Abstract
In this study, we cloned and sequenced a DNA fragment from an ordered cosmid library of Helicobacter pylori NCTC 11638 which confers to a siderophore synthesis mutant of Escherichia coli (EB53 aroB hemA) the ability to grow on iron-restrictive media and to reduce ferric iron. Sequence analysis of the DNA fragment revealed the presence of an open reading frame with high homology to the ribA gene of Bacillus subtilis. This gene encodes a bifunctional enzyme with the activities of both 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) synthase and GTP cyclohydrolase II, which catalyze two essential steps in riboflavin biosynthesis. Expression of the gene (designated ribBA) resulted in the formation of one translational product, which was able to complement both the ribA and the ribB mutation in E. coli. Expression of ribBA was iron regulated, as was suggested by the presence of a putative FUR box in its promotor region and as shown by RNA dot blot analysis. Furthermore, we showed that production of riboflavin in H. pylori cells is iron regulated. E. coli EB53 containing the plasmid with H. pylori ribBA excreted riboflavin in the culture medium, and this riboflavin excretion also appeared to be iron regulated. We postulate that the iron-regulated production of riboflavin and ferric-iron-reduction activity by E. coli EB53 transformed with the H. pylori ribBA gene is responsible for the survival of EB53 on iron-restrictive medium. Because disruption of ribBA in H. pylori eliminates its ferric-iron-reduction activity, we conclude that ribBA has an important role in ferric-iron reduction and iron acquisition by H. pylori.
Iron plays an essential role in many enzymatic processes in most living cells. The role of iron in microbial infections has been largely investigated, since pathogenic bacteria encounter an iron-limiting environment when they attempt to colonize or invade a mammalian host (19, 26). In the presence of oxygen, iron is found in the ferric state (Fe3+) and may form insoluble ferric hydroxides not readily available for living organisms (31). Furthermore, intracellular, iron is predominantly found bound to heme, iron-sulfur proteins, or ferritin. The small quantities of extracellular iron are withheld from invading organisms in serum by transferrin and by lactoferrin on mucosal surfaces. It is believed that the various amounts of iron available in the host largely influence the establishment and extent of microbial infection (38).
Two general mechanisms have been developed by microorganisms to survive in the iron-limited environment of the mammalian host (26). First, host iron-binding compounds like transferrin, lactoferrin, and heme can be used directly by binding of these iron-containing compounds to the bacterial cell surface. Subsequently, the iron is released at the cell surface or the host iron compound is taken up as a whole. Second, the pathogen can synthesize and excrete high-affinity iron chelators called siderophores or ferric reductants to dissociate iron from host iron complexes. The ferrisiderophore complex is subsequently bound by specific receptors and internalized. Ferric reductants keep iron in its soluble ferrous form (Fe2+), and in this form iron may diffuse through the porin channels. Subsequently, the iron can be transported by a specific ferrous-iron-uptake system.
Helicobacter pylori, the causative agent of human gastric and duodenal ulcerations, resides in the mucosa of the human stomach (6). Both hemin and lactoferrin can be used as sole iron sources (21), and a lactoferrin-binding protein has been described for H. pylori (12). We have previously described iron-repressible outer membrane proteins potentially involved in heme uptake by H. pylori (39). These iron-repressible outer membrane proteins were also expressed in vivo (40). In another previous report, we could not detect siderophores but we did find iron-regulated ferric-iron-reduction activity in culture supernatants of H. pylori (41).
In the human stomach the ingested iron from the food is present partially as heme iron (10 to 20%) but mostly as nonheme iron (80%) which might form insoluble ferric hydroxide complexes (23). Both for the host and for H. pylori, reduction of ferric iron to the ferrous form is essential to permit membrane transport. The H. pylori-associated ferric-iron-reduction activity may be involved in the mobilization of the iron from insoluble ferric complexes present in the more neutral environment underneath the mucus layer of the human stomach mucosa. Furthermore, this ferric-iron-reduction activity may increase the accessibility of free iron from several bound iron sources (heme compounds, ferritin, lactoferrin, transferrin, and ferrisiderophores), as has been described for Listeria monocytogenes (11).
In this study we describe a gene from H. pylori which could confer to an iron-uptake mutant of E. coli the ability to liberate bound iron from its environment through ferric-iron-reduction activity. The gene responsible for this, designated ribBA, encodes one bifunctional enzyme catalyzing two essential steps in riboflavin biosynthesis. The transcription of ribBA was iron regulated and resulted in the iron-regulated production of the ferric reductant riboflavin in H. pylori.
MATERIALS AND METHODS
Culture media, bacterial strains, and growth conditions.
The H. pylori type strain ATCC 43504 was routinely cultured on Columbia Agar supplemented with 5% lysed horse blood and Dent supplement (Dent plates; Oxoid, Hampshire, United Kingdom). Liquid culture of H. pylori was performed in 50-ml Erlenmeyer flasks that were placed in a microaerobic jar fixed on a gently shaking platform at 37°C for 2 days. At the onset of each experiment, cultures were inoculated to a final optical density at 650 nm of 0.01. Iron-restrictive liquid culture conditions were achieved by addition of 20% newborn calf serum (Gibco Ltd., Paisly, Scotland) to brucella broth (BS20 [39]). Iron repletion was achieved by addition of 1 mM Fe(III) nitrate to BS20 medium.
Escherichia coli DH5α, which harbors the ordered cosmid library of H. pylori (7), was grown in Luria-Bertani broth (LB) containing 25 μg of kanamycin per ml. E. coli EB53 (aroB hemA [14]) was grown in LB supplemented with 50 μg of 5-aminolevulinic acid (Sigma Chemical Co., St. Louis, Mo.) per ml. The iron-restricted medium used for selection of EB53 ferric-iron-reduction transformants was nutrient broth (Oxoid) with 0.3 mM dipyridyl (BDH Chemicals), solidified with 1.5% Bacto Agar (Difco) (NBD plates [32]). E. coli strains were grown under aerobic conditions at 37°C.
Recombinant DNA and RNA techniques.
The ordered cosmid library of H. pylori NCTC 11638 (7) was used for transformation of E. coli EB53. This library contains 68 cosmids with DNA fragments of approximately 40 kb that had been partially digested with Sau3AI and cloned in the low-copy-number vector Lorist6. The cosmids were extracted and purified with plasmid spin columns as described by the manufacturer (Qiagen Ltd., Crawly, United Kingdom). EB53 was transformed by a standard electroporation protocol for E. coli (24). For Southern blot hybridization analysis, HindIII-digested cosmids were electrophoresed through a 0.8% agarose gel, transferred to a nylon filter (Boehringer Mannheim, Mannheim, Germany) by capillary blotting, and cross-linked to the filter by UV irradiation for 3 min.
For RNA spot blot analysis, total RNA was isolated from iron-restricted and iron-replete cultures of H. pylori (109 cells) with an RNeasy kit (Qiagen). To ensure complete removal of residual DNA, the RNA was treated with RNase-free DNase I according to the manufacturer’s (Promega, Madison, Wis.) instructions. Amounts of 100, 10, 1, and 0.1 ng of RNA in a total volume of 50 μl of 20× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) were spotted on nylon filters (Boehringer Mannheim) with a spot blot apparatus (Bio-Rad, Veenendaal, The Netherlands). To check whether equal amounts of RNAs of the iron-restricted and iron-replete cells were spotted, we used an 850-bp PCR fragment derived from the 23S rRNA gene of H. pylori as a probe.
DNA probes were labelled with [α-32P]dATP (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) with a random primer labelling kit (Prime-it II; Stratagene, La Jolla, Calif.). Hybridizations were performed overnight at 68°C in hybridization solution (1% [wt/vol] sodium dodecyl sulfate [SDS], 0.1% [wt/vol] sodium lauryl sarcosinate [Sarkosyl; CIBA-GEIGY Pharmaceutical Co.], 1% [wt/vol] blocking reagent [Boehringer Mannheim], 6× SSPE). Washings were performed at room temperature (twice for 10 min each time) with 2× SSPE–1% (wt/vol) SDS and at 68°C (twice for 10 min each time) with 0.1× SSPE–1% SDS (wt/vol).
RNA spot hybridization was quantified with a PhosphorImage screen and PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Radioactivity counts of the cosmids hybridizing with the RNA spots were determined by scanning the blot strips with Imagequant (Molecular Dynamics). By integration of the areas of the count peaks, total hybridization counts of the RNA spots were determined, and these counts were used to calculate the hybridization ratio.
Sequence analysis.
Nucleotide sequences were determined by the dideoxy chain termination method with an automated DNA sequencer (Applied Biosystems model 371 A) and fluorescent-dye-labelled terminators (30). Homology searches were performed by making use of the National Center for Biotechnology Information BLAST Network service. Alignments were performed with the Lasergene program (DNAstar, Madison, Wis.).
PCR analysis and expression of H. pylori ribBA as a glutathione S-transferase (GST) fusion protein.
H. pylori ribBA and flanking regions from clone 1B and genomic DNA from H. pylori NCTC 11638 were amplified under standard conditions by PCR (Perkin-Elmer) with the sense primer 5′-TAAGCGGTTTGCTAAATGCGG-3′ and the antisense primer 5′-CGCAAACTATCCAAATCTTTGGG-3′. The amplified DNA was cloned in the pGEM-T PCR cloning vector from Promega.
To introduce an EcoRI and a SalI site at the 5′ and 3′ ends of H. pylori ribBA, respectively, we used PCR with the sense primer 5′-TAGGGAaTTcGAATGATCTTAAAACGAGTTACTGAA-3′ and the antisense primer 5′-ATCGTgTCgACAAGCTTAAGCCCAAACCCGCTCAAAGCC-3′. Lowercase letters indicate changes from the original sequence for correct restriction sites. The amplified DNA was cloned in frame into EcoRI- and SalI-digested pGEX-4T-2 (Pharmacia) to generate an ribBA-GST fusion construct. E. coli DH5α was transformed with this construct, and induction of the recombinant protein with isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) was performed according to standard protocols. Cells were lysed by sonication, and aliquots of the pellets (insoluble fusion protein fraction) were analyzed on an SDS–12% polyacrylamide gel.
Complementation of E. coli ribA and ribB mutants.
The original plasmid isolated from clone 1B, containing the ribBA gene, was tested for its ability to complement ribA (GTP cyclohydrolase II) and ribB (3,4-dihydroxy-2-butanone 4-phosphate [DHBP] synthase) mutations in E. coli E. coli ribA::Tn5 (BSV18) and ribB::Tn5 (BSV11) used for the complementation studies were kindly provided by Barbara Bachmann (E. coli Genetic Stock Center, Yale University). E. coli strains were cultured in LB supplemented with 20 μg of riboflavin (Sigma) per ml. The mutant strains were transformed with the ribBA-containing plasmid by a standard electroporation protocol for E. coli (24). Complementation of the E. coli mutations was determined by restoration of the ability of BSV18 and BSV11 to grow on M9 minimal medium plates in the absence of riboflavin.
Construction of an ribBA disruption in H. pylori.
The 1,346-bp ribBA PCR product (see Fig. 2) cloned in the PCR cloning vector pGEM-T (Promega) was used. A kanamycin resistance gene, aphA (36), was cloned in the unique XmaI site of this construct. The final construct with the disrupted ribBA gene, which is not able to replicate in H. pylori, was introduced directly into H. pylori by natural transformation as described previously (37). Transformants were selected on Dent plates containing 10 μg of kanamycin per ml and 20 μg of riboflavin (Sigma) per ml. Single colonies were isolated and restreaked several times on Dent plates with kanamycin and riboflavin. To select for possible ribBA mutants, the transformants were subsequently tested for growth on plates without riboflavin. Transformants not growing on these plates were isolated, and insertion of the kanamycin cassette into ribBA was confirmed by PCR analysis.
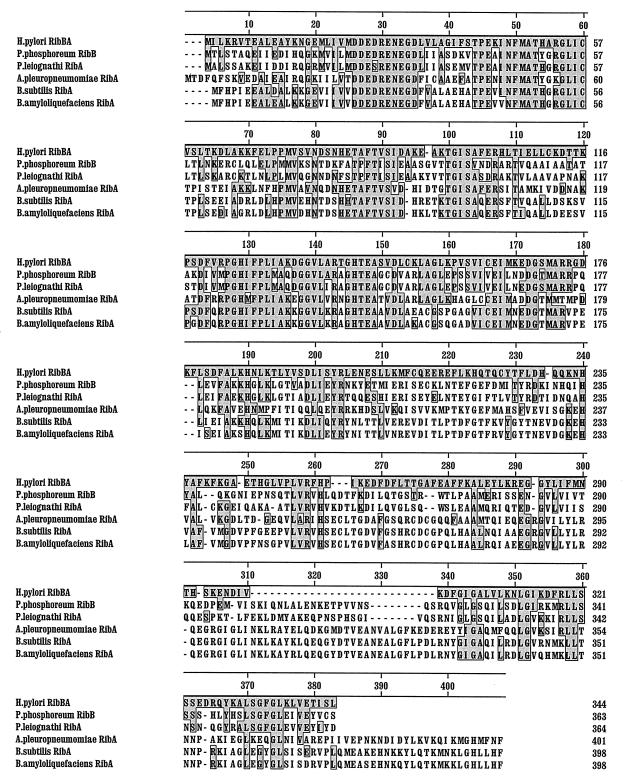
FIG. 2.
Alignment of H. pylori RibBA with homologous proteins from other bacteria by the Clustal method. Boxed residues are amino acids that match exactly the residues of the H. pylori RibBA protein.
Reduction assays.
For the determination of ferric-iron-reduction activity, we used bathophenantroline-disulfonate (BPDS; Sigma) as a chromogenic ferrous-iron chelator. E. coli EB53 transformants were cultured overnight (O/N) in 10 ml of LB containing 25 μg of kanamycin per ml and 50 μg of 5-aminolevulinic acid per ml. H. pylori transformants were cultured O/N in 10 ml of brucella broth containing 1 g of cyclodextrins per liter, 10 μg of kanamycin per ml, and 20 μg of riboflavin per ml. Medium not inoculated with bacteria was incubated under the same conditions for use as a reference. BPDS was added to the O/N cultured cells and inoculated media to a final concentration of 1 mM. As a ferric-iron source, Fe(III) ammonium-sulfonate was added to a final concentration of 50 μM. Subsequently, cells and media were incubated at 37°C for 1 h. One-milliliter aliquots were taken, the cells were removed by centrifugation, and the optical densities of the Fe(II)-BPDS complexes in the supernatants were measured at 535 nm. The Fe(III)-reduction activity was quantified as nanomoles of Fe(II)-BPDS formed per hour per 109 cells. For the determination of the amount of Fe(II)-BPDS, a molar extinction coefficient of 22,140 at 535 nm was used (25).
Fluorescence emission spectra.
Fluorescence emission spectra from 480 to 650 nm were measured in a Perkin-Elmer model LS50B spectrofluorometer with an excitation beam of 450 nm. Cell-free culture supernatants (800 μl) from overnight cultures were analyzed in a quartz cuvette. For determination of intracellular riboflavin, 108 H. pylori cells were washed with phosphate-buffered saline and lysed by solubilizing the pellets in 1 ml of a 5% (wt/vol) Sarkosyl solution. This extract (800 μl) was analyzed in the spectrofluorometer.
RESULTS
Selection of E. coli EB53 transformants with ferric-iron-reduction activity.
In order to find genes in H. pylori which are involved in ferric-iron reduction, we transformed an iron-uptake mutant of E. coli (EB53 aroB hemA) with the ordered cosmid library of Bukanov and Berg (7). All 68 cosmids were pooled and electroporated in EB53. Upon transformation the bacteria were grown O/N on iron-restrictive NBD medium containing kanamycin and 5-aminolevulinic acid to select only for aroB complementation, and many transformants were observed. Transformation of EB53 with the Lorist6 cosmid not containing an insert did not result in growth on this medium. Transformant colonies varied in colony diameter (0.5 to 1 mm), and some of the larger colonies showed a pronounced red color. This red color indicated that in these colonies more Fe(II) was present, since Fe(II) being bound by the ferrous-iron chelator 2,2 dipyridyl present in the NBD plates results in a red color. The accumulation of Fe(II) may possibly have been the result of ferric-iron-reduction activity in these transformed colonies. To investigate this, we selected the colony, designated clone 1B, that had the most pronounced growth and red color for further analysis and tested the bacteria from this colony for ferric-iron-reduction activity. We could not detect any significant ferric-iron-reduction activity by untransformed EB53 or EB53 containing the Lorist6 cosmid without the H. pylori insert. Clone 1B, however, did show significant ferric-iron-reduction activity; 2.25 (±0.3) nmol of Fe(II)-BPDS/h/109 cells.
Sequence analysis of clone 1B.
Clone 1B contained the Lorist6 cosmid with an insert of approximately 3 kb. Obviously, a deletion or recombination occurred in the original ∼40-kb cosmid. Such deletions of cosmid clones are, however, not uncommon in Rec+ strains like EB53. Sequence analysis of the 3-kb insert revealed the presence of two partial open reading frames (ORFs) at the borders of the insert and only one complete ORF, which encodes a polypeptide of 38 kDa. To rule out the possibility that this ORF was the result of a deleted or recombinant DNA fragment, we performed PCR analysis with primers flanking the 38-kDa ORF and with both clone 1B plasmid and H. pylori genomic DNA as templates. The same 1,346-bp fragment was amplified from both clone 1B plasmid DNA and genomic DNA.
We cloned the PCR product amplified from the genomic template in the PCR cloning vector pGEM-T and confirmed the correctness of the DNA fragment by sequence analysis. Figure 1 shows the PCR product with the corresponding genomic ferric-iron-reduction gene. The putative translation product of the gene has high homology with polypeptides encoded by ribA from Bacillus subtilis and several other bacteria (Fig. 2). The ribA gene from B. subtilis is part of the rib operon and encodes a bifunctional enzyme with both DHBP synthase (5′ end) and GTP cyclohydrolase II activities (3′ end) (27).
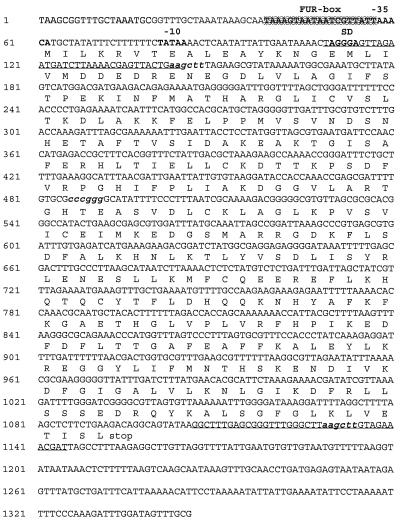
FIG. 1.
Complete nucleotide sequence of the 1,346-bp PCR product amplified from H. pylori NCTC 11638. The Shine-Dalgarno (SD) and the −10 and −35 promotor sequences are indicated in boldface type, and the putative FUR box is shaded. Primers used for PCR amplification and subsequent cloning in the pGEX-4T expression vector are underlined. HindIII restriction sites and the unique XmaI site (used in the construction of the ribBA mutants) are in boldface, italic, lowercase letters.
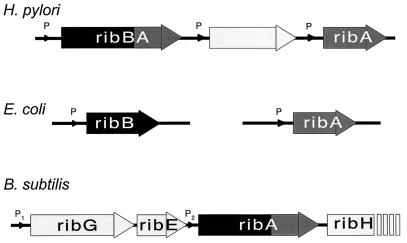
These enzyme activities catalyze essential, rate-limiting steps in riboflavin biosynthesis. In order to investigate whether the H. pylori gene indeed encodes a single, possibly bifunctional polypeptide as in B. subtilis ribA, we cloned the gene in the expression vector pGEX-4T. IPTG induction of this construct resulted in the expression of a 65-kDa fusion protein consisting of the 27-kDa GST and a polypeptide of 38 kDa (Fig. 3, lane 2). This result indicated that both the ribB (5′ end) and ribA (3′ end) homologs are encoded by one single gene, transcription of which results in a single translation product. For these reasons, we designated the H. pylori gene ribBA and the encoded protein RibBA. In E. coli, DHBP synthase and GTP cyclohydrolase activities are encoded by two genes (ribB and ribA, respectively) that are separated and not organized in an operon (2) (Fig. 4). A separate ribA gene in H. pylori has already been cloned by Bereswill et al. (4), and we confirmed the presence of this ribA in our H. pylori strain by PCR analysis (data not shown). Furthermore, in the recently published genomic sequence of H. pylori 26695, both ribA and ribBA are present, separated by only one unidentified ORF (35) (Fig. 4). There was no significant homology between RibA and RibBA, except for some similarities of the C-terminal parts of RibBA and RibA. In the genomic sequence of H. pylori 26695, no separate ribB gene was identified.
FIG. 3.
Schematic representations of the genomic organizations of the H. pylori, E. coli, and B. subtilis genes encoding GTP cyclohydrolase (black) and DHBP synthase (dark gray) activities. The B. subtilis ribA gene encodes a bifunctional enzyme with both enzymatic activities. Promotors are indicated by P’s.
FIG. 4.
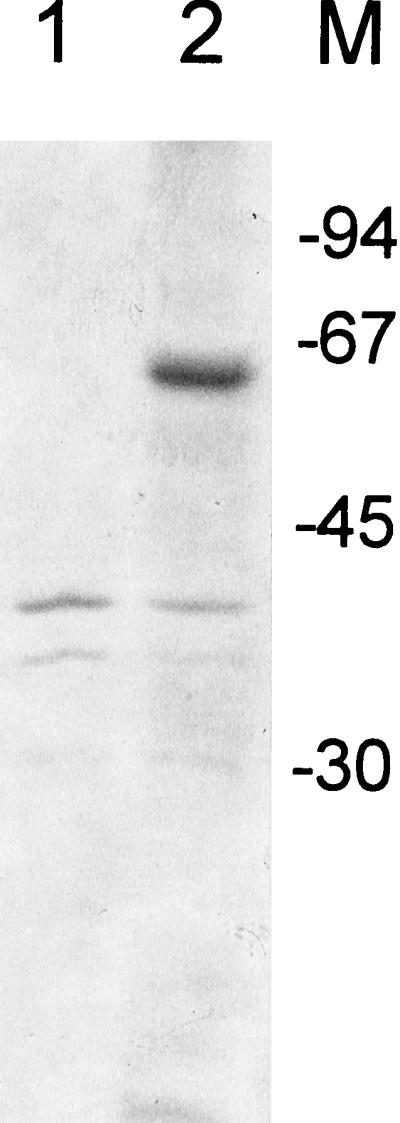
Insoluble protein fraction of E. coli DH5α transformed with pGEX-4T (lane 1) and pGEX-4T with the H. pylori ribBA insert (lane 2). Molecular size markers in kilodaltons are shown at the right.
Complementation of E. coli ribA and ribB mutants by H. pylori ribBA.
In order to determine whether the RibBA protein encoded by ribBA indeed contains both GTP cyclohydrolase II and DHBP synthase activity, we tested the ribBA-containing plasmid derived from clone 1B for its ability to complement an E. coli ribA and ribB mutant. Only upon transformation with the H. pylori ribBA-containing plasmid were these E. coli ribA and ribB mutants able to grow on M9 minimal medium.
Genomic mapping of the H. pylori ribBA gene.
A Southern blot that contained the HindIII-digested DNAs of the 68 cosmids from the ordered cosmid library of strain NCTC 11638 (7) and genomic DNAs from H. pylori ATCC 43504 and NCTC 11638 was hybridized with the 1,346-bp ribBA PCR product shown in Fig. 1. The probe hybridized strongly with a 1-kb HindIII fragment and faintly with bands of approximately 900 and 600 bp present on cosmid 5 and in the genomic DNA digestions of both strains. HindIII restriction sites within ribBA are indicated in Fig. 1.
Iron-regulated transcription of ribBA in H. pylori.
The complete nucleotide sequence of H. pylori ribBA and flanking regions is shown in Fig. 3. The promotor region preceding ribBA contains typical Shine-Dalgarno and −10 and −35 sequences. Furthermore, a putative FUR-binding site (FUR box) could be localized at the −35 promotor region (Fig. 1) (5′-TAAAGTAATAATCGTTATT-3′). Thirteen of these 19 nucleotides match exactly the E. coli FUR consensus sequence: 5′-GATAATGATAATCATTATC-3′. The presence of a FUR consensus sequence at this position indicates that transcription of ribBA is iron regulated. To find evidence for iron-regulated transcription of the ribBA gene, we performed an RNA spot blot analysis with the 1,346-bp ribBA PCR product as a probe. When compared to results with RNA from iron-replete cells, results of hybridization of ribBA with RNA from iron-restricted cells indicated a 1.8-fold (±0.2-fold) increase in hybridization counts.
Iron-regulated excretion of riboflavin by clone 1B.
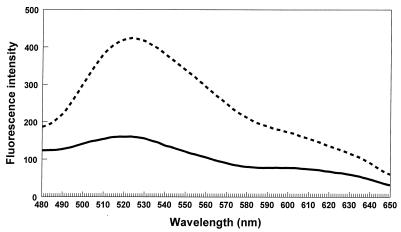
Because cloning of the H. pylori ribBA gene in E. coli might result in an increase of the extracellular riboflavin concentration, we analyzed culture supernatants of the EB53 clone 1B for the presence of riboflavin. The fluorescence emission spectrum of an aqueous riboflavin standard solution showed a single, broad structureless band with a maximum around 523 nm (Fig. 5A).
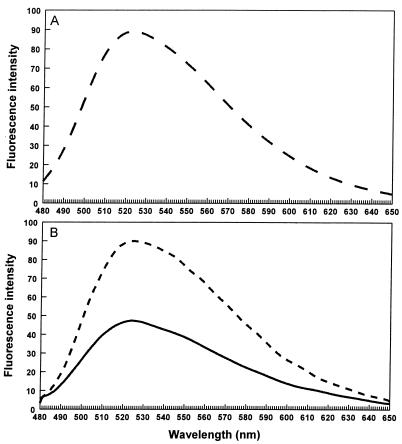
FIG. 5.
(A) Fluorescence emission spectra of an aqueous riboflavin standard solution; (B) culture supernatants from EB53 clone 1B cultured in iron-rich (——) and iron-poor (–––) media.
We measured fluorescence emission spectra of cell-free culture supernatants harvested from an O/N culture of clone 1B cultured in NB medium and used cell-free culture supernatants from an O/N culture of untransformed EB53 in NB medium as a reference. The culture supernatant of clone 1B contained significantly more riboflavin (Fig. 5B). When clone 1B was cultured in iron-restrictive medium (NB with 0.3 mM 2,2-bipyridyl), higher amounts of riboflavin in the culture supernatants were detected than in culture supernatants from clone 1B grown in iron-rich NB medium (Fig. 5B).
Iron-regulated hemolytic activity of clone 1B.
Riboflavin can cause hemolysis of erythrocytes (34). It has been shown that H. pylori contains hemolytic genes (13). These genes were mapped in the H. pylori genome by making use of the same ordered cosmid library from Bukanov and Berg (7) that we used for this study. One of the hemolytic genes was mapped to cosmid 5, the same cosmid that contains H. pylori ribBA. To investigate whether expression of H. pylori ribBA in E. coli results in hemolytic activity, we cultured E. coli clone 1B and an unrelated EB53 transformant which was able to grow on iron-restrictive medium but which did not show any ferric-iron-reduction activity (clone 1A) on blood agar plates containing kanamycin, aminolevulinic acid, and 0.3 mM 2,2 dipyridyl. After 1 day of growth, colonies of clone 1B showed clearly β-hemolysis whereas clone 1A did not. When both transformants were grown on the same plates without 2,2 dipyridyl (non-iron restrictive), neither clone 1A nor clone 1B showed hemolytic activity.
Iron-regulated production of riboflavin in H. pylori.
H. pylori exhibits both extracellular and cell-associated ferric-iron-reduction activities that are clearly iron regulated (41). This reduction activity may be due to the extracellular secretion of reduced riboflavin. To test this hypothesis, we measured the fluorescence of culture supernatants from H. pylori cultured in iron-restricted (BS20) and iron-replete [BS20 plus 1 mM Fe(III) nitrate] media. We could not detect any significant differences in the riboflavin concentrations in the culture supernatants. Possibly, the fluorescence emission spectrum was disturbed by the high background fluorescence caused by the large amount of serum in the culture medium. To circumvent this problem, we decided to test washed H. pylori cells that were cultured in iron-restrictive medium for a potential accumulation of riboflavin in the cell or at the cell surface. We could hardly detect riboflavin in iron-replete cells, but we detected large amounts of riboflavin in iron-restricted cells (Fig. 6).
FIG. 6.
Fluorescence emission spectra of Sarkosyl extracts from 109 washed H. pylori cells cultured in iron-rich (——) and iron-poor (–––) media.
Ferric-iron-reduction activity of H. pylori ribBA mutants.
To study the possible role of ribBA in H. pylori ferric-iron-reduction activity, we sought to inactivate ribBA. To inactivate ribBA, a kanamycin resistance gene was cloned into an ribBA-containing vector (pGEM-T) and this construct was introduced into H. pylori by natural transformation. Because this vector is unable to replicate in H. pylori, selection of transformants on kanamycin-containing plates resulted in insertion of the kanamycin cassette by homologous recombination of flanking sequences within ribBA. This result was confirmed by PCR analysis and the inability of these mutants to grow on plates which do not contain riboflavin. These mutants were tested for their ability to reduce ferric iron. Transformants which were given the same treatments but grew normally on plates not containing riboflavin were used as a positive control in these ferric-iron-reduction experiments. Uninoculated media were used as negative controls. We could not detect any difference between the amounts of BPDS-Fe(II) formed in 1 h in the culture supernatant of broth inoculated with the ribBA mutants and our negative control (uninoculated broth). However, the H. pylori transformants without an ribBA mutation showed significant ferric-iron-reduction activity: 41.3 (±6.5) nmol of BPDS-Fe(II)/h/109 cells.
DISCUSSION
We have cloned and sequenced a gene from H. pylori that has high homology to the ribA gene of B. subtilis, which encodes a bifunctional enzyme with GTP cyclohydrolase and DHBP synthase activities (27). This bifunctional gene is in contrast to the gene arrangement in E. coli, which has two separate genes (ribA and ribB) for these activities. These two enzymatic activities catalyze the initial steps in each of the two converging branches of the riboflavin synthesis pathway, and it has been suggested that these reaction steps are rate limiting for the biosynthesis of riboflavin (1, 28, 29). We confirmed that transcription of the H. pylori gene results in a single translational product. This translational product must contain both enzyme activities, as was determined by complementation studies of E. coli ribA and ribB mutants. Therefore, we designated the H. pylori gene ribBA.
Riboflavin (vitamin B2) is a precursor of the coenzymes flavine adenine dinucleotide and flavin mononucleotide, and it is essential for basic metabolism. It is synthesized by plants and by most microorganisms but not by higher animals (1). The flavins are well-known as the key prosthetic groups of several redox enzymes called flavoproteins. Protein-free flavins play roles as electron transfer mediators in important biological functions, such as activation of ribonucleotide reductase (15), bioluminescence (20), oxygen activation (18), and ferric-iron reduction (8, 16).
Reduced free flavins can increase the availability of iron from a wide range of iron-containing compounds by reduction of their iron to the ferrous form, for which the different ligands have less affinity (8, 11, 42). Several flavin reductases (NAD[P]H:flavin oxidoreductases) have been reviewed by Fontecave et al. (16). Reduced free flavins are well-suited for ferric-iron reduction probably because (i) they are small molecules (when compared with flavoproteins), (ii) they have very low redox potentials (below −0.2 V), and (iii) they are able to transfer their two electrons stepwise, due to the relative stability of the semiflavin state (16). It is well-known that riboflavin synthesis is induced under low-iron conditions in fungi and bacteria (10). In some of these organisms, riboflavin might be used in a ferric iron reduction system. However, both iron-regulated riboflavin synthesis and ferric-iron-reduction activity do not normally occur in E. coli.
Cloning of H. pylori ribBA in E. coli EB53 (aroB hemA) restored H. pylori’s ability to grow on iron-restrictive media. Furthermore, the ribBA-containing transformant (clone 1B) showed higher ferric-iron-reduction activity than untransformed EB53. Because ferric-iron reduction has been described as one of several iron-uptake mechanisms pathogenic bacteria use (26), this might explain growth of clone 1B on iron-restrictive medium. Clone 1B showed a marked increase in extracellular riboflavin production compared to that of EB53. A similar riboflavin excretion has also been described for the cloning of Actinobacillus pleuropneumoniae rib genes in E. coli (17).
The ribBA promotor region contained a putative FUR box within the −35 region, and we have evidence for iron-regulated transcription of the H. pylori ribBA gene. First, total RNA from H. pylori cultured in iron-restricted medium hybridized stronger with ribBA than RNA from H. pylori cultured in iron-replete medium. Second, both in EB53 and in H. pylori, more riboflavin is produced when the bacteria are cultured under iron-restrictive conditions. This iron-regulated production of riboflavin indicates a role in iron uptake of H. pylori through ferric-iron reduction.
Recently, we have described the existence of both extracellular and cell-associated ferric-iron-reduction activity in H. pylori (41). These reduction activities were also induced by low concentrations of iron in the culture media. This is not unique for H. pylori, because recently, a secreted ferric reductase enzyme and a membrane-bound reductase have been described for L. monocytogenes (3, 11). Extracellular secreted ferric reductants 3-hydroxyanthranilate (3HAA) have also been characterized for yeasts (9, 22, 25). Interestingly, it is known that plant roots exhibit ferric-iron-reduction activity (5) and excrete and/or accumulate riboflavins when they are cultured under iron-restrictive conditions. Therefore, it has been proposed that riboflavins have a role in plant iron acquisition (33).
We were unable to establish whether riboflavin is excreted by H. pylori under iron-restrictive conditions, because our iron-restricted medium (BS20) had an extremely high background fluorescence that disturbed our detection method. However, we were able to detect larger amounts of H. pylori-associated riboflavin when H. pylori was cultured in iron-restricted medium than in iron-replete medium. This accumulation of riboflavin in the cell or at the cell surface, in combination with a flavin reductase to generate reduced flavins, might form a potent iron-reduction system. ribBA plays an important role in this iron-reduction system, since disruption of the gene results not only in the inability of H. pylori to survive on media lacking riboflavin but also in elimination of its ferric-iron-reduction activity.
We postulate that under iron-poor conditions, the iron-reduction system of H. pylori is activated, which implies an increased demand for riboflavin. Because the initial, rate-limiting reaction steps in riboflavin synthesis are catalyzed by the bifunctional ribBA gene product, higher expression of the ribBA gene would increase the amount of riboflavin. Expression of the other rib genes remains necessary, but under iron restriction an increase in the expression of ribBA is enough to elevate riboflavin levels. Bereswill et al. cloned a separate ribA gene from H. pylori (4), which was also present in our ribBA-containing H. pylori strain. Furthermore, in the recently published genomic sequence of H. pylori 26695, both ribA and ribBA are present, separated by only one unidentified ORF (35) (Fig. 3). There was no significant homology between RibA and RibBA, except for some similarities in the C-terminal parts of RibBA and RibA.
Interestingly, both ribA and ribBA conferred hemolytic activity to E. coli (reference 4 and this paper). This hemolytic activity probably results from overproduction of riboflavin mediated by these genes, which can result in lysis of erythrocytes (34). In contrast with ribA, however, the hemolytic activity caused by ribBA was iron regulated. The ribA gene was not iron regulated and probably serves as a housekeeping gene necessary for a cell’s metabolic riboflavin demands. In the genomic sequence of strain 26695, a separate ribB gene was not identified. From this we conclude that ribBA, which can provide a sudden increased demand for riboflavin under iron-poor conditions, also serves as an ribB housekeeping gene.
In bacteria that need more riboflavin in certain stress situations, the separate ribB and ribA may have evolved to the combined ribBA form to allow fast, tightly coregulated, and high expression of DHBP synthase and GTP cyclohydrolase.
ACKNOWLEDGMENTS
We thank J. Maaskant for her technical assistance with experiments and G. Koningstein for his assistance with the determination of nucleotide sequences.
REFERENCES
- 1.Bacher A. Biosynthesis of flavins. In: Muller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 1. Boca Raton, Fla: Chemical Rubber Co.; 1991. pp. 215–259. [Google Scholar]
- 2.Bandrin S V, Rabinovich P M, Stepanov A I. Three linkage groups of genes involved in riboflavin biosynthesis in Escherichia coli. Sov Genet. 1983;19:1103–1109. [PubMed] [Google Scholar]
- 3.Barchini E, Cowart R E. Extracellular iron reductase activity produced by Listeria monocytogenes. Arch Microbiol. 1996;166:51–57. doi: 10.1007/s002030050354. [DOI] [PubMed] [Google Scholar]
- 4.Bereswill S, Fassbinder F, Covacci A, Völzing C, Ries H, Bacher A, Kist M. Characterization of the Helicobacter pylori ribA gene that confers haemolytic activity to Escherichia coli. Ir J Med Sci. 1997;166:30. [Google Scholar]
- 5.Bienfait H F, Bino R J, van der Bliek A M, Duivenvoorden J F, Fontaine J M. Characterization of ferric reducing activity in roots of Fe-deficient Phaseolus vulgaris. Physiol Plant. 1983;59:196–202. [Google Scholar]
- 6.Blaser M J. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 7.Bukanov N O, Berg D E. Ordered cosmid library and high-resolution physical-genetic map of Helicobacter pylori strain NCTC 11638. Mol Microbiol. 1994;11:509–523. doi: 10.1111/j.1365-2958.1994.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 8.Covès J, Fontecave M. Reduction and mobilization of iron by a NAD(P)H:flavin oxidoreductase from Escherichia coli. Eur J Biochem. 1993;211:635–641. doi: 10.1111/j.1432-1033.1993.tb17591.x. [DOI] [PubMed] [Google Scholar]
- 9.Dancis A, Klausner R D, Hinnebusch A G, Barriocanal J G. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demain A L. Oversynthesis of riboflavin. Annu Rev Microbiol. 1972;26:369–388. doi: 10.1146/annurev.mi.26.100172.002101. [DOI] [PubMed] [Google Scholar]
- 11.Deneer H G, Healey V, Boychuk I. Reduction of exogenous ferric iron by a surface-associated ferric reductase of Listeria spp. Microbiology. 1995;141:1985–1992. doi: 10.1099/13500872-141-8-1985. [DOI] [PubMed] [Google Scholar]
- 12.Dhaenens L, Szczebara F, Husson M O. Identification, characterization, and immunogenicity of the lactoferrin-binding protein from Helicobacter pylori. Infect Immun. 1997;65:514–518. doi: 10.1128/iai.65.2.514-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drazek E S, Dubois A, Holmes R K, Kersulyte D, Akopyants N S, Berg D E, Warren R L. Cloning and characterization of hemolytic genes from Helicobacter pylori. Infect Immun. 1995;63:4345–4349. doi: 10.1128/iai.63.11.4345-4349.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberspächer B, Braun V. The involvement of cytochromes in the uptake of ferrichrome by Escherichia coli K-12. FEMS Microbiol Lett. 1980;7:61–64. [Google Scholar]
- 15.Fontecave M, Eliasson R, Reichard P. Enzymatic regulation of the radical content of the small subunit of Escherichia coli ribonucleotide reductase involving reduction of its redox centers. J Biol Chem. 1989;264:9164–9170. [PubMed] [Google Scholar]
- 16.Fontecave M, Covès J, Pierre J L. Ferric reductases or flavin reductases. Biometals. 1994;7:3–8. doi: 10.1007/BF00205187. [DOI] [PubMed] [Google Scholar]
- 17.Fuller T E, Mulks M H. Characterization of Actinobacillus pleuropneumoniae riboflavin biosynthesis genes. J Bacteriol. 1995;177:7265–7270. doi: 10.1128/jb.177.24.7265-7270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudu P, Touati D, Nivière V, Fontecave M. The NAD(P)H:flavin oxidoreductase from Escherichia coli as a source of superoxide radicals. J Biol Chem. 1994;269:8182–8188. [PubMed] [Google Scholar]
- 19.Griffiths E. The iron-uptake systems of pathogenic bacteria. In: Bullen J J, Griffiths E, editors. Iron and infection: molecular, physiological and clinical aspects. Chichester, United Kingdom: John Wiley & Sons; 1987. pp. 69–138. [Google Scholar]
- 20.Hastings J W, Potrikus C J, Gupta S C, Kurfurst M, Makenson J C. Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol. 1985;26:235–291. doi: 10.1016/s0065-2911(08)60398-7. [DOI] [PubMed] [Google Scholar]
- 21.Husson M A, Legrand D, Spik G, Leclerc H. Iron acquisition by Helicobacter pylori: importance of human lactoferrin. Infect Immun. 1993;61:2694–2697. doi: 10.1128/iai.61.6.2694-2697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesuisse E, Simon M, Klein R, Labbe P. Excretion of anthranilate and 3-hydroxyanthranilate by Saccharomyces cerevisiae: relationship to iron metabolism. J Gen Microbiol. 1992;138:85–89. doi: 10.1099/00221287-138-1-85. [DOI] [PubMed] [Google Scholar]
- 23.Lombard M, Chua E, O’Toole P. Regulation of intestinal non-haem iron absorption. Gut. 1997;40:435–439. doi: 10.1136/gut.40.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 25.Nyhus K J, Wilborn A T, Jacobson E S. Ferric iron reduction by Cryptococcus neoformans. Infect Immun. 1997;65:434–438. doi: 10.1128/iai.65.2.434-438.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne S M. Iron and virulence in the family Enterobacteriaceae. Crit Rev Microbiol. 1988;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 27.Perkins J B, Pero J G. Biosynthesis of riboflavin, biotin, folic acid, and cobalamin. In: Sonenshein A, editor. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 319–334. [Google Scholar]
- 28.Richter G, Ritz H, Katzenmeier G, Volk R, Kohnle A, Lottspeich F, Allendorf D, Bacher A. Biosynthesis of riboflavin: cloning, sequencing, mapping, and expression of the gene coding for GTP cyclohydrolase II in Escherichia coli. J Bacteriol. 1993;175:4045–4051. doi: 10.1128/jb.175.13.4045-4051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter G, Volk R, Krieger C, Lahm H-W, Röthlisberger U, Bacher A. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J Bacteriol. 1992;174:4050–4056. doi: 10.1128/jb.174.12.4050-4056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiro T G, Saltman P. Polynuclear complexes of iron and their biological implications. Struct Bonding. 1969;6:116–120. [Google Scholar]
- 32.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Susin S, Abian J, Sanchez-Baeza F, Luisa Peleato M, Abadia A, Gelpi E, Abadia J. Riboflavin 3"- and 5′-sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris) J Biol Chem. 1993;268:20958–20965. [PubMed] [Google Scholar]
- 34.Suzuki Y, Miura T, Ogiso T. Riboflavin photosensitized hemolysis of rat erythrocytes in the presence of serum. J Pharmacobio-Dyn. 1982;5:568–575. doi: 10.1248/bpb1978.5.568. [DOI] [PubMed] [Google Scholar]
- 35.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Petreson J D, Kelley J M, Cotton M D, Weidman J M, Fuji C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J G. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–546. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 36.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Roos K P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 38.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worst D J, Otto B R, De Graaff J. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect Immun. 1995;63:4161–4165. doi: 10.1128/iai.63.10.4161-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worst D J, Sparrius M, Kuipers E J, Kusters J G, De Graaff J. Human serum antibody response against iron-repressible outer membrane proteins of Helicobacter pylori. FEMS Microbiol Lett. 1996;144:29–32. doi: 10.1111/j.1574-6968.1996.tb08504.x. [DOI] [PubMed] [Google Scholar]
- 41.Worst, D. J., C. M. J. E. Vandenbroucke-Grauls and J. G. Kusters. Siderophore-like activity and ferric iron reduction in Helicobacter pylori. Submitted for publication.
- 42.Yubisui T, Maksuki T, Tanishima K, Takeshita K, Yoneyama M. NADPH-flavin reductase in human erythrocytes and the reduction of methemoglobin through flavin by the enzyme. Biochem Biophys Res Commun. 1977;76:174–182. doi: 10.1016/0006-291x(77)91683-7. [DOI] [PubMed] [Google Scholar]