Abstract
The characterization of a hyd gene cluster encoding the stable, bidirectional [NiFe]hydrogenase 1 enzyme in Thiocapsa roseopersicina BBS, a purple sulfur photosynthetic bacterium belonging to the family Chromatiaceae, is presented. The heterodimeric hydrogenase 1 had been purified to homogeneity and thoroughly characterized (K. L. Kovacs et al., J. Biol. Chem. 266:947–951, 1991; C. Bagyinka et al., J. Am. Chem. Soc. 115:3567–3585, 1993). As an unusual feature, a 1,979-bp intergenic sequence (IS) separates the structural genes hydS and hydL, which encode the small and the large subunits, respectively. This IS harbors two sequential open reading frames (ORFs) which may code for electron transfer proteins ISP1 and ISP2. ISP1 and ISP2 are homologous to ORF5 and ORF6 in the hmc operon, coding for a transmembrane electron transfer complex in Desulfovibrio vulgaris. Other accessory proteins are not found immediately downstream or upstream of hydSL. A hup gene cluster coding for a typical hydrogen uptake [NiFe]hydrogenase in T. roseopersicina was reported earlier (A. Colbeau et al. Gene 140:25–31, 1994). The deduced amino acid sequences of the two small (hupS and hydS) and large subunit (hupL and hydL) sequences share 46 and 58% identity, respectively. The hup and hyd genes differ in the arrangement of accessory genes, and the genes encoding the two enzymes are located at least 15 kb apart on the chromosome. Both hydrogenases are associated with the photosynthetic membrane. A stable and an unstable hydrogenase activity can be detected in cells grown under nitrogen-fixing conditions; the latter activity is missing in cells supplied with ammonia as the nitrogen source. The apparently constitutive and stable activity corresponds to hydrogenase 1, coded by hydSL, and the inducible and unstable second hydrogenase may be the product of the hup gene cluster.
Hydrogenases are widely spread metalloenzymes in bacteria catalyzing the uptake or production of molecular hydrogen. Hydrogenases have been classified into two groups on the basis of their metal content at the active center: Fe-only hydrogenases, and [NiFe]hydrogenases (some [NiFe]hydrogenases contain also selenium) (2, 14, 30, 38). Hydrogenases are located in the cytoplasm, in the periplasm, or in the membrane of the various bacteria.
Most microorganisms apparently contain a designated hydrogenase for the diverse physiological tasks. Therefore, for a number of species more than one hydrogenase has been described (for reviews, see references 14, 17, and 38). The hydrogenases may differ in electron carrier specificity, in subcellular localization, and in regulation of expression.
Several purple and green photosynthetic bacteria have been reported to contain hydrogenase, and all known examples belong to the typical heterodimer [NiFe]hydrogenase family (for a recent review, see reference 37). In spite of the metabolic versatility of these organisms, only one membrane-associated enzyme has been reported for Rhodobacter capsulatus (10, 11, 35) and for Chromatium vinosum (reviewed in reference 2). R. rubrum also contains a membrane-bound [NiFe]hydrogenase (1), although recent observations (16, 19) are compatible with the occurrence of more than one distinct hydrogenases in this organism.
In Thiocapsa roseopersicina BBS, a remarkably stable hydrogenase activity has been found (18) and characterized (20–22, 24). In this report, we present molecular biology evidence that in the photosynthetic bacterium T. roseopersicina BBS there are at least two [NiFe]hydrogenases, encoded by the hup and hyd gene clusters. The two enzymes differ in sequence, gene organization, and regulation of biosynthesis. The genes encoding the two enzymes are located at distinct sites on the chromosome, and two hydrogenase activities of dissimilar stability properties can be separated from cells containing active nitrogenase. The two hydrogenases of similar subunit composition and amino acid sequence but dissimilar stability properties offer a unique possibility for studying molecular structural parameters that stabilize proteins in a single bacterium.
MATERIALS AND METHODS
Growth conditions.
T. roseopersicina BBS, obtained originally from E. N. Kondratieva (Moscow State University, Moscow, Russia), was grown at 28 to 30°C under continuous illumination from a bank of light bulbs for 2 to 4 days in Pfennig’s mineral medium containing either 0.1% NH4Cl or 0.1% sodium glutamate as the nitrogen source (29).
Determination of amino acid sequences from the stable hydrogenase.
Hydrogenase (hydrogenase 1 [see Results and Discussion]) was purified from ammonia-grown cell paste as described by Kovacs et al. (20), from the soluble fraction following cell sonication, or from the Triton X-100-solubilized membrane fraction. The pure protein was identified by activity staining of non-heat-denatured samples after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (24) and by reaction with antibodies raised against various [NiFe]hydrogenases (22). The active hydrogenase band was recovered from the crushed gel slices by soaking in 20 mM sodium phosphate buffer (pH 7.0). The large and small subunits of the pure hydrogenase 1 were isolated after separation of the completely heat-denatured enzyme on a second sodium dodecyl sulfate-polyacrylamide gel. For determination of N-terminal amino acid sequences, the protein bands were transferred onto Problott (Applied Biosystems) membranes. The isolated subunits were treated with aspartate endopeptidase (sequencing grade; Boehringer), and internal peptides were analyzed as described by Toussaint et al. (36).
DNA manipulations.
Preparation of total DNA and plasmids and cloning were done as described by Colbeau et al. (12) or according to general practice (32).
PCR conditions.
Several oligonucleotides were designed (Table 1) by using the determined hydrogenase 1 peptide sequences and the codon usage of T. roseopersicina BBS (12). PCR was performed in a thermal cycler (Techne/Gene E), using an annealing temperature of 50 to 55°C and an elongation step at 72°C. The products were analyzed in 1% agarose gels or in 5% acrylamide gels (32), typically after 25 PCR cycles. In most cases, several PCR products were obtained. Fragments of the expected sizes were isolated from the gel and cloned into the cloning vectors (e.g., pBluescript [Stratagene]). DNA sequencing was done manually by the dideoxy-chain termination method (33) with a Sequenase version 2.0 kit (U.S. Biochemicals) or by automatic sequencing (Applied Biosystems model 373 Stretch DNA sequencer).
TABLE 1.
Oligonucleotide sequences used in PCRs to identify the hydS and hydL genes of T. roseopersicina
| Name | Sequencea |
|---|---|
| ACX12/N | ATCGAGGTSCARATGGAYGG |
| ACX13/R | TCYTGGATCTGSCCSGCRTC |
| ACX14/R | TGSGCSACRTACTCYTGGAT |
| ACX15/R | TGSCCSGCSACSGTSSWRTASCC |
| TRSA/N | GCNGCNTTNGAGCAGGCNAAGMGNCCNCC |
M, A or C; N, A, C, G, or T; R, A or G; S, C or G; Y, C or T.
Hybridization conditions.
Total genomic DNA samples of T. roseopersicina BBS digested with restriction enzymes were separated in a 1.0% agarose gel in TAE buffer (32). DNA fragments were denatured and then transferred onto a Hybond nylon membrane (Amersham) by capillary transfer or in a semidry vacuum apparatus (Pharmacia-LKB) in the presence of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (32). Southern or colony hybridization experiments were carried out as recommended by the manufacturer (Boehringer).
Separation of hydrogenase activities.
Photosynthetically grown T. roseopersicina BBS culture (2 liters) was harvested at the late exponential phase by centrifugation. The cell paste was broken by sonication (Branson Sonifier) on ice. Almost all hydrogenase 1 activity can be detached from the membrane (20); however, it was not known how strongly hydrogenase 2 was associated with the membrane. Therefore, various solubilization methods were tested. The total cell extract or the Triton X-100-solubilized membrane fraction separated after ultracentrifugation (100,000 × g, 45 min, 15°C) was immediately applied onto a DEAE-cellulose (DE52; Whatman) column (3.5 by 12 cm) and washed with 20 mM Tris-HCl buffer containing 20 mM sodium phosphate (pH 8.0). The column was washed with 1 column volume of the loading buffer, and after elution of the loosely bound protein fraction with 40 mM phosphate in the same buffer (3 column volumes), a linear phosphate gradient of between 40 and 500 mM (5 column volumes) was applied. The conductivity and hydrogen-evolving activity (4) of the collected fractions were determined immediately after elution and following overnight incubation under air at 4°C. All of these operations were done under air.
Nucleotide sequence.
The 7,873-bp DNA sequence of T. roseopersicina determined in this study has been deposited at GenBank under accession no. AF002817.
RESULTS AND DISCUSSION
Comparison of the purified hydrogenase 1, HydSL, with the predicted products of the hupSL genes.
A gene cluster, encoding the hupSLC hydrogenase structural genes and various hydrogenase accessory genes, had been identified in T. roseopersicina BBS by hybridization with the homologous hup genes from R. capsulatus and sequenced (12). The N-terminal amino acid sequences of the predicted HupS and HupL structural gene products differed from the subunits of the protein routinely isolated based on its hydrogenase activity (12, 24). To determine whether the difference originated in protein modification during purification, in this study we used hydrogenase samples obtained from various cell fractions of cultures grown in the presence of ammonia (see Materials and Methods) to redetermine the N-terminal amino acid sequences. The N termini of the hydrogenase subunits from these preparations were identical and differed from those predicted for HupS and HupL. This was also the case for an internal peptide sequence determined for each subunit. The results strongly suggested the occurrence of at least two hydrogenases in T. roseopersicina BBS.
Identification of hydSL genes.
Oligonucleotides corresponding to the N-terminal and the internal peptide sequences of the large subunit of hydrogenase 1 yielded PCR products ACRG4 and TSUP2 (Fig. 1). PCR using ACX12 and ACX14 followed by a second PCR on the product with ACX12 and ACX13 gave the 0.9-kb ACRG4 fragment (oligonucleotide sequences are listed in Table 1).
FIG. 1.
Cloning, restriction map, and arrangement of hyd genes. The locations of PCR primer sequences, designed on the basis of the N-terminal and internal peptide sequences, are indicated. The principal PCR products have been cloned as pTSUP and pACRG4 for sequencing. The positive cosmid was identified by hybridization with the PCR product ACRG4. This cosmid insert was digested with BamHI and PstI, and the 5-kb BamHI and 4.5-kb PstI fragments, giving positive hybridization, were subcloned into the corresponding sites of pBluescribe vector (Stratagene), yielding clones pTSH2/8 and pTSH4/5, respectively. The overlapping clones were further subcloned into pBluescribe and pBluescriptKS and -SK and then sequenced by using specific synthetic primers and/or T3 and T7 primers. The hydS and hydL genes code for the small and large subunits, respectively, of hydrogenase 1. IS is the 2-kb IS harboring two ORFs, isp1 and isp2. Abbreviations for restriction endonucleases: BHI, BamHI; BHII, BssHII; BII, BglII; BXI, BstXI; E, EcoRI; H, HincII; P, PstI; S, SphI; X, XhoI.
The ACRG4 PCR product was identified by the following criteria. (i) The main PCR product was a fragment of approximately 0.9 kb, the size expected from the distance between the N-terminal and internal peptides. (ii) The translated amino acid sequence of the cloned PCR product matched exactly the known hydrogenase 1 peptide sequences adjacent to the amino acids coded by the PCR primers. The overall DNA sequence of the PCR product showed homology with other [NiFe]hydrogenase large-subunit sequences. (iii) A highly specific Southern hybridization response, showing only one strongly hybridizing band in the T. roseopersicina genome, was obtained with ACRG4 (data not shown). These bands did not correspond to hupSL (12).
Cloning and sequencing of hydSL.
Positive clones have been identified in a cosmid library (12) of T. roseopersicina by colony hybridization with labeled ACRG4 or TSUP2. The arrangement of the corresponding hydrogenase genes is summarized in Fig. 1.
The hydS and hydL genes encode proteins of 370 and 577 amino acids, respectively, which are processed to form the mature HydS (34,062-Da) and HydL (63,892-Da) proteins. These data are in good agreement with the stable hydrogenase 1 subunit sizes determined by denaturating electrophoresis (34 and 64 kDa [24]). The translated HydS and HydL amino acid sequences and the N termini and internal peptides from hydrogenase 1 subunits are in perfect match.
We found significant homology between HydSL and other [NiFe]hydrogenase coding sequences, including HupSL of T. roseopersicina. The HupS-HydS and HupL-HydL amino acid sequence identities are 46 and 58%, respectively.
Compared with the translated HupSL sequences, large blocks of identical amino acids characterize the HupL and HydL homologies, whereas identity is restricted to short amino acid sequences within the small subunit outside the conserved motifs. The positions of cysteine residues and those of recognized conserved amino acid motifs for class I [NiFe]hydrogenases (39) are well preserved both in HupSL and in HydSL. Protein database search revealed that the HydL subunit shows strongest homology with the corresponding subunit sequences of Bradyrhizobium japonicum (34), R. capsulatus (26), Azotobacter chroococcum (15), Desulfovibrio gigas (27), and D. vulgaris (13). These results corroborate earlier immunological findings (22).
At the N terminus of HydS, a typical signal peptide sequence, containing the consensus RRXFXK sequence motif (38), is found. The putative signal peptidase cleavage site is alanine. The presence of the signal peptide coding region at the 5′ end of hydS, and a similar sequence in hupS (12), suggests that both hydrogenases are exported into or through the photosynthetic membrane.
Immediately upstream and downstream from the gene coding for the large subunit, a set of accessory genes are located in a typical [NiFe]hydrogenase gene cluster (17, 37). Apparently, the hyd genes in T. roseopersicina BBS represent an exception, as no sequence homologous with any hydrogenase accessory genes has been detected within 1 kb upstream and 2 kb downstream from hydSL. The region downstream from hydSL contains sequences characteristic of Rho-independent terminators (8). Therefore, genes coding for products necessary for the maturation of hydrogenase 1 of T. roseopersicina BBS are not organized as in other [NiFe]hydrogenase gene clusters. Either the accessory genes are located elsewhere in the genome or the accessory genes around hupSL participate in the biosynthesis of HydSL.
The 2-kb IS between hydS and hydL.
hydS and hydL are separated by a 1,979-bp intergenic sequence (IS). Such an arrangement of [NiFe]hydrogenase structural genes is rather unusual. In cyanobacteria, a DNA fragment is intercalated inside the hupL gene. This element plays a significant role in heterocyst differentiation (9). The structural gene cluster coding for one of the hydrogenase isoenzymes in Escherichia coli (28) is the only known example where the two [NiFe]hydrogenase structural genes are separated. In this case, the hybAB genes are reported to be intercalated between the structural genes hybO and hybC (7, 28).
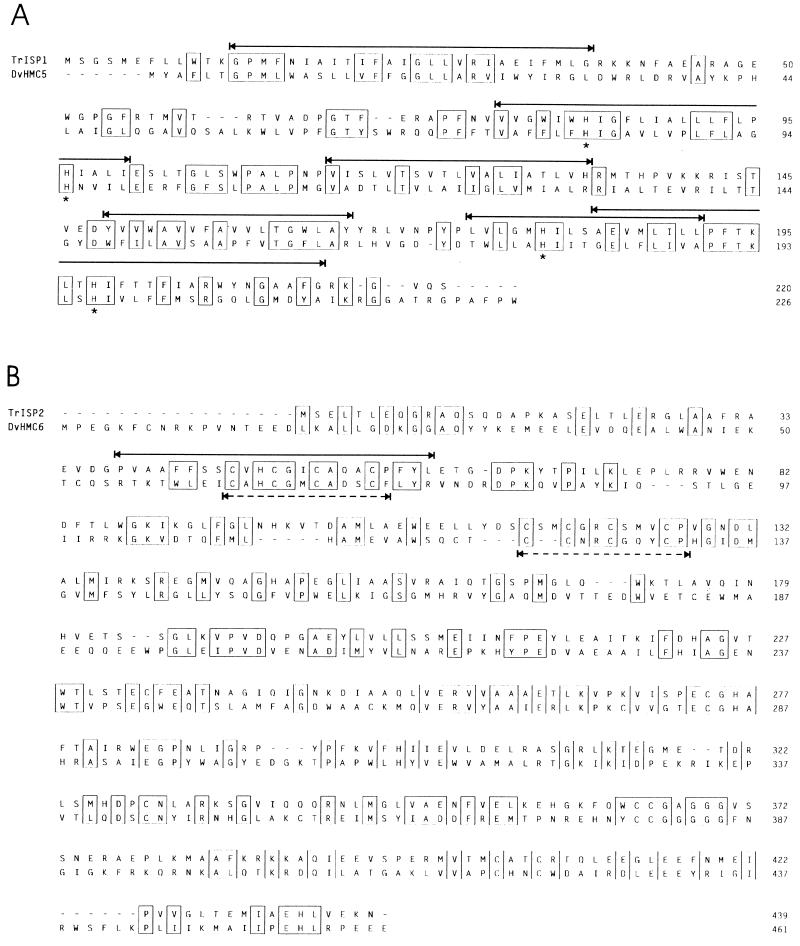
Two open reading frames (ORFs) which may code for the ISP1 and ISP2 protein products have been identified within the IS fragment (Fig. 2). ISP1 contains five putative membrane-spanning domains and a cytochrome heme-binding site, it shows 28% sequence identity to the predicted product of ORF5, found in D. vulgaris as part of a large transmembrane electron transport protein cluster encoded by the hmc operon (31). Adjacent to two of the transmembrane domains there are at least three amino acid residues with strong positive charge which may anchor the protein at the interior surface of the membrane. ISP2 is unlikely an integral membrane protein, it contains two ferredoxin-type iron-sulfur cluster-binding sites, as ORF6 does in the same Desulfovibrio hmc operon, and there is 26% amino acid sequence identity between the two putative proteins. Genes showing significant homology to ORF5 and ORF6 of the Desulfovibrio hmc operon have been found in methanogens, and the gene products have been identified recently as a b-type cytochrome (MbhdrD) and heterodisulfide reductase (MbhdrE), (25). Interestingly, homologous genes have also been discovered in the dsr locus of C. vinosum D (DSMZ180), which encodes enzymes participating in sulfur metabolism (12a). The presence of the two ORFs intercalated between the hydrogenase 1 structural genes of T. roseopersicina BBS is intriguing in light of the involvement of homologous gene products in the sulfur metabolism of several phylogenetically distant species. Preliminary Northern blot analysis indicates that the IS region is transcribed together with the hydSL hydrogenase structural genes (data not shown). It is also noted that although hybAB in E. coli is about the same size as the IS in T. roseopersicina, the homology between ISP1 of T. roseopersicina and HybB of E. coli does not seem significant outside the heme-binding motif, and ISP2 and HybA are apparently unrelated.
FIG. 2.
Alignment of translated ORFs ISP1 and ISP2 (TrISP1 and TrISP2) from the DNA region intercalated between hydS and hydL of T. roseopersicina with the predicted products of ORF5 and ORF6 of the hmc operon of D. vulgaris (DvHMC5). The multiple alignment was done by using the CLUSTALW and PRETTYPLOT programs of the Genetics Computer Group program package. Homologous amino acids are boxed, the putative transmembrane hydrophobic domains in the T. roseopersicina sequence are marked with continuous lines between arrows, the heme (ORF5)-binding conserved histidines (at positions 83, 96, 180, and 198) are shown marked with asterisks, and the iron-sulfur cluster (ORF6)-binding domains are indicated with dashed lines between arrows. As evident from the homologies, ORF5 and ORF6 contain the corresponding sequences and therefore are not marked separately.
The structural genes of the two hydrogenases are at least 15 kb apart.
Two deoxyoligonucleotides which corresponded to the N-terminal sequences of the two hydrogenases were synthesized. To design a probe specific for hydrogenase 1, the N-terminal four amino acids of the small subunit were selected since they differed from the predicted sequence of HupS (12). For a HupSL-specific probe, the deoxyoligonucleotide that coincided to the first 21 nucleotides of the hupL sequence was chosen since the corresponding amino acid sequence was missing from the large subunit of the isolated hydrogenase 1.
The two oligonucleotides hybridized to dissimilar DNA regions. The oligonucleotide specific for hydrogenase 1 did not hybridize with the cosmid containing the hupSL genes, suggesting that hydSL was separated from hupSL by at least 15 kb, the distance of hupSL from the ends of its cosmid (data not shown). The hybridization results corroborated the existence of two distinct sets of hydrogenase genes in T. roseopersicina BBS and indicated that the genes encoding the two hydrogenases were not contiguous.
Separation of two hydrogenase activities by ion-exchange chromatography.
The synthesis of hydrogenase in R. capsulatus is induced under nitrogenase derepressed conditions by the hydrogen gas produced through the nitrogenase complex (reviewed in reference 37). In T. roseopersicina, however, the hydrogenase activity is apparently constitutively synthesized (18). To detect a hydrogen-inducible hydrogenase activity in T. roseopersicina, cells were grown either in ammonia-containing medium (nitrogenase repressed) or in a medium where glutamate served as nitrogen source (nitrogenase active). The cell paste was disrupted by sonication and was applied onto a DEAE-cellulose DE52 ion-exchange column within 2 h. Fractions eluted by a linear sodium phosphate gradient were assayed by the hydrogen evolution assay (4).
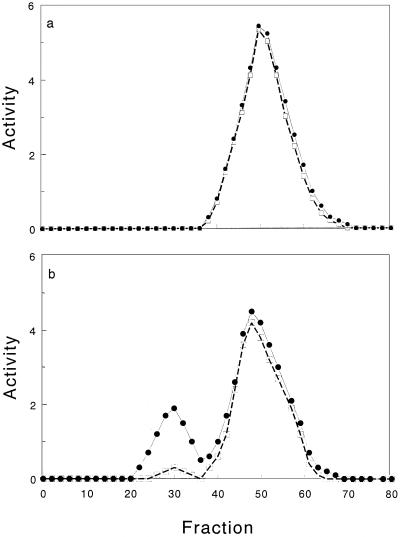
A single hydrogenase peak, eluting at about 0.3 M sodium phosphate, was recovered from ammonia-grown cells (Fig. 3). This corresponded to the stable hydrogenase activity currently identified as hydrogenase 1 and purified routinely from nitrogenase-repressed T. roseopersicina cells (20).
FIG. 3.
DEAE chromatogram of total cell extract prepared from ammonia-grown cells (a; nitrogenase repressed) and cells grown with glutamate as the nitrogen source (b; nitrogenase active). The hydrogen evolution activity of each fraction was determined after elution (•) and following incubation at 4°C under air for 24 h (□). Activities are expressed as micromoles of H2 produced per hour per milligram of protein.
In contrast to the ammonia-grown cells, two peaks displaying hydrogenase activity were separated from cells supplied with glutamate as the nitrogen source. The new hydrogenase peak, present in T. roseopersicina cells synthesizing nitrogenase, was tentatively assigned to hydrogenase 2. This hydrogenase peak emerged at a salt concentration of about 0.1 M, comparable to the elution profile of the hydrogenase of R. capsulatus (10). The hydrogen-evolving activity in the peak containing the hydrogenase 2 represented 5 to 30% of the activity in the hydrogenase 1 peak. This may be due either to a low synthesis of the protein or to a significantly lower hydrogen-evolving activity of hydrogenase 2 than of hydrogenase 1; alternatively, perhaps hydrogenase 2 was simply less stable than hydrogenase 1.
Indeed, after storage of the hydrogenase-containing fractions at cold room temperature for 24 h, about 90% of the hydrogen-evolving activity in the hydrogenase 2 peak was lost, while the hydrogenase 1 peak remained fairly unaltered (Fig. 3). This observation is in good agreement with the facts that the solubilized hydrogenase 1 is reversibly inactivated only during purification under air (20), while other hydrogenases (e.g., that of R. capsulatus [10]) are irreversibly inactivated by oxygen and/or in the cold under similar conditions.
The oxygen and cold sensitivity, combined with the apparently inducible nature of hydrogenase 2, explains why the enzyme has not been detected in earlier studies. Hydrogenase 1 has been routinely purified under air and from nitrogenase-repressed cells (20). The two subunits of the hydrogenase 1 protein have molecular masses of 34 and 64 kDa, respectively (24). The estimated molecular masses of the small (HupS, 36 kDa; HydS, 34 kDa) and large (HupL, 65 kDa; HydL, 64 kDa) subunits are in the same range; therefore, the two hydrogenases cannot be definitively distinguished on the basis of size. However, it is to be noted that the biochemical and molecular biology data do not rigorously exclude the possibility of the presence of more than two hydrogenases in T. roseopersicina.
Conclusion.
In this study, a 7,873-bp nucleotide sequence from T. roseopersicina BBS that includes the hyd genes was cloned and sequenced. The results provide conclusive evidence for the occurrence of at least two distinct hydrogenases in T. roseopersicina BBS. Two hydrogenase fractions with distinct oxygen and cold sensitivity could be isolated from cells grown under conditions where nitrogenase was synthesized, and two sets of genes capable of encoding two distinct [NiFe]hydrogenases have been identified in T. roseopersicina BBS genomic DNA.
The main source of hydrogenase activity, i.e., hydrogenase 1, has been characterized as a membrane-associated protein (3, 23). Indeed, hydS contains a signal peptide coding sequence, similarly to the signal peptide sequence motif upstream from hupSLC (12). However, once detached from the membrane, hydrogenase activity remained soluble.
Taking the various observations together, we conclude that the two T. roseopersicina hydrogenases belong to the class I of [NiFe]hydrogenases as defined by Wu and Mandrand (39). They are both heterodimers, they possess subunits of similar size, and they are both associated with the membrane. The deduced amino acid sequences of the two small (hupS and hydS) and large subunit (hupL and hydL) sequences show 46 and 58% identity, respectively.
Differences between HupSL and HydSL are as follows. HydSL, which has been implicated in membrane bioenergetic functions (23), is apparently a constitutive enzyme, whereas HupSL appears to be an inducible enzyme. Hydrogenase 1 shows outstanding stability against irreversible heat, oxygen, and protease inactivation (21, 24), while hydrogenase 2 is quickly inactivated under air and/or by the cold (Fig. 3). Mutagenesis studies will be required to establish unequivocally that the products of the hupSL genes correspond to the hydrogenase 2 activity.
An intriguing question is the processing and role of the intergenic 2-kb IS between hydS and hydL. This IS is composed of two ORFs, which may code for electron transfer proteins. Future experiments are needed to establish the putative link between sulfur metabolism and hydrogenase 1. The accessory genes required to process and mature hydrogenase 1 in T. roseopersicina are also yet to be located on the chromosome.
It was noted that under stringent Southern hybridization conditions, labeled probes of hydSL gave positive hybridization signals only with genomic DNA fragments containing hyd-specific sequences. As the identification of hydrogenase-encoding genes is usually done through heterologous hybridization with hup-related gene probes, any hyd-related sequence in the genome may have been easily overlooked in other organisms. A systematic analysis for the occurrence and distribution of this [NiFe]hydrogenase subfamily is therefore warranted. Preliminary data show that a hyd-related gene cluster, including the IS, is present in C. vinosum, an other photosynthetic bacterium (6).
ACKNOWLEDGMENTS
The collaboration was partially supported by travel grants through the COST Action 8.18 and the French-Hungarian Bilateral Exchange Program “Balaton” (project 29). The Hungarian laboratory thanks the financial support of OTKA, OMFB, PHARE-TDQM, PHARE-TEMPUS, UNDP HUN/95/002, and the U.S.-Hungarian Joint Fund (JF221), and the Laboratoire de Biochimie et Biophysique des Systemes Integres of the French CEA/CNRS UMR314.
REFERENCES
- 1.Adams M W W, Hall D O. Physical and catalytic properties of the hydrogenase of Rhodospirillum rubrum. In: Schlegel H G, Schneider K, editors. Hydrogenases: their catalytic activity, structure and function. Göttingen, Germany: Goltze K. G.; 1979. pp. 159–169. [Google Scholar]
- 2.Albracht S P J. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 3.Bagyinka C, Kovacs K L, Rak E. Localization of hydrogenase in Thiocapsa roseopersicina photosynthetic membrane. Biochem J. 1982;202:255–258. doi: 10.1042/bj2020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagyinka C, Zorin N A, Kovacs K L. Unconsidered factors affecting hydrogenase activity measurement. Anal Biochem. 1984;142:7–15. doi: 10.1016/0003-2697(84)90509-8. [DOI] [PubMed] [Google Scholar]
- 5.Bagyinka C, Whitehead J P, Maroney M J. An X-ray absorption study of nickel redox chemistry in hydrogenase. J Am Chem Soc. 1993;115:3567–3585. [Google Scholar]
- 6.Barta F, Rakhely G, Kovacs K L. Proceedings of the 5th International Conference on the Molecular Biology of Hydrogenases. Grenoble, France: CEA Publishers; 1997. Molecular biology of hydrogenase from Chromatium vinosum; p. 110. [Google Scholar]
- 7.Boxer D H, Sargent F, Richard D. Proceedings of the COST Action 818 Workshop on Regulation of Hydrogenase Biosynthesis. 1996. Biosynthesis of Escherichia coli hydrogenases; p. 8. [Google Scholar]
- 8.Carafa Y A, Brody E, Thermes C. Prediction of Rho-independent Escherichia coli transcription terminators. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco C D, Buettner J A, Golden J W. Programmed rearrangement of a cyanobacterial hupL gene in heterocysts. Proc Natl Acad Sci USA. 1995;92:791–795. doi: 10.1073/pnas.92.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colbeau A, Chabert J, Vignais P M. Purification, molecular properties and localization in the membrane of the hydrogenase of Rhodopseudomonas capsulata. Biochim Biophys Acta. 1983;748:116–127. [Google Scholar]
- 11.Colbeau A, Richaud P, Toussaint B, Caballero F J, Elster C, Delphin C, Smith R L, Chabert J, Vignais P M. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993;8:15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 12.Colbeau A, Kovacs K L, Chabert J, Vignais P M. Cloning and sequences of the structural (hupSLC) and accessory (hupDHI) genes for hydrogenase biosynthesis in Thiocapsa roseopersicina. Gene. 1994;140:25–31. doi: 10.1016/0378-1119(94)90726-9. [DOI] [PubMed] [Google Scholar]
- 12a.Dahl, C. (University of Bonn). Personal communication.
- 13.Deckers H M, Wilson F R, Voordouw G. Cloning and sequencing of a [NiFe] hydrogenase operon from Desulfovibrio vulgaris Miyazaki F. J Gen Microbiol. 1990;136:2021–2028. doi: 10.1099/00221287-136-10-2021. [DOI] [PubMed] [Google Scholar]
- 14.Fauque G, Peck H D, Jr, Moura J J G, Huynh B H, Berlier Y, DerVartanian D V, Teixeira M, Przybyla A E, Lespinat P A, Moura I, LeGall J. The three classes of hydrogenases from sulfate reducing bacteria of the genus Desulfovibrio. FEMS Microbiol Rev. 1988;54:299–344. doi: 10.1111/j.1574-6968.1988.tb02748.x. [DOI] [PubMed] [Google Scholar]
- 15.Ford C M, Garg N, Garg R P, Tibelius K H, Yates M G, Arp D J, Seefeldt L C. The identification, characterization, sequencing and mutagenesis of the genes (hupSL) encoding the small and large subunits of the H2—uptake hydrogenase of Azotobacter chroococcum. Mol Microbiol. 1990;4:999–1008. doi: 10.1111/j.1365-2958.1990.tb00672.x. [DOI] [PubMed] [Google Scholar]
- 16.Fox J D, Kerby R L, Roberts G P, Ludden P W. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J Bacteriol. 1996;178:1515–1524. doi: 10.1128/jb.178.6.1515-1524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich B, Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- 18.Gogotov I N. Hydrogenases of phototrophic microorganisms. Biochimie. 1986;68:181–187. doi: 10.1016/s0300-9084(86)81082-3. [DOI] [PubMed] [Google Scholar]
- 19.Kern M, Klipp W, Klemme J-H. Increased nitrogenase-dependent H2 photoproduction by hup mutants of Rhodospirillum rubrum. Appl Environ Microbiol. 1994;60:1768–1774. doi: 10.1128/aem.60.6.1768-1774.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacs K L, Tigyi G, Alfonz H. Purification of hydrogenase by fast protein liquid chromatography and by conventional separation techniques: a comparative study. Prep Biochem. 1985;15:321–334. doi: 10.1080/00327488508062449. [DOI] [PubMed] [Google Scholar]
- 21.Kovacs K L, Bagyinka C, Tigyi G. Proteolytic resistance and its utilization in purification of hydrogenase from Thiocapsa roseopersicina. Biochim Biophys Acta. 1988;935:166–172. [Google Scholar]
- 22.Kovacs K L, Seefeldt L C, Tigyi G, Doyle C M, Mortenson L E, Arp D J. Immunological relationship among hydrogenases. J Bacteriol. 1989;171:430–435. doi: 10.1128/jb.171.1.430-435.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacs K L, Bagyinka C. Structural properties, functional states and physiological roles of hydrogenase in photosynthetic bacteria. FEMS Microbiol Rev. 1990;87:407–412. [Google Scholar]
- 24.Kovacs K L, Tigyi G, Thanh L T, Lakatos S, Kiss Z, Bagyinka C. Structural rearrangements in active and inactive forms of hydrogenase from Thiocapsa roseopersicina. J Biol Chem. 1991;266:947–951. [PubMed] [Google Scholar]
- 25.Künkel A, Vaupel M, Heim S, Thauer R K, Hedderich R. Heterodisulfide reductase from methanol-grown cells of Methanosarcina barkeri is not a flavoenzyme. Eur J Biochem. 1997;244:226–234. doi: 10.1111/j.1432-1033.1997.00226.x. [DOI] [PubMed] [Google Scholar]
- 26.Leclerc M, Colbeau A, Cauvin B, Vignais P M. Cloning and sequencing of the genes encoding the large and small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol Gen Genet. 1988;214:97–107. doi: 10.1007/BF00340186. . (Erratum, 215:368, 1989.) [DOI] [PubMed] [Google Scholar]
- 27.Li C, Peck H D, Jr, LeGall J, Przybyla A E. Cloning, characterization, and sequencing of the genes encoding the large and small subunits of the periplasmic [NiFe] hydrogenase of Desulfovibrion gigas. DNA. 1987;6:539–551. doi: 10.1089/dna.1987.6.539. [DOI] [PubMed] [Google Scholar]
- 28.Menon N K, Chatelus C Y, Dervartanian M, Wendt J C, Shanmugam K T, Peck H D, Jr, Przybyla A E. Cloning, sequencing and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J Bacteriol. 1994;176:4416–4423. doi: 10.1128/jb.176.14.4416-4423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfennig N. Photosynthetic bacteria. Annu Rev Microbiol. 1967;21:286–324. doi: 10.1146/annurev.mi.21.100167.001441. [DOI] [PubMed] [Google Scholar]
- 30.Przybyla A E, Robbins J, Menon N, Peck H D., Jr Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev. 1992;88:109–136. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- 31.Rossi M, Brent W, Pollack R, Reij M W, Keon R G, Fu R, Voordouw G. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J Bacteriol. 1993;175:4699–4711. doi: 10.1128/jb.175.15.4699-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayavedra-Soto L A, Powell G K, Evans H J, Morris R O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci USA. 1988;85:8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seefeldt L C, McCollum L C, Doyle C M, Arp D J. Immunological and molecular evidence for a membrane-bound, dimeric hydrogenase in Rhodopseudomonas capsulata. Biochim Biophys Acta. 1987;914:299–303. [Google Scholar]
- 36.Toussaint B, Delic-Attree I, De Sury D’Aspermont R, David L, Vincon M, Vignais P M. Purification of the integration host factor homolog of Rhodobacter capsulatus: cloning and sequencing of the hip gene, which encodes the b subunit. J Bacteriol. 1993;175:6499–6504. doi: 10.1128/jb.175.20.6499-6504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vignais P M, Toussaint B, Colbeau A. Regulation of hydrogenase gene expression. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1175–1190. [Google Scholar]
- 38.Voordouw G. Evolution of hydrogenase genes. Adv Inorg Chem. 1992;38:397–422. [Google Scholar]
- 39.Wu L F, Mandrand M A. Microbial hydrogenases: primary structure, classification, signatures and phylogeny. FEMS Microbiol Rev. 1993;104:243–270. doi: 10.1111/j.1574-6968.1993.tb05870.x. [DOI] [PubMed] [Google Scholar]