Abstract
Background
miR-4270 is a regulatory factor has been linked with the progression of various cancers, such as nasopharyngeal carcinoma, hepatocellular carcinoma (HCC), and gastric cancer. However, the underlying mechanisms through which miR-4270 modulates HCC development are not fully understood.
Methods
miR-4270 expression levels were analyzed in various HCC cell lines and tissue samples. An online bioinformatics tool was then utilized to predict the miR-4270 target gene. The binding relationship between miR-4270 and its target gene DNMT3A was verified using dual-luciferase reporter and Ago2–RIP assays. Then, co-immunoprecipitation (Co-IP) and chromatin immunoprecipitation (ChIP) assays were conducted to investigate the association between DNMT3A and the hepatocyte growth factor activator (HGFAC) promoter region. To assess the methylation level of the HGFAC promoter, methylation-specific PCR (MSP) was employed. Furthermore, rescue analyses were carried out to evaluate the functional relevance of miR-4270 and HGFAC in the modulation of the malignant properties of HCC cells. Finally, HepG2 cells overexpressing miR-4270 were subcutaneously injected into nude mice to estimate the impact of miR-4270 on the xenograft tumor growth of HCC.
Results
A substantial miR-4270 downregulation was revealed in HCC patient samples and cell lines. miR-4270 upregulation suppressed both cell proliferation and invasion while promoting apoptosis. At the molecular level, miR-4270 was found to bind to the 3′untranslated region (3′UTR) of DNMT3A, thereby inhibiting DNMT3A-mediated methylation of the HGFAC promoter. Functional assays indicated that inhibition of miR-4270 stimulated HCC cell growth, an effect counteracted by overexpression of HGFAC. In vivo assays further verified that miR-4270 effectively suppressed the progression of HCC xenograft tumors.
Conclusions
miR-4270 was found to mitigate the malignant characteristics of HCC by inhibiting DNMT3A-mediated methylation of the HGFAC promoter, suggesting a potential therapeutic avenue for the management of HCC.
Keywords: Hepatocellular carcinoma, miR-4270, DNMT3A, HGFAC
Introduction
Liver cancer is among the leading causes of cancer-associated mortality throughout the world. Hepatocellular carcinoma (HCC) is the most prevalent type of primary liver cancer (Shen et al., 2022; Ong, Huey & Shelat, 2022). The incidence of HCC is highest in Asia and Africa, primarily due to prevalent underlying conditions such as liver disease and cirrhosis. Infections by the hepatitis B and C viruses are the most common causes of HCC, followed by cirrhosis (Xu et al., 2019). Additional risk factors for liver cancer include alcohol abuse, obesity, excess iron, environmental pollutants, and exposure to aflatoxins. Surgical resection stands as the primary treatment modality for HCC at present. Nonetheless, the high propensity for metastasis and recurrence in HCC often undermines post-surgical prognosis, with the accompanying surgical trauma presenting further complications (Piñero, Dirchwolf & Pessôa, 2020). With advancements such as the completion of human genome sequencing, therapies targeting specific molecular pathways are emerging as breakthrough approaches. Therefore, identifying target genes that can suppress HCC cell migration and invasion has become an important research direction in this field.
MicroRNAs (miRNAs) are approximately endogenous RNAs between 19 and 25 base pairs in length. They are non-coding and interact primarily with the 3′ untranslated regions (3′UTRs) of target genes, resulting in RNA degradation or the suppression of translation, and thus post-transcriptionally downregulating gene expression (Lu & Rothenberg, 2018). More than 2,500 miRNAs have been discovered to date, each capable of affecting the expression of hundreds of genes. MiRNAs are essentially associated with various cellular biological mechanisms, such as development, differentiation, growth, and apoptosis (Ni & Leng, 2016; Wang et al., 2022b). Therefore, changes in miRNA expression levels can disrupt these cellular functions, potentially leading to various diseases. Decades of research focusing on the qualitative and quantitative evaluation of miRNA levels have revealed significant alterations in their expression profiles across different diseases (Oura, Morishita & Masaki, 2020; Yan et al., 2022b). These findings underscore the potential of miRNA expression profiling as a valuable tool in disease diagnosis and cure. Some miRNAs are highly expressed in HCC and function as oncogenes, while others are under-expressed, serving as tumor suppressors (Wong, Tsang & Ng, 2018; Komoll et al., 2021). For instance, decreased miR-744-5p expression has been observed in HCC tissues and cells, whereas its upregulation significantly reduced HCC cell and tissue growth (Huang et al., 2021; Yu et al., 2022). Research studies have indicated the downregulation of miR-515-5p in HCC tissues, with its upregulation significantly suppressing malignant behaviors in HCC cells (Ni et al., 2020). Dong et al. (2021) discovered that downregulated expression of miR-369 in HCC tissues predicted poor prognosis in patients with HCC (Dong et al., 2021; Wang et al., 2022a). miR-4270, a recently identified miRNA, is under-expressed in different cancers, such as nasopharyngeal carcinoma, retinoblastoma, and gastric cancer, where it acts as a tumor suppressor influencing processes such as tumor growth, invasion, and metastasis (Hao et al., 2021; Feng et al., 2021; Shen et al., 2020). Nevertheless, there are few reports on the relationship between HCC and miR-4270. Our analysis revealed the downregulation of miR-4270 in HCC clinical tissue samples within the GEO datasets, indicating a need for comprehensive investigation into its functional roles and molecular mechanisms in HCC.
Epigenetic modifications, encompassing RNA and non-coding RNA modifications, histone modifications, DNA methylation, and chromatin remodeling, are crucial for regulating gene expression and maintaining cellular identity. Numerous studies have shown that abnormal epigenetic reprogramming is a key driver in the pathogenesis of various human malignant tumors. DNA methylation is a typical form of epigenetic regulation, that is primarily catalyzed by DNA methyltransferases (DNMTs), including DNMT3A, DNMT1, and DNMT3B (Long et al., 2019; Liu et al., 2022; Cheng et al., 2018). Methylation typically occurs within CpG islands in the promoter regions of genes, often leading to durable and heritable gene silencing. CpG islands, rich in CpG sequences and ranging from 200 to several thousand base pairs, are predominantly situated near gene promoters. It has been suggested that abnormal promoter DNA methylation is related to tumor occurrence. DNMT3A has a molecular weight of 130 kDa and is localized on the human 2p23 chromosome. In mammals, the protein is significantly conserved with 98% homology between humans and mice. Numerous studies have reported that aberrant overexpression of DNMT3A contributes to the progression of various cancers, including colorectal cancer (Zhou et al., 2022), ovarian cancer (He et al., 2019), pancreatic ductal adenocarcinoma (Jing et al., 2019), and breast cancer (Man et al., 2022). While the precise mechanisms behind aberrant DNA methylation remain unclear, multiple studies have shown a significant upregulation of DNMT3A mRNA levels in cirrhotic tissues in comparison with normal hepatic tissues, with even increased expression in primary HCC tissues compared to surrounding non-tumorous tissues (Wang et al., 2021).
Based on these insights, our study employed both in vivo and in vitro analyses to elucidate the effects of miR-4270 on the biological behavior of HCC cells. Bioinformatics was then utilized to predict and analyze the potential relationship and underlying mechanisms between miR-4270 and DNMT3A, offering a novel perspective for gene therapy approaches in HCC.
Materials and Methods
HCC tissue specimens
Data were collected from 114 HCC patients treated at our hospital between October 2017 and January 2020, comprising 35 females and 79 males between the ages of 31 and 78 years (average age = 48.55 ± 10.17 years). Inclusion parameters were patients with confirmed diagnosis through surgical pathology, imaging, and other relevant examinations, complete admission data, no history of prior antitumor treatments, aged over 20 years, and who had consented to laparoscopic radical hepatectomy for HCC. The exclusion criteria were the presence of neurological diseases, other malignant tumors, a combination of secondary HCC or liver abscess hemangioma, abnormal function of the heart, liver, kidney, and other important organs, inability to communicate normally, involvement in other clinical studies, or withdrawal from the present study. This investigation was authorized by the Ethics Committee of Tianjin Medical University Cancer Hospital, and all the participants provided written informed consent.
Bioinformatics assay
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) was accessed, and a search using ‘hepatocellular carcinoma’ as the keywords was conducted. ‘Homo sapiens’ and ‘expression profiling by array’ were selected, and the gene data matrix files for GSE108724 were downloaded. The GSE108724 microarray files, sourced from the GPL20712 platform (HuGene-1_0-st Affymetrix Human Gene 1.0 ST Array (transcript (gene) version), comprised 7 HCC samples and seven corresponding adjacent non-tumor tissue samples. All microarray data were processed for batch effects with the support of the R package “sva.” Following batch effect processing, we further standardized the data. The normalization process encompassed three steps: (1) correcting the background data, which involved cleaning unbound probe impurities to detect genes with low differential expression; (2) conducting normalization to remove systematic measurement errors, ensuring comparability of measurements across experimental conditions; (3) utilizing robust array average algorithm processing to derive consistent average fluorescence intensity values from the probe group, preparing the GEO data for subsequent comparative analyses. Differentially expressed genes (DEGs) were determined through differential analysis using the ‘Limma’ package in R software. The GEPIA database (http://gepia.cancer-pku.cn/) was utilized to assess DNMT3A and HGFAC expression levels in 369 HCC tumor samples and 160 paired normal tissues spanning various stages of HCC. Additionally, the correlations between the levels of DNMT3A and HGFAC and patient outcomes, specifically overall survival (OS) and disease-free survival (DFS), were evaluated.
Cell lines and culture
The HCC cell lines SMMC-7721, Hep3B, MHCC97H, HepG2, and Huh7, and the human normal hepatocyte L02 cell line, were cultured in DMEM containing penicillin-streptomycin (1%; Sigma-Aldrich, St. Louis, MI, USA) and 10% fetal bovine serum (FBS; Thermo Fisher, Waltham, MA, USA) in a 5% CO2 incubator at 37 °C. Cells were grown until 80% confluent before detachment with trypsin and passaging at a ratio of 1:2 to 1:3.
Cell transfection
HCC cells in logarithmic growth were plated at 2 × 103cells/well in 96-well plates. An miR-4270 mimic, miR-4270 inhibitor, and pcDNA-HGFAC (GenePharma Co., Ltd., Shanghai, China) were transfected into cells at 70% confluence using Lipofectamine 3000, according to the provided protocol. After 48 h, the cells were harvested for subsequent analyses.
RT-qPCR
Total RNA was extracted from HCC tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The reverse transcription reaction (20 μL) comprised 10 μL total RNA, 1.2 μL miR-RT primer (1 μmol/L), MMLV (0.2 μL, 200 U/μL), 5 × RT buffer (4 μL), dNTP (0.75 μL, 10 mmol/L), and RNase-free ddH2O (3.85 μL). We assessed the levels of miR-4270, DNMT3A, and HGFAC in HCC tissues and cells using the miScript PCR starter kit (Qiagen, Hilden, Germany) and amplified the mRNA on a Bio-Rad CFX90 Real-time PCR system. The 2−ΔΔCT method was utilized for assessing the relative expression levels. The primers for miR-4270, DNMT3A, and HGFAC used in this investigation were as follows: miR-4270 forward, 5′-GCC GAG TCA GGG AGT CAG GG-3′; and reverse, 5′-CTC AAC TGG TGT CGT GGA-3′; DNMT3A forward, 5’-GTG GAT GTT GAT GGG AGG CA-3′; and reverse, 5′-CGC ACC ACT GTT TTC ACC AG-3′; HGFAC forward: 5′-AGG GAG CTA GAA AGA GGG GG-3′; and reverse, 5′-AGG CTC CAG GGG TCT CTT AG-3′.
Western blotting
Briefly, proteins were separated on SDS-PAGE gels comprising 10% separating gel and 5% concentrating gel. The proteins were then transferred to PVDF membranes according to the molecular weights of the target proteins after electrophoresis. After electrotransfer, 5% BSA was used to block the membranes for 3 h at room temperature. The membrane was then incubated on a shaker at 4 °C in diluted primary antibodies against DNMT3A and HGFAC overnight. The membranes were then washed three times with 1 × TBST, each wash lasting 10 min, and were then incubated in a diluted secondary antibody (goat anti-rabbit IgG) solution for 1 h at ambient temperature and washed three times with 1 × TBST, with each wash lasting 10 min. Finally, the luminescent substrate was prepared per the reagent instructions and applied to the PVDF membrane, and the membrane was observed using an automatic imager.
Cell proliferation assay
HCC cells (1 × 103/mL) were seeded into 96-well plates in 100 μL cell suspension/well. After 24, 48, and 72 h, CCK-8 solution (10 μL) was added to the cells for 1 h. The absorbances (OD values) at 450 nm were measured using a microplate reader.
Cell invasion assay
Following the Transwell chamber protocol, 200 μl of each differently transfected HCC cell suspension (serum-free) were placed in the Matrigel-coated upper chamber while 500 μl of DMEM containing 10% FBS was added to the lower chamber. After 24 h, the cells were stained with crystal violet and visualized by microscopy (IX53; Olympus Corp, Tokyo, Japan) and counted. Each assay was performed three times.
Cell cycle assay
Exponentially growing cells were seeded in six-well plates and then processed and collected using a Cell Cycle Staining kit, following standardized procedures. Subsequently, cell cycle distribution was evaluated by flow cytometry and assessed via the FlowJo 7.6 software.
Cell apoptosis assay
HCC cells from each treatment group were collected and centrifuged at 2,000 rpm for 5 min, followed by three rinses with PBS. Subsequently, the cells were resuspended in binding buffer (500 μL). Then propidium iodide (PI) and Annexin V/FITC (Pricella, Wuhan, China) were added (5 μL, respectively). After incubation for 15 min in the dark, a FACScan flow cytometer (Attune NxT; Thermo Fisher, Waltham, MA, USA) was used to analyze the proportions of apoptotic cells with Annexin V or PI-stained cells identified as apoptotic cells.
Methylation-specific PCR
DNA bisulfite modification relies on the ability of bisulfite and hydroquinone to convert cytosine into uracil on a DNA strand, a reaction prevented by cytosine methylation. In this way, methylated vs unmethylated DNA sequences can be distinguished by sequence-specific primers after bisulfite modification. Here, 1‒2 μL of genomic DNA was diluted to 30 μL and denatured by adding 20 μL 0.5 mol/L NaOH at 37 °C. Then, 520 μL of freshly prepared 3 mol/L NaHSO3 (pH 5.0) and 30 μL of 10 mmol/L hydroquinone were added, mixed, and incubated for 16 h at 50 °C. The sodium bisulfite-modified DNA was recovered and purified by dialysis against 1% agarose, and then 50 μL of 0.6 mol/L NaOH was added at 37 °C to terminate the reaction. DNA was recovered by ethanol precipitation, dissolved in 30 μL sterile double-distilled water. Finally, the collected DNA was used immediately for PCR amplification or stored at −80 °C.
Co-immunoprecipitation (Co-IP)
The Pierce™ Direct IP Kit 26148 was used, following the manufacturers’ guide (Takara Biotechnology, Dalian, China). Twenty microliters of Amino Link PlusResin and Pierce Control Agarose Resin (a negative control to eliminate nonspecific protein binding) were added with 4 μg of an anti-HGFAC rabbit monoclonal antibody for 100 min. Subsequently, the protein sample (500 μL) was added and incubated overnight, followed by four rinses with IP lysis/wash buffer and one with conditioning buffer. HGFAC protein complexes were then eluted with solution buffer, and 10% SDS-PAGE electrophoresis (voltage 110 V, electrophoresis time 25 min) was performed, followed by western blotting, as described above. The membranes were incubated with rabbit anti-human HGFAC (1:1,000 dilution) and DNMT3A (1:1,000 dilution) primary antibodies overnight at 4 °C. After three washes in TBST, the membranes were developed using an enhanced chemiluminescence (ECL) reagent.
Chromatin immunoprecipitation (ChIP)
HCC cells from each group were fixed with 1% formaldehyde for 10 min, and the membranes and nuclei were sequentially lysed. Chromatin DNA was fragmented ultrasonically into fragments between 200 and 1,000 BP. The chromatin suspension was diluted five-fold with dilution buffer, and 5% was reserved as control (input). We then aliquoted the remainder, adding an anti-DNMT3A antibody to one portion and a normal rabbit IgG antibody to another, incubating on a shaker overnight at 4 °C. Protein A/G magnetic beads were utilized for collection of the immunoprecipitated complexes, followed by extensive washing in a 65 °C water bath overnight. DNA was extracted using a PCR product extraction kit (Qiagen, Hilden, Germany) with the total DNA (input) and each immunoprecipitated DNA was used as a template. The results were calculated as the relative expression level (% input) using the following formula:
This represents the ratio of immunoprecipitated DNA to total DNA (input). The relative expression level (% input) of rabbit normal IgG precipitated DNA vs input DNA was taken as a negative control.
Xenograft tumor mouse models
BALB/c-nu nude mice (age = 4 weeks, male, n = 24) were allowed to acclimatize for 2 days in a specific pathogen-free (SPF) facility. HepG2 cells (5 × 106 in a 200 μL suspension) were inoculated into the right groin area of the mice, after which the animals were randomly divided into the NC mimic and miR-4270 mimic groups (each n = 12). The tumor growth was monitored and the long and short diameters of the tumors were measured every 5 days. The tumor volumes were calculated using the formula:
The mice were euthanized after 30 days and the tumors were harvested and stored for later use.
Statistical measurements
Statistical analyses were conducted using GraphPad Prism 7.0 software. Three replicates were used for all assays. Data distributions were verified using the Shapiro-Wilk test, and the homogeneity of variances was verified by Levene’s test. Normally distributed and variance-homogenous data were analyzed by one-way analysis of variance (ANOVA) or t-tests followed by post hoc analysis (Tukey-Kramer correction). Non-normally distributed or non-homogenous data were analyzed using Kruskal-Wallis non-parametric tests. P < 0.05 was considered statistically significant.
Results
miR-4270 levels were downregulated in HCC tissues and cell lines
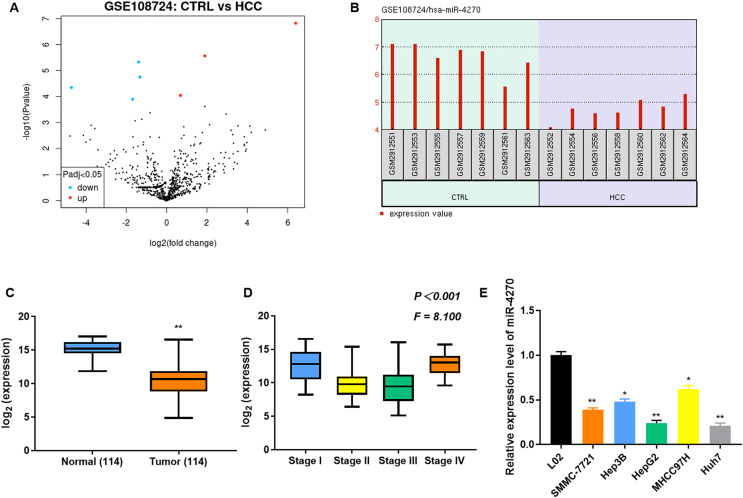
Seven differentially expressed miRNAs (three upregulated and four downregulated) were identified in the GSE108724 dataset (Fig. 1A). Among them, miR-4270 showed significant downregulation in HCC tissues compared with normal tissues (Fig. 1B). Furthermore, to confirm the expression of miR-4270 in HCC, the miR-4270 levels in HCC and normal tissues at different stages, as well as in the HCC cell lines (SMMC-7721, Hep3B, HepG2, MHCC97H, and Huh7) were measured via RT-qPCR. The results indicated that miR-4270 was markedly downregulated in HCC, and its expression correlated with various clinical stages of HCC (Figs. 1C and 1E).
Figure 1. Downregulation of miR-4270 in HCC tissues and cell lines.
(A) Volcano plot illustrating the differential expression of genes in the GSE108724 dataset, comprising 7 HCC and seven adjacent non-tumor tissue samples. The x-axis represents the log2 (fold change). (B) Expression levels of miR-4270 in individual samples from the GSE108724 dataset. (C) Comparative expression of miR-4270 in 114 paired HCC tumor and adjacent non-tumor tissues. **P < 0.01 vs Normal, non-paired t-test. (D) miR-4270 expression analysis across different HCC clinical stages, one-way ANOVA, P < 0.001, F = 8.100. (E) miR-4270 levels in different HCC cell lines. *P < 0.05, **P < 0.01 vs L02. N = 6.
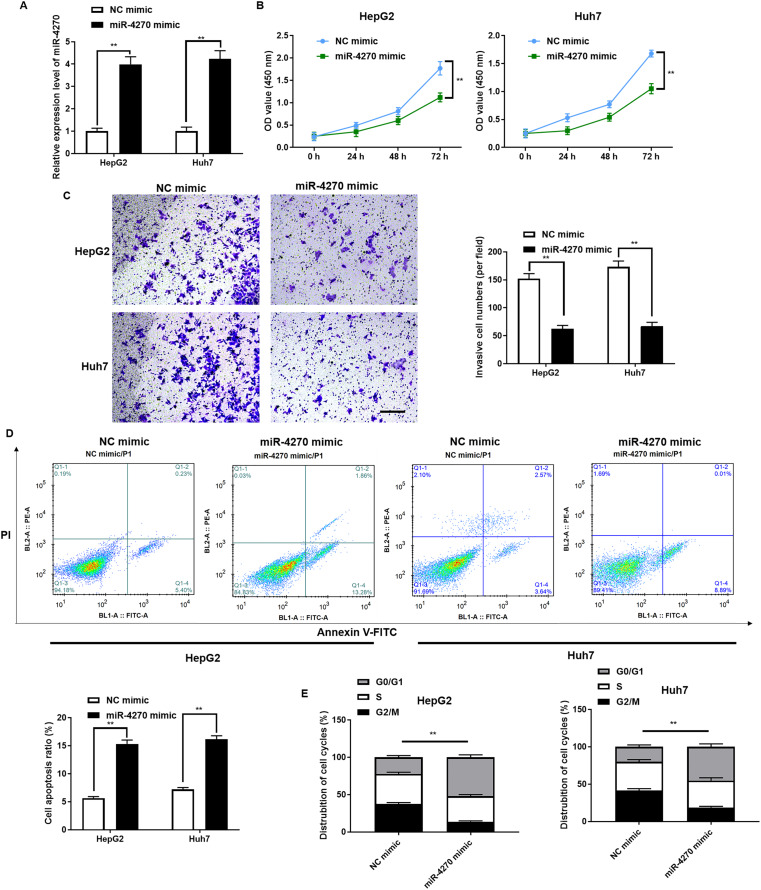
miR-4270 mimic repressed HCC cell proliferation and invasion
To elucidate miR-4270’s biological function in HCC, HepG2, and Huh7 cells were transfected with the miR-4270 mimic, achieving transient overexpression. Figure 2A demonstrates the efficiency of miR-4270 mimic overexpression. miR-4270 overexpression markedly reduced both growth and invasion in HepG2 and Huh7 cells and induced apoptosis (Figs. 2B and 2D). Furthermore, miR-4270 overexpression substantially increased the number of cells in the G0/G1 phase while significantly decreasing those in the G2/M phase (Fig. 2E).
Figure 2. miR-4270 suppresses malignant characteristics in HCC cells.
(A) RT-qPCR elucidation of miR-4270 levels in miR-4270 mimic transfected HCC cells, t-test. (B) miR-4270 mimic transfected HCC cells’ ability to proliferate was assessed via the CCK-8 assay, t-test and two-way ANOVA. (C) Transwell analysis evaluated the miR-4270 mimic transfected HCC cells’ invasion capability. Scale bar: 100 µm, t-test. (D) miR-4270 mimic transfected HCC cells was evaluated by Annexin V-FITC/PE assay, t-test. (E) Flow cytometry evaluation of the cycle distribution of miR-4270 mimic transfected HCC cells, t-test. **P < 0.01. N = 6.
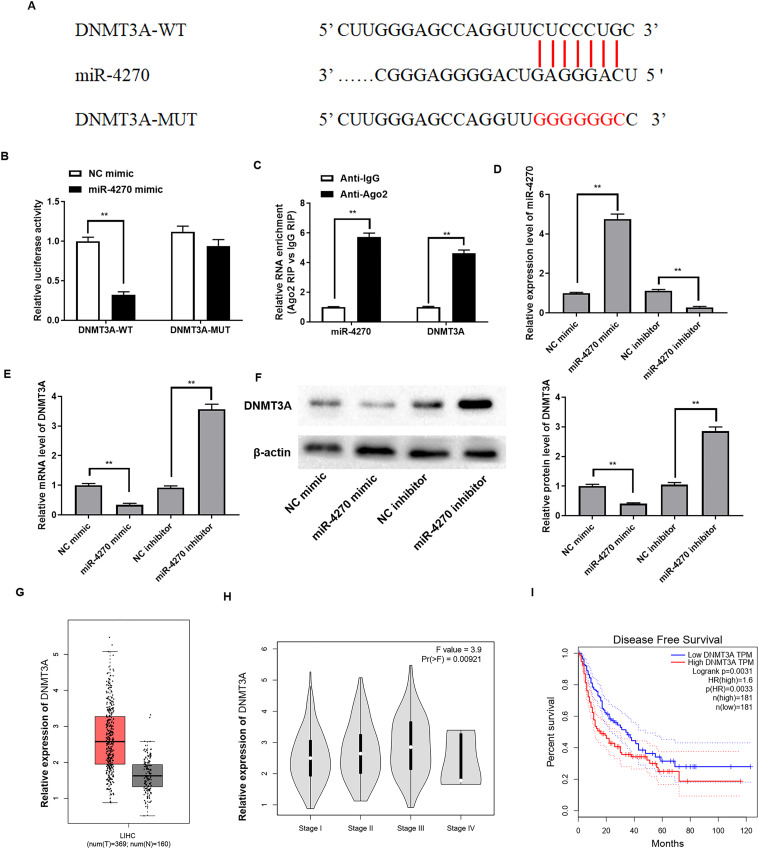
miR-4270 targets the DNMT3A 3’UTR
DNMT3A was identified as a potential target gene of miR-4270 through StarBase online database analysis. Figure 3A illustrates the binding site of miR-4270 on the 3′UTR of DNMT3A. In addition, we further verified the binding relationship by dual-luciferase reporter and Ago2-RIP assays. The results indicated that binding of the miR-4270 mimic to DNMT3A-WT resulted in significantly less luciferase activity than the DNMT3A-MUT (Fig. 3B). Furthermore, the RNA enrichment of Ago2 on miR-4270 and DNMT3A was increased compared with IgG (Fig. 3C).
Figure 3. miR-4270 directly targets the 3′UTR of DNMT3A.
(A) Binding sequences of miR-4270 and the 3′UTR of DNMT3A. (B) Relative luciferase activities of miR-4270 and DNMT3A were tested by luciferase reporter gene assay, t-test. (C) The RIP test was conducted to assess the relative enrichment level of miR-4270 and DNMT3A in HCC cells, t-test. (D) The mRNA levels of miR-4270, and (E) DNMT3A in miR-4270 mimic or miR-4270 inhibitor transfected HCC cells were assessed by RT-qPCR, t-test. (F) Western blot evaluation of DNMT3A levels in miR-4270 mimic or miR-4270 inhibitor transfected HCC cells, t-test. (G) Online analysis of DNMT3A levels in HCC tissues through the GEPIA website. (H) Online analysis of DNMT3A in different clinical stages of HCC patients through the GEPIA website. (I) Online analysis of disease-free survival curves of DNMT3A patients through the GEPIA website. **P < 0.01. N = 6.
Subsequently, to explore whether miR-4270 could regulate DNMT3A expression, the miR-4270 mimic, and miR-4270 inhibitor were individually transfected into HepG2 cells. Figure 3D shows the interference efficiency of the miR-4270 inhibitor. DNMT3A protein and mRNA levels were markedly reduced by miR-4270 upregulation and increased by miR-4270 knockdown (Figs. 3E and 3F). Interestingly, we discovered that DNMT3A levels were substantially elevated in HCC patients compared to controls, shown by Sanger prediction, and were correlated with HCC clinical stage (Figs. 3G and 3H). Additionally, patients with elevated DNMT3A expression had notably poorer disease-free survival than those with low DNMT3A levels (Fig. 3I).
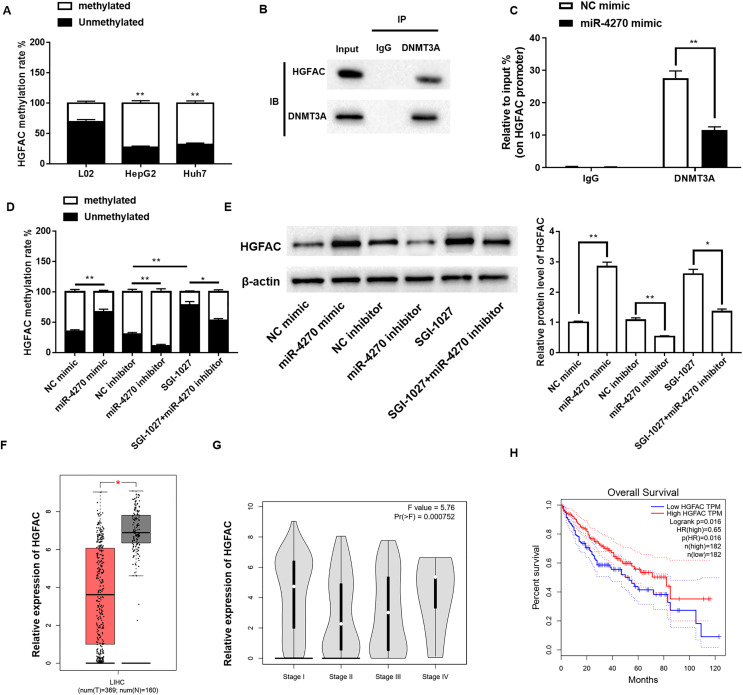
miR-4270 inhibits DNMT3A-mediated HGFAC promoter methylation
Previous studies have indicated a negative correlation between hepatocyte growth factor activator (HGFAC) expression and methylation of its promoter region, implying that DNA methylation may control HGFAC expression. Accordingly, we further analyzed whether HGFAC is regulated by DNMT3A methylation. Using the MSP, a significant increase was observed in HGFAC promoter methylation levels in HCC cells (Fig. 4A). Furthermore, Co-IP and ChIP assays demonstrated that DNMT3A could bind to HGFAC and was significantly enriched in its promoter region (Figs. 4B and 4C). Additionally, the methylation levels of the HGFAC promoter decreased significantly following miR-4270 overexpression and increased after miR-4270 inhibition. However, treatment with the DNMT3A inhibitor SGI-1027 markedly reversed the repressive effect of miR-4270 inhibition on HGFAC promoter methylation levels (Fig. 4D). Moreover, miR-4270 upregulation increased HGFAC protein levels, a trend reversed by miR-4270 downregulation of HGFAC and SGI-1027 treatment (Fig. 4E). Subsequently, we found that the HGFAC levels were significantly lower in HCC patients compared to controls, with Sanger prediction, and were correlated with the HCC clinical stage (Figs. 4F and 4G). Furthermore, patients with low HGFAC expression had significantly poorer disease-free survival than those with high HGFAC expression (Fig. 4H).
Figure 4. miR-4270 mitigates DNMT3A-induced methylation of the HGFAC promoter.
(A) Methylation-specific PCR was employed to detect HGFAC methylation rate in L02, HepG2, and Huh7 cells, t-test and one-way ANOVA. (B) Co-IP evaluation of the protein interaction of HGFAC and DNMT3A. (C) The enrichment of DNMT3A in the HGFAC promoter region was elucidated by te ChIP assay, t-test. (D) Methylation-specific PCR was employed to detect HGFAC methylation rate in HepG2 cells transfected with miR-4270 mimic, miR-4270 inhibitor, or treated with SGI-1027, t-test and one-way ANOVA. (E) Western blot assessment of HGFAC levels in miR-4270 mimic and miR-4270 inhibitor transfected or SGI-1027 treated HepG2 cells, t-test and one-way ANOVA. SGI-1027: DNMT3A inhibitor (8 µM). (F) Online analysis of HGFAC levels in HCC tissues through the GEPIA website. (G) Online analysis of DNMT3A in different clinical stages of HCC patients through the GEPIA website. (H) Online analysis of disease-free survival curves of DNMT3A patients through the GEPIA website. *P < 0.05, **P < 0.01. N = 6. For image analysis, an inverted fluorescence microscope (#IX53; Olympus Corp, Tokyo, Japan) was utilized and Image-ProPlus 5.1 Chinese was employed for capturing images.
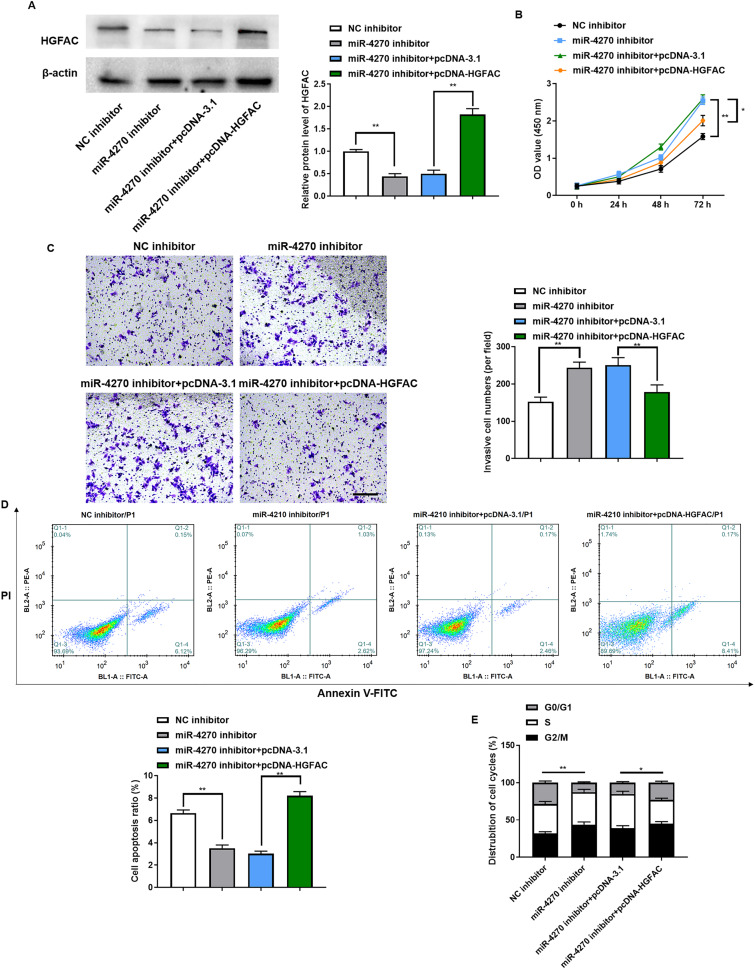
HGFAC overexpression antagonizes the promotion of malignancy by the miR-4270 inhibitor
To determine if miR-4270 mediates its tumor-suppressive role in HCC cells through HGFAC regulation, we conducted rescue experiments involving co-transfection with the miR-4270 inhibitor and pcDNA-HGFAC plasmid. The efficiency of pcDNA-HGFAC transfection in HepG2 cells was verified by Western blotting (Fig. 5A). Transfection with pcDNA-HGFAC reversed the promotion of cell proliferation by the miR-4270 inhibitor (Fig. 5B), as well as invasion (Fig. 5C) and its inhibition of apoptosis (Fig. 5D). Moreover, HGFAC upregulation counteracted the miR-4270 inhibitor’s effect on the cell cycle, leading to an increased percentage of cells in the G0/G1 phase (Fig. 5E). Collectively, these data demonstrate that HGFAC inhibits HCC cell growth and serves an essential role in the pathway downstream of miR-4270.
Figure 5. HGFAC overexpression could antagonize the facilitative effect of miR-4270 inhibitor on the malignant behavior of HCC cells.
(A) Western blot assessment of HGFAC levels in HepG2 cells transfected with miR-4270 inhibitor or/and pcDNA-HGFAC, t-test. (B) Proliferation of miR-4270 inhibitor or/and pcDNA-HGFAC transfected HCC cells by CCK-8 assay, t-test and two-way ANOVA. (C) The invasion ability of miR-4270 inhibitor or/and pcDNA-HGFAC transfected HCC cells was assessed by transwell assay. Scale bar: 100 µm, t-test. (D) Annexin V-FITC/PE assay evaluation of the apoptotic ratio of HCC cells transfected with miR-4270 inhibitor or/and pcDNA-HGFAC, t-test. (E) Flow cytometry evaluation of the cycle distribution of miR-4270 inhibitor or/and pcDNA-HGFAC transfected HCC cells t-test. *P < 0.05, **P < 0.01. N = 6.
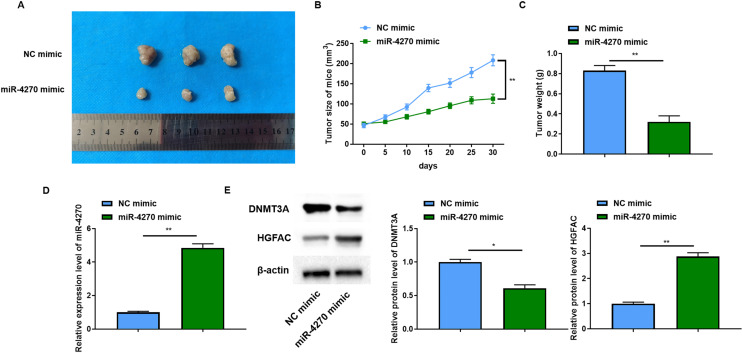
miR-4270 reduced xenograft tumor growth in mice
Following the initial focus of the in vitro tumor-suppressive effects of miR-4270, in vivo experiments were then performed. The results revealed a significant reduction in the size of the xenograft tumors in mice following miR-4270 overexpression (Fig. 6A). Measurements of tumor volume and mass also showed that miR-4270 overexpression retarded tumor growth in mice (Figs. 6B and 6C). Consistent with the in vitro findings, HGFAC protein levels increased significantly, and DNMT3A levels were decreased in tumor tissues from nude mice with increased miR-4270 expression (Figs. 6D and 6E). Taken together, these results confirmed that the upregulation of miR-4270 hindered the progression of HCC in vivo.
Figure 6. miR-4270 impeded xenograft tumor development in mice.
(A) Representative images of each groups’ tumors, two-way ANOVA, and t-test. (B) Each groups’ tumor volumes, t-test. (C) Each groups’ tumor weight, t-test. (D) RT-qPCR evaluation of miR-4270 expression in each group of tumors, t-test. (E) Western blot evaluation of DNMT3A and HGFAC protein levels in each group of tumors, t-test. *P < 0.05, **P < 0.01. N = 12.
Discussion
HCC is a global health problem and is one of the most common malignancies, ranking second in the causes of death due to cancer (Jiří, Igor & Mba, 2020; Elms, Badami & Dhanarajan, 2022). Given HCC’s high rate of metastasis and postoperative recurrence, understanding the molecular mechanisms underlying HCC initiation and progression is crucial, necessitating an in-depth search for key markers and gene targets pertinent to hepatocarcinogenesis and metastasis (Nagy et al., 2018; Qadir & Rizvi, 2017). Beyond common etiological factors such as hepatitis virus infection, cirrhosis, alcohol abuse, and fatty liver disease, genomic instability, and mutations also contribute significantly to the HCC predisposition. However, the precise pathological mechanism of HCC has not been fully elucidated.
Numerous studies on miRNAs have demonstrated their critical roles in regulating tumor cell proliferation, apoptosis, the cell cycle, and angiogenesis, processes of immense clinical significance in tumor development and progression (Hill & Tran, 2021; He et al., 2020). Current literature on miR-4270, although limited, indicates that its expression varies across various cancers, suggesting tissue-specific roles. Downregulation of its expression is associated with the progression of many tumors, and the mechanism may be to participate in gene regulation as a tumor suppressor (Wang et al., 2020). Wang et al. (2022a) reported reduced miR-4270 levels in nasopharyngeal cancer cell lines, associating its downregulation with increased cellular radiosensitivity. Additionally, reduced expression of miR-4270 in lung cancer tissues and cell lines underscores its potential role as a tumor suppressor (Zou et al., 2019). Sartorius et al. (2018) found that the levels of 10 miRNAs in HCC tissues were lower than those in matched non-tumor tissues, including miR-4270. This study used bioinformatics and tissue microarray data, providing evidence that miR-4270 levels are significantly downregulated in HCC tissues. Further, the miR-4270 mimic significantly reduced HCC cell proliferation and invasion and promoted cell apoptosis, compared with the NC mimic group.
MiRNAs play indispensable roles in HCC development, predominantly through their regulation of downstream target genes (Papanicolau-Sengos & Aldape, 2022). Based on the understanding that miR-4270 is a tumor suppressor in HCC, our study aimed to elucidate its underlying mechanisms by identifying downstream targets influenced by miR-4270. DNMT3A was found to be a target of miR-4270 by Sanger prediction as well as by dual-luciferase reporter assay and RIP assay verification. One of the early manifestations of many tumors is increased methylation of tumor suppressor genes, suggesting that changes in DNA methylation patterns may be among the first detectable tumor-specific changes associated with tumorigenesis (Hernandez-Meza et al., 2021).
Epigenetic alterations and modifications are important components of tumor initiation and progression, with DNA methylation being the most studied (Zhang et al., 2016). Previous studies have observed a correlation between altered gene methylation patterns and poorer prognoses, underscoring the significant role of epigenetic modifications in tumor development (Zhu et al., 2018). Recently reported differential methylation of promoters associated with the activation of oncogenes and premetastatic genes has been revealed to play a crucial role in cancer initiation and metastasis (Nishiyama & Nakanishi, 2021). DNA methylation, characterized by the addition of a methyl group to adenine or cytosine nucleotides within DNA sequences, is catalyzed by DNA methyltransferases (DNMTs) and constitutes a fundamental epigenetic imprint (Bestor & Verdine, 1994). In DNA methylation, s-adenosylmethionine serves as a methyl donor, facilitating a reaction catalyzed by three distinct DNMTs, namely, DNMT1, DNMT3A, and DNMT3B (Tajima et al., 2016). Among these, DNMT3A and DNMT3B are de novo methyltransferases that recognize methylated, hemimethylated, and unmethylated DNA (Okano et al., 1999). DNMT3A can introduce a methyl group into the demethylated CpG site to remethylate, regulating gene expression (Zhang et al., 2018). DNMT3A has been verified to be significantly up-regulated in HCC (Zhao et al., 2010). A study found that lncRNA SNHG5 decreased DNMT3A expression to suppress the methylation level of SPATS2 in HCC (Yan et al., 2022a). This study demonstrated that miR-4270 effectively repressed DNMT3A-mediated methylation of the HGFAC promoter.
HGFAC is a novel serine protease that activates the precursor of hepatocyte growth factor and promotes angiogenesis, tumorigenesis, and regeneration in the tumor microenvironment (Fukushima et al., 2018). Three endogenous protease inhibitors of HGFAC, HAI-1, HAI-2, and protein C, regulate HGFAC activity. Notably, Guerra et al. (2019) initially reported the critical role of HAI-1 and HAI-2 as HGFAC repressors in gastric cancer, highlighting their significance in tumor progression. Research has indicated a correlation between reduced HGFAC levels in HCC and poor survival outcomes, with DNA hypermethylation contributing to this reduction in HGFAC expression, thereby proposing HGFAC as a prognostic biomarker for HCC patients (Yin et al., 2019). Similarly, we discovered that HGFAC promoter methylation levels were markedly increased in HCC cells, which were counteracted by miR-4270 upregulation. In contrast, downregulation of miR-4270 suppressed HGFAC protein expression and increased methylation of the HGFAC promoter, effects that were counteracted by treatment with SGI-1027. Furthermore, HGFAC overexpression antagonized the promotion of malignant behavior by the miR-4270 inhibitor in HCC cells.
In conclusion, this study, by in vivo and in vitro analyses, revealed that miR-4270 can reduce growth, invasion, and migration in HCC cells. Mechanistically, miR-4270 inhibited the malignant behavior of HCC by suppressing DNMT3A-mediated methylation of the HGFAC promoter. Nevertheless, a more comprehensive mechanism of miR-4270 in HCC cells still needs further investigation. A limitation of our study was the omission of control cells and mice from the experimental design, preventing us from ruling out potential effects attributable to the NC mimic or NC inhibitor treatments. Moreover, our findings indicate a relative under-expression of miR-4270 in HCC cells. However, determining whether similar patterns of miR-4270 expression occur in tumor tissues and sera of HCC patients requires verification through comprehensive clinical trials.
Supplemental Information
Funding Statement
The authors received no funding for this work.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Qiang Zou conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Shasha Cao conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Human Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
All samples obtained in this study were approved by the ethics committee of the Tianjin Medical University Cancer Institute and Hospital and abided by the ethical guidelines of the Declaration of Helsinki.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
Animal experiments were approved and supervised by the Animal Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. All methods were carried out in accordance with relevant guidelines and regulations.
Data Availability
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.
References
- Bestor & Verdine (1994).Bestor TH, Verdine GL. DNA methyltransferases. Current Opinion in Cell Biology. 1994;6(3):380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Cheng et al. (2018).Cheng D, Deng J, Zhang B, He X, Meng Z, Li G, Ye H, Zheng S, Wei L, Deng X, Chen R, Zhou J. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;36:159–170. doi: 10.1016/j.ebiom.2018.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong et al. (2021).Dong Y, Li F, Wang J, Hu J, Li Z, Gu Y, Feng Y. miR-369 inhibits liver cancer progression by targeting ZEB1 pathway and predicts the prognosis of HCC patients. Journal of Cancer. 2021;12(10):3067–3076. doi: 10.7150/jca.54759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elms, Badami & Dhanarajan (2022).Elms D, Badami A, Dhanarajan A. Systemic therapy in metastatic hepatocellular carcinoma. Current Gastroenterology Reports. 2022;24:65–71. doi: 10.1007/s11894-022-00842-9. [DOI] [PubMed] [Google Scholar]
- Feng et al. (2021).Feng W, Zhu R, Ma J, Song H. LncRNA ELFN1-AS1 promotes retinoblastoma growth and invasion via regulating miR-4270/SBK1 axis. Cancer Management and Research. 2021;13:1067–1073. doi: 10.2147/CMAR.S281536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima et al. (2018).Fukushima T, Uchiyama S, Tanaka H, Kataoka H. Hepatocyte growth factor activator: a proteinase linking tissue injury with repair. International Journal of Molecular Sciences. 2018;19(11):3435. doi: 10.3390/ijms19113435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra et al. (2019).Guerra MT, Florentino RM, Franca A, Filho ACL, dos Santos ML, Fonseca RC, Lemos FO, Fonseca MC, Kruglov E, Mennone A, Njei B, Gibson J, Guan F, Cheng YC, Ananthanarayanam M, Gu J, Jiang J, Zhao H, Lima CX, Vidigal PT, Oliveira AG, Nathanson MH, Leite MF. Expression of the type 3 InsP(3) receptor is a final common event in the development of hepatocellular carcinoma. Gut. 2019;68(9):1676–1687. doi: 10.1136/gutjnl-2018-317811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao et al. (2021).Hao W, Zhu Y, Wang H, Guo Y. miR-4270 modulates the irradiation-sensitivity of nasopharyngeal carcinoma cells through modulation of p53 in vivo. Tohoku Journal of Experimental Medicine. 2021;254(2):63–70. doi: 10.1620/tjem.254.63. [DOI] [PubMed] [Google Scholar]
- He et al. (2019).He D, Wang X, Zhang Y, Zhao J, Han R, Dong Y. DNMT3A/3B overexpression might be correlated with poor patient survival, hypermethylation and low expression of ESR1/PGR in endometrioid carcinoma: an analysis of the cancer genome atlas. Chinese Medical Journal. 2019;132(2):161–170. doi: 10.1097/CM9.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2020).He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, Xie H, Peng X, Yin W, Tao Y, Wang X. miRNA-based biomarkers, therapies, and resistance in cancer. International Journal of Biological Sciences. 2020;16(14):2628–2647. doi: 10.7150/ijbs.47203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Meza et al. (2021).Hernandez-Meza G, Von felden J, Gonzalez Kozlova EE, Garcia-lezana T, Peix J, Portela A, Craig AJ, Sayols S, Schwartz M, Losic B, Mazzaferro V, Esteller M, Llovet JM, Villanueva A. DNA Methylation profiling of human hepatocarcinogenesis. Hepatology. 2021;74(1):183–199. doi: 10.1002/hep.31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill & Tran (2021).Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Disease Models & Mechanisms. 2021;14(4):dmm047662. doi: 10.1242/dmm.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2021).Huang W, Chen Q, Dai J, Zhang Y, Yi Y, Wei X, Wu Z. miR-744-5p suppresses tumor proliferation and metastasis by targeting transforming growth factor-beta 1 (TGF-β1) in hepatocellular carcinoma (HCC) Journal of Gastrointestinal Oncology. 2021;12(4):1811–1822. doi: 10.21037/jgo-21-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing et al. (2019).Jing W, Song N, Liu YP, Qu XJ, Qi YF, Li C, Hou KZ, Che XF, Yang XH. DNMT3A promotes proliferation by activating the STAT3 signaling pathway and depressing apoptosis in pancreatic cancer. Cancer Management and Research. 2019;11:6379–6396. doi: 10.2147/CMAR.S201610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiří, Igor & Mba (2020).Jiří T, Igor K, Mba Hepatocellular carcinoma future treatment options. Klinicka Onkologie. 2020;33(Supplementum 3):26–29. doi: 10.14735/amko20203s26. [DOI] [PubMed] [Google Scholar]
- Komoll et al. (2021).Komoll RM, Hu Q, Olarewaju O, von Döhlen L, Yuan Q, Xie Y, Tsay HC, Daon J, Qin R, Manns MP, Sharma AD, Goga A, Ott M, Balakrishnan A. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. Journal of Hepatology. 2021;74(1):122–134. doi: 10.1016/j.jhep.2020.07.039. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2022).Liu Y, Cheng H, Cheng C, Zheng F, Zhao Z, Chen Q, Zeng W, Zhang P, Huang C, Jiang W, Liu X, Liu G. ZNF191 alters DNA methylation and activates the PI3K-AKT pathway in hepatoma cells via transcriptional regulation of DNMT1. Cancer Medicine. 2022;11(5):1269–1280. doi: 10.1002/cam4.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long et al. (2019).Long J, Chen P, Lin J, Bai Y, Yang X, Bian J, Lin Y, Wang D, Yang X, Zheng Y, Sang X, Zhao H. DNA methylation-driven genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Theranostics. 2019;9(24):7251–7267. doi: 10.7150/thno.31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu & Rothenberg (2018).Lu TX, Rothenberg ME. MicroRNA. Journal of Allergy and Clinical Immunology. 2018;141(4):1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man et al. (2022).Man X, Li Q, Wang B, Zhang H, Zhang S, Li Z. DNMT3A and DNMT3B in breast tumorigenesis and potential therapy. Frontiers in Cell and Developmental Biology. 2022;10:916725. doi: 10.3389/fcell.2022.916725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy et al. (2018).Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni & Leng (2016).Ni WJ, Leng XM. miRNA-dependent activation of mRNA translation. MicroRNA. 2016;5(2):83–86. doi: 10.2174/2211536605666160825151201. [DOI] [PubMed] [Google Scholar]
- Ni et al. (2020).Ni JS, Zheng H, Ou YL, Tao YP, Wang ZG, Song LH, Yan HL, Zhou WP. miR-515-5p suppresses HCC migration and invasion via targeting IL6/JAK/STAT3 pathway. Surgical Oncology. 2020;34:113–120. doi: 10.1016/j.suronc.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Nishiyama & Nakanishi (2021).Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends in Genetics. 2021;37(11):1012–1027. doi: 10.1016/j.tig.2021.05.002. [DOI] [PubMed] [Google Scholar]
- Okano et al. (1999).Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases DNMT3A and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ong, Huey & Shelat (2022).Ong Y, Huey CWT, Shelat VG. Paraneoplastic syndromes in hepatocellular carcinoma: a review. Expert Review of Gastroenterology & Hepatology. 2022;16(5):449–471. doi: 10.1080/17474124.2022.2085556. [DOI] [PubMed] [Google Scholar]
- Oura, Morishita & Masaki (2020).Oura K, Morishita A, Masaki T. Molecular and functional roles of microRNAs in the progression of hepatocellular carcinoma—a review. International Journal of Molecular Sciences. 2020;21(21):8362. doi: 10.3390/ijms21218362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolau-Sengos & Aldape (2022).Papanicolau-Sengos A, Aldape K. DNA methylation profiling: an emerging paradigm for cancer diagnosis. Annual Review of Pathology: Mechanisms of Disease. 2022;17(1):295–321. doi: 10.1146/annurev-pathol-042220-022304. [DOI] [PubMed] [Google Scholar]
- Piñero, Dirchwolf & Pessôa (2020).Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6):1370. doi: 10.3390/cells9061370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir & Rizvi (2017).Qadir MI, Rizvi SZ. miRNA in hepatocellular carcinoma: pathogenesis and therapeutic approaches. Critical Reviews in Eukaryotic Gene Expression. 2017;27(4):355–361. doi: 10.1615/CritRevEukaryotGeneExpr.2017019539. [DOI] [PubMed] [Google Scholar]
- Sartorius et al. (2018).Sartorius K, Sartorius B, Winkler C, Chuturgoon A, Makarova J. The biological and diagnostic role of miRNA’s in hepatocellular carcinoma. Frontiers in Bioscience. 2018;23(9):1701–1720. doi: 10.2741/4668. [DOI] [PubMed] [Google Scholar]
- Shen et al. (2022).Shen J, Shen H, Ke L, Chen J, Dang X, Liu B, Hua Y. Knowledge mapping of immunotherapy for hepatocellular carcinoma: a bibliometric study. Frontiers in Immunology. 2022;13:815575. doi: 10.3389/fimmu.2022.815575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen et al. (2020).Shen D, Zhao H, Zeng P, Song J, Yang Y, Gu X, Ji Q, Zhao W. Circular RNA hsa_circ_0005556 accelerates gastric cancer progression by sponging miR-4270 to increase MMP19 expression. Journal of Gastric Cancer. 2020;20(3):300–312. doi: 10.5230/jgc.2020.20.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima et al. (2016).Tajima S, Suetake I, Takeshita K, Nakagawa A, Kimura H. Domain structure of the Dnmt1, DNMT3A, and Dnmt3b DNA methyltransferases. Yeast Membrane Transport. 2016;945:63–86. doi: 10.1007/978-3-319-43624-1_4. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2020).Wang Y, Li CF, Sun LB, Li YC. microRNA-4270-5p inhibits cancer cell proliferation and metastasis in hepatocellular carcinoma by targeting SATB2. Human Cell. 2020;33(4):1155–1164. doi: 10.1007/s13577-020-00384-0. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2022a).Wang L, Ma X, Chen Y, Zhang J, Zhang J, Wang W, Chen S. MiR-145-5p suppresses hepatocellular carcinoma progression by targeting ABHD17C. Oncologie. 2022a;24(4):897–912. doi: 10.32604/oncologie.2022.025693. [DOI] [Google Scholar]
- Wang et al. (2021).Wang J, Wang Z, Yuan J, Wang Q, Shen X. Upregulation of miR-137 expression suppresses tumor growth and progression via interacting with DNMT3A through inhibiting the PTEN/Akt signaling in HCC. OncoTargets and Therapy. 2021;14:165–176. doi: 10.2147/OTT.S268570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2022b).Wang X, Xu J, Gu Q, Tang DX, Ji HY, JU SQ, Wang F, Chen L, Yuan RY. A UHPLC/MS/MS assay based on an isotope-labeled peptide for sensitive miR-21 detection in HCC serum. Oncologie. 2022b;24(3):513–526. doi: 10.32604/oncologie.2022.024373. [DOI] [Google Scholar]
- Wong, Tsang & Ng (2018).Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nature Reviews Gastroenterology & Hepatology. 2018;15(3):137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- Xu et al. (2019).Xu XF, Xing H, Han J, Li ZL, Lau WY, Zhou YH, Gu WM, Wang H, Chen TH, Zeng YY, Li C, Wu MC, Shen F, Yang T. Risk factors, patterns, and outcomes of late recurrence after liver resection for hepatocellular carcinoma. JAMA Surgery. 2019;154(3):209–217. doi: 10.1001/jamasurg.2018.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2022a).Yan J, Huang QY, Huang YJ, Wang CS, Liu PX. SPATS2 is positively activated by long noncoding RNA SNHG5 via regulating DNMT3A expression to promote hepatocellular carcinoma progression. PLOS ONE. 2022a;17(1):e0262262. doi: 10.1371/journal.pone.0262262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2022b).Yan D, Li C, Zhou Y, Yan X, Zhi W, Qian H, Han Y. Exploration of combinational therapeutic strategies for HCC based on TCGA HCC database. Oncologie. 2022b;24(1):101–111. doi: 10.32604/oncologie.2022.020357. [DOI] [Google Scholar]
- Yin et al. (2019).Yin L, Mu Y, Lin Y, Xia Q. HGFAC expression decreased in liver cancer and its low expression correlated with DNA hypermethylation and poor prognosis. Journal of Cellular Biochemistry. 2019;120(6):9692–9699. doi: 10.1002/jcb.28247. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2022).Yu S, Lv H, Zhang J, Zhang H, Ju W, Jiang Y, Lin L. Heparanase/Syndecan-1 axis regulates the grade of liver cancer and proliferative ability of hepatocellular carcinoma cells. Oncologie. 2022;24(3):539–551. doi: 10.32604/oncologie.2022.024882. [DOI] [Google Scholar]
- Zhang et al. (2016).Zhang C, Li J, Huang T, Duan S, Dai D, Jiang D, Sui X, Li D, Chen Y, Ding F, Huang C, Chen G, Wang K. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. OncoTarget. 2016;7(49):81255–81267. doi: 10.18632/oncotarget.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018).Zhang ZM, Lu R, Wang P, Yu Y, Chen D, Gao L, Liu S, Ji D, Rothbart SB, Wang Y, Wang GG, Song J. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554(7692):387–391. doi: 10.1038/nature25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2010).Zhao Z, Wu Q, Cheng J, Qiu X, Zhang J, Fan H. Depletion of DNMT3A suppressed cell proliferation and restored PTEN in hepatocellular carcinoma cell. Journal of Biomedicine and Biotechnology. 2010;2010:737535. doi: 10.1155/2010/737535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2022).Zhou Y, Yang Z, Zhang H, Li H, Zhang M, Wang H, Zhang M, Qiu P, Zhang R, Liu J. DNMT3A facilitates colorectal cancer progression via regulating DAB2IP mediated MEK/ERK activation. Biochimica et Biophysica Acta—Molecular Basis of Disease. 2022;1868(4):166353. doi: 10.1016/j.bbadis.2022.166353. [DOI] [PubMed] [Google Scholar]
- Zhu et al. (2018).Zhu HR, Huang RZ, Yu XN, Shi X, Bilegsaikhan E, Guo HY, Song GQ, Weng SQ, Dong L, Janssen HLA, Shen XZ, Zhu JM. Microarray expression profiling of microRNAs reveals potential biomarkers for hepatocellular carcinoma. World Journal of Experimental Medicine. 2018;245(2):89–98. doi: 10.1620/tjem.245.89. [DOI] [PubMed] [Google Scholar]
- Zou et al. (2019).Zou A, Liu X, Mai Z, Zhang J, Liu Z, Huang Q, Wu A, Zhou C. LINC00472 acts as a tumor suppressor in NSCLC through KLLN-Mediated p53-signaling pathway via MicroRNA-149-3p and MicroRNA-4270. Molecular Therapy—Nucleic Acids. 2019;17:563–577. doi: 10.1016/j.omtn.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.