the identification of individual cells in the AP sections of amylin-treated Wistar rats (Figure 2a). Cells were randomly selected and individually collected by LCM (Figure 2a inset). A presence/absence assay was performed; GAPDH was used as endogenous control and the analyzed neuronal population was divided into CTRa+ and CTRa− cells, respectively. The resulting neuronal landscape showed a complex scenario (Figure 2b). Almost 40% of the collected neurons were CTRa+ cells; importantly, all CTRa+ cells co-expressed CTRa and RAMP1 (3%), CTRa and RAMP3 (10%) or CTRa and both RAMP1 and RAMP3 (25%). 62% of the randomly collected cells were CTRa negative. Moreover, most of the CTRa− cells were also RAMP1− and RAMP3− (53%); only 9% of CTRa− cells showed either RAMP1 (6%) or RAMP3 (3%) expression. Hence, RAMP1 and RAMP3 mRNAs were preferentially expressed in CTRa+ versus CTRa− cells (Chi-square test (30.13,3), P < 0.0001). The key finding of this study was that CTRa+ cells co-expressed RAMP1 and/or RAMP3 transcript, which are the necessary subunits required for a specific and functional AMY1 or AMY3.
Figure 2.
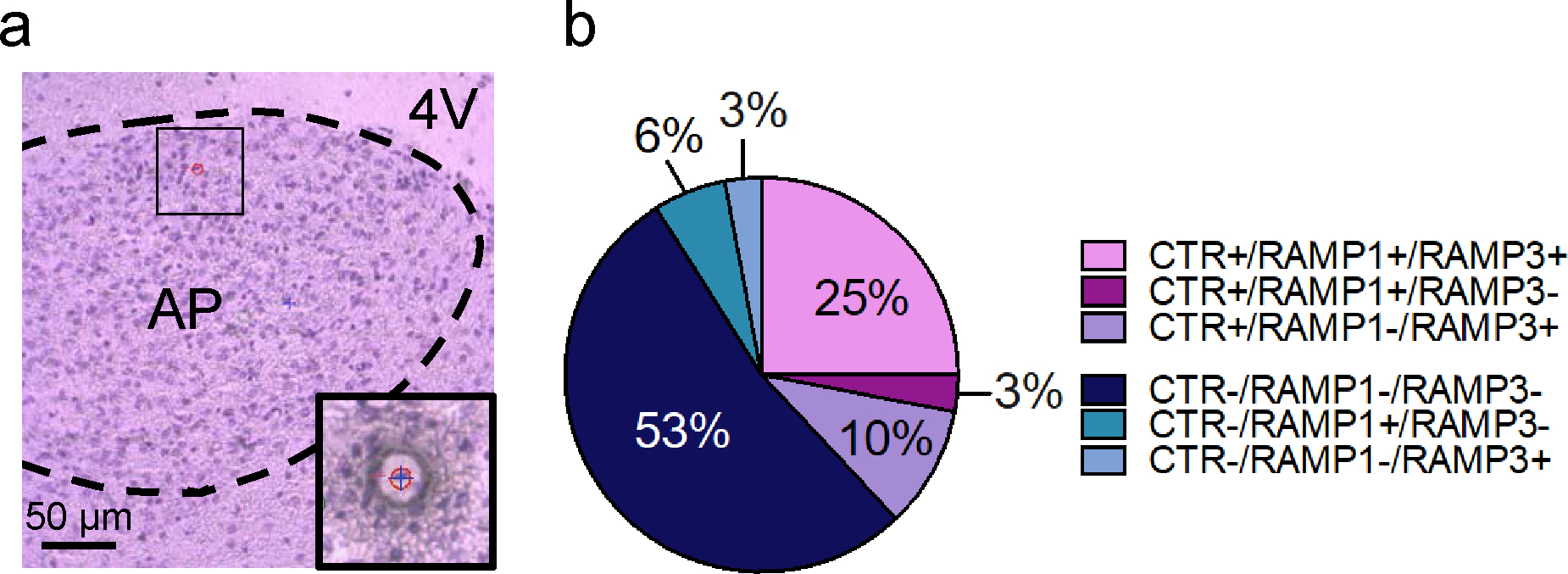
Distribution of CTR, RAMP1 and RAMP3 mRNAs in single cell of amylin-treated rats. (a) Nissl staining of rat AP at 20x. Representative image showing the LCM laser (blue cross) in the act of collecting a single neuron (red circle with number 1). The green circle represents the ultraviolet laser cutting spot. (b) In randomly picked individual neurons (n=8–10 cells per rat) from the AP of amylin-treated (20 μg/kg) male Wistar rats (n=4), 62% of the randomly collected single cells are CTRa−; most of the CTRa− cells are also RAMP1− and RAMP3− (53%); 6% of all cells are CTRa−, RAMP1+ and RAMP3− and 3% of all cells are CTRa−, RAMP1− and RAMP3+. The remaining 38% of individual neurons are CTRa+, and the 25% of them co-express CTRa and both RAMP1 and RAMP3. The remaining cells co-expressed CTRa and only RAMP1 (3%) or CTRa and only RAMP3 (10%). RAMP1 and RAMP3 transcripts are preferentially expressed in CTRa positive vs CTRa negative neurons; Chi-square test, *** P < 0.001.
Experiment 3: CTRa, RAMP1 and RAMP3 mRNAs are all co-expressed in the same single amylin-activated AP neurons of Fos-GFP rats.
Results from the second experiment clearly showed that in the neuronal population defined as CTRa+, single cells co-expressed RAMP1, RAMP3 or both RAMP1 and RAMP3 mRNAs in addition to CTRa. We reasoned that only this pool of cells expressed a biologically functional AMY1 or AMY3 and, therefore, can be activated by amylin. To test this hypothesis, we combined the use of Fos-GFP transgenic rats and single-cell qPCR to better characterize the transcripts of CTRa, RAMP1 and RAMP3 in amylin-activated individual neurons.
Transgenic rats were treated with vehicle or amylin (Figure 3a and 3d, respectively). In the control group, single cells were visualized with Nissl staining and randomly collected. Results obtained from the randomly collected single cells treated with vehicle replicated the scenario of Experiment 2; we confirmed that CTRa− cells are also normally RAMP1− and RAMP3− (50% of all cells investigated in male and 33% in female rats respectively), whereas CTRa+ cells are also RAMP1+, RAMP3+ or both RAMP1+ and RAMP3+ (see Figure 3b and 3c for the detailed distribution in male and female rats, respectively). Amylin treatment induced strong Fos-GFP expression in the AP (Figure 3d), allowing the specific collection of single amylin-activated neurons. In male rats, 64% of the cell population showed co-expression of CTRa, RAMP1 and RAMP3 in the same single neuron; 9% of the collected single neurons co-expressed CTRa with either RAMP1 (1%) or RAMP3 (8%). Only 27% of the neuronal population expressed one of the RAMPs but not the CTR (see Figure 3e for details). Results obtained from female transgenic rats displayed a pattern similar to the males. In the amylin-treated female rats, the entire population of amylin-responsive cells showed co-expression with CTRa and RAMPs; 70% of the collected single cells were CTRa+, RAMP1+ and RAMP3+ while the remaining 30% showed co-expression of CTRa with either RAMP1(10%) or RAMP3 (20%) (Figure 3f).
Figure 3.
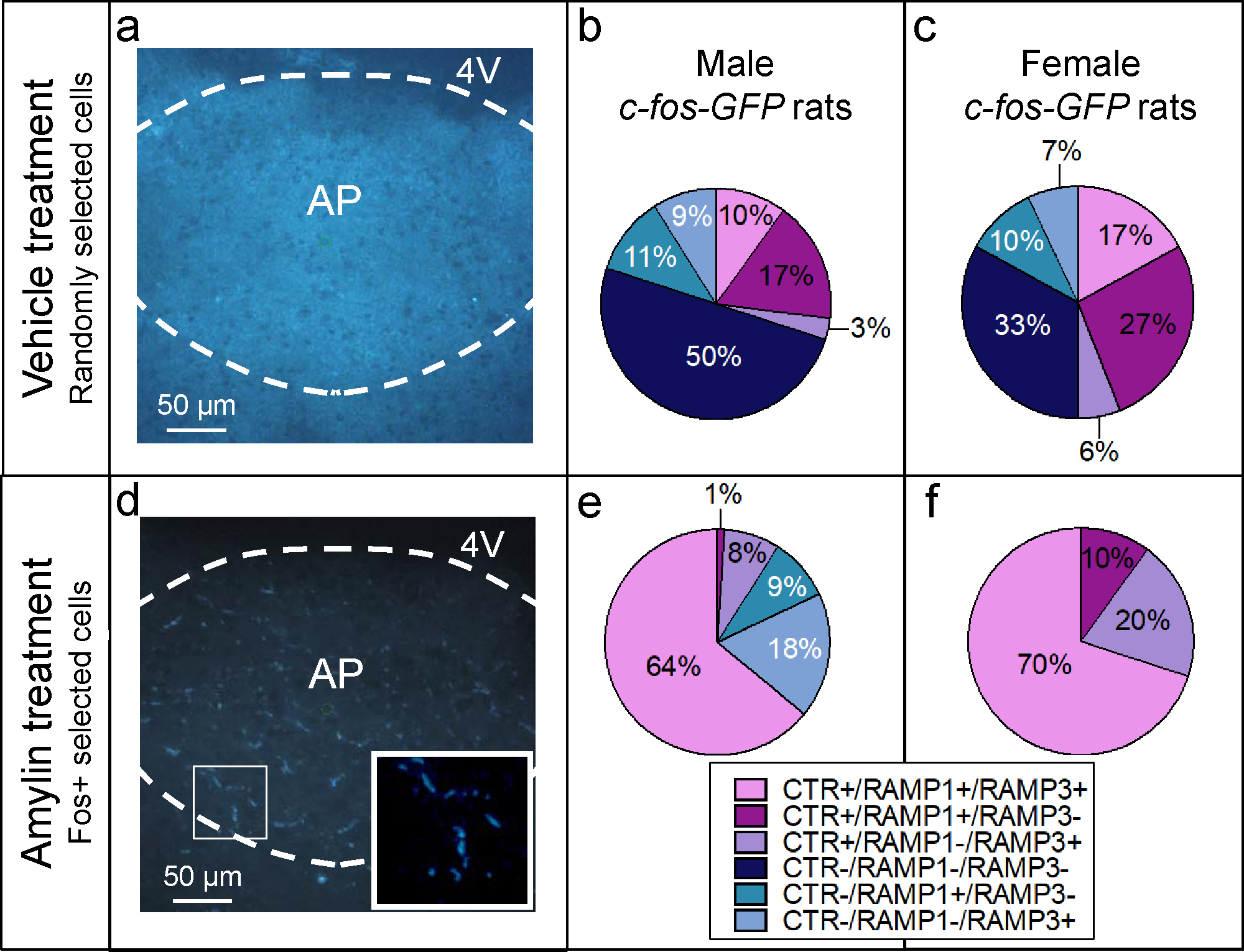
Analysis of single cells captured from vehicle and amylin-treated Fos-GFP rats. AP sections of Fos-GFP rats. (a) In vehicle-treated male rats (n=4 per group/per gender) (b), the randomly collected single-cells (n=9–10 cells per rat) show a mixed distribution: 30% of the single cells were CTRa+/RAMPs+; specifically 10% was CTRa+/RAMP1+/RAMP3+ and 17% was CTRa+/RAMP1+/RAMP3− and 3% was CTRa+/RAMP1−/RAMP3+. In the remaining cell population, 50% of the neurons were CTRa−/RAMP1−/RAMP3−, 11% were CTRa−/RAMP1+/RAMP3− and 9% were CTRa−/RAMP1−/RAMP3+. (c) In Fos-GFP female rats, the results in the vehicle-treated group was consistent with what was shown in males; 50% of the collected single cells were CTRa+/RAMPs+; in particular, 17% was CTRa+/RAMP1+/RAMP3−, 27% was CTRa+/RAMP1+/RAMP3− and 6% was CTRa+/RAMP1−/RAMP3+. The remaining cell population was characterized as CTRa− : 33% being CTRa−/RAMP1−/RAMP3−, 10% being CTRa−/RAMP1+/RAMP3− and 7% being CTRa−/RAMP1−/RAMP3+. (d) Amylin-induced Fos expression in AP neurons. The inset shows examples of single amylin- Fos-GFP activated cells. The green circle represents the ultraviolet cutting spot. (e) In c-fos-GFP male rats, the 73% of neurons that showed Fos-GFP expression after amylin injection (50 μg/kg) were CTRa+. Most of CTRa+ cells (64%) co-expressed the mRNA of CTRa, RAMP1 and RAMP3, 8% was defined as of CTRa+/RAMP1−/RAMP3+ and 1% was CTRa+/RAMP1+/RAMP3−. The remaining neuronal population was characterized as CTR−/RAMP1+/RAMP3− (9%) and CTR−/RAMP1−/RAMP3+ (18%). (f) In amylin-treated Fos-GFP female rats, all Fos-GFP activated neurons were CTRa+/RAMPs+; 70% of the population co-expressed CTRa, RAMP1 and RAMP3 in the same single cell; the remaining 30%, co-expressed CTRa and RAMP1 (10%) or CTRa and RAMP3 (20%).
Experiment 4: Amylin down-regulated RAMP1 and RAMP3 transcripts in single AP neurons; whereas RAMP2 mRNA is up-regulated.
To provide an absolute quantification of the expression levels of the mRNAs of CTRa and RAMPs, we selected all the CTRa+ cells and compared the relative expression of RAMP1, RAMP2 and RAMP3 in control-treated and amylin-treated Fos-GFP transgenic rats. Hence, RAMP2 mRNA was also included in this experiment to test specifically if identified CTRa+ cells also express RAMP2, in addition to RAMP1 and RAMP3.
Our findings demonstrate that while CTRa expression seems to be un-affected by acute amylin treatment (CTRa; t79 = 0.3573, P = 0.3573;), RAMP1 and RAMP3 mRNAs were down-regulated in amylin-activated single neurons (RAMP1; t62 = 5.409, P < 0.0001; RAMP3; t67 = 6.640, P < 0.0001; Figure 4b and 4d). Interestingly, amylin up-regulated RAMP2 mRNA in single AP cells (RAMP2; t34 =3.832; P < 0.0001, Figure 4c).
Figure 4.
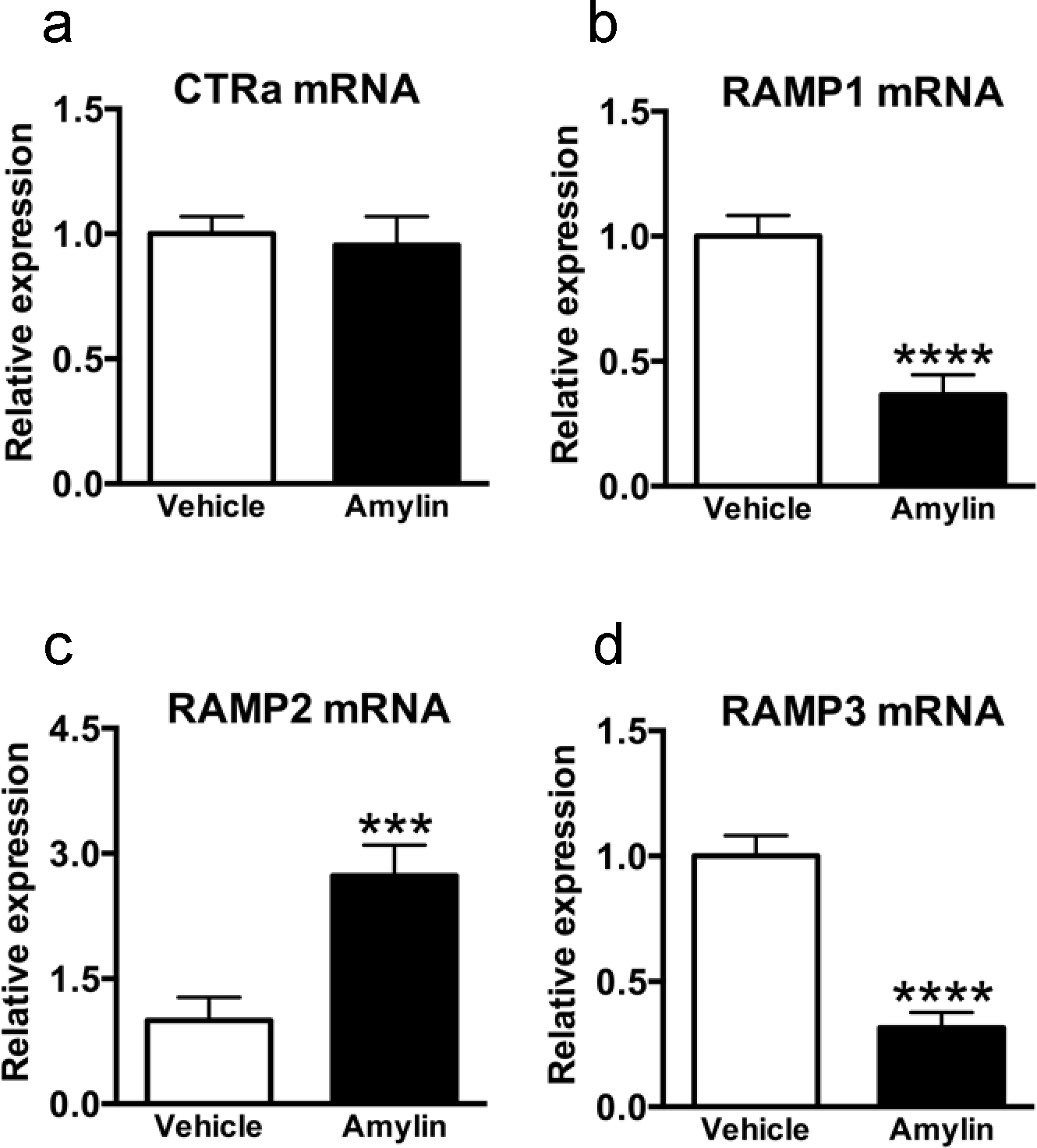
Effect of amylin on the transcriptional expression of CTRa, RAMP1, RAMP2 and RAMP3 mRNA in area postrema-single-cells. In Fos-GFP females rats (n=4 per group), AP single neurons (n=9–10 cells per rat), that showed co-expression of CTRa and RAMPs in both vehicle-treated and amylin-treated animals were analysed by qPCR. mRNA expression is shown as fold change; rat GAPDH was used as housekeeping gene to normalize the data. (a) CTRa mRNA levels were unaffected by the experimental condition, whereas (b) RAMP1 and (d) RAMP3 transcripts were significantly down-regulated in amylin-treated animals compared to control-treated rats (RAMP1, ***P < 0.001; RAMP3, ***P <0.001). Amylin up-regulated RAMP2 mRNA compared to control (RAMP2, ***P < 0.001); Data shown as mean±SEM.
To evaluate the co-expression of RAMP2 mRNA relatively to RAMP1 and RAMP3, a presence/absence analysis was performed in all the CTRa+ cells. In vehicle-treated animals, 70% of single CTRa+ AP-neurons expressed RAMP2+ in addition to RAMP1 (2%), RAMP3 (13%) or both RAMP1 and 3 (55%; Figure 5a); none of the CTRa+ cells expressed RAMP2 exclusively. The RAMP2 mRNA distribution was relatively similar in amylin-treated animals (see Figure 5b for details). These findings provide additional information on the distribution of RAMP2 mRNA relatively to the other RAMPs.
Figure 5.
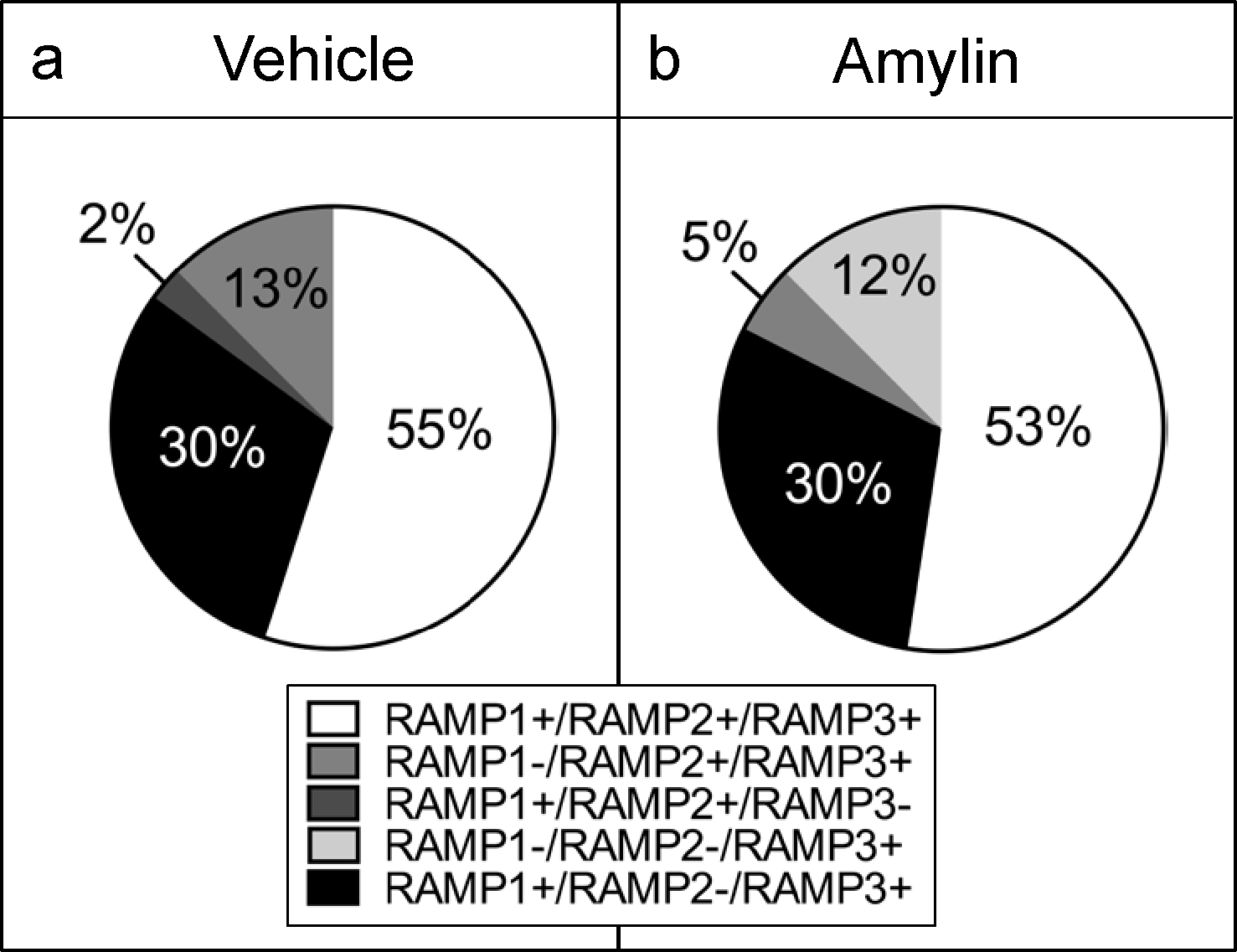
Distribution of RAMP2 mRNA respectively to the other RAMPs. All the selected cells were CTRa+. (a) In vehicle Fos-GFP females rats (n=4 per group), the collected AP single neurons (n=9–10 cells per rat), were mainly characterized as RAMP2+, being RAMP1+/RAMP2+/RAMP3+ (55%), RAMP1−/RAMP2+/RAMP3+ (13%) and RAMP1+/RAMP2+/RAMP3− (2%); 30% being RAMP1+/RAMP2−/RAMP3+). (b) RAMP2 mRNA distribution in amylin-treated animals was the following: RAMP1+/RAMP2+/RAMP3+ (53%), RAMP1−/RAMP2+/RAMP3+ (5%); RAMP1−/RAMP2−/RAMP3+ (12%) and RAMP1+/RAMP2−/RAMP3+ (30%).
Experiment 5: 30% of amylin-treated single AP neurons that co-expressed all the necessary components to form a functional AMY also co-expressed LepRb mRNA.
The presence of LepRb mRNA was investigated in the rats from Experiment 3 in single cells that were found to express all the necessary components to form a functional AMY (CTRa+ and either individual or combined RAMPs was defined as AMY+). Single, AMY+ cells from both vehicle and amylin-treated groups were analyzed. Under vehicle conditions, 52% of single AMY+ cells also co-expressed LepRb mRNA (Figure 6a). In rats treated with amylin, 30% of the AMY+ single cells also co-expressed LepRb (Figure 6b). Interestingly, the relative mRNA level of LepRb in AMY+ cells was significantly up-regulated by amylin treatment (t9 = 2.286, P < 0.05; Figure 6c).
Figure 6.
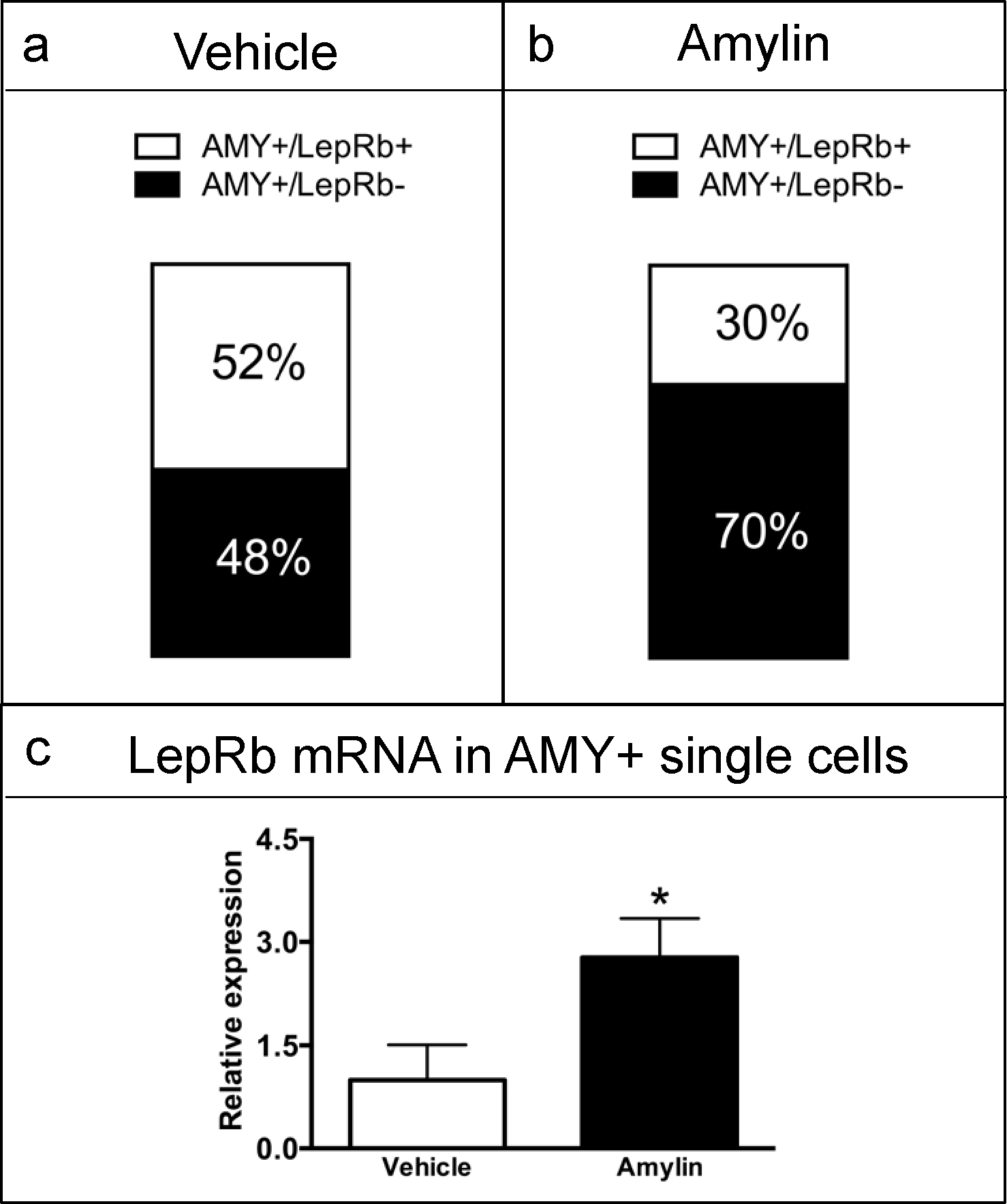
Quantitative analysis of LepRb mRNA levels in area postrema single neurons. Single cells (n=4 rat per group; 9–10 cells per rat) that were positive for all components of the functional AMY (AMY+) were analysed for the presence or absence of LepRb mRNA. (a) In vehicle-treated rats, 52% of the single-cells co-expressed AMY and LepR mRNAs. (b) In amylin-treated rats, 30% of the Fos positive single-cells that were AMY+ also co-expressed LepRb mRNA. (c) Quantitative analysis of LepRb mRNA levels; GAPDH was used as housekeeping gene and mRNA expression is shown as fold change. LepRb transcript levels were significantly up-regulated in AMY+ cells from amylin-treated compared to vehicle treated rats (*P < 0.05). Data shown as mean±SEM.
Discussion
CTR and the RAMPs generate receptors with high affinity for amylin (Christopoulos et al., 1999), with the precise nature of these receptors depending on the CTR splice variant and the cellular background. In the present study we provide evidence that the mRNA for all components of the AMY complex is co-expressed in the same single neurons in the rat AP, suggesting intracellular interactions between these subunits to mediate amylin signaling in the brain. This is the first demonstration in native tissue that single cells express all components of the functional AMY that allows them to be activated directly by amylin. We confirmed that only the CTRa isoform of the CTR is expressed in the AP, therefore the AMY complex in this brain nucleus results from a dimer of one or more RAMPs exclusively with CTRa (Barth et al., 2004; Becskei et al., 2004).
By applying the LCM technique, we were able to fully characterize the presence of functional AMY at the single cell level. Our results indeed support a scenario where in basal conditions AP neurons stochastically express zero, one, two, three or four components of the AMY. However, only the neurons that concurrently express CTRa and at least one of the RAMPs bear a functional AMY, and have therefore the full potential to be directly activated by amylin.
Previous findings demonstrated that RAMP1, RAMP2 and RAMP3 all interact with CTRa and CTRb to generate functional AMY1–3 in vitro. Different CTR/RAMPs complexes have distinct pharmacology, which may determine the rate of amylin binding to a specific AMY subtype. AMY1 and AMY3 have high affinities for amylin and salmon calcitonin (sCT) and both have the potential to bind the calcitionin gene-related peptide (CGRP). However AMY1 has a 30-fold higher affinity for CGRP than AMY3 (Christopoulos et al., 1999), implying that CGRP is most likely to target AMY1 than AMY3. These findings might suggest that amylin binds predominantly to AMY3 than AMY1; supporting the role of AMY3 as the main AMY. It is important to mention that in particular the ability of RAMP2 to form functional AMY2 is clearly dependent on the cellular background and on the receptor isoform (Tilakaratne et al., 2000). In fact, CTRb displayed greater capacity to dimerize with RAMP2 to generate a functional AMY2 than CTRa (Morfis et al., 2008). Given that CTRb is not present in the AP, our work mainly focused on the characterization and co-expression of CTRa, RAMP1 and RAMP3.
Our findings showed that in both male and female rats, the majority (over 60%) of the amylin-activated, Fos-GFP-positive single cells co-expressed CTRa, RAMP1 and RAMP3 and a smaller percentage co-expressed CTRa and either RAMP1 or RAMP3. This suggests that amylin mainly activates cells that express mRNA for CTRa, RAMP1 and RAMP3. When CTRa was co-expressed with only one of the RAMPs, the accessory subunit was preferentially RAMP3 (see Figure 3e and 3f for details), suggesting that AMY3 might be the primary AMY at the cellular level for AP mediated amylin actions. The preferential expression of CTRa with RAMP1 and/or RAMP3 in these activated cells also supports the idea that AMY1 and AMY3 are the most physiologically relevant receptors mediating amylin actions in the AP, and indicates that both subtypes can be present in an individual AP-neuron. In Fos-GFP male animals, 27% of amylin-activated, Fos-GFP positive neurons were characterized as CTR- but expressed either RAMP1 (9%) or RAMP3 (18%); it is unlikely that these cells were directly activated by amylin but because they expressed GFP after amylin administration, they possibly represent second-order neurons.
The congruent results obtained in males and females suggest that the sex of the rat does not seem to be an important factor influencing the genetic expression of the components of the AMY. However, sex hormones may still influence the relative expression of the AMY components. Estradiol strongly upregulates mRNA levels of RAMP3 in rat uterus (Watanabe et al., 2006) but it has not been tested whether estradiol may have similar effects in the brain. Interestingly, our previous work indicates that estradiol-treated ovariectomized rats demonstrate enhanced suppression of eating after acute amylin (Asarian et al., 2011), suggesting that amylin sensitivity fluctuates over the course of the estrous cycle, which could in part be the result of estrogen-induced changes in RAMPs expression.
We also provide evidence that RAMP2 mRNA is co-expressed with both RAMP1 and RAMP3 transcripts in over 50% of single AP-cells; this highlights the possibility of co-expression of all RAMP1, RAMP2 and RAMP3 along with CTRa in individual AP-neurons. However, none of the individually collected AP-neurons co-expressed CTRa and only RAMP2 mRNA; either one or both RAMP1 and RAMP3 transcripts were co-expressed with RAMP2 in single CTRa+ AP-cells. These results further support the notion that amylin acts predominantly via AMY1,3 in the AP, rather than AMY2.
Whether these different AMY have different physiological relevance is difficult to answer at present; further studies are required to investigate the contribution of more than one RAMP (i.e; CTRa/RAMP1, CTRa/RAMP3, CTRa/RAMP1/RAMP3 or CTRa/RAMP1/RAMP2/RAMP3 amylin receptors) to the binding and physiological effect of amylin. Our current work does not discern whether different variations of the AMY complex might contribute to the discrete physiological actions of amylin, such as the control of eating, the increase in energy expenditure, the slowing of gastric empting or the inhibition of glucagon secretion, all of which seem to be AP-mediated effects (Lutz, 2012; Hay et al., 2015). Knockout or knockdown models of either one or more RAMPs would be an interesting experimental approach to answer such questions.
Our results also indicate that amylin has the potential to modify individual components of its own receptors. The relative quantification of the mRNA levels revealed that expression of RAMPs but not of CTRa was differentially regulated by amylin. Precisely, the mRNA of the primary contributors necessary to form the functional AMY1,3 (RAMP1 and RAMP3) was consistently and significantly down-regulated by amylin while CTRa mRNA seemed to be unaffected. In contrast to RAMP1 and RAMP3, RAMP2 mRNA was up-regulated after amylin administration. The functional consequences of the latter effect are unknown at present. RAMP2 transcript level could be elevated to allow the dimerization with the calcitionin-like receptor (CLR) in the rat AP, and therefore generate an adrenomedullin-responsive receptor over an AMY2. However, this was not the purpose of our study.
The functional relevance of reduced RAMP1 and RAMP3 expression was an unexpected finding. Interestingly, supporting evidence has shown that when amylin was infused chronically at high doses (i.e. 50 μg/kg/d), its eating inhibitory effect was stronger in the first few days of infusion compared to later time points (Lutz et al., 2001), suggesting a decrease in amylin sensitivity. However, our recent work showed that when rats are chronically treated with more physiological doses of amylin (5–10 μg/kg/d), the animals’ responsiveness to acute amylin injections was maintained over time (Boyle et al., 2011). Based on these data, we therefore hypothesize that amylin, especially when administered at supraphysiological concentrations, acts on its own primary receptors (AMY1,3) to modulate RAMP1 and RAMP3 mRNAs via a potential negative feedback regulation mechanism. RAMP1 and RAMP3 may therefore act as the dynamic and regulatory component of the functional AMY1,3 in single AP-neurons, while CTRa may preferentially play a structural role as the core AMY subunit.
A number of studies have shown that amylin increases the leptin sensitivity in leptin- resistant animals (Trevaskis et al., 2010b). Leptin exerts its eating and weight lowering effects primarily by binding to the LepRb. The LepRb mediated effects on energy balance seem to rely on a wide network of brain areas, including the VMH and the arcuate nucleus, but possibly also regions in the brainstem, such as the nucleus of the solitary tract (NTS) which is also rich in LepRb (Myers et al., 2009). Even though the VMH has been proposed to mediate the interactions between leptin and amylin (Trevaskis et al., 2010b), the involvement of the caudal brainstem, and specifically the AP, has not been fully examined. Interestingly, the LepRb has also been found within the AP, even though in low abundance (Wada et al., 2014). Moreover, chronic amylin treatment in rats was shown to elevate both basal and leptin-induced activation of pSTAT3 in the AP (Roth et al., 2008), thus supporting the idea that amylin might also interact with leptin at the level of the AP (Trevaskis et al., 2010b). Here, we showed that after acute amylin treatment, 30% of the single, Fos-GFP-positive AP cells co-expressed the transcripts of LepRb and AMY. Moreover, amylin up-regulated AP LepRb expression at the single cell level. Our results therefore suggest the presence of a first-order neuronal population in the AP that is responsive to both amylin and leptin. This idea is consistent with recent findings that amylin and leptin are able to excite the same neurons isolated from the rat AP, using a whole cell current clamp recording technique (Smith et al., 2015). Further, our observation that acute amylin treatment increased LepRb expression, provides a possible mechanism by which amylin enhances leptin sensitivity, as was previously speculated (Trevaskis et al., 2010a). The specific contribution of AP neurons with the LepRb to leptin’s effects on eating and to the interaction between amylin and leptin will require additional investigation.
The results from our current study provide the first concrete evidence that CTRa, RAMP1, RAMP2, RAMP3 and LepRb mRNAs, are all co-expressed in single, amylin-activated neurons of native rat AP. Our data reported transcriptional changes at the mRNA level only. This limitation was mainly due to the lack of availability of commercial antibodies against the RAMPs and the LepRb; such studies would be important to show that the amylin receptor components are also co-expressed at the protein level. Regardless, our data support the possibility to have more than one AMY subtypes in the same amylin-activated AP-neuron. In addition, amylin has the potential to self-regulate its own receptors by modifying the transcriptional expression of the RAMPs. Finally, we showed that the LepRb mRNA is co-expressed along with CTRa and at least one of the RAMPs, in 30% of individual amylin-activated AP-neurons. These data indicate that the AP may directly contribute to the interaction between amylin and leptin.
Acknowledgement
This work was supported by the Swiss National Science Foundation and the Center for Integrative Human Physiology of the University of Zurich. The authors gratefully acknowledge the Center for Clinical Studies, the Center for Microscopy and Image Analysis (University of Zurich, Zurich, Switzerland) and Dr. Savina Adamo for technical support.
Abbreviations
- AP
area postrema
- AMY
amylin receptor
- CGRP
calcitonin gene-related peptide
- CLR
calcitonin-like receptor
- CTR
calcitonin receptor
- CTRa
calcitonin receptor isoform a
- CTRb
calcitonin receptor isoform b
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescence protein
- HYP
hypothalamus
- LCM
laser capture microdissection
- LepR
leptin receptor
- LepRb
leptin receptor isoform beta
- NTS
nucleus of the solitary tract
- pSTAT3
phospho-signal transducer and activator of transcription 3
- qPCR
quantitative polymerase chain reaction
- RAMP1
receptor activity modifying protein 1
- RAMP2
receptor activity modifying protein 2
- RAMP3
receptor activity modifying protein 3
- sCT
salmon calcitonin
- VMH
ventro-medial hypothalamus
- VTA
ventral tegmental area
- SFO
subfornical organ
References
- Alexander SP, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, Peters JA, Harmar AJ & Collaborators C (2013) The Concise Guide to PHARMACOLOGY 2013/14: G protein-coupled receptors. Br J Pharmacol, 170, 1459–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asarian L, Boyle CN & Lutz TA (2011) Estradiol (E2) increases the acute eating-inhibitory effect of amylin in ovariectomized (OVX) rats. Appetite, 57, Supplement 1, S2. [Google Scholar]
- Barth SW, Riediger T, Lutz TA & Rechkemmer G (2004) Peripheral amylin activates circumventricular organs expressing calcitonin receptor a/b subtypes and receptor-activity modifying proteins in the rat. Brain Res, 997, 97–102. [DOI] [PubMed] [Google Scholar]
- Becskei C, Riediger T, Zund D, Wookey P & Lutz TA (2004) Immunohistochemical mapping of calcitonin receptors in the adult rat brain. Brain Res, 1030, 221–233. [DOI] [PubMed] [Google Scholar]
- Boyle CN, Rossier MM & Lutz TA (2011) Influence of high-fat feeding, diet-induced obesity, and hyperamylinemia on the sensitivity to acute amylin. Physiol Behav, 104, 20–28. [DOI] [PubMed] [Google Scholar]
- Christopoulos G, Paxinos G, Huang XF, Beaumont K, Toga AW & Sexton PM (1995) Comparative distribution of receptors for amylin and the related peptides calcitonin gene related peptide and calcitonin in rat and monkey brain. Canadian journal of physiology and pharmacology, 73, 1037–1041. [DOI] [PubMed] [Google Scholar]
- Christopoulos G, Perry KJ, Morfis M, Tilakaratne N, Gao Y, Fraser NJ, Main MJ, Foord SM & Sexton PM (1999) Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol, 56, 235–242. [DOI] [PubMed] [Google Scholar]
- Cifani C, Koya E, Navarre BM, Calu DJ, Baumann MH, Marchant NJ, Liu QR, Khuc T, Pickel J, Lupica CR, Shaham Y & Hope BT (2012) Medial prefrontal cortex neuronal activation and synaptic alterations after stress-induced reinstatement of palatable food seeking: a study using c-fos-GFP transgenic female rats. J Neurosci, 32, 8480–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautron L & Elmquist JK (2011) Sixteen years and counting: an update on leptin in energy balance. The Journal of clinical investigation, 121, 2087–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DL, Chen S, Lutz TA, Parkes DG & Roth JD (2015) Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacological reviews, 67, 564–600. [DOI] [PubMed] [Google Scholar]
- Hilton JM, Chai SY & Sexton PM (1995) In vitro autoradiographic localization of the calcitonin receptor isoforms, C1a and C1b, in rat brain. Neuroscience, 69, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Kadar A, Wittmann G, Liposits Z & Fekete C (2009) Improved method for combination of immunocytochemistry and Nissl staining. J Neurosci Methods, 184, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Foll C, Johnson MD, Dunn-Meynell AA, Boyle CN, Lutz TA & Levin BE (2015) Amylin-induced central IL-6 production enhances ventromedial hypothalamic leptin signaling. Diabetes, 64, 1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz TA (2009) Control of food intake and energy expenditure by amylin-therapeutic implications. Int J Obes (Lond), 33 Suppl 1, S24–27. [DOI] [PubMed] [Google Scholar]
- Lutz TA (2010) The role of amylin in the control of energy homeostasis. Am J Physiol Regul Integr Comp Physiol, 298, R1475–1484. [DOI] [PubMed] [Google Scholar]
- Lutz TA (2012) Control of energy homeostasis by amylin. Cellular and molecular life sciences : CMLS, 69, 1947–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz TA, Mollet A, Rushing PA, Riediger T & Scharrer E (2001) The anorectic effect of a chronic peripheral infusion of amylin is abolished in area postrema/nucleus of the solitary tract (AP/NTS) lesioned rats. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity, 25, 1005–1011. [DOI] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG & Foord SM (1998) RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature, 393, 333–339. [DOI] [PubMed] [Google Scholar]
- Mietlicki-Baase EG, Olivos DR, Jeffrey BA & Hayes MR (2015) Cooperative interaction between leptin and amylin signaling in the ventral tegmental area for the control of food intake. Am J Physiol Endocrinol Metab, ajpendo 00087 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietlicki-Baase EG, Rupprecht LE, Olivos DR, Zimmer DJ, Alter MD, Pierce RC, Schmidt HD & Hayes MR (2013) Amylin receptor signaling in the ventral tegmental area is physiologically relevant for the control of food intake. Neuropsychopharmacology, 38, 1685–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfis M, Tilakaratne N, Furness SG, Christopoulos G, Werry TD, Christopoulos A & Sexton PM (2008) Receptor activity-modifying proteins differentially modulate the G protein-coupling efficiency of amylin receptors. Endocrinology, 149, 5423–5431. [DOI] [PubMed] [Google Scholar]
- Mori I, Ishii A, Nakamura A, Nakamura M, Nakagomi N, Takeda K & Kakudo K (2006) Expression and cellular localization of calcitonin receptor: RT-PCR and in situ hybridization studies. Cellular and molecular biology, 52, 9–13. [PubMed] [Google Scholar]
- Myers MG Jr., Munzberg H, Leinninger GM & Leshan RL (2009) The geometry of leptin action in the brain: more complicated than a simple ARC. Cell Metab, 9, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G & Watson C (2007) The rat brain in stereotactic coordinates. Academic press/ELSEVIER, Amsterdam; Boston. [Google Scholar]
- Potes CS, Boyle CN, Wookey PJ, Riediger T & Lutz TA (2012) Involvement of the extracellular signal-regulated kinase 1/2 signaling pathway in amylin’s eating inhibitory effect. Am J Physiol Regul Integr Comp Physiol, 302, R340–351. [DOI] [PubMed] [Google Scholar]
- Potes CS & Lutz TA (2010) Brainstem mechanisms of amylin-induced anorexia. Physiol Behav, 100, 511–518. [DOI] [PubMed] [Google Scholar]
- Qi YF, Shi YR, Bu DF, Pang YZ & Tang CS (2003) Changes of adrenomedullin and receptor activity modifying protein 2 (RAMP2) in myocardium and aorta in rats with isoproterenol-induced myocardial ischemia. Peptides, 24, 463–468. [DOI] [PubMed] [Google Scholar]
- Riediger T, Schmid HA, Lutz T & Simon E (2001) Amylin potently activates AP neurons possibly via formation of the excitatory second messenger cGMP. Am J Physiol Regul Integr Comp Physiol, 281, R1833–1843. [DOI] [PubMed] [Google Scholar]
- Riediger T, Zuend D, Becskei C & Lutz TA (2004) The anorectic hormone amylin contributes to feeding-related changes of neuronal activity in key structures of the gut-brain axis. Am J Physiol Regul Integr Comp Physiol, 286, R114–122. [DOI] [PubMed] [Google Scholar]
- Roth JD, Erickson MR, Chen S & Parkes DG (2012) GLP-1R and amylin agonism in metabolic disease: complementary mechanisms and future opportunities. Br J Pharmacol, 166, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JD, Roland BL, Cole RL, Trevaskis JL, Weyer C, Koda JE, Anderson CM, Parkes DG & Baron AD (2008) Leptin responsiveness restored by amylin agonism in diet-induced obesity: evidence from nonclinical and clinical studies. Proc Natl Acad Sci U S A, 105, 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton PM, Albiston A, Morfis M & Tilakaratne N (2001) Receptor activity modifying proteins. Cellular signalling, 13, 73–83. [DOI] [PubMed] [Google Scholar]
- Sexton PM, Paxinos G, Kenney MA, Wookey PJ & Beaumont K (1994) In vitro autoradiographic localization of amylin binding sites in rat brain. Neuroscience, 62, 553–567. [DOI] [PubMed] [Google Scholar]
- Skofitsch G, Wimalawansa SJ, Jacobowitz DM & Gubisch W (1995) Comparative immunohistochemical distribution of amylin-like and calcitonin gene related peptide like immunoreactivity in the rat central nervous system. Canadian journal of physiology and pharmacology, 73, 945–956. [DOI] [PubMed] [Google Scholar]
- Smith P, Brzezinska P, Hubert F, Maurice D & Ferguson A (2015) Leptin and Amylin Influence the Excitability of Area Postrema Neurons, Possibly via cAMP-Mediated Mechanisms. The FASEB Journal, vol. 29 no. 1 Supplement 988.4.25466899 [Google Scholar]
- Takahashi Y, Smith P, Ferguson A & Pittman QJ (1997) Circumventricular organs and fever. The American journal of physiology, 273, R1690–1695. [DOI] [PubMed] [Google Scholar]
- Tilakaratne N, Christopoulos G, Zumpe ET, Foord SM & Sexton PM (2000) Amylin receptor phenotypes derived from human calcitonin receptor/RAMP coexpression exhibit pharmacological differences dependent on receptor isoform and host cell environment. The Journal of pharmacology and experimental therapeutics, 294, 61–72. [PubMed] [Google Scholar]
- Trevaskis JL, Parkes DG & Roth JD (2010a) Insights into amylin-leptin synergy. Trends Endocrinol Metab, 21, 473–479. [DOI] [PubMed] [Google Scholar]
- Trevaskis JL, Parkes DG & Roth JD (2010b) Insights into amylin-leptin synergy. Trends Endocrinol Metab, 21, 473–479. [DOI] [PubMed] [Google Scholar]
- Turek VF, Trevaskis JL, Levin BE, Dunn-Meynell AA, Irani B, Gu G, Wittmer C, Griffin PS, Vu C, Parkes DG & Roth JD (2010) Mechanisms of amylin/leptin synergy in rodent models. Endocrinology, 151, 143–152. [DOI] [PubMed] [Google Scholar]
- Ueda T, Ugawa S, Saishin Y & Shimada S (2001) Expression of receptor-activity modifying protein (RAMP) mRNAs in the mouse brain. Brain Res Mol Brain Res, 93, 36–45. [DOI] [PubMed] [Google Scholar]
- Wada N, Hirako S, Takenoya F, Kageyama H, Okabe M & Shioda S (2014) Leptin and its receptors. Journal of chemical neuroanatomy, 61–62, 191–199. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Takahashi E, Kobayashi M, Goto M, Krust A, Chambon P & Iguchi T (2006) The estrogen-responsive adrenomedullin and receptor-modifying protein 3 gene identified by DNA microarray analysis are directly regulated by estrogen receptor. Journal of molecular endocrinology, 36, 81–89. [DOI] [PubMed] [Google Scholar]
- Wielinga PY, Alder B & Lutz TA (2007) The acute effect of amylin and salmon calcitonin on energy expenditure. Physiol Behav, 91, 212–217. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Morgan DA, Kuburas A, Thedens DR, Russo AF & Rahmouni K (2011) Neuronal receptor activity-modifying protein 1 promotes energy expenditure in mice. Diabetes, 60, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]


