Highlights
-
•
Prevalence of the Omicron subvariants during the flu season and beyond.
-
•
Gradual emergence and dominance of the recombinant XBB and its sub-lineages.
-
•
XBB.1.5 was the most common circulating variant XBB.1.5.
-
•
Timely detection and characterization of SARS-CoV-2 variants is important to reduce transmission through established disease control measures.
-
•
Avoid introductions into animal populations that could lead to serious public health implications.
Keywords: SARS-CoV-2, XBB, Omicron, XBB.1.5
Abstract
Background
Early SARS-CoV-2 variant detection relies on testing and genomic surveillance. The Omicron variant (B.1.1.529) has quickly become the dominant type among the previous circulating variants worldwide. Several subvariants have emerged exhibiting greater infectivity and immune evasion. In this study we aimed at studying the prevalence of the Omicron subvariants during the flu season and beyond in Lebanon through genomic screening and at determining the overall standing and trajectory of the pandemic in the country.
Methods
A total of 155 SARS-CoV-2 RNA samples were sequenced, using Nanopore sequencing technology.
Results
Nanopore sequencing of 155 genomes revealed their distribution over 39 Omicron variants. XBB.1.5 (23.29 %) was the most common, followed by XBB.1.9.1 (10.96 %) and XBB.1.42 (7.5 %). The first batch collected between September and November 2022, included the BA.2.75.2, BA.5.2, BA.5.2.20, BA.5.2.25 and BQ.1.1.5 lineages. Between December 2022 and January 2023, those lineages were replaced by BA.2.75.5, BN.1, BN.1.4, BQ.1, BQ.1.1, BQ.1.1.23, CH.1.1, CM.4 and XBK. Starting February 2023, we observed a gradual emergence and dominance of the recombinant XBB and its sub-lineages (XBB.1, XBB.1.5, XBB.1.5.2, XBB.1.5.3, XBB.1.9, XBB.1.9.1, XBB.1.9.2, XBB.1.16, XBB.1.22 and XBB.1.42).
Conclusions
The timely detection and characterization of SARS-CoV-2 variants is important to reduce transmission through established disease control measures and to avoid introductions into animal populations that could lead to serious public health implications.
1. Introduction
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a betacoronavirus (Gralinski and Menachery, 2020), is a single-stranded positive-sense RNA virus (Zhou et al., 2020). Its genome is arranged into a 5′-leader sequence-ORF1/ab-S-ORF3a-E-M-ORF6a-ORF7a-ORF7b-ORF8-N-ORF10–3′ sequence (Parra-Lucares et al., 2022). Viral replication is mediated by RNA-dependent-RNA polymerase (RdRp) and associated with proofreading activity through exoribonuclease (ExoN) (Kim et al., 2020; Moeller et al., 2022), both located within ORF1ab. The accumulation of mutations is common in ssRNA viruses and is largely linked to the high error rate of the RdRp (Eskier et al., 2020).
The persistence of the COVID-19 pandemic was mainly attributed to the emergence of variants with distinct phenotypic characteristics including transmissibility, severity, and immune evasion. Several genetically distinct lineages have evolved, and the occurrence of the new variants raised concerns regarding the potential impact on vaccination programs (McLean et al., 2022). Adaptation to host and the evasion of the immune response, including that triggered through immunization, are the two common forces that significantly shaped the SARS-CoV-2 genome. SARS-CoV-2 variant carrying the spike protein amino acid change D614G, for example, has become the most prevalent form in the global pandemic with the emergence of a large number of viral lineages worldwide, including several variants of concern (VOC) (Korber et al., 2020). Following Alpha, Beta, Gamma, and Delta SARS-CoV-2 variants, the WHO has identified and classified Omicron (B.1.1.529) as a highly mutated VOC which became dominant in several countries owing to its very high transmissibility (Nie et al., 2022; Parums, 2022). The B.1.1.529 Omicron variant expanded into a series of sub-lineages with variable and slightly distinct phenotypes. Several Omicron sub-lineages were identified (BA.1; BA.2; BA.4; BA.5; BA.2.12.1; BA.2.75; BQ.1; XBB), and which have spread globally. BA.1 and BA.2 initially predominated, but then replaced by BA.4, BA.5, and sub-lineage BA.2.12.1 in Europe and the USA (Parums, 2022). BQ.1 is a BA.5 sub-lineage with additional spike mutations L452R, N460K, and K444T. BA.5 and its sub-lineages reached high prevalence and became globally prevalent (UK Health Security Agency, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1115077/Technical-Briefing-47.pdf).
Multiple genetic recombination events have been also reported in coinfection cases, and the earliest detected inter-lineage SARS-CoV-2 recombinant was the XA, first appearing in the United Kingdom (Markov et al., 2023; Gangavarapu et al., 2023; Khare et al., 2021). The X indicates that the subvariant emerged from the recombination of at least two sub-lineages (Pango Network, https://www.pango.network/The-pango-nomenclature-system/statement-of-nomenclature-rules/). The circulating recombinants of B.1.631 and B.1.634 were designated as lineage XB, according to Pango nomenclature, and showed more extensive circulation compared to XA. The recombinant variant XBB, originated by the recombination of two BA.2 descendants, BJ.1 and BM.1.1.1. The XBB has 13 additional mutations compared to BA.2 (including 9 on the RBD) and XBB.1.5 has two additional spike mutations compared to its ancestor XBB, with a similar spike profile as XBB.1.9.1. The two additional spike mutations (F486P and G252V) increased its binding affinity to ACE-2 receptors (Gangavarapu et al., 2023; Khare et al., 2021; Mahase, 2023). As of October 2022, XBB.1.5 was reported from 38 countries, and became the most transmissible variant. XBB.1.5 had a growth advantage compared to other circulating Omicron sub-lineages and evaded antibodies in humans (WHO; https://www.who.int/activities/tracking-SARS-CoV-2-variants).
COVID-19 was first detected in Lebanon on the 21st of February 2020 (Feghali et al., 2021), and throughout the pandemic, SARS-CoV-2 cases, and subsequent hospitalizations, driven by different variants, have been on a rise. As of 5 July 2023, more than 750 million COVID-19 cases were reported and over 6.9 million deaths (WHO; https://covid19.who.int/). SARS-CoV-2 variants possessing unique combinations of mutations will continue to emerge and circulate and without continuous surveillance intermediate sequences will be missed altogether. The under sampling and limited genomic surveillance in Lebanon and elsewhere increase the possibility of circulating viral lineages being undetected until the emergence of a dominating VOC. Accordingly, we aimed at providing an updated status during the flu season for the dominant variants in Lebanon through genomic screening and reviewed the overall standing and trajectory.
2. Materials and methods
Between September 26, 2022, and August 17, 2023, samples were collected from LAU Medical Center-Rizk Hospital (LAUMCRH), Serhal Hospital and Mashrek Medical Diagnostic Center. Samples collected from LAUMCRH were designated as LAU-MC-1–199, those collected from Serhal Hospital were designated as SH-1–109, and those collected from Mashrek Medical Diagnostic Center were designated as MDC-1–16. A total of 155 genomes were sequenced.
2.1. Viral genome amplification
cDNA was reverse transcribed from 11 μL of viral RNA using the SuperScript IV reverse transcriptase and RNaseOUT (Thermo) with random hexamers. Polymerase chain reaction was performed using Q5 Hot Start Master Mix (NEB, USA) and the Artic V4.1 NCOV-2019 Panel primer pools A and B (Integrated DNA Technologies, IDT), designated in the ARTIC network SARS-CoV-2 sequencing protocol (Tyson et al., 2020). Tiled amplicons were purified using the Agencourt AMPure XP beads (Beckman Coulter, USA).
2.2. Genome sequencing
Purified amplicon libraries were prepared using the SQK-LSK109 Ligation Sequencing kit according to the ARTIC sequencing protocol (Tyson et al., 2020). End preparation was performed with NEBNext Ultra II buffer and enzyme mix (NEB, USA). Unique barcodes were assigned and ligated to each sample with the Native barcoding expansion kits (EXP-NBD104 and EXP-NBD114; Oxford Nanopore Technologies, UK). Barcoded samples were pooled together, and library concentration was determined with the Qubit 2.0 fluorometer using the dsDNA HS Assay Kit (Thermo Fisher Scientific, United States). Sequencing was performed with the MinION Mk1B device using a FLO-MIN106D (R9.4.1) flow cell (Oxford Nanopore Technologies, UK). 75 µl (15 ng of amplicon DNA) of the final library mixture were loaded on the primed flow cell and the sequencing run was set for 10 h.
2.3. Consensus sequence assembly
Fast base calling of the raw fast5 files was performed in real time through guppy v3.1.5 (https://community.nanoporetech.com/protocols/Guppy-protocol/v/gpb_2003_v1_revu_14dec2018/guppy-software-overview) as part of the MinKNOW software suit (Oxford Nanopore Technologies, UK). Read pass selection criteria was based on a Phred value score > 7 and barcode identification on both ends. Barcode trimming was also performed through guppy barcoder integrated in the MinKNOW software.
Read filtering was performed with the artic guppyplex script (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html) with the “—skip-quality-check” flag, a minimum read length of 350 and maximum length of 700. Subsequently, amplicon mapping, primer trimming, and consensus genomes were generated through submitting the filtered reads to the artic minion Nanopolish pipeline (https://github.com/jts/nanopolish). Samples with less than 10,000 Ns were used to proceed with the analysis.
2.4. Lineage assignment
Lineage assignment was completed using the Phylogenetic Assignment of Named Global Outbreak LINeages ‘Pangolin’ web server tool (https://pangolin.cog-uk.io/) (O'Toole et al., 2021).
2.5. Phylogenetic analysis
We downloaded 1031 genome sequences of SARS-CoV-2 from GSAID recovered from Lebanon and used them along with samples sequenced in this study for phylogenetic analysis. The phylogenetic tree was generated using Ultrafast Sample placement on Existing tRees (UShER) (Turakhia et al., 2021), and the tree file was obtained through Nextstrain (Hadfield et al., 2018). The tree was then visualized and colored-coded using the interactive Tree Of Life (iTOL) online tool (Letunic and Bork, 2021).
2.6. Mutation analysis
Nextclade platform was used to detect non-synonymous mutations and their relative amino acid variations (https://clades.nextstrain.org) (Aksamentov et al., 2021).
3. Results
3.1. Sequencing and variant detection during the flu season-2023
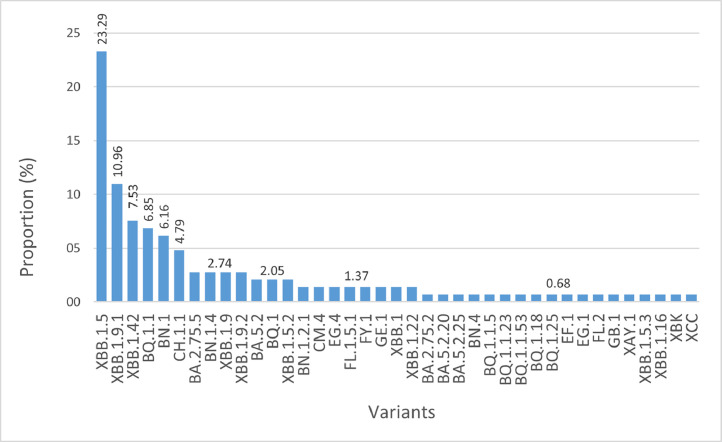
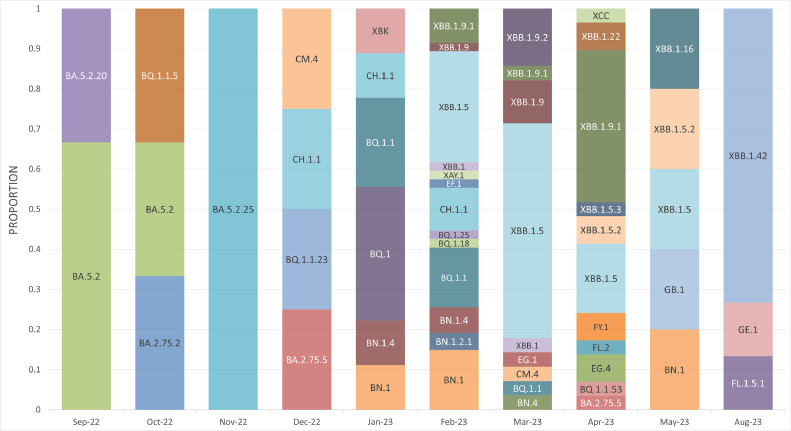
The samples, included in this study, were collected from three healthcare settings in Lebanon between September 26, 2022, and August 17, 2023, to cover the flu season (2022–2023), and assess frequency and abundance. A total of 155 samples were sequenced and deposited on GISAID (Table S1). Lineage assignment through Pangolin was applied to the 155 sequenced genomes, and results obtained showed their distribution over 39 Omicron variants. XBB.1.5, representing 23.29 % of the sequenced genomes, was the most common, followed by XBB.1.9.1 (10.96 %) and XBB.1.42 (7.5 %) (Fig. 1). The first batch collected between September and November 2022, contained several lineages including BA.2.75.2, BA.5.2, BA.5.2.20, BA.5.2.25 and BQ.1.1.5, which, between December 2022 and January 2023, were replaced by BA.2.75.5, BN.1, BN.1.4, BQ.1, BQ.1.1, BQ.1.1.23, CH.1.1, CM.4 and XBK. Starting February 2023, we observed a gradual emergence and dominance of the recombinant XBB lineage and its sub lineages (XBB.1, XBB.1.5, XBB.1.5.2, XBB.1.5.3, XBB.1.9, XBB.1.9.1, XBB.1.9.2, XBB.1.16, XBB.1.22 and XBB.1.42). XBB.1.42 was detected during mid-August and the high number doesn't reflect a change in the dominance but more of a group of related people who got infected through exposure to a primary case. Other lineages and sub lineages were also detected starting February 2023, including some that were not previously reported such as BQ.1.1.53, BQ.1.18, BQ.1.25, BN.1.2.1, BN.4, EF.1, EG.1, EG.4, FL.1.5.1, FL.2, FY.1, GB.1, GE.1, XAY.1 and XCC (Fig. 2).
Fig. 1.
Proportion (%) of detected variants during the study period.
Fig. 2.
Monthly distribution of variants detected during the study period.
3.2. Phylogenetic analysis
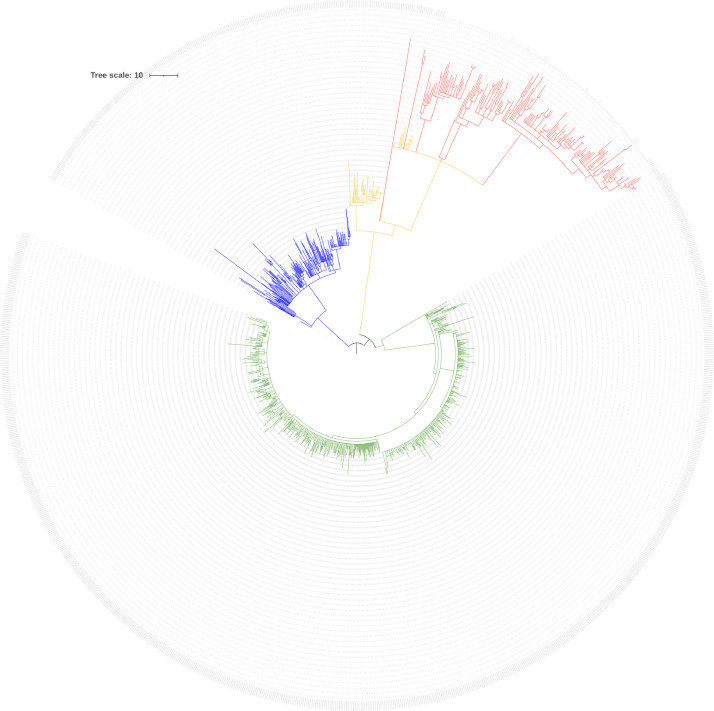
The 155 sequenced genomes in this study along with 1031 others recovered from Lebanon and deposited on GISAID were aligned using Wuhan_WIV04 (EPI_ISL_402,124) as a reference. All the genomes were distributed over the three main distinct clades representing the Alpha, Delta, and Omicron variants. Each cluster was further divided into subclades based on intra-lineage differences. The Alpha, Delta, Omicron, and the genomes sequenced in this study were all color coded (Fig. 3).
Fig. 3.
Phylogenetic placement of the sequenced genomes along with all sequences recovered from Lebanon and deposited on GSAID. GSAID others recovered and sequenced from Lebanon Clades representing the different variants including the sample from this study were color coded (Alpha: green; Delta: blue; Omicron: orange; Sequenced genomes from this study: red).
3.3. Mutations in the S gene
The highest number of mutations in the S gene were found in the RBD region. The most common mutation was A27S, detected in 153/155 samples, followed by D796Y then N679K and P681H. Several other mutations were also detected in the RBD but with less than 10 occurrences and were classified as uncommon. All the detected mutations along with the number of occurrences were included in Table S2.
4. Discussion
Overall, 2755 SARS-CoV-2 genomes have been submitted to GISAID from Lebanon as of 11 July 2023 (Khare et al., 2021). B.1, B.1.1, and B.4 variants, part of the 20A, 20B, and 19A clades respectively, were the first detected variants in Lebanon (Feghali et al., 2021), following which Fayad et al. (2021) revealed the detection of B.1.398 and B.1.1.7. The largest scale sequencing attempt however was the one reported by Merhi et al., 2022 which was the transition period (December 2020 and June 2021) marked by the worldwide gradual replacement of the Alpha variant and the dominance of the Delta. Other sporadic studies were also conducted with different focus groups such as the one by AlKalamouni et al., 2023 which revealed the prevalence of BA.1, BA.1.1, and BA.2, Omicron lineages in healthcare workers, between December 2021 and January 2022.
The Omicron variant was first detected on the 9th of November 2021 (Vieillard-Baron et al., 2022). It has 37 mutated amino acids, 15 of which are part of the RBD (Cameroni et al., 2021; European Center for Disease prevention and Control, https://www.ecdc.europa.eu/sites/default/files/documents/Implications-emergence-spread-SARS-CoV-2%20B1.1.529-variant-concern-Omicron-for-the-EU-EEA-Nov2021.pdf). The ones with key contact sites with the human ACE2 included the N501Y. N501Y causes a sixfold increase in the binding affinity to ACE-2. The association of N501Y with Q498R significantly increased the affinity to the ACE2 receptor and the transmissibility of this variant (Araf et al., 2022), having a 10-fold higher infectivity compared to the wild-type strain (Riediker et al., 2022). We have detected the co-occurrence of those two mutations in 148/155 of the variants sequenced in this study.
E484K, a mutation found in the Beta and Gamma variants, was previously linked to the higher reinfection rate observed with the Gamma variant. The Omicron, however, has the E484A substitution instead, which possibly changes the interaction between the RBD and the ACE2 (Kannan et al., 2021; Resende et al., 2021), and alters the shape of the spike protein site which interacts with neutralizing antibodies (Ren et al., 2022). Furthermore, the RBD exposure and ACE2 binding are facilitated through the furin cleavage site in the S protein (Peacock et al., 2021; Wrobel et al., 2020). N679K mutation, located next to the furin cleavage site in Omicron, had reduced spike protein in purified virions compared to wild-type and with a replication advantage in the upper airway over wild-type SARS-CoV-2 in hamsters (Vu et al., 2023). On the other hand, the R203K and G204R mutations, in the nucleocapsid gene, played a role in increasing the infectivity of the Omicron variant by increasing the proliferation of the virus, and were also detected in Alpha and Gamma variants (Johnson et al., 2022).
BA.2.75.2, BQ.1, and BQ.1.1, were among the detected variants in this study and which exhibited strong neutralization resistance (Qu et al., 2023). In BQ.1 and BQ.1.1, resistance was mostly conferred by the N460K, and to a lesser extent by R346T, K444T, and N658S mutations (Bruel et al., 2022; Sheward et al., 2022; Qu et al., 2022) (WHO, https://www.who.int/news/item/27–10–2022-tag-ve-statement-on-omicron-sublineages-bq.1-and-xbb). R346T and K444T alter the antibody recognition epitope, destabilize the affinity between RBD and related antibodies, and mediate antibody recognition by affecting the stability of hydrogen bonds and salt bridges (Qu et al., 2023). Nonetheless, R346T and F486S mutations contributed to the significantly enhanced resistance of BA.2.75.2 (Sheward et al., 2022).
XBB, was detected for the first time in August 2022, in India, and between 3 and 9 October 2022, it constituted more than half of the cases in Singapore (Kurhade et al., 2022). XBB.1, XBB.1.5, known as “Kraken”, and XBB.1.9 were among the detected variants in this study. XBB.1 carried the R346T mutation, which was linked to neutralization evasion (Bruel et al., 2022; Sheward et al., 2022; Kurhade et al., 2022), whereas the XBB.1.5 and XBB.1.9 increased infectivity was attributed to the F486P spike mutation and its role in increasing the binding affinity to ACE2 (Parums, 2023; Yamasoba et al., 2023). Furthermore, XBB.1.16, compared to XBB.1.5 and XBB.1.9.1, has two substitutions in the spike protein: E180V in the N-terminal domain and T478R in the RBD. XBB.1.16 showed a greater growth advantage compared to XBB.1 and XBB.1.5, but similar immune evasion. The increased fitness of XBB.1.16 was probably linked to the mutations in non-spike proteins (Yamasoba et al., 2023).
CH.1.1, which was among the detected variants in this study and also known as “Orthus”, was first detected in July 2022 in India (Bazzani et al., 2023). CH.1.1 harbored the R346T and K444T mutations similar to BA.2.75.2 and BQ.1, respectively. Compared with BA.2.75, however, CH.1.1 had additional mutations such as L452R and F486S. These mutations were linked to reduced CH.1.1 antibody neutralization (Uraki et al., 2023). F486S mutation hindered the binding to ACE2 and exerted a negative impact on viral replication (Bazzani et al., 2023).
XAY.1 a Delta-Omicron recombinant (Liu et al., 2023; Focosi and Maggi, 2022), was also detected in this study confirming previous results with the Delta variant backbone still existing in several regions and spreading as a Delta-Omicron hybrid.
Finally, it's noteworthy, that among the variants detected in this study, XBB.1.9.1, XBB.1.9.2, BA.2.75 and CH.1.1 remain classified by the WHO as variants under monitoring (VUMs), whereas XBB.1.5 and XBB.1.16 as variants of interest (VOIs) (as of August 17, 2023).
5. Conclusion
In this study, we performed a genome-based survey to assess genetic variability and provide an updated epidemiological trajectory. Our study revealed that the Omicron (B.1.1.529) has quickly replaced the previous circulating variants in Lebanon. Omicron wave was associated with the emergence of several subvariants exhibiting greater infectivity and immune evasion. Early variant detection relies on testing and genomic surveillance, with timely sequence reporting. Therefore, continuous genomic surveillance is important to trace and map the prevalence and emergence of the variants, reduce transmission through established disease control measures, and avoid introductions into animal populations that could lead to serious public health implications.
Ethics statement
All experiments were performed in compliance with relevant laws and institutional guidelines and in accordance with the ethical standards of the Declaration of Helsinki. The study protocol was ethically approved by the Lebanese American University, Institutional Review Board (LAU IRB) (reference LAU.SAS.ST2.19/May/2020). Authors had access to limited de-identified information on each patient.
CRediT authorship contribution statement
Ibrahim Al Kodsi: Investigation, Data curation, Visualization, Writing – original draft. Douaa El Rayes: Investigation, Data curation, Visualization, Writing – original draft. Jad Koweyes: Investigation, Data curation. Charbel Al Khoury: Data curation, Visualization, Writing – review & editing. Kelven Rahy: Data curation, Visualization. Sergio Thoumi: Data curation, Visualization. Marc Chamoun: Data curation, Resources. Hoda Haddad: Investigation, Data curation. Jacques Mokhbat: Data curation, Resources, Supervision, Writing – review & editing. Sima Tokajian: Conceptualization, Methodology, Resources, Supervision, Data curation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199289.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6(67):3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- AlKalamouni H., Abou Hassan F.F., Bou Hamdan M., Page A.J., Lott M., Matthews M., Ghosn N., Rady A., Mahfouz R., Araj G.F., Dbaibo G., Zaraket H., Melhem N.M., Matar G.M. Genomic surveillance of SARS-CoV-2 in COVID-19 vaccinated healthcare workers in Lebanon. BMC Med Genomics. 2023;16(1):14. doi: 10.1186/s12920-023-01443-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araf Y., Akter F., Tang Y.D., Fatemi R., Parvez M.S.A., Zheng C., Hossain M.G. Omicron variant of SARS-CoV-2: genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzani L., Imperia E., Scarpa F., Sanna D., Casu M., Borsetti A., Pascarella S., Petrosillo N., Cella E., Giovanetti M., et al. SARS-COV CH.1.1 variant: genomic and Structural insight. Infect. Dis. Rep. 2023;15(3):292–298. doi: 10.3390/idr15030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel T., Stefic K., Nguyen Y., Toniutti D., Staropoli I., Porrot F., Guivel-Benhassine F., Bolland W.H., Planas D., Hadjadj J., et al. Longitudinal analysis of serum neutralization of SARS-CoV-2 Omicron BA.2, BA.4, and BA.5 in patients receiving monoclonal antibodies. Cell Rep. Med. 2022;3(12) doi: 10.1016/j.xcrm.2022.100850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021;602(7898):664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskier D., Karakülah G., Suner A., Oktay Y. RdRp mutations are associated with SARS-CoV-2 genome evolution. PeerJ. 2020;8:e9587. doi: 10.7717/peerj.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayad N., Habib W.A., Kandeil A., El-Shesheny R., Kamel M.N., Mourad Y., Mokhbat J., Kayali G., Goldstein J., Abdallah J. SARS-COV-2 variants in Lebanon: evolution and current situation. Biology. 2021;10(6):531. doi: 10.3390/biology10060531. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghali R., Merhi G., Kwasiborski A., Hourdel V., Ghosn N., Tokajian S. Genomic characterization and phylogenetic analysis of the first SARS-CoV-2 variants introduced in Lebanon. PeerJ. 2021;9:e11015. doi: 10.7717/peerj.11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., Maggi F. Recombination in coronaviruses, with a focus on SARS-CoV-2. Viruses. 2022;14(6):1239. doi: 10.3390/v14061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu K., Latif A.A., Mullen J.L., Alkuzweny M., Hufbauer E., Tsueng G., Haag E., Zeller M., Aceves C.M., Zaiets K., et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. Methods. 2023;20(4):512–522. doi: 10.1038/s41592-023-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Zhou Y., Lokugamage K.G., Vu M.N., Bopp N., Crocquet-Valdes P.A., Kalveram B., Schindewolf C., Liu Y., Scharton D., et al. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLOS Pathog. 2022;18(6) doi: 10.1371/journal.ppat.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S., Ali S., Sheeza A. Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review. PubMed. 2021;25(24):8019–8022. doi: 10.26355/eurrev_202112_27653. [DOI] [PubMed] [Google Scholar]
- Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T.C., Yeo W., et al. GISAID's role in pandemic response. China CDC Wkly. 2021;3(49):1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurhade C., Zou J., Xia H., Liu M., Chang H.C., Ren P., Xie X., Shi P.Y. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med. 2022;29(2):344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.T., Tsai J.J., Chu J.J.H., Chen C.H., Chen L.J., Lin P.C., Tsai C.Y., Hsu M.C., Chuang W.L., Hwang S.J., et al. The identification and phylogenetic analysis of SARS-CoV-2 delta variants in Taiwan. Kaohsiung J. Med. Sci. 2023;39(6):624–636. doi: 10.1002/kjm2.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. COVID-19: what do we know about XBB.1.5 and should we be worried? BMJ. 2023;380:p153. doi: 10.1136/bmj.p153. [DOI] [PubMed] [Google Scholar]
- Markov P.V., Ghafari M., Beer M., Lythgoe K., Simmonds P., Stilianakis N.I., Katzourakis A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023;21(6):361–379. doi: 10.1038/s41579-023-00878-2. [DOI] [PubMed] [Google Scholar]
- McLean G., Kamil J., Lee B., Moore P., Schulz T.F., Muik A., Sahin U., Türeci Ö., Pather S. The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. MBio. 2022;13(2) doi: 10.1128/mbio.02979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhi G., Trotter A.J., de Oliveira Martins L., Koweyes J., Le-Viet T., Abou Naja H., Al Buaini M., Prosolek S.J., Alikhan N.F., Lott M., Tohmeh T., Badran B., Jupp O.J., Gardner S., Felgate M.W., Makin K.A., Wilkinson J.M., Stanley R., Sesay A.K., Webber M.A., Davidson R.K., Ghosn N., Pallen M., Hasan H., Page A.J., Tokajian S. Replacement of the Alpha variant of SARS-CoV-2 by the Delta variant in Lebanon between April and June 2021. Microb Genom. 2022;8(7):mgen000838. doi: 10.1099/mgen.0.000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller N.H., Shi K., Demir Ö., Belica C., Banerjee S., Yin L., Durfee C., Amaro R.E., Aihara H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. U. S. A. 2022;119(9) doi: 10.1073/pnas.2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie C., Sahoo A., Herrmann A., Ballauff M., Netz R.R., Haag R. Charge matters: mutations in Omicron variant favor binding to cells. ChemBioChem. 2022;23(6) doi: 10.1002/cbic.202100681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2):veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Lucares A., Segura P., Rojas V., Pumarino C., Saint-Pierre G., Toro L. Emergence of SARS-CoV-2 variants in the world: how could this happen? Life. 2022;12(2):194. doi: 10.3390/life12020194. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parums D.V. Editorial: world health organization (WHO) variants of concern lineages under monitoring (VOC-LUM) in response to the global spread of lineages and sublineages of Omicron, or B.1.1.529, SARS-COV-2. Med. Sci. Monit. 2022;28 doi: 10.12659/msm.937676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parums D.V. Editorial: the XBB.1.5 (‘Kraken’) subvariant of Omicron SARS-CoV-2 and its rapid global spread. Med. Sci. Monit. 2023;29 doi: 10.12659/msm.939580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Qu P., Evans J.P., Zheng Y.M., Carlin C., Saif L.J., Oltz E.M., Xu K., Gumina R.J., Liu S.L. Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe. 2022;30(11):1518–1526. doi: 10.1016/j.chom.2022.09.015. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P., Evans J.P., Faraone J.N., Zheng Y.M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe. 2023;31(1):9–17. doi: 10.1016/j.chom.2022.11.012. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases. 2022;10(1):1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Bezerra J.F., Vasconcelos R.H.T., Arantes I., Appolinario L., Mendonça A.C., Paixao A.C., Duarte A.C., Silva T., Rocha A.S., et al. Severe acute respiratory syndrome Coronavirus 2 P2 lineage associated with reinfection case, Brazil, June–October 2020. Emerg. Infect. Dis. 2021;27(7):1789–1794. doi: 10.3201/eid2707.210401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riediker M., Briceno-Ayala L., Ichihara G., Albani D., Poffet D., Tsai D.H., Iff S., Monn C. Higher viral load and infectivity increase risk of aerosol transmission for delta and Omicron variants of SARS-CoV-2. Schweiz. Med. Wochenschr. 2022;152(0102):w30133. doi: 10.4414/smw.2022.w30133. [DOI] [PubMed] [Google Scholar]
- Sheward D.J., Kim C.I., Fischbach J., Sato K., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect. Dis. 2022;22(11):1538–1540. doi: 10.1016/s1473-3099(22)00663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhia Y., Thornlow B., Hinrichs A.S., De Maio N., Gozashti L., Lanfear R., Haussler D., Corbett-Detig R. Ultrafast sample placement on existing tRees (UshER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat. Genet. 2021;53(6):809–816. doi: 10.1038/s41588-021-00862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R., Ito M., Kiso M., Yamayoshi S., Iwatsuki-Horimoto K., Sakai-Tagawa Y., Furusawa Y., Imai M., Koga M., Yamamoto S., et al. Efficacy of antivirals and bivalent mRNA vaccines against SARS-CoV-2 isolate CH.1.1. Lancet Infect. Dis. 2023;23(5):525–526. doi: 10.1016/s1473-3099(23)00132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieillard-Baron A., Flicoteaux R., Salmona M., Chariot A., De Maupeou D'Ableiges B., Darmon M., Batteux F. APHP reality research group. Omicron variant in the critical care units of the Paris metropolitan area: the reality research group. Am. J. Respir. Crit. Care Med. 2022;206(3):349–363. doi: 10.1164/rccm.202202-0411le. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson, J.R., James, P., Stoddart, D., Sparks, N., Wickenhagen, A., Hall, G., Choi, J.H., Lapointe, H., Kamelian, K., Smith, A.D., Prystajecky, N., Goodfellow, I., Wilson, S.J., Harrigan, R., Snutch, T.P., Loman, N.J., Quick, J. 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv [Preprint]. 2020.09.04.283077. doi:10.1101/2020.09.04.283077.
- Vu, M.N., Alvarado, R.E., Morris, D.R., Lokugamage, K.G., Zhou, Y., Morgan, A.L., Estes, L.K., McLeland, A.M., Schindewolf, C., Plante, J.A., Ahearn, Y.P., Meyers, W.M., Murray, J.T., Crocquet-Valdes, P.A., Weaver, S.C., Walker, D.H., Russell, W.K., Routh, A.L., Plante, K.S., Menachery, V. 2023. Loss-of-function mutation in Omicron variants reduces spike protein expression and attenuates SARS-CoV-2 infection. bioRxiv [Preprint]. 2023.04.17.536926. doi: 10.1101/2023.04.17.536926.
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27(8):763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D., Uriu K., Plianchaisuk A., Kosugi Y., Pan L., Zahradnik J. Genotype to phenotype Japan (G2P-Japan) consortium; Ito J, Sato K. Virological characteristics of the SARS-CoV-2 omicron XBB.1.16 variant. Lancet Infect. Dis. 2023;23(6):655–656. doi: 10.1016/s1473-3099(23)00278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.