Abstract
Nitrogen fixation occurs in two domains, Archaea and Bacteria. We have characterized a nif (nitrogen fixation) gene cluster in the methanogenic archaeon Methanococcus maripaludis. Sequence analysis revealed eight genes, six with sequence similarity to known nif genes and two with sequence similarity to glnB. The gene order, nifH, ORF105 (similar to glnB), ORF121 (similar to glnB), nifD, nifK, nifE, nifN, and nifX, was the same as that found in part in other diazotrophic methanogens and except for the presence of the glnB-like genes, also resembled the order found in many members of the Bacteria. Using transposon insertion mutagenesis, we determined that an 8-kb region required for nitrogen fixation corresponded to the nif gene cluster. Northern analysis revealed the presence of either a single 7.6-kb nif mRNA transcript or 10 smaller mRNA species containing portions of the large transcript. Polar effects of transposon insertions demonstrated that all of these mRNAs arose from a single promoter region, where transcription initiated 80 bp 5′ to nifH. Distinctive features of the nif gene cluster include the presence of the six primary nif genes in a single operon, the placement of the two glnB-like genes within the cluster, the apparent physical separation of the cluster from any other nif genes that might be in the genome, the fragmentation pattern of the mRNA, and the regulation of expression by a repression mechanism described previously. Our study and others with methanogenic archaea reporting multiple mRNAs arising from gene clusters with only a single putative promoter sequence suggest that mRNA processing following transcription may be a common occurrence in methanogens.
Nitrogen fixation, an exclusively prokaryotic process whereby molecular dinitrogen is transformed into ammonia, is not limited to the primary domain Bacteria but is also observed in several methanogenic members of the domain Archaea (5, 6, 31). In Bacteria, the organization and regulated expression of nitrogen fixation (nif) genes have been described for a variety of species (15, 22, 35, 36). The genes nifH, nifD, and nifK are typically found together in a single operon and are physically adjacent to other nif genes as part of a larger nif regulon. The genes nifD and nifK encode the structural subunits of dinitrogenase, an α2β2 heterotetramer, which is the site of N2 reduction. The nifH gene codes for the protein dinitrogenase reductase, which provides electrons to the nitrogenase complex. Often found downstream of nifK, in a separate operon, are the genes nifE, nifN, and nifX. The genes nifE and nifN encode a scaffold-like structure in which an essential cofactor for the nitrogenase complex is assembled (14). The function of nifX is unclear, but it may play a role in cofactor biosynthesis (29) or in regulation (20).
The fact that nitrogen fixation also occurs in methanogenic members of the Archaea has stimulated a comparison of the processes in the two domains. The discovery in methanogens of genes homologous to nifH, nifD, and nifK suggests that the basic mechanism of nitrogen fixation is similar (9, 37, 39). Biochemical analysis of nitrogen fixation in Archaea supports this view (27, 28). In addition, most nitrogenases of methanogens seem to have a molybdenum-containing cofactor, as do the primary nitrogenases of Bacteria (25). There are several interesting differences between the domains as well. First, phylogenetic analysis of nifD and nifK places the nitrogenases of methanogens near the center of the tree, separate from most bacterial nitrogenases (8, 9, 25). Second, although nifH, nifD, and nifK occur in the same order in methanoarchaea as in the Bacteria, all diazotrophic methanogens contain two open reading frames (ORFs) inserted between nifH and nifD that show strong similarity to glnB. In enteric bacteria, glnB encodes the PII protein that participates in the nitrogen regulatory cascade (23). Third, transcription of the nif genes in methanogens occurs from typically archaeal promoters, rather than from promoters resembling those of Bacteria; this difference reflects the difference between the archaeal and the bacterial transcription apparatuses (the transcriptional apparatus in Archaea is similar to that of Eucarya [4]). Finally, we have studied transcriptional regulation from the nifH promoter in Methanococcus maripaludis and have found an operator sequence that mediates repression by binding an as yet unidentified factor (12). This regulatory mechanism contrasts with those of bacterial nif genes, which are regulated by activation.
In order to learn more about the arrangement and expression of nif genes in methanogenic Archaea, we identified and characterized an operon in M. maripaludis that is required for nitrogen fixation and that contains eight genes, six with sequence similarity to known nif genes and two with similarity to glnB. Although the order of the six nif genes resembles that found in many Bacteria members, their inclusion in a single operon and the presence of the glnB-like genes are unique to Archaea. In addition, an unusual pattern of multiple mRNAs arises from within the operon due to internal termination, intramolecular processing, or both. The implementation of transposon insertion mutagenesis, developed in our laboratory for M. maripaludis (6), was instrumental in establishing the operon nature of the gene cluster as well as its role in diazotrophic growth.
MATERIALS AND METHODS
Strains and plasmids.
In a previous report (6), we have indicated our use of the type strain of M. maripaludis, JJ. Recently, we detected differences between our laboratory strain and JJ, which led us to compare the 16S ribosomal DNA (rDNA) sequences of the two strains. The 16S rDNA of our strain was PCR amplified (Gene Amp PCR kit; Perkin-Elmer) with the conserved reverse primer 1492RPL (5′ GGCTCGAGCGGCCGCCCGGGTTACCTTGTTACGACTT 3′). Sequences published in GenBank for several M. maripaludis strains were used to generate a forward primer (5′ ATAAGAATGCGGCCGCGATCCCGCCGGAGGCCACTG 3′). Both of these primers have flanking NotI restriction sites. The resulting 1,392-bp PCR product was digested with NotI, cloned into pBluescript KS+ (Stratagene), and sequenced on both strands. Our strain, now designated LL, was 99.85% identical in 16S rDNA sequence to strain JJ. This difference places LL well within the species M. maripaludis (26, 43). All LL strains of M. maripaludis used here are listed in Table 1.
TABLE 1.
Plasmids, phages, and strains used in this study
| Plasmid, phage, or strain | Characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pBluescript | KS+, cloning vector; Amr | Stratagene |
| pGEM-7 | Z+, cloning vector; Amr | Promega |
| pBluePur | pac (puromycin resistance) EcoRI cassette in pBluescript; Amr | 6 |
| pMMP1.3 | HindIII subclone of pMMP1.0 with nifH and 5′ end of ORF105; Amr | This study |
| pMMP1.4 | HindIII/XbaI subclone of pMMP1.0 with nifE; Amr | This study |
| pMMP1.6 | HindIII subclone of pMMP1.0 with nifD, nifK, and 5′ end of nifE; Amr | This study |
| pMMP1.8 | HindIII subclone of pMMP1.0 with nifH, ORF105, ORF121, and 5′ end of nifD; Amr | This study |
| pMMP2.1 | HindIII/XbaI subclone of pMMP2.0 with nifN; Amr | This study |
| pMMP2.8.1 | HindIII/EcoRI subclone of pMMP2.0 with 3′ portion of nifN, nifX, and 140 bp 3′ to nifX; Amr | This study |
| pMMP2.8.1.1 | pMMP2.8.1 with pac gene at the SacI site of nifX; Amr | This study |
| pMMP2.8.2 | EcoRI/HincII subclone of pMMP2.0 with nifX and 140 bp 3′ to nifX; Amr | This study |
| pMMP2.9 | HindIII fragment of pMMP2.0 with 3′ end of nifN, nifX, and 1.9 kb 3′ to nifX in pGEM-7(Z+) vector; Amr | This study |
| pMMP2.9ΔEcoΩpac | pMMP2.9 with a deletion of an internal 1.2-kb EcoRI fragment and the insertion of the pac EcoRI cassette; Amr | This study |
| Phages | ||
| Mmpλ-1 | M. maripaludis λ genomic library clone containing nif gene cluster | 6 |
| Mmpλ-1-4 | Mmpλ-1 nifD4::Mudpur Cmr | This study |
| Mmpλ-1-5 | Mmpλ-1 Ω5::Mudpur Cmr | This study |
| Mmpλ-1-10 | Mmpλ-1 Ω10::Mudpur Cmr | This study |
| Mmpλ-1-12 | Mmpλ-1 Ω12::Mudpur Cmr | This study |
| Mmpλ-1-23 | Mmpλ-1 nifE23::Mudpur Cmr | This study |
| Mmpλ-1-31 | Mmpλ-1 nifK31::Mudpur Cmr | This study |
| Mmpλ-1-32 | Mmpλ-1 nifN32::Mudpur Cmr | This study |
| Mmpλ-1-41 | Mmpλ-1 Ω41::Mudpur Cmr | This study |
| M. maripaludis strains | ||
| LL | Wild type | W. Whitman |
| Mm4 | LL nifD4::Mudpur (Purr) | This study |
| Mm5 | LL Ω5::Mudpur (Purr) | This study |
| Mm10 | LL Ω10::Mudpur (Purr) | This study |
| Mm12 | LL Ω12::Mudpur (Purr) | This study |
| Mm18 | LL nifH18::Mudpur (Purr) | 6 |
| Mm20 | LL nifH20::Mudpur (Purr) | 6 |
| Mm23 | LL nifE23::Mudpur (Purr) | This study |
| Mm29 | LL Ω29::Mudpur (Purr) | 6 |
| Mm31 | LL nifK31::Mudpur (Purr) | This study |
| Mm32 | LL nifN32::Mudpur (Purr) | This study |
| Mm33 | LL Ω33::Mudpur (Purr) | 6 |
| Mm41 | LL Ω41::Mudpur (Purr) | This study |
| Mm51 | LL nifX51::pac with pac gene inserted at the SacI site in nifX (Purr) | This study |
| Mm52 | LL with a deletion of an internal 1.2-kb EcoRI fragment and the insertion of the pac EcoRI cassette (Purr) | This study |
Media and growth conditions.
Media were prepared as previously described (6). Techniques for growing methanogens were those of Balch et al. (3). M. maripaludis was routinely grown at 31°C in McN, McC (43), or nitrogen-free medium. McN is a minimal medium that contains 10 mM NH4+, while McC contains in addition yeast extract, sodium acetate, and vitamins. Nitrogen-free medium is McN modified so that all forms of combined nitrogen are lacking and Na2WO4 · 2H2O is omitted from the trace minerals (6). Puromycin was added as needed to a final concentration of 2.5 μg/ml. For diazotrophic growth, N2-CO2 (80/20 ratio) to 10 lb/in2 was added to the headspace of tubes containing nitrogen-free medium. After the addition of amendments and inoculum, the headspace was filled with H2-CO2 (80/20 ratio) to 40 lb/in2. For the phenotypic analysis of chromosomal insertion mutants, cultures were grown in McN to an optical density at 660 nm (OD660) of 0.4, and 0.1 ml of this culture was used to inoculate 5.0 ml of nitrogen-free medium. Cultures were incubated for 5 to 7 days on their sides without shaking at 31°C. H2-CO2 gas was added every 48 h to a final pressure of 40 lb/in2.
Transposon insertion mutagenesis and transformation of M. maripaludis.
Identification of a recombinant lambda clone with sequence homology to the nifH gene of M. maripaludis, Mmpλ-1, and transposon insertion mutagenesis of that clone have been described previously (6). Transformation of M. maripaludis with 7 μg of mutagenized phage DNA was accomplished by a recently developed polyethylene glycol-protoplast procedure (41). Transformants were selected on McN plates containing 2.5 μg of puromycin per ml.
Construction of directed insertions.
Two strains of M. maripaludis were created by directed insertions of the puromycin resistance gene (18). Mm51 contains an insertion into the coding region of nifX. For this construct, pMMP2.8.1, a pBluescript KS+-derived plasmid with a 1,079-bp HindIII/EcoRI fragment containing the 3′ end of nifN, all of nifX, and 140 bp 3′ to nifX, was digested with SacI, which cuts 180 bp 3′ to the putative translational start site of nifX. The ends of this plasmid digest were blunted with Klenow enzyme. The puromycin resistance cassette was cloned into the EcoRI site of pBluescript KS+, digested with PvuII, and ligated into pMMP2.8.1. This new construct, pMMP2.8.1.1, was used to transform wild-type M. maripaludis. To create Mm52, which contains a deletion and insertion downstream of nifX, a 3.0-kb HindIII fragment of M. maripaludis including the 3′ end of nifN, all of nifX, and 2.0 kb 3′ to nifX was cloned into a pGEM-7(Z+)-derived plasmid with the EcoRI site deleted from its polycloning region (12). From this plasmid, pMMP2.9, an EcoRI fragment that begins 140 bp downstream of nifX and continues downstream for 1.2 kb, was deleted. This region was replaced with the puromycin resistance cassette to create pMMPΔEcoΩpac, which was transformed into M. maripaludis.
Sequence analysis.
DNA sequencing was performed by cycle sequencing with dye terminators (Perkin-Elmer), and gels were run by the Biochemistry Sequence Facility and by the DNA core facility of the Molecular Pharmacology Unit at the University of Washington. Templates were plasmid subclones of MMPλ-1. The junctions of all subclones were checked by sequencing directly from the larger parental plasmids or from PCR-amplified genomic DNA. The results were assembled with SeqApp 1.9 (19), and the final sequences were organized and analyzed with the sequence analysis package of the University of Wisconsin’s Genetics Computer Group (16). The FASTA program of the Genetics Computer Group was used for the initial identification of the translated ORFs.
Southern analysis.
A total of 2.5 μg of DNA from wild-type M. maripaludis and chromosomal insertion mutants was isolated as described previously (6), digested with PvuII, run on a 1% agarose gel, transferred to a nylon membrane (Bio-Rad Laboratories), and probed with an oligonucleotide specific for the internal portion of nifH. The probe was end labeled with [γ-32P]ATP with polynucleotide kinase (New England Biolabs), and unincorporated radionucleotides were removed with a spin column of DNA-grade Sephadex G-50 (Pharmacia Biotech).
Northern analysis.
Four 5-ml tubes of McN medium were inoculated with 0.1 ml of M. maripaludis and grown overnight to an OD660 of 0.5 to 0.8. The tubes were centrifuged for 10 min at 750 × g, the supernatant was removed, and the cell pellet was resuspended in 1 ml of nitrogen-free medium. This was then added to 20 ml of prereduced nitrogen-free medium in a 120-ml serum vial and incubated for 4 to 5 h on an orbital shaker at 31°C. The culture was then transferred to a 35-ml centrifuge tube and spun aerobically at 10,000 × g for 10 min at 4°C. The pellet was resuspended in 250 μl of 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) followed by RNA extraction with guanidinium thiocyanate-phenol-chloroform (11). The RNA was resuspended in deionized formamide (10). RNA ladders were radiolabeled according to the manufacturer’s instructions (Gibco BRL). Five micrograms of total cellular RNA was run on 1% formaldehyde-agarose gels and transferred to nylon membranes which were then cross-linked in a UV Stratalinker (Stratagene). To verify that equal amounts of RNA were loaded in each lane, the membranes were stained with methylene blue. Quantitative analysis of the methylene blue staining images was done with NIH Image v1.54. The calculated amounts of RNA loaded and the amount of staining observed in the 16S and 23S rRNA bands agreed within 10% (results not shown). Hybridizations were done in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–5× Denhardt’s medium–1% sodium dodecyl sulfate (SDS)–10% dextran sulfate (Pharmacia Biotech; molecular weight, 500,000)–50% formamide. Washes were done twice for 5 min at room temperature with 2× SSC–0.1% SDS and then twice more for 10 min at 42°C with 0.2× SC–0.1% SDS. Blots were exposed to phosphor screens and processed on a phosphorimager (Molecular Dynamics).
Northern probes.
Probes were radiolabeled with [α-32P]dATP by random priming (Boehringer Mannheim), and unincorporated nucleotides were removed with a spin column. The following DNA fragments were used: for nifH, the HindIII/EcoRI fragment of pMMP1.3; for ORF105 and ORF121, the BspEI/BseRI fragment of pMMP1.8; for nifD, an 861-bp HindIII/SspI fragment of pMMP1.6; for nifK, an 849-bp BsrDI fragment of pMMP1.6; for nifE, an 853-bp HindIII/XbaI fragment of pMMP1.4; for nifN, a 644-bp HindIII/EcoRI fragment of pMMP2.1; and for nifX, a 563-bp EcoRI/PstI fragment of pMMP2.8.2.
Nucleotide sequence accession number.
The nucleotide sequence of the nif operon and the 16S rDNA sequence of M. maripaludis LL have been deposited in GenBank under accession no. U75887 and AF005049, respectively.
RESULTS
Identification of a nif gene cluster.
In our initial examination of nitrogen fixation in M. maripaludis (6), we identified a recombinant λ clone from a genomic library of M. maripaludis, Mmpλ-1, that contained the nifH gene. We mutagenized this clone by a novel system for transposon insertion mutagenesis of recombinant λ DNA. The transposon we created, Mudpur, carries selectable markers that function in both Escherichia coli (chloramphenicol resistance) and M. maripaludis (puromycin resistance). Using this technique, we obtained 19 independent transposition events distributed across the 15.3-kb insert of Mmpλ-1. DNA from four mutagenized λ clones was used to transform M. maripaludis, generating four mutants, three of which were Nif− (6). Here we describe eight additional mutants. In total, we have chosen for analysis 12 representative mutants with transposon insertions evenly distributed across the 15.3-kb insert of Mmpλ-1.
Mutagenized Mmpλ-1 DNA was used to create eight additional chromosomal insertion mutants of M. maripaludis. DNA containing Mudpur in the cloned λ insert was introduced into M. maripaludis by transformation. As before (6), we expected double homologous recombination to lead to replacement of the wild-type locus with mutagenized DNA. We verified this by Southern hybridization (results not shown). The sizes of hybridizing fragments in chromosomal digests confirmed that the placement of the transposons was the same in the chromosomal insertion mutants as in mutagenized Mmpλ-1 clones (6). Additional restriction mapping was used to localize the transposon insertions within 200 bp. The positions of these mutations, as well as those previously determined (6), are shown in Fig. 1.
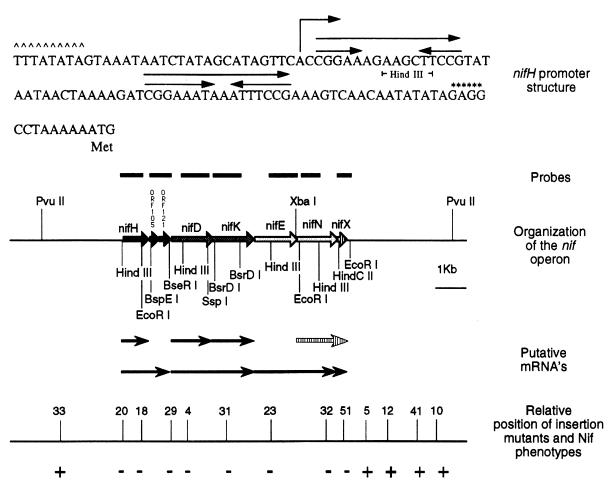
FIG. 1.
Organization of the nif operon. The nifH promoter (indicated by ∧∧∧∧) and the transcription initiation site (indicated by bent arrow) precede the first of two palindromes. The putative ribosome binding site is indicated by ∗∗∗∗. Restriction sites on the operon map were used to create probes for Northern analysis. A summary of the transcripts observed from the Northern analysis is also included. The shaded arrow indicates the heavily processed message that may include nifN, nifX, and downstream regions. The positions of 13 chromosomal insertions are shown, and the Nif phenotypes of the corresponding strains are indicated (results from this study and reference 6).
We tested the effects of these chromosomal insertions on diazotrophic growth. All strains grew equally well in N-free medium supplemented with NH4+. Mutants Mm33, Mm5, Mm12, Mm41, and Mm10 grew like the wild type under diazotrophic conditions, while mutants Mm18, Mm4, Mm31, Mm23, and Mm32 were unable to grow (results not shown). We also constructed two additional insertion mutants. The first, Mm51, placed the puromycin resistance gene into nifX, while the second, Mm52, deleted 1.2 kb of DNA beginning 140 bp 3′ to nifX. The nifX insertion mutant was Nif−, while the deletion of the region 3′ to nifX resulted in a Nif+ phenotype. This analysis combined with our previous results (6) identified a region of 8 kb containing elements necessary for nitrogen fixation (Fig. 1).
Sequence analysis of 7,955 bp corresponding to the region required for nitrogen fixation revealed eight genes: nifH, ORF105, ORF121, nifD, nifK, nifE, nifN, and nifX (Fig. 1; Table 2). Overlapping coding regions were observed between nifD and nifK and between nifN and nifX. Each coding region was preceded by a putative ribosome binding site. All nif genes showed sequence homology with the bacterial counterparts over their entire length. Phylogenetic analysis of nifH, nifD, and nifK is described elsewhere (25). Both ORF105 and ORF121 showed considerable sequence homology to the glnB gene of Klebsiella pneumoniae and other Bacteria spp. They differed from bacterial glnB in the region just N terminal to and including a conserved uridylylation site found in Bacteria spp. but were aligned over their entire length with the corresponding genes from the nif regions of other methanogens (37, 39). It is notable that the six genes with homology to known nif genes are contained within the region shown by transposon insertion to be required for diazotrophic growth. However, due to the polar effects of the transposon insertions, these results do not eliminate the formal possibility that only the downstream genes are necessary for nitrogen fixation.
TABLE 2.
Summary of nif gene cluster sequence
| Gene | Nucleotide sequence positiona | Gene size (bp) | Product size (kDa) | % Amino acid identityb |
|---|---|---|---|---|
| nifH | 139–966 | 828 | 30.1 | 64.3 |
| ORF105 | 1009–1326 | 318 | 11.6 | 38.7 |
| ORF121 | 1336–1701 | 366 | 13.2 | 36.9 |
| nifD | 1751–3184 | 1,434 | 53.3 | 44.0 |
| nifK | 3177–4565 | 1,389 | 50.5 | 39.7 |
| nifE | 4588–6039 | 1,452 | 53.6 | 44.8 |
| nifN | 6049–7425 | 1,377 | 49.9 | 28.0, 42.6c |
| nifX | 7391–7711 | 321 | 11.9 | 29.7 |
Nucleotide positions correspond to those in the GenBank database.
nifH, nifD, nifK, and nifE were compared with genes from Clostridium pasteurianum. ORF105 and ORF121 were compared with glnB from K. pneumoniae. Homologous sequences for glnB, nifN, and nifX are not available for C. pasteurianum; nifN and nifX were compared with genes from K. pneumoniae.
Percent identity with nifK from C. pasteurianum.
Transcriptional organization of the nif genes and characterization of the cluster as an operon.
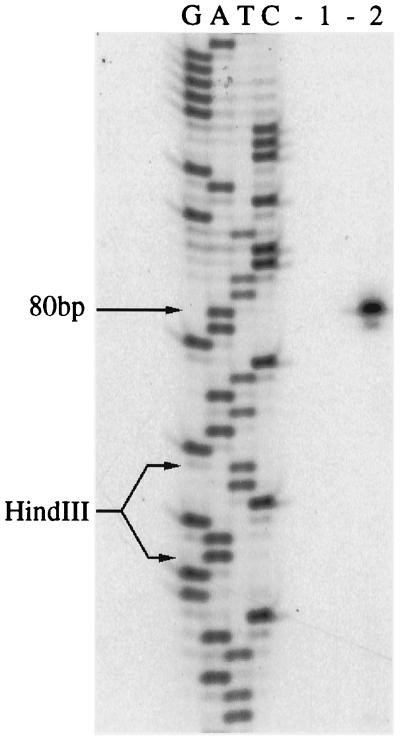
Primer extension analysis of RNA prepared from cultures grown in diazotrophic conditions revealed a 5′ end 80 bp upstream of the putative translational start site of nifH (Fig. 2). Similar analysis with nifD and nifE failed to show discrete 5′ ends (results not shown). A consensus Box A promoter sequence of methanogenic Archaea [TTTA(T/A)ATA] (33) was centered 24 bp 5′ to the end of the nifH mRNA (Fig. 1). No similar promoter sequences could be found at appropriate locations 5′ of the other genes.
FIG. 2.
Primer extension analysis of nifH. RNA was used from cells grown in nitrogen-free medium supplemented with 10 mM NH4+ (lane 1) or nitrogen-free medium alone (lane 2). The same primer was used in both the primer extension and the DNA sequence ladder. The indicated HindIII site shows the junction between the M. maripaludis genomic sequence and the vector sequence.
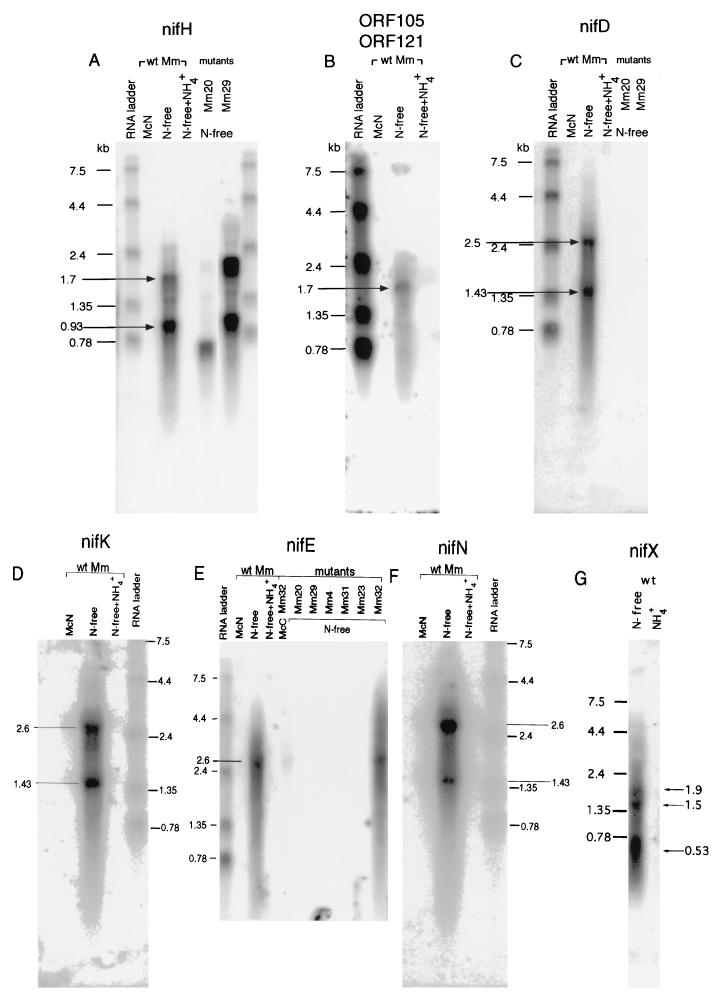
Using specific probes for each gene (Fig. 1), we detected multiple nif mRNAs. Transposon insertions consistently eliminated the expression of the downstream genes. Probing with an internal fragment of nifH revealed the presence of two transcripts, one of 0.93 kb and another of 1.7 kb (Fig. 3A, lane 3). The sizes of the observed transcripts agreed with the predicted length of a nifH and a nifH-ORF105-ORF121 mRNA. To verify that the larger transcript included both nifH and ORF105-ORF121, a similar blot was probed with a fragment of ORF105-ORF121. The only signal detected with this probe was one of 1.7 kb (Fig. 3B, lane 3), consistent with the notion that the larger transcript is derived from both ORF105-ORF121 and nifH. Two mutants were also analyzed for their nifH expression, Mm20 and Mm29 (Fig. 3A, lanes 5 and 6). In Mm20, which contains a Mudpur insertion at the 5′ end of nifH, only a shortened transcript was observed. In Mm29, with an insertion at the end of ORF121, signals for both nifH and nifH-ORF105-ORF121 messages were observed. These results suggest the presence of two mRNAs as shown in Fig. 1.
FIG. 3.
Northern analysis of the nif operon of M. maripaludis. Probes are designated above each figure. wt, wild type.
Analysis of nifD and nifK revealed three distinct mRNAs. Transcripts of 1.4 and 2.6 kb were seen for both nifD (Fig. 3C, lane 3) and nifK (Fig. 3D, lane 2). The observed sizes were consistent with those expected for nifD, nifK, and nifDK. Expression of nifD was not observed in mutants containing upstream insertions, Mm20 and Mm29 (Fig. 3C, lanes 5 and 6). Similarly, expression of nifK was not seen in mutant Mm20, Mm29, or Mm4 (results not shown).
Our sequence analysis extended to three additional genes located 3′ to nifK, the genes nifE, nifN, and nifX. Probes for nifE and nifN revealed two transcripts, a single 2.6-kb signal for nifE (Fig. 3E, lanes 3 and 11), and 1.43- and 2.6-kb signals for nifN (Fig. 3F, lane 2). The 2.6-kb signal apparently contains nifE and nifN, while the 1.43-kb signal could contain nifN alone or parts of nifN and nifX (see below). Significantly, no signal for nifE was observed in mutants with upstream insertions extending from nifH to nifE itself (Fig. 3E).
A probe for nifX produced three signals, of 0.53, 1.5, and 1.9 kb (Fig. 3G). The smallest transcript could contain nifX alone. The larger transcripts obviously contain additional regions. However, the 1.9-kb transcript does not appear to contain nifN as well because its size does not correspond to any signal detected with the nifN probe. This transcript may contain nifX with additional regions downstream of nifX. The 1.5-kb transcript could correspond to the 1.43-kb transcript detected with the nifN probe and could contain parts of nifN and nifX, or it could contain nifX with downstream regions. The presence of multiple bands hybridizing to nifX suggests that this portion of the message may be heavily processed.
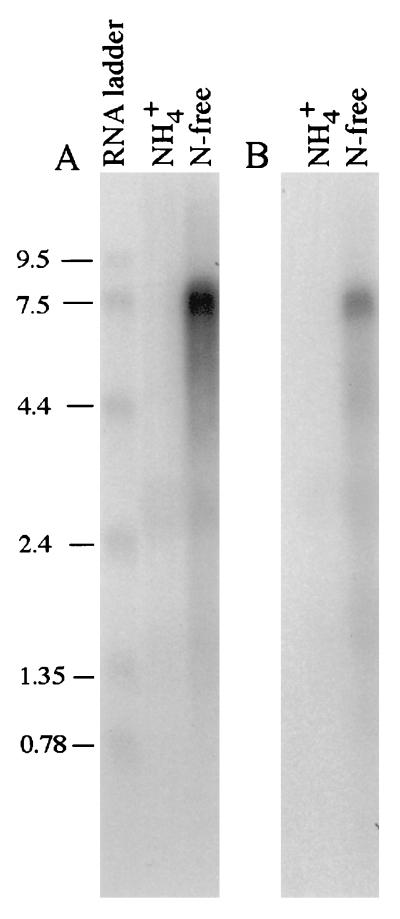
Interestingly, when RNA was prepared from cells initially grown in McC instead of McN before transfer to nitrogen-free medium, a single 7.5-kb transcript was observed for the operon. This was verified as a full-length nif transcript by probes for both nifH and nifX (Fig. 4).
FIG. 4.
Northern analysis with probes for nifH (A) and nifX (B). Four 5-ml cultures of M. maripaludis LL (wild type) were grown in McC overnight at 37°C to an OD660 of 0.8 to 0.9. These cultures were spun down, resuspended in N-free medium, and incubated for 4 to 5 h on an orbital shaker at 31°C before RNA was extracted. Numbers at left are molecular sizes in kilobases.
All transcripts were detected only under diazotrophic conditions, in the absence of ammonia. The sole exception was in mutant Mm32. A weak but consistent signal for nifE was observed in Mm32 when it was grown in McN, which contains NH4+ (Fig. 3E, lane 5). Indeed, weak transcripts for all of the nif genes of the cluster were observed for this mutant (results not shown), with the exception of nifX, which lies downstream of the insertion. One possible explanation for this unusual result is that insertion 32 stabilizes nif mRNA by altering its structure at the 3′ end. Alternatively, insertion 32 could exert its effect by eliminating the expression of nifN or nifX. Indeed, nifX may play a regulatory role in K. pneumoniae (20). However, other insertions that should eliminate nifN and nifX expression did not have the same effect.
DISCUSSION
In this study we have identified the gene cluster nifH-ORF105-ORF121-nifD-nifK-nifE-nifN-nifX in M. maripaludis. This cluster appears to constitute the minimum extent of a single operon, despite the fact that the mRNAs detected in Northern blots were fragmented in most experiments. Thus, transposon insertions across the cluster (generated by a system that we recently developed [6]) were uniformly polar on downstream mRNAs. The detection of a single 7.6-kb nif transcript in one experiment (Fig. 4) confirmed that all of the genes in the cluster are expressed as a single operon. Transcription initiation occurred 80 bp 5′ to the putative translation start site for nifH, 24 bp downstream of a promoter sequence identical to the consensus for methanogenic Archaea spp. In intergenic regions downstream of nifH, the lack of appropriately positioned sequences resembling promoters, and our failure to observe discrete 5′ mRNA ends, tended to eliminate the possibility of transcription initiation other than that from the nifH promoter. The entire operon was regulated by the cellular nitrogen status.
We cannot eliminate the possibility that additional genes on either side of the nif cluster are also involved in nitrogen fixation or are coregulated with the genes studied here. However, a preliminary BLAST analysis of approximately 1 kb flanking each end of the cluster failed to reveal the presence of ORFs with homology to known nif genes (24). In addition, transposon insertions 2 to 3 kb on each side of the nif gene cluster, as well as the deletion mutation in Mm52 downstream from nifX, failed to identify any more genes required for nitrogen fixation. It will be interesting to determine whether genes unlinked to those studied here correspond in sequence or function to the additional nif genes that are found in Bacteria spp. (15).
Compared to Bacteria, several distinctive features are observed in the nif gene cluster in M. maripaludis. Although the nif genes are in the same order as that usually found in Bacteria (15, 40), their organization into a single operon is unparalleled in the Bacteria. K. pneumoniae, for example, contains nifH, nifD, and nifK in an operon with the genes nifT and nifY, while nifE, nifN, and nifX are in a separate, though adjacent, operon. Another distinction is the presence of two genes with homology to glnB, positioned between nifH and nifD. Although their sequence suggests that these genes should function in some way in nitrogen sensing and signaling, their function is as yet unestablished (see below). In a wide range of other methanogenic Archaea spp., the same gene order is observed, including the presence of the glnB-like genes. Thus, sequencing through nifK in Methanococcus thermolithotrophicus (39), through nifE in Methanosarcina barkeri (9), and through nifX in Methanobacterium thermoautotrophicum (GenBank accession no. X87971) shows the same gene order. However, it has not been rigorously established in the other methanogenic species that the genes are in a single operon. Primer extension experiments in M. thermolithotrophicus revealed 5′ mRNA ends upstream of nifH and nifD, but a corresponding consensus promoter was found only for nifH (39).
Another distinction of the M. maripaludis nif gene cluster is its pattern of expression. We observed 10 different mRNAs corresponding to subsets of genes within the cluster. Multiple overlapping nif mRNAs are not uncommon and have been reported for Bacteria spp. In Azotobacter vinelandii (22) and Azospirillum brasilense (17), mRNA containing just nifH and mRNA containing nifH with nifD or with nifD and nifK are found, and it has been suggested that intergenic termination of transcription occurs at inverted repeats. It was noted that the NifH protein is more abundant than the NifDK protein, and termination of transcription after nifH could be a mechanism to bring this about (17). In Rhodobacter capsulatus, a similar composition of nif mRNAs is observed, with the additional presence of a nifDK message (44). In this instance, intramolecular RNA processing at inverted repeats was invoked. In M. maripaludis, a combination of intergenic termination and intramolecular processing could be going on, but we have not detected any intergenic inverted repeats to explain how either process would be directed. Whatever the mechanism, the pattern of mRNAs could be a way to assure higher levels of NifH, NifD, and NifK than of the other gene products.
It should be noted that there is no evidence for a similar pattern of fragmentation in the nif mRNA of other methanogens studied. In M. barkeri, probing for nifH and nifK revealed a single transcript corresponding in length to nifH-ORF105-ORF125-nifD-nifK (9), while in M. thermolithotrophicus, a single transcript corresponding to nifH-ORF105-ORF128 was observed (39). However, we did notice that the fragmentation pattern appeared to depend on the growth conditions leading up to the preparation of the mRNA. On the other hand, examples in which apparent operons give rise to multiple mRNAs appear to be common among the methanogenic Archaea spp. Thus, for the “spectinomycin operon” of Methanococcus vannielii (2), the pta and ack genes (38) and the CO dehydrogenase-acetyl coenzyme A synthase operon (30) of Methanosarcina thermophila, and the fdhCAB genes of Methanobacterium formicicum (42), mRNA processing has been invoked as a possible mechanism to explain the appearance of smaller transcripts. Our observation of a similar phenomenon, together with our demonstration that the genes are in a single operon, suggests that intramolecular mRNA processing may be a common occurrence in the methanogens.
In addition to defining the physical and transcriptional organization of the nif gene cluster in M. maripaludis, this study establishes the background against which a detailed study of regulation may take place. Some aspects of nif gene regulation in M. maripaludis are already known. At the level of transcription initiation, we previously found two palindromic sequences (inverted repeats) just downstream of the transcription start site for the nif gene cluster. The first of these palindromes functions in repression by NH4+ and specifically binds a factor found in extracts prepared from ammonia-grown cells (12). Two similar repeats are also found in the nifH promoter region of M. thermolithotrophicus (39), and one similar repeat sequence is also found upstream of the glnA gene, encoding glutamine synthetase, of M. maripaludis (13), Methanococcus voltae (32), and Methanococcus jannaschii (7). This observation suggests a possible shared mechanism for the nitrogen-regulated transcription of nif genes and glnA in methanococci.
Our identification of nifX and of two ORFs with homology to glnB suggests other facets of regulation. The observation that all methanogenic nif gene clusters maintain two glnB-like genes is striking. In enteric bacteria, glnB encodes the PII protein, part of the nitrogen-sensing regulatory cascade that controls the transcription and activity of glutamine synthetase (34). In addition, glnB-like genes are involved in the regulation of nif gene transcription in several proteobacteria (1, 21). However, the presence of glnB-like genes within a nif gene cluster as found in methanogens is unusual. The role of these potential regulatory genes in methanogens is still uncertain, but they do not appear to function at the level of transcription initiation. Thus, no glnB message is found in our mutant Mm20, yet expression of a truncated nifH mRNA in this mutant is still regulated by nitrogen, as is the case for the wild-type strain (results not shown). nifX, too, could play a minor regulatory role, as it appears to in K. pneumoniae (20). Additional work should lead to an understanding of how all of these factors function to regulate nitrogen fixation and assimilation in methanogenic Archaea spp.
ACKNOWLEDGMENTS
This work was supported by U.S. Department of Agriculture grant 92-37305-7965. P.S.K. was supported by a fellowship from the Office of Naval Research.
REFERENCES
- 1.Arsene F, Kaminski P A, Elmerich C. Modulation of NifA activity in Azospirillum brasilense: evidence for a regulatory role of the NifA N-terminal domain. J Bacteriol. 1996;178:4830–4838. doi: 10.1128/jb.178.16.4830-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auer J, Spicker G, Böck A. Organization and structure of the Methanococcus transcriptional unit homologous to the Escherichia coli “spectinomycin operon”: implications for the evolutionary relationship of 70S and 80S ribosomes. J Mol Biol. 1989;209:21–36. doi: 10.1016/0022-2836(89)90167-8. [DOI] [PubMed] [Google Scholar]
- 3.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P, Qureshi S A, Jackson S P. Transcription: new insights from the studies on Archaea. Trends Genet. 1995;11:279–283. doi: 10.1016/s0168-9525(00)89075-7. [DOI] [PubMed] [Google Scholar]
- 5.Beley N, Sparling R, Daniels L. Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature. 1984;312:286–288. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- 6.Blank C E, Kessler P S, Leigh J A. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J Bacteriol. 1995;177:5773–5777. doi: 10.1128/jb.177.20.5773-5777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Chien Y, Zinder S H. Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD1 from the eubacterium Clostridium pasteurianum. J Bacteriol. 1994;176:6590–6598. doi: 10.1128/jb.176.21.6590-6598.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien Y, Zinder S H. Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archaeon Methanosarcina barkeri. J Bacteriol. 1996;178:143–148. doi: 10.1128/jb.178.1.143-148.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski P. Solubilization in formamide protects RNA from degradation. Nucleic Acids Res. 1992;20:3791–3792. doi: 10.1093/nar/20.14.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Cohen-Kupiec R, Blank C, Leigh J A. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc Natl Acad Sci USA. 1997;94:1316–1320. doi: 10.1073/pnas.94.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen-Kupiec, R., and J. A. Leigh. 1997. Unpublished results.
- 14.Dean D R, Bolin J T, Zheng L. Nitrogenase metalloclusters: structures, organization, and synthesis. J Bacteriol. 1993;175:6737–6744. doi: 10.1128/jb.175.21.6737-6744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834. [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Zamaroczy M, Delorme F, Elmerich C. Regulation of transcription and promoter mapping of the structural genes for nitrogenase (nifHDK) of Azospirillum brasilense Sp7. Mol Gen Genet. 1989;220:88–94. doi: 10.1007/BF00260861. [DOI] [PubMed] [Google Scholar]
- 18.Gernhardt P, Possot O, Foglino M, Sibold L, Klein A. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol Gen Genet. 1990;221:273–279. doi: 10.1007/BF00261731. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert D G. SeqApp 1.9a169. A biological sequence editor and analysis program for Macintosh computers. 1992. Available via anonymous ftp to ftp.bio.indiana.edu. [Google Scholar]
- 20.Gosink M M, Franklin N M, Roberts G P. The product of the Klebsiella pneumoniae nifX gene is a negative regulator of the nitrogen fixation (nif) regulon. J Bacteriol. 1990;172:1441–1447. doi: 10.1128/jb.172.3.1441-1447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Institut Pasteur. Proceedings of the 11th International Congress on Nitrogen Fixation. Paris, France: Institut Pasteur; 1997. [Google Scholar]
- 22.Jacobson M R, Brigle K E, Bennett L T, Setterquist R A, Wilson M S, Cash V L, Newton W E, Dean D R. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J Bacteriol. 1989;171:1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keener J, Wong P, Popham D, Wallis J, Kustu S. A sigma factor and auxiliary proteins required for nitrogen-regulated transcription in enteric bacteria. In: Reznikoff W S, editor. RNA polymerase and regulation of transcription. New York, N.Y: Elsevier; 1987. pp. 159–175. [Google Scholar]
- 24.Kessler, P. S., and J. L. Leigh. 1997. Unpublished results.
- 25.Kessler P S, McLarnan J, Leigh J A. Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J Bacteriol. 1997;179:541–543. doi: 10.1128/jb.179.2.541-543.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keswani J, Orkand S, Premachandran U, Mandelco L, Franklin M J, Whitman W B. Phylogeny and taxonomy of mesophilic Methanococcus spp. and comparison of rRNA, DNA hybridization, and phenotypic methods. Int J Syst Bacteriol. 1996;46:727–735. doi: 10.1099/00207713-46-3-727. [DOI] [PubMed] [Google Scholar]
- 27.Lobo A L, Zinder S H. Diazotrophy and nitrogenase activity in the archaebacterium Methanosarcina barkeri 227. Appl Environ Microbiol. 1988;54:1656–1661. doi: 10.1128/aem.54.7.1656-1661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobo A L, Zinder S H. Nitrogenase in the archaebacterium Methanosarcina barkeri 227. J Bacteriol. 1990;172:6789–6796. doi: 10.1128/jb.172.12.6789-6796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludden P, Shah V, Roberts G, Rangaraj C, Ruttimann-Johnson T, Foulger T, Allen R, Homer M, Chatterjee R. Proceedings of the 11th International Congress on Nitrogen Fixation. Paris, France: Institut Pasteur; 1997. Biosynthesis of the iron-molybdenum cofactor and iron vanadium cofactor of nitrogenases, abstr. L005; p. 10. [Google Scholar]
- 30.Maupin-Furlow J A, Ferry J G. Analysis of the CO dehydrogenase/acetyl-coenzyme A synthase operon of Methanosarcina thermophila. J Bacteriol. 1996;178:6849–6856. doi: 10.1128/jb.178.23.6849-6856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray P A, Zinder S H. Nitrogen fixation by a methanogenic archaebacterium. Nature. 1984;312:284–286. [Google Scholar]
- 32.Possot O, Sibold L, Aubert J P. Nucleotide sequence and expression of the glutamine synthetase gene, glnA, of the archaebacterium Methanococcus voltae. Res Microbiol. 1989;140:355–371. doi: 10.1016/0923-2508(89)90012-0. [DOI] [PubMed] [Google Scholar]
- 33.Reeve J N. Structure and organization of genes. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry and genetics. New York, N.Y: Chapman and Hall; 1993. pp. 493–527. [Google Scholar]
- 34.Reitzer L J, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1987. pp. 302–320. [Google Scholar]
- 35.Roberts G P. Genetics and regulation of nitrogen fixation. Annu Rev Microbiol. 1981;35:207–235. doi: 10.1146/annurev.mi.35.100181.001231. [DOI] [PubMed] [Google Scholar]
- 36.Robson R, Kennedy C, Postgate J. Progress in comparative genetics of nitrogen fixation. Can J Microbiol. 1983;29:954–967. doi: 10.1139/m83-152. [DOI] [PubMed] [Google Scholar]
- 37.Sibold L, Henriquet M, Possot O, Aubert J P. Nucleotide sequence of nifH regions from Methanobacterium ivanovii and Methanosarcina barkeri 227 and characterization of glnB-like genes. Res Microbiol. 1991;142:5–12. doi: 10.1016/0923-2508(91)90091-n. [DOI] [PubMed] [Google Scholar]
- 38.Singh-Wissmann K, Ferry J G. Transcriptional regulation of the phosphotransacetylase-encoding and acetate kinase-encoding genes (pta and ack) from Methanosarcina thermophila. J Bacteriol. 1995;177:1699–1702. doi: 10.1128/jb.177.7.1699-1702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souillard N, Sibold L. Primary structure, functional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol Microbiol. 1989;3:541–551. doi: 10.1111/j.1365-2958.1989.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 40.Thiel T, Lyons E M, Erker J C. Characterization of genes for a second Mo-dependent nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol. 1997;179:5222–5225. doi: 10.1128/jb.179.16.5222-5225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tumbula D L, Makula R A, Whitman W B. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. Microbiol Lett. 1994;121:309–314. [Google Scholar]
- 42.White W B, Ferry J G. Identification of formate dehydrogenase-specific mRNA species and nucleotide sequence of the fdhC gene of Methanobacterium formicicum. J Bacteriol. 1992;174:4997–5004. doi: 10.1128/jb.174.15.4997-5004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitman W B, Shieh J, Seehyang S, Caras D S, Premachandran U. Isolation and characterization of 22 mesophilic methanococci. Syst Appl Microbiol. 1986;7:235–240. [Google Scholar]
- 44.Willison J C, Pierrard J, Hübner P. Sequence and transcript analysis of the nitrogenase structural gene operon (nifHDK) of Rhodobacter capsulatus: evidence for intramolecular processing of nifHDK mRNA. Gene. 1993;133:39–46. doi: 10.1016/0378-1119(93)90222-o. [DOI] [PubMed] [Google Scholar]