Abstract
The 20S proteasome from the methanoarchaeon Methanosarcina thermophila was produced in Escherichia coli and characterized. The biochemical properties revealed novel features of the archaeal 20S proteasome. A fully active 20S proteasome could be assembled in vitro with purified native α ring structures and β prosubunits independently produced in Escherichia coli, which demonstrated that accessory proteins are not essential for processing of the β prosubunits or assembly of the 20S proteasome. A protein complex with a molecular mass intermediate to those of the α7 ring and the 20S proteasome was detected, suggesting that the 20S proteasome is assembled from precursor complexes. The heterologously produced M. thermophila 20S proteasome predominately catalyzed cleavage of peptide bonds carboxyl to the acidic residue Glu (postglutamyl activity) and the hydrophobic residues Phe and Tyr (chymotrypsinlike activity) in short chromogenic and fluorogenic peptides. Low-level hydrolyzing activities were also detected carboxyl to the acidic residue Asp and the basic residue Arg (trypsinlike activity). Sodium dodecyl sulfate and divalent or monovalent ions stimulated chymotrypsinlike activity and inhibited postglutamyl activity, whereas ATP stimulated postglutamyl activity but had little effect on the chymotrypsinlike activity. The results suggest that the 20S proteasome is a flexible protein which adjusts to binding of substrates. The 20S proteasome also hydrolyzed large proteins. Replacement of the nucleophilic Thr1 residue with an Ala in the β subunit abolished all activities, which suggests that only one active site is responsible for the multisubstrate activity. Replacement of β subunit active-site Lys33 with Arg reduced all activities, which further supports the existence of one catalytic site; however, this result also suggests a role for Lys33 in polarization of the Thr1 N, which serves to strip a proton from the active-site Thr1 Oγ nucleophile. Replacement of Asp51 with Asn had no significant effect on trypsinlike activity, enhanced postglutamyl and trypsinlike activities, and only partially reduced lysozyme-hydrolyzing activity, which suggested that this residue is not essential for multisubstrate activity.
The 26S proteasome is a high-molecular-weight proteinase which, in the Eucarya domain, is responsible for the rapid proteolysis of central metabolic enzymes, regulatory proteins, transcription factors, and misfolded or damaged proteins (reviewed in reference 4). The 26S eucaryal proteasome is also an essential component of the ATP-dependent, nonlysosomal proteolytic pathway and, in higher vertebrates, generates peptides which are presented to the immune system on major histocompatibility complex class I molecules. The 20S catalytic core of the 26S eucaryal proteasome is highly conserved with the archaeal 20S proteasomes from Thermoplasma acidophilum (5) and Methanosarcina thermophila (14). A 20S proteasome has been purified from the hyperthermophilic archaeon Pyrococcus furiosus (2), and novel forms of proteasomes have also been reported in microbes from the Bacteria domain (reviewed in reference 4), signifying the broad importance of these high-molecular-weight proteases in procaryotes.
The 20S proteasome from T. acidophilum is a cylinder of four stacked rings, each comprised of seven subunits in an α7β7β7α7 configuration with the α subunits localized to the outer rings and the β subunits forming the inner rings (10, 13). The active sites are localized to the β subunits on the inner surface of the cylinder. The walls of the cylinder have no openings, which limits the entry of substrates into the two openings at the ends of the cylinder. The recent crystal structure of the eucaryal 20S proteasome from yeast is similar (9), except that there are seven different types of α and β subunits. Studies with the T. acidophilum enzyme suggest that α subunits form seven-membered rings early in assembly, presumably to provide a scaffolding for the formation of β rings from monomeric β prosubunits (26), which contain a propeptide that is autocatalytically processed during assembly to expose an N-terminal threonine (Thr1), forming the active-site nucleophile for peptide hydrolysis (13, 22). A relatively stable half-proteasome intermediate has been detected during assembly of the mammalian 20S proteasome (8, 21). A chaperone is proposed to assist in assembly of the two half-proteasomes into the eucaryal 20S proteasome. The eucaryal 20S proteasome is multicatalytic, displaying CL (chymotrypsinlike), TL (trypsinlike), and PG (postglutamyl) activities (16, 19). Three distinct β-type subunits (Pre3, Pup1, and Pre2), each with an N-terminal Thr1 active site, are responsible for PG, TL, and CL activities of the yeast 20S proteasome (11). The eucaryal enzyme also catalyzes cleavage after branched-chain amino acids (BRAAP activity) and small neutral amino acids (SNAAP activity) (17). Understanding of the assembly and multisubstrate activity of the archaeal 20S proteasome is not as advanced.
The 20S proteasome from T. acidophilum is the only archaeal 20S proteasome characterized biochemically (1, 6). T. acidophilum is a thermoacidophile which grows aerobically by utilizing glucose as an energy source and is both phylogenetically and metabolically distant from M. thermophila, which is an obligately anaerobic methanoarchaeon obtaining energy for growth by converting simple one- and two-carbon substrates to methane at neutral pH (15). Here we report biochemical features of the M. thermophila 20S proteasome which extend an understanding of the 20S proteasome in general for the Archaea and specifically for the methanoarchaea.
MATERIALS AND METHODS
Materials.
Biochemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). Other organic and inorganic chemicals were from Fisher Scientific and were analytical grade. Restriction endonucleases and DNA-modifying enzymes were from New England Biolabs (Beverly, Mass.), Promega (Madison, Wis.), or US Biochemical (Cleveland, Ohio). Oligonucleotides were from the University of Florida-Interdisciplinary Center for Biotechnology (Gainesville, Fla.) and Integrated DNA Technologies (Coralville, Iowa).
Cloning of psmA and psmB into expression vector pT7-7.
The psmB gene, encoding the β prosubunit, was amplified by PCR with the high-fidelity thermophilic Vent DNA polymerase to minimize the error rate in DNA synthesis. The template DNA (1 ng) was a pUC19-based plasmid containing the complete psmB gene on a 3.0-kb HindIII-to-Sau3A1 fragment from the M. thermophila genome (14). Flanking oligonucleotides with either an NdeI or an EcoRI restriction enzyme site (underlined), 5′-CTGCCCATATGGATAATGACAAATA-3′ and 5′-GGCTAGAATTCAACAGTTAAATGAT-3′, were used for the amplification reaction. The NdeI and EcoRI sites were used to clone the amplification product into expression vector pT7-7 (25). The resulting plasmid, pMT7β, has the psmB translational start codon located 8 bp downstream of the T7 ribosome binding site (Table 1). The psmA gene, encoding the α subunit, was isolated from a previously reported 17-kb Sau3A1 M. thermophila genomic fragment (14) by using restriction enzymes EcoRV and XmnI. The resulting 0.98-kb psmA-specific fragment was cloned into the HincII site of pUC19, and then, by using the restriction enzymes EcoRI and HindIII, psmA was subcloned into both pT7-7 and pMT7β. The resulting plasmids, pMT7α and pMT7βα (Table 1), have 165 bp of genomic DNA included upstream of psmA.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant genotype or phenotype |
|---|---|
| pT7-7 | Apr; contains T7 RNA polymerase promoter φ10 and translation start site for T7 gene 10 protein inserted between PvuII and ClaI sites of pT7-5 (27) |
| pMT7β | Apr; contains 656-bp NdeI-EcoRI fragment with complete psmB gene immediately downstream of translation start site for T7 gene 10 protein in pT7-7 |
| pMT7α | Apr; contains 980-bp EcoRI-HindIII fragment with complete psmA gene and 165 bp of genomic DNA upstream of psmA; fragment is positioned downstream of T7 RNA polymerase promoter φ10 in pT7-7 |
| pMT7βα | Apr; contains 980-bp EcoRI-HindIII fragment with complete psmA gene positioned downstream of psmB gene in pMT7β |
| pMT7βΔ | Apr; contains 632-bp NdeI-EcoRI fragment with psmB gene truncated at 5′ end to delete coding region of 8-residue β propeptide |
| pMT7βΔα | Apr; contains 980-bp EcoRI-HindIII fragment with complete psmA gene positioned downstream of truncated psmB gene in pMT7βΔ |
Heterologous production and purification of the 20S proteasome and α or β subunits.
The psmB and psmA genes were coexpressed and independently expressed in Escherichia coli BL21(DE3) by using the bacteriophage T7 RNA polymerase-promoter system (25). Freshly transformed cells were inoculated into Luria-Bertani medium supplemented with ampicillin (100 mg/liter) and grown at 30°C and 200 rpm until the cells reached an A600 of about 0.7. T7 RNA polymerase-dependent transcription was then induced by incubation with 0.4 mM isopropyl-γ-d-thiogalactopyranoside for 3 h. Cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C and then stored at −70°C. The typical cell yield was about 4.5 g (wet weight)/liter of culture. Cells (4.5 g) were thawed in 6 volumes (wt/vol) of 20 mM Tris buffer (pH 7.2) containing 1 mM dithiothreitol and passed through a French pressure cell at 20,000 lb/in2. This was followed by centrifugation at 16,000 × g for 30 min at 4°C.
The 20S proteasome and α subunit were purified by first adding NH4(SO4)2 to cell extract at a final concentration of 60%. The samples were then equilibrated for 30 min and centrifuged at 10,000 × g for 15 min at 4°C. The resulting supernatant solutions were equilibrated at 85% NH4(SO4)2 for 1 h, and the proteins were centrifuged at 10,000 × g for 30 min at 4°C. The β subunits were purified by first adding NH4(SO4)2 to cell extract at a final concentration of 30%. The sample was then equilibrated for 30 min and centrifuged at 10,000 × g for 15 min at 4°C. The resulting supernatant solution was equilibrated at 60% NH4(SO4)2 for 1 h and centrifuged at 10,000 × g for 30 min at 4°C. The resulting protein pellets were resuspended in 5 to 10 ml of Tris buffer and dialyzed twice against 2 liters of Tris buffer at 4°C for 18 h. Each sample was then applied to a Q-Sepharose (Pharmacia) column (2.5 by 28.5 cm). For the β subunit, the column was equilibrated with Tris buffer and then developed with a linear NaCl gradient (0 to 50 mM NaCl in 20 ml of Tris buffer). For the α subunit and the 20S proteasome, the Q-Sepharose column was equilibrated with Tris buffer containing 200 mM NaCl and then developed with a linear NaCl gradient (200 to 350 mM NaCl in 80 ml of Tris buffer). Q-Sepharose fractions of interest were pooled, precipitated with 85% NH4(SO4)2, and resuspended in 0.5 ml of Tris buffer. The samples were applied to a Superose 6 HR 10/30 column equilibrated with Tris buffer containing 150 mM NaCl. Protein fractions were assayed by measuring CL activity with the substrate Suc-LLVY-Amc (succinyl-LLVY-7-amido-4-methyl-coumarin) (see below). Neither the α nor β subunits alone catalyzed detectable peptide-hydrolyzing activity; thus, fractions were preincubated for 30 min at 37°C in the presence of either β or α subunits prior to assay. About 12 mg of 20S proteasome, 20 mg of α subunit, and 10 mg of β subunit were purified from 1.0 liter of culture. Protein concentrations were determined by the bicinchoninic acid method with bovine serum albumin as the standard as previously described (14).
Activity assays and electrophoretic techniques.
Peptide-hydrolyzing activities were assayed by monitoring the release of β-naphthylamine colorimetrically or that of 7-amino-4-methylcoumarin fluorometrically as previously described (14). Specific activities are reported as nanomoles of product per minute per milligram of protein. Protein hydrolysis activity was assayed by measuring the generation of ninhydrin-reactive products (18, 20). Reaction mixtures (150-μl final volume) contained 150 μg of substrate and 10 μg of 20S proteasome in 10 mM Tris-HCl (pH 8.0) and were incubated at 65°C. The reaction was terminated after 80 min by addition of 50 μl of 10% trichloroacetic acid. Ninhydrin-reactive products were determined from a 100-μl aliquot of the supernatant after trichloroacetic acid precipitation of protein. Specific activity is reported as nanomoles of leucine equivalents per minute per milligram of protein. Control reactions included the enzyme and the substrate protein incubated separately and then precipitated with trichloroacetic acid after 80 min at 65°C. The irreversible inhibition was determined by incubating the purified 20S proteasome at 0.01 mg/ml with 20 μM 3,4-dichloroisocoumarin for 30 min at 22°C. A Spectra/Por cellulose ester membrane with a molecular mass cutoff of 15,000 Da (Spectrum, Houston, Tex.) was used to dialyze a 5-ml sample twice against 4 liters of Tris buffer at 4°C for 18 h. After dialysis, the protein concentration and peptide-hydrolyzing activity of the sample were reassessed.
Nondenaturing electrophoresis was performed by using a horizontal submarine gel of 2.0% MetaPhor XR agarose (FMC BioProducts, Rockland, Maine) in 500 mM Tris-HCl–160 mM boric acid–1 M urea (pH 8.5). Proteins were electrophoresed with a 90 mM Tris-HCl–90 mM boric acid buffer (pH 8.5) and then stained with Coomassie blue R-250. The proteasome proteins and subunits were also separated by denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (12) using 12 or 14% polyacrylamide. N-terminal sequencing by Edman degradation was done as previously described (14).
In vitro assembly.
A subunit equimolar ratio of α subunits and β prosubunits at a final concentration of 1.3 mg of protein/ml of Tris buffer was incubated at temperatures ranging from 0 to 65°C for 90 min and then electrophoresed under nondenaturing and denaturing conditions. The sample incubated at 37°C was also equilibrated at 80% NH4(SO4)2 and then centrifuged at 10,000 × g for 15 min at 4°C. The protein pellet (2 mg) was resuspended in 0.1 ml of Tris buffer and applied to a Superose 6 HR 10/30 column equilibrated with Tris buffer containing 150 mM NaCl. Fractions eluting at about 645 kDa were analyzed for peptide-hydrolyzing activity and electrophoresed by denaturing SDS–14% PAGE.
Site-specific amino acid replacements in PsmB and purification of the altered proteins.
Amino acids DNDKYLKG of the N terminus of the β prosubunits were deleted by PCR amplification of the psmB gene as described above, except that the oligonucleotide CTGCCCATATGACAACTACCGTAGG (oligo βΔ8aa) was used for annealing to the 5′ end of psmB to generate plasmid pMT7βΔ. Alterations in the N-terminal region of the mature form of the β subunit were generated by using derivations of oligo βΔ8aa for PCR amplification in which A12→G, A12→T, and A15→G were introduced to generate βΔThr1Ala, βΔThr1Ser, and βΔThr2Ala, respectively. Mutations were confirmed by DNA sequence analysis.
For site-directed replacements of amino acids in the central region of the mature β subunit, oligonucleotide-directed, site-specific mutagenesis of the psmB gene was performed by using the Morph site-specific plasmid DNA mutagenesis kit as recommended by the supplier (5 Prime → 3 Prime, Boulder, Colo.). Double-stranded plasmid pMT7βΔ was used as a template for annealing of the mutagenic oligonucleotide. Mutations were confirmed by DNA sequence analysis of plasmids which had been selected in E. coli BMH71-18. Plasmids with the desired mutation were transformed into E. coli TB-1, and the DNA sequence of the altered psmB gene was reconfirmed. psmA was then subcloned into the plasmids with the confirmed site-directed mutation in the psmB gene. The mutant 20S proteasome proteins were produced in E. coli and purified as described above.
Transmission electron microscopy.
Isolated 20S proteasomes produced in E. coli and authentic 20S proteasomes purified from M. thermophila were placed on 200-mesh grids coated with either Formvar or carbon films and briefly stained with 1% aqueous uranyl acetate. Some preparations were prefixed with 2% cacodylate-buffered glutaraldehyde before staining. Samples were viewed and photographed on a Zeiss EM-10CA transmission electron microscope operated at 80 kV.
RESULTS AND DISCUSSION
Heterologous production of the M. thermophila 20S proteasome and independent subunits.
The 20S proteasome was produced in E. coli by coproduction of α subunits and β prosubunits with the T7 RNA polymerase-promoter system (25). The same system was used to independently produce either α subunits or β prosubunits containing the nine-residue propeptide which is absent in the authentic 20S proteasome purified directly from M. thermophila (Fig. 1). The plasmids used are shown in Table 1. The 20S proteasome and independently produced subunits each comprised 5 to 10% of the total cell protein of E. coli, which facilitated purification to homogeneity, as judged by SDS-PAGE (Fig. 2). The in vivo-assembled, heterologously produced 20S proteasome catalyzed hydrolysis of 20 μM Suc-AAF-Amc and 300 μM Cbz-LLE-β-Na (carbobenzoxy-LLE-β-naphthylamide) at 42°C with specific activities (1.3 ± 0.2 and 9.1 ± 0.2 nmol/min/mg of protein) commensurate with those previously reported (14) for the authentic M. thermophila proteasome (1.2 and 8.9 nmol/min/mg of protein) when assayed at the same concentrations of substrates and temperature. Electron microscopy revealed a cylindrical structure comprised of four stacked rings indistinguishable from the quaternary structure of the authentic 20S proteasome (Fig. 3C and D). The results indicate that the 20S proteasome produced in E. coli is similar to the previously described authentic proteasome isolated from M. thermophila (14).
FIG. 1.
Deduced and experimentally determined N-terminal sequences of the M. thermophila 20S proteasome α and β subunits. PsmA and PsmB, α subunit and β prosubunit sequences deduced from the psmA and psmB gene sequences shown in uppercase letters; α-1 and α-2, experimentally determined sequences of the α subunit from the authentic 20S proteasome purified directly from M. thermophila; α-3, experimentally determined sequence of the α subunit produced in E. coli either independently or in the 20S proteasome; β-1, experimentally determined sequence obtained for either the mature β subunit from the authentic 20S proteasome or the mature β subunits from the heterologously produced 20S proteasomes assembled either in vivo or in vitro; β-2, experimentally determined sequence of the β prosubunit independently produced in E. coli. X, uncertain amino acid assignment. The bases in boldface are putative ribosome binding sites.
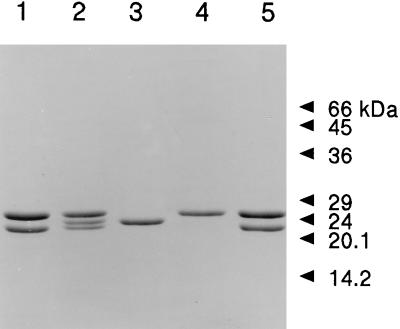
FIG. 2.
SDS-PAGE of the authentic and heterologously produced M. thermophila 20S proteasome and individual α or β subunits. Lanes: 1, 20S proteasome produced in E. coli; 2, α subunit and β prosubunit independently produced in E. coli, combined in vitro, and incubated for 90 min at 37°C; 3, β prosubunit independently produced in E. coli; 4, α subunit independently produced in E. coli; 5, authentic 20S proteasome purified from M. thermophila. Proteins were stained with Coomassie blue R-250. Molecular size standards are indicated on the right.
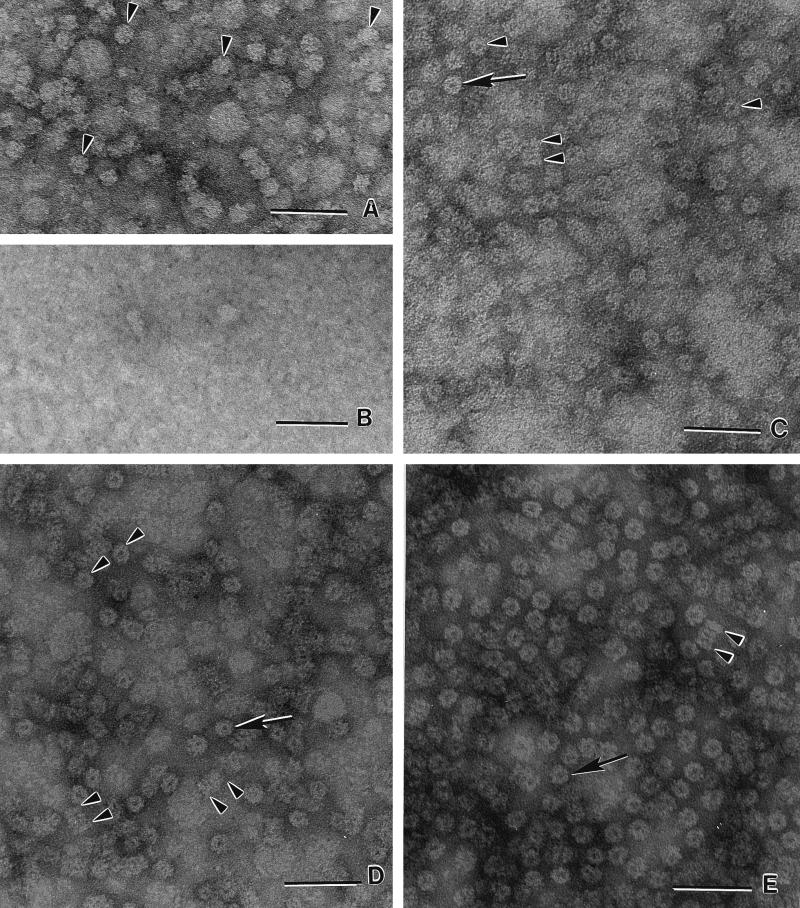
FIG. 3.
Transmission electron micrographs of negatively stained M. thermophila 20S proteasomes and subunits. (A) α subunit produced in E. coli independently of the β prosubunit. Seven-membered ring structures (α7) with no apparent central pore are visible (arrowheads). (B) β prosubunit produced in E. coli independently of the α subunit. (C) Authentic 20S proteasome purified from M. thermophila. End-on views (single arrow and arrowheads) and lateral views (double arrowheads) of the cylindrical 20S proteasome comprised of four stacked (α7β7β7α7) rings. (D) 20S proteasome produced in E. coli. The structures are indistinguishable from those in C. (E) The 20S proteasome assembled in vitro from α7 and β prosubunits independently produced in E. coli. The structures are indistinguishable from those in C and D. Bars, 50 nm.
The β subunit from the 20S proteasome produced in E. coli migrated (22 kDa) similarly to the mature β subunit from the authentic 20S proteasome (Fig. 2); however, the independently produced β prosubunit migrated significantly slower, suggesting a slightly larger molecular mass. N-terminal sequencing (Fig. 1) confirmed that the nine-residue propeptide was still present. N-terminal sequencing also indicated that the β subunits from the 20S proteasome produced in E. coli were processed to expose the N-terminal threonine as previously shown for mature β subunits from the authentic 20S proteasome (Fig. 1). No evidence for a β ring structure was obtained in electron micrographs of purified, independently produced β prosubunits (Fig. 3B). These results, and results obtained during studies on in vitro assembly (see below), provide further evidence that maturation and assembly of the archaeal β prosubunit require the α subunit as a chaperone (22).
Electron micrographs (Fig. 3A) show that the independently produced α subunits formed α ring structures. Analysis of the α subunit from the authentic M. thermophila 20S proteasome revealed two N-terminal sequences which are identical except in length (14) (Fig. 1). DNA analysis indicated three potential translational start sites encoded in psmA yielding three potential N termini (Fig. 1); thus, the two different N termini in the authentic 20S proteasome could be ascribed to either two translational start sites or processing of a single N terminus. The functional significance of two N termini for the α subunit of the authentic 20S proteasome is unknown. Elimination of the first 34 residues of the T. acidophilum α subunit prevents formation of α7 ring structures, signifying the importance of N-terminal residues in the assembly of the 20S proteasome (26). Furthermore, the N terminus of the T. acidophilum 20S proteasome is located at the entrance of the α subunit rings (13), indicating a potential role in translocation of the substrate into the central chamber or interaction with regulatory complexes. The heterologously produced α subunit, whether from the 20S proteasome or produced independently, appeared to migrate (24 kDa) similarly to the α subunit of the authentic 20S proteasome purified from M. thermophila (Fig. 2); however, the N-terminal sequences of the α subunits produced in E. coli were two residues longer than the longest α subunit in the authentic 20S proteasome, suggesting that translation in E. coli started at the first site (Fig. 1). The presence of these two additional residues in the α subunit did not affect α ring assembly (Fig. 3A). The two additional residues also did not affect β prosubunit maturation or the assembly and catalytic activity of the 20S proteasome produced in E. coli (see below), which suggests that the residues are inconsequential for these essential properties of the 20S proteasome. The results also suggest that either the α subunit was processed by an M. thermophila protease not present in the eubacterial host or the translational start sites encoded by the psmA gene are not recognized by E. coli.
In vitro assembly of the 20S proteasome.
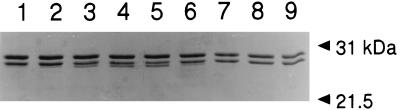
When independently produced α ring structures and β prosubunits were purified and reconstituted in subunit equimolar amounts at pH 7.2, approximately 50% of the β prosubunit was processed, as determined by SDS-PAGE (Fig. 2). Sequencing identified an N-terminal threonine for the processed β subunits that was identical to the β subunit in the authentic 20S proteasome (Fig. 1). Mature β subunits were not detected in the absence of the α subunit rings (Fig. 2), which indicates that processing is dependent on the α ring. Gel filtration chromatography resolved a 645-kDa protein from the reconstitution mixture, which was judged homogeneous by SDS-PAGE. The purified protein catalyzed hydrolysis of 20 μM Suc-AAF-Amc and 300 μM Cbz-LLE-βNa with specific activities (1.3 ± 0.2 and 9.1 ± 0.2 nmol/min/mg of protein) identical to those of the in vivo-assembled, heterologously produced 20S proteasome reported here and similar to those of the previously reported (14) authentic 20S proteasome purified from M. thermophila (1.2 and 8.9 nmol/min/mg of protein). Electron microscopy revealed a cylindrical structure comprised of four stacked rings indistinguishable from the quaternary structure of the authentic 20S proteasome (Fig. 3E). These results indicate that a fully active 20S proteasome was assembled in vitro. Both maturation of the β prosubunit (Fig. 4) and assembly of the 20S proteasome (Fig. 5) were optimal between 37 and 42°C.
FIG. 4.
Temperature-dependent processing of the M. thermophila β prosubunit. Purified α7 rings and β prosubunits were combined in subunit equimolar amounts (15 μM, final concentration) and incubated for 60 min at 16°C (lane 1), 21°C (lane 2), 30°C (lane 3), 37°C (lane 4), 42°C (lane 5), 50°C (lane 6), 55°C (lane 7), 60°C (lane 8), or 70°C (lane 9). A sample of the protein mixture (1.1 μg) was analyzed by SDS-PAGE by using 12% polyacrylamide. Proteins were stained with Coomassie blue R-250. Molecular size standards are indicated on the right.
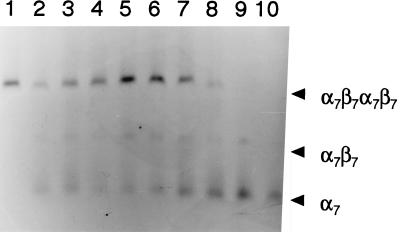
FIG. 5.
Temperature-dependent assembly of the M. thermophila 20S proteasome. The purified 20S proteasome produced in E. coli is shown in lane 1. Purified α7 rings and β prosubunits independently produced in E. coli were combined in subunit equimolar amounts (15 μM) and incubated for 60 min at 16°C (lane 2), 21°C (lane 3), 30°C (lane 4), 37°C (lane 5), 42°C (lane 6), 50°C (lane 7), 55°C (lane 8), or 60°C (lane 9). Purified α7 rings produced in E. coli independently of β prosubunits are in lane 10. Samples (1.1 μg) were analyzed by nondenaturing electrophoresis using 2.0% agarose. Proteins were stained with Coomassie blue R-250. The α7β7β7α7 20S proteasome, a putative α7β7 structure, and α7 rings are indicated by arrowheads (right). The unincorporated β prosubunits eluted from the gel and were not detectable.
Neither processing of β prosubunits (Fig. 2) nor assembly into β subunit ring structures (Fig. 3B) was observed in the absence of α rings. A protein complex with a molecular mass intermediate to those of the α ring structure and the 20S proteasome was observed (Fig. 5) which was not present if either α rings or β prosubunits were omitted from the reconstitution mixture. The results are consistent with the proposed mechanism of in vivo assembly for the mouse and yeast 20S proteasome in which an α7 ring structure chaperones assembly of β prosubunits, leading to the formation of two half-proteasome precursor structures which assemble to form the 20S proteasome (3, 8, 21). Assembly intermediates have yet to be reported for in vivo or in vitro synthesis of the 20S proteasome from T. acidophilum.
The in vitro assembly of a catalytically active T. acidophilum 20S proteasome has only been accomplished by combining individually purified α subunits and β prosubunits at pH 2.6 and then adjusting the pH to 7.5 (26); thus, it is was not possible to rule out additional proteins or other factors essential for in vivo assembly. It was recently suggested that a chaperone may be essential for final assembly of the mammalian 20S proteasome from half-proteasome complexes (21). The results presented here suggest that accessory proteins, such as chaperones or proteases, are not essential for assembly of the M. thermophila 20S proteasome or processing of the β prosubunit. However, only 50% of the β prosubunits were processed and incorporated into fully active 20S proteasomes; thus, accessory proteins which enhance the efficiency or rate of assembly cannot be ruled out. Nonetheless, the ability to assemble a highly active M. thermophila 20S proteasome in vitro under physiological conditions and isolate a potential precursor complex will facilitate future experiments designed to probe the mechanisms of β prosubunit autoprocessing and assembly of the archaeal 20S proteasome which were not previously possible with the T. acidophilum enzyme. For example, it may be possible to determine if β prosubunit processing occurs in precursor complexes or during final assembly of the 20S archaeal proteasome, as proposed for the eucaryal enzyme (3).
Multisubstrate activity.
Heterologous production of the M. thermophila 20S proteasome in vivo by coproduction of α subunits and β prosubunits in E. coli facilitated the isolation of amounts which enabled an examination of the multisubstrate activity with a variety of fluorogenic and chromogenic small-peptide substrates (Table 2). The greatest activity was obtained with Cbz-LLE-βNa, in which the peptide bond was cleaved carboxyl to the acidic glutamate residue (PG activity). Substantial activity was also obtained with Suc-LLVY-Amc and Suc-AAF-Amc, in which the peptide bonds carboxyl to the aromatic tyrosine and phenylalanine residues were cleaved (CL activity) at 73 and 38% of the PG activity. This result contrasts with that obtained with the 20S proteasome from T. acidophilum, which cleaves Cbz-LLE-βNa at only 7 to 8% of the rate of Suc-LLVY-Amc hydrolysis (1), suggesting significant differences in the active sites between these two archaeal 20S proteasomes. Minor TL activity was measurable with the substrates Benz-FVR-Amc (N-benzoyl-FVR-Amc) and Boc-FSR-Amc (N-t-butoxycarbonyl)-FSR-Amc (Table 2); however, the activity was only 2% of the PG activity. Significant SNAAP activity was not detected. The M. thermophila 20S proteasome also catalyzed hydrolysis of a variety of proteins (see Table 4); however, proteolysis was only measurable at temperatures above 50°C (data not shown), which suggests that substrate proteins must be at least partially unfolded prior to degradation.
TABLE 2.
Multiple peptide-hydrolyzing activities of the M. thermophila 20S proteasome
| Activity | Substrate | Sp acta (nmol/min/mg of protein) |
|---|---|---|
| PG | Cbz-LLE-βNa | 115.0 ± 2.3 |
| Ac-YVAD-Amc | 2.2 ± 0.5 | |
| AE-Amc | UDb | |
| CL | Suc-LLVY-Amc | 83.7 ± 1.2 |
| Suc-AAF-Amc | 43.7 ± 1.5 | |
| Suc-IIW-Amc | 2.4 ± 0.7 | |
| Suc-LY-Amc | UD | |
| GPL-βNa | UD | |
| TL | Benz-FVR-Amc | 2.5 ± 0.4 |
| Boc-FSR-Amc | 1.5 ± 0.3 | |
| GGR-Amc | UD | |
| SNAAP | GG-βNa | UD |
| Suc-IA-Amc | UD |
The 20S proteasome was heterologously produced in E. coli. The assay mixtures contained 150 μM substrate and 0.027 mg of protein per ml. The reaction temperature was 60°C.
UD, undetectable.
TABLE 4.
Protein degradation activity of the 20S proteasome from M. thermophila
| Substrate | Sp acta |
|---|---|
| β-Casein | 31.0 |
| Dephosphorylated β-casein | 31.0 |
| Lysozyme | 52.0 |
| Bovine serum albumin | 2.0 |
| Phosphorylase b | 0.1 |
Expressed as nanomoles of leucine equivalents per hour per milligram of protein. The assay (60°C) contained 1.0 mg of substrate/ml and 0.067 mg of the 20S proteasome produced in E. coli per ml.
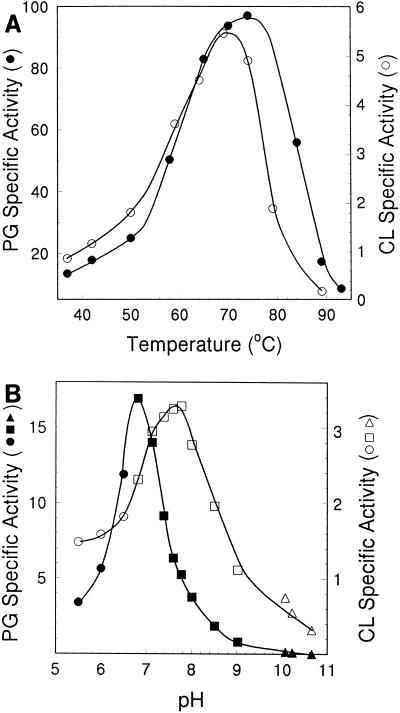
In contrast to that of the T. acidophilum proteasome, the PG activity of the M. thermophila enzyme was substantial (Table 2), which permitted a comparison with CL activity. The temperature optima (70 to 75°C) for both PG and CL activities (Fig. 6) were similar and compatible with the thermophilic nature of M. thermophila; however, the pH optima for PG (6.8) and CL (7.8) activities were significantly different (Fig. 6). Differences in PG and CL activities were also apparent when the M. thermophila proteasome was preincubated with various compounds (Table 3). Preincubation with ATP stimulated PG activity but had no apparent effect on CL activity. Low levels of SDS stimulated CL activity and inhibited PG activity. The divalent cations Mg2+, Mn2+, Zn2+, and Ca2+ and the monovalent cation K+ increased CL activity significantly; however, the same concentrations of these cations significantly reduced PG activity. The different behaviors of PG and CL activities on perturbation of the M. thermophila 20S proteasome suggest that the archaeal 20S proteasome is a conformationally flexible protein which adjusts to the binding of ligands.
FIG. 6.
Effects of temperature and pH on the activity of the M. thermophila 20S proteasome. (A) The heterologously produced 20S proteasome was assayed for PG (•) or CL (○) activity at the indicated temperatures by using (final concentrations) 250 μM Cbz-LLE-βNa and 6% (vol/vol) dimethyl sulfoxide (•) or 20 μM Suc-LLVT-Amc and 0.4% dimethyl sulfoxide (○). Specific activities are expressed as nanomoles of product per minute per milligram of protein. (B) The heterologously produced 20S proteasome was assayed for PG (•, ▪, ▴) or CL (○, □, ▵) activity at 37°C and the indicated pH by using (final concentrations) 150 μM Cbz-LLE-βNa and 6% (vol/vol) dimethyl sulfoxide (•, ▪, ▴) or 20 μM Suc-LLVY-Amc and 0.4% (vol/vol) dimethyl sulfoxide (○, □, ▵). Reaction mixtures contained the following buffers (final concentrations): 50 mM morpholineethanesulfonic acid (•, ○), 100 mM Tris-Cl (▪, □), or 50 mM ••• (▴, ▵). Specific activities are expressed as nanomoles of product per minute per milligram of protein.
TABLE 3.
Effects of various compounds on the peptide hydrolyzing activities of the 20S proteasome from M. thermophila
| Compound | Concn (mM) | Activitya
|
|
|---|---|---|---|
| CL | PG | ||
| None | 100 ± 0.5 | 100 ± 0.2 | |
| ATP | 1 | 100 ± 1.1 | 131 ± 2.1 |
| GTP | 1 | 100 ± 1.0 | 98 ± 2.0 |
| MgCl2 | 10 | 151 ± 9.8 | 54 ± 1.0 |
| MnCl2 | 10 | 129 ± 2.2 | 51 ± 1.2 |
| ZnCl2 | 10 | 140 ± 5.6 | 3 ± 0.5 |
| CaCl2 | 100 | 204 ± 3.0 | 45 ± 1.0 |
| KCl | 300 | 177 ± 6.0 | 33 ± 0.5 |
| NaCl | 100 | 150 ± 9.5 | 99 ± 1.0 |
| SDS | 0.7 | 208 ± 14 | 37 ± 0.5 |
| Spermidine | 1 | 103 ± 7.9 | 94 ± 4.0 |
| EDTA | 5 | 98 ± 3.8 | 103 ± 2.1 |
| Cysteine | 1 | 100 ± 2.6 | 96 ± 3.0 |
| 3,4-Dichloroisocoumarin | 0.022 | 0 ± 0.1 | 3 ± 0.5 |
| Ac-Leu-Leu-Nle-alb | 0.13 | 7 ± 1.0 | 26 ± 2.0 |
Expressed as relative activity, where activity with no added effector molecule is 100%. CL activity was assayed by using 20 μM Suc-LLVY-Amc, where 100% specific activity was 0.7 nmol of product/min/mg. PG activity was measured by using 250 μM Cbz-LLE-βNa, where 100% specific activity was 12.0 nmol of product/min/mg. The 20S proteasome used was produced in E. coli. The enzyme was preincubated with the indicated compound in 20 mM Tris-HCl (pH 7.2) at 21°C for 10 min prior to assay at 37°C. The final concentration of the 20S proteasome in the assay was 0.01 mg/ml.
Ac-Leu-Leu-Nle-al; acetyl-Leu-Leu-norleucine-al.
Active-site analysis of multisubstrate activity.
The N-terminal Thr1 Oγ of the 20S proteasome β subunit is presumed to act as the nucleophile in hydrolysis of peptide bonds, as determined by both the crystal structure of the T. acidophilum 20S proteasome in complex with an inhibitor (13) and replacement of β subunit Thr1 with Ala, which abolishes CL activity (23). The discovery that the natural inhibitor lactacystin becomes covalently linked to N-terminal Thr1 of β subunits of the eucaryal 20S proteasome supports this mechanism (7). Recent genetic analysis of the yeast 20S proteasome shows that three distinct β-type subunits (Pre3, Pup1, and Pre2), each with an N-terminal Thr1 active site, are responsible for PG, TL, and CL activities (11). Clearly, with only one type of β subunit, this basis for multisubstrate activity cannot apply to the archaeal 20S proteasome. The CL, PG, TL, and protein-hydrolyzing activities of the M. thermophila 20S proteasome permitted an investigation of the basis for the multisubstrate activity of the archaeal proteasome by site-directed replacement of potential active-site residues in the β subunit. The experiments were performed with 20S proteasomes assembled in vivo by coproduction of α and βΔ subunits in E. coli. The βΔ subunit lacks the propeptide, except for an N-terminal methionine (Met−1Thr1Thr2Thr3Val4Gly5Val6Val7…). The activities of the unaltered 20S proteasome (see Table 5) were similar to those reported in Tables 2 and 4, which indicated that the N-terminal methionine of the βΔ subunit was removed by E. coli aminopeptidase, exposing the essential N-terminal threonine (Thr1). Electron micrographs (not shown) of αβΔ 20S proteasomes altered by amino acid replacements revealed quaternary structures for each which resembled those of the authentic 20S proteasome purified from M. thermophila. Thus, the effects of amino acid substitutions on the multisubstrate activity were independent from effects on processing of the β prosubunit and assembly of the 20S proteasome.
TABLE 5.
Small-peptide- and protein-hydrolyzing activities of M. thermophila 20S proteasomes altered by amino acid replacements in the βΔ subunit
| Alterationb | Sp acta
|
|||
|---|---|---|---|---|
| CL | PG | TL | Proteolysis | |
| None | 95 ± 0.83 | 106 ± 2.1 | 2.30 ± 0.38 | 54.0 ± 0.97 |
| Thr1→Ala | UDc | UD | UD | UD |
| Thr2→Ala | 40 ± 1.4 | 32 ± 2.8 | 0.48 ± 0.07 | 10.0 ± 0.40 |
| Thr1→Ser | 72 ± 3.4 | 24 ± 0.84 | 0.35 ± 0.02 | 9.1 ± 0.09 |
| Asp51→Asn | 204 ± 3.0 | 155 ± 0.75 | 2.30 ± 0.08 | 35.0 ± 1.4 |
| Lys33→Ala | UD | UD | 7.10 ± 0.36 | 1.7 ± 0.03 |
| Lys33→Arg | 27 ± 1.2 | 9 ± 0.08 | 0.30 ± 0.04 | 8.6 ± 0.68 |
CL activity was determined with 150 μM Suc-LLVY-Amc, PG activity was determined with 150 μM Cbz-LLE-βNa, and TL activity was determined with 300 μM Benz-FVR-Amc. CL, PG, and TL activities are expressed as nanomoles per minute per milligram. Proteolysis activity was determined with lysozyme and is expressed as nanomoles of leucine equivalents per minute per milligram of protein. The proteolysis assay contained 1.0 mg of substrate/ml and 0.17 mg of 20S proteasome/ml. All assays were done at 60°C.
Amino acid replacements were in the βΔ subunit of 20S proteasomes heterologously produced by coproduction of α and βΔ subunits in E. coli. The βΔ subunit has the propeptide deleted.
UD, undetectable.
When Thr1 of the βΔ subunit was changed to Ala, all activities were abolished (Table 5), which suggests that this residue is essential for the multiple peptide-hydrolyzing activities of the M. thermophila 20S proteasome and supports the existence of one catalytic site. Replacement of Thr2 with Ala yielded a 20S proteasome all of whose activities were reduced relative to those of the unaltered enzyme; however, in each case, a significant amount of activity was preserved, suggesting that this residue is not essential for catalysis. When Thr1 of the βΔ subunit was replaced with Ser, the altered 20S proteasome catalyzed hydrolysis of peptide bonds; however, all activities were significantly lower than those of the unaltered 20S proteasome (Table 5), further supporting the idea of the existence of one catalytic site. Apparently, the side-chain hydroxyl group of Ser only partially substitutes for Thr1 Oγ. The pH optima for the CL and PG activities of the αβΔ Thr1→Ser enzyme increased 0.8 and 0.3 pH unit relative to those of the unaltered enzyme (data not shown), a result consistent with an expected increase in the pKa for the Ser hydroxyl group compared with the Thr hydroxyl group. The pH increased for both CL and PG activities, which suggests that the same nucleophile functions for both activities, further supporting the idea that only one catalytic site is responsible for the multisubstrate activity of the M. thermophila 20S proteasome. It follows that this active site must accommodate all of the short chromogenic and fluorogenic substrates tested. This conclusion is consistent with the results reported here, which suggest that the active site of the M. thermophila 20S proteasome is a conformationally flexible protein that is able to adjust to the binding of different ligands. Pairs of adjacent β subunits of different types are necessary for the CL and PG activities of the yeast 20S proteasome, suggesting that the proteolytic centers are formed by cooperation between specific subunit pairs (3, 11). The crystal structure of the T. acidophilum 20S proteasome suggests that the active-site residues of the β subunit are in close contact with residues of the adjacent β subunit and therefore could interact. Although the archaeal 20S proteasomes contain only one type of β subunit, it may be possible that ligand-induced conformational changes produce specific subunit interactions required for multisubstrate activity. The more sluggish PG activity of the T. acidophilum 20S proteasome relative to that of the M. thermophila enzyme likely arises from structural differences in the β subunits which affect either substrate binding or substrate-induced subunit interactions.
Asp51 is conserved among all α- and β-type subunits from members of the Archaea and Eucarya domains and is not essential for Suc-LLVY-Amc hydrolysis (CL activity) by the T. acidophilum proteasome (24). In fact, the CL activity of the T. acidophilum 20S proteasome is threefold greater when β subunit Asp51 is replaced with Asn; however, hydrolysis of larger proteins was not investigated. The same modification in the β subunit of the M. thermophila proteasome had no significant effect on TL activity and enhanced PG and CL activities compared with those of the unaltered enzyme. The lysozyme-hydrolyzing activity was partially reduced (Table 5). These results demonstrate that Asp51 is not essential for multisubstrate activity with short peptides but may have an indirect role in protein hydrolysis. A second β subunit site (in addition to Thr1) has been proposed for protein-hydrolyzing activity of the yeast 20S proteasome (9). This second site is postulated to be a water molecule acting as the general base and acid required for internal peptide bond hydrolysis of the protein substrate. In the mechanism, peptide binding and acyl-enzyme formation occur at the Thr1 site to position protein substrates for internal peptide cleavage at the active-site water. Acyl-enzyme hydrolysis then releases the short peptide product. This mechanism is similar to that of aspartate proteases, in which the carboxyl group of aspartate activates a water molecule. Clearly, it is essential to determine if this proposed mechanism also applies to protein hydrolysis by the archaeal proteasome and if Asp51 has any role.
The function of Lys33 in catalysis.
The crystal structure of the T. acidophilum 20S proteasome indicates that Lys33 is in close proximity to Thr1, where it is proposed to function either directly or indirectly to strip a proton from the initial attacking nucleophile (Thr1 Oγ). The Lys33→Arg replacement in the M. thermophila enzyme led to significant reduction of all activities (Table 5), further supporting the idea of one active site for multisubstrate activity. Although the Lys33→Ala replacement eliminated CL and PG activities and greatly reduced proteolysis, TL activity was threefold greater. The reason for increased TL activity is unknown; however, it is possible that Lys33 prevents active-site access of substrates with basic P1 residues. The crystal structure of the T. acidophilum 20S proteasome suggests that the side-chain amino group of Lys33 and the NH2 terminus of the β subunit (Thr1 N) are in close proximity to the attacking Thr1 Oγ and that, therefore, either one is positioned for the proton acceptor-donor function (13). It was proposed that Lys33 is positively charged and unlikely to accept a proton; however, direct evidence has not been reported for this hypothesis. Although the quaternary structure was preserved, replacement of Lys33 with Arg in the T. acidophilum 20S proteasome completely abolished CL activity (23). Unfortunately, the complete loss of activity did not resolve the question of whether Lys33 is directly involved in proton transfer and, therefore, essential for catalysis or whether it functions only indirectly to polarize the Thr1 N. The Lys33→Arg replacement preserved a substantial amount of all of the activities of the M. thermophila enzyme (Table 5). The side chain of Arg (pKa, 12.5) is more likely to be protonated than Lys33 (pKa, 10.5); thus, these results support the idea that the positive charge of the side-chain amino group of Lys33 only assists in catalysis by polarization of the Thr1 N, which functions as the direct proton acceptor-donor in catalysis.
Conclusions.
The 20S proteasome from M. thermophila is only the second to be characterized from the Archaea domain. This 20S proteasome, the first from a methanoarchaeon, has features which distinguish it from the T. acidophilium 20S proteasome. The ability to assemble the M. thermophila 20S proteasome in vitro demonstrated that accessory proteins are not essential for processing or assembly. Investigation of the multisubstrate activity has extended an understanding of the archaeal 20S proteasome in general and the active site in particular. Our results support the previously proposed role for Lys33 in polarization of the active-site Thr1 N.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Individual National Research Service Award 1F32GM15877-03 and National Science Foundation award MCB95-13863.
We are grateful to Mark Ou for technical assistance in purification of the in vitro-assembled 20S proteasome, Claudia Jakubzick for technical assistance in nondenaturing electrophoresis, and Donna Williams for technical assistance in electron microscopy.
REFERENCES
- 1.Akopian T N, Kisselev A F, Goldberg A L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- 2.Bauer M W, Bauer S H, Kelly R M. Purification and characterization of a proteasome from the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 1997;63:1160–1164. doi: 10.1128/aem.63.3.1160-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26s proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, Hegerl R, Baumeister W. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989;251:125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- 6.Dahlmann B, Kuehn L, Grziwa A, Zwickl P, Baumeister W. Biochemical properties of the proteasome from Thermoplasma acidophilum. Eur J Biochem. 1992;208:789–797. doi: 10.1111/j.1432-1033.1992.tb17249.x. [DOI] [PubMed] [Google Scholar]
- 7.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 8.Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel P M. 20S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13-16 S preproteasome complexes. J Mol Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 9.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 10.Grziwa A, Baumeister W, Dahlmann B, Kopp F. Localization of subunits in proteasomes from Thermoplasma acidophilum by immunoelectron microscopy. FEBS Lett. 1991;290:186–190. doi: 10.1016/0014-5793(91)81256-8. [DOI] [PubMed] [Google Scholar]
- 11.Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf D H. The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lowe J, Stock D, Jap R, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 14.Maupin-Furlow J A, Ferry J G. A proteasome from the methanogenic archaeon Methanosarcina thermophila. J Biol Chem. 1995;270:28617–28622. doi: 10.1074/jbc.270.48.28617. [DOI] [PubMed] [Google Scholar]
- 15.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlowski M. The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochemistry. 1990;29:10289–10297. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- 17.Orlowski M, Cardozo C, Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry. 1993;32:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 18.Orlowski M, Michaud C. Pituitary multicatalytic proteinase complex. Specificity of components and aspects of proteolytic activity. Biochemistry. 1989;28:9270–9278. doi: 10.1021/bi00450a006. [DOI] [PubMed] [Google Scholar]
- 19.Rivett A J. The multicatalytic proteinase. Multiple proteolytic activities. J Biol Chem. 1989;264:12215–12219. [PubMed] [Google Scholar]
- 20.Rosen H. A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys. 1957;67:10–15. doi: 10.1016/0003-9861(57)90241-2. [DOI] [PubMed] [Google Scholar]
- 21.Schmidtke G, Schmidt M, Kloetzel P M. Maturation of mammalian 20 s proteasome: purification and characterization of 13s and 16s proteasome precursor complexes. J Mol Biol. 1997;268:95–106. doi: 10.1006/jmbi.1997.0947. [DOI] [PubMed] [Google Scholar]
- 22.Seemuller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–470. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 23.Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 24.Seemuller E, Lupas A, Zuhl F, Zwickl P, Baumeister W. The proteasome from Thermoplasma acidophilum is neither a cysteine nor a serine protease. FEBS Lett. 1995;359:173–178. doi: 10.1016/0014-5793(95)00036-9. [DOI] [PubMed] [Google Scholar]
- 25.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat Struct Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]