Abstract
rRNA transcription in Escherichia coli is activated by the FIS protein, which binds upstream of rrnp1 promoters and interacts directly with RNA polymerase. Analysis of the contribution of FIS to rrn transcription under changing physiological conditions is complicated by several factors: the wide variation in cellular FIS concentrations with growth conditions, the contributions of several other regulatory systems to rRNA synthesis, and the pleiotropy of fis mutations. In this report, we show by in vivo footprinting and Western blot analysis that occupancy of the rrnBp1 FIS sites correlates with cellular levels of FIS. We find, using two methods of measurement (pulse induction of a FIS-activated hybrid promoter and primer extension from an unstable transcript made from rrnBp1), that the extent of transcription activation by FIS parallels the degree of FIS site occupancy and therefore cellular FIS levels. FIS activates transcription throughout exponential growth at low culture density, but rrnp1 transcription increases independently of FIS immediately following upshift, before FIS accumulates. These results support the model that FIS is one of a set of overlapping signals that together contribute to transcription from rrnp1 promoters during steady-state growth.
The seven rRNA (rrn) operons in Escherichia coli each have two promoters, p1 and p2 (Fig. 1). The rate of rRNA transcription increases with increasing steady-state growth rate and begins to change within 1 min of an improvement in the nutritional quality of the medium (upshift) (19, 35). The rrnp1 promoters are three- to fivefold more active than the p2 promoters during rapid growth (1, 12, 34) and are responsible for growth rate-dependent regulation of rRNA transcription (13, 24).
FIG. 1.
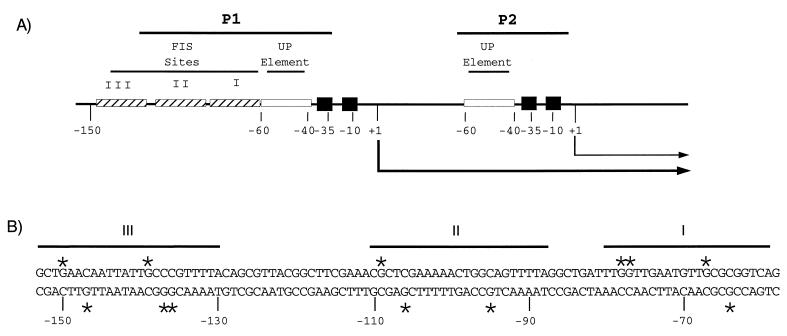
The rrnB promoter region. (A) FIS sites I, II and III, UP elements, −10 and −35 hexamers, transcription start sites (+1), and transcripts from the p1 and p2 promoters (arrows) are indicated. (B) DNA sequence of the region containing FIS sites I, II, and III in rrnBp1. G residues protected by FIS against methylation by DMS (Fig. 2 and reference 8) are indicated with asterisks. Lines indicate sequences protected by FIS against DNase I cleavage (48).
Several factors contribute to the regulation and extremely high activity of the p1 promoters (11, 25, 32). In rrnBp1, the −10 and −35 hexamers (recognized by the sigma subunit of RNA polymerase [RNAP]) are near consensus. The core promoter (−40 to +1) is stimulated 30- to 60-fold by an upstream (UP) element (the binding site for the RNAP alpha subunit [Fig. 1]) (7, 15, 47, 49). Additionally, rrnBp1 is stimulated by the 11.2-kDa transcription factor FIS (factor for inversion stimulation). FIS binds to three sites, centered at −71 (site I), −102 (site II), and −143 (site III) relative to the transcription start site, and stimulates transcription approximately fivefold in vivo (Fig. 1) (8, 9, 21, 22, 48). FIS dimers bind noncooperatively at adjacent sites in rrnBp1 (8). FIS bound at site I is sufficient to account for most of the effect of FIS on rrnBp1, primarily through direct interactions with the RNAP alpha subunit (8, 9, 22, 39, 48). Deletion of an A:T base pair at position −72 (the Δ72 mutation) (i) eliminates FIS binding at site I and activation in vitro and (ii) reduces promoter activity in strains with a wild-type fis gene but not in fis::kan strains (47, 48). Because of other regulatory systems acting on rrnp1 core promoters that can compensate for the loss of activation (18, 27), transcription from a wild-type rrnBp1 containing FIS sites is not reduced in a fis::kan strain (48). The presence of overlapping regulatory systems has made it difficult to determine the extent of the contribution of FIS to rrnp1 promoter activity at different times and under different growth conditions.
FIS is active in many cellular processes in addition to activation of rrnp1 promoters, although the fis gene is not essential for growth at 37°C (28). FIS plays a role in Hin- and Gin-mediated DNA inversion (28, 33), Tn5 transposition (52), oriC-directed DNA replication (16), transcriptional repression (53), and activation of transcription of tRNA operons and the proP promoter (14, 40–43, 54), as well as rrnp1 promoters (30, 48), and in many other systems (17, 20). Strains deleted for fis show slightly reduced growth rates in rich media and have altered morphology at high temperatures, and some fis strains have a longer lag phase before attaining logarithmic growth upon dilution of stationary-phase cultures (16, 42, 45). It is not certain which function(s) of FIS is responsible for the pleiotropic effects of fis mutations.
It was proposed that FIS may account for the rapid increase in rrnp1 transcription during transition from stationary- to exponential-phase growth (41, 48). In agreement with this proposal, intracellular FIS concentrations vary widely with changing growth phase and rate (3, 43). FIS is present in very low or undetectable quantities in stationary-phase cells but accumulates to more than 50,000 molecules per cell within two generations of growth after stationary-phase cultures are diluted into fresh rich medium. Production of fis mRNA ceases and cellular levels of FIS decrease sharply as cells enter stationary phase. FIS concentrations are lower in cells grown in poor medium (3, 43). However, previous studies have not established whether the variation in FIS levels is important to rRNA transcription activation; e.g., it is conceivable that FIS site occupancy is complete at low FIS concentrations and that high FIS levels are required only for other cell functions.
We have examined the relationship between FIS concentration, FIS site occupancy, and FIS-dependent activation of rrnBp1 at different stages of growth, using a variety of experimental approaches to circumvent the complications described above. We find that FIS increases rrnBp1 transcription during exponential growth but is not responsible for the increase in rrnBp1 transcription at the earliest times following dilution of cells from stationary phase. This finding supports a model in which FIS is one of several systems that influence rrn promoter function to different extents depending on nutritional conditions and allows efficient production of rRNA during rapid logarithmic growth. The approaches used here should be generally applicable to the study of transcription in other systems where the interpretation of experimental conclusions is complicated by redundancy, pleiotropy, or transiency of expression.
MATERIALS AND METHODS
Plasmids and strains.
Bacterial strains and plasmids are listed in Table 1. The fis::kan767 allele has been described elsewhere (28). Lysogens carried single copy λ prophages containing promoter-lacZ constructs in one of two fusion systems (system I or II, as described in reference 47). The plasmid vectors pSL6 and pRLG770 have been described previously (21, 48). pRLG770 has rrnB T1 and T2 transcription terminators approximately 170 bp downstream of a site for promoter fragment insertion (48).
TABLE 1.
Strains, λ lysogens, and plasmids used
| Strain, λ lysogen, or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| MG1655 | 2 | |
| RLG851 | MG1655 ΔlacX74 (=CAG4000) | 48 |
| LL309 | F′ lacIq ZΔM15Y+pro+/Δ(lac/pro) thi nalA supE Bfer | 24 |
| RLG1863 | RLG851 fis::kan-767 | This work |
| RLG911 | RLG851(pSL7) | This work |
| RLG912 | RLG851(pAJ4) | 8 |
| RLG921 | RLG1863(pSL7) | This work |
| RLG1819 | RLG851(pRLG1819) | This work |
| RLG1820 | RLG851(pRLG1820) | This work |
| λ lysogens | ||
| RLG1350 | RLG851/λ system I rrnBp1 −88 to +1-lacZ | 48 |
| RLG1816 | LL309/λ system II rrnBp1 −88 to −37, lac −36 to +57-lacZ | This work |
| RLG1817 | LL309/λ system II rrnBp1 −88 to −37 Δ72, lac −36 to +57-lacZ | 47 |
| RLG1823 | LL309 fis::kan 767/λ system II rrnBp1 −88 to −37, lac −36 to +57-lacZ | This work |
| RLG1824 | LL309 fis::kan 767/λ system II rrnBp1 −88 to −37 Δ72, lac −36 to +57-lacZ | This work |
| RLG1829 | RLG851/λ system I rrnBp1 −88 to +50-lacZ | This work |
| RLG1831 | MG1655/λ system I rrnBp1 −88 to +1 Δ72-lacZ | This work |
| Plasmids | ||
| pSL6 | 21 | |
| pRLG770 | 48 | |
| pSL7 | pSL6 (rrnBp1 −154 to +50) | 21 |
| pAJ4 | pSL6 (rrnBp1 −154 to +50 Δ72) | 8 |
| pRLG1819 | pRLG770 (rrnBp1 −88 to −37, lac −36 to +57) | This work |
| pRLG1820 | pRLG770 (rrnBp1 −88 to −37 Δ72, lac −36 to +57) | This work |
In vivo DMS footprinting.
The in vivo methylation protection procedure was modified from that of Thompson et al. (51). Saturated cultures grown for 13 to 24 h were diluted into prewarmed LB containing 100 μg of ampicillin per ml at a starting optical density at 600 nm (OD600) of 0.03 to 0.1 and grown at 30°C with aeration (doubling time of ∼40 min). Aliquots of culture sufficient to yield plasmid DNA for analysis (e.g., ∼100 ml of culture at an OD600 of ∼0.1 to 0.3) were removed at intervals to an Erlenmeyer flask, and dimethyl sulfate (DMS) was added to a final concentration of 0.2 to 0.5% (vol/vol) and swirled for 30 s. (All work with DMS was carried out in a fume hood except centrifugation of cells, which was carried out in bottles sealed with O-rings). The cells were poured immediately over a half volume of ice (prechilled to −20°C) and mixed to melt the ice, and EDTA was added to a final concentration of 40 mM. For subsequent steps, all containers, centrifuge bottles, rotors, media, and buffers were prechilled. The chilled culture was centrifuged to pellet cells, and cells were washed with LB, centrifuged again, resuspended in 2.5 ml of 20% sucrose–0.01 M Tris-Cl (pH 8.1), and kept on ice for 30 min. Lysozyme (0.25 ml of freshly dissolved 10-mg/ml solution in 0.25 M Tris-Cl [pH 8.1]) was added, and cells were incubated for 5 min on ice, followed by addition of 0.3 ml of 0.2 M EDTA and further incubation for 10 min on ice. A solution of 0.2% Triton X-100–25 mM EDTA–50 mM Tris-Cl (pH 8.1) was added slowly with stirring on ice (∼20 min). The cleared cell lysate was centrifuged at 25,000 × g at 5°C for 1 h. Sodium acetate was added to the supernatant to 0.2 M, followed by two extractions with an equal volume of phenol and two extractions with phenol-chloroform (1:1). Plasmid DNA was precipitated with 2 volumes of ethanol, pelleted, washed with ethanol, and resuspended in 10 mM Tris-Cl (pH 8.1).
Plasmid DNA was digested with HindIII (at +50 of rrnBp1), phenol extracted, 3′-end labeled with Sequenase (U.S. Biochemical) and [α32P]dATP (38), and digested with XhoI to generate a ∼300-bp fragment. The labeled fragment was isolated from an acrylamide gel and further purified and concentrated by using benzolated naphthylated DEAE-cellulose (23). Strand cleavage at G residues methylated by DMS was carried out in 100 μl of piperidine at 95°C for 30 min, and then piperidine was thoroughly evaporated in a vacuum centrifuge. Samples were resuspended in distilled water and dried before resuspension in gel loading buffer (8 M urea, 0.5× TBE [0.1 M Tris, 0.1 M boric acid, 2.5 mM EDTA], 0.05% xylene cyanol, 0.05% bromophenol blue). Samples containing equivalent amounts of radioactivity were electrophoresed on 8% acrylamide–7 M urea gels containing 0.5× TBE, dried, and autoradiographed. DMS footprints with purified FIS were carried out in vitro as described elsewhere (8). This procedure precluded analyzing all time points in one experiment. Three experiments were used to cover the range of times indicated in Fig. 3.
FIG. 3.
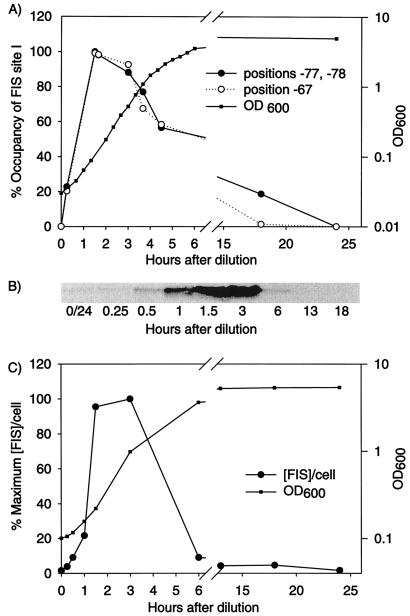
Occupancy of FIS site I and comparison with FIS levels in vivo. (A) rrnBp1 FIS site I occupancy after dilution from stationary phase. Percent occupancy was derived from protection of positions −67, −77, and −78 against DMS methylation (see the legend to Fig. 2B and Materials and Methods for details). Growth curve of a representative experiment is included. (B) FIS levels at various stages of growth. Extracts were prepared from RLG911 at the indicated times, and FIS levels were visualized on Western blots as described in Materials and Methods. The bands shown comigrated with purified FIS protein (data not shown). (C) Amounts of FIS in each band were quantified by using ImageQuant software (Molecular Dynamics) and normalized so that the maximum observed band intensity was equivalent to 100%. Growth curve of RLG911 cells sampled is shown.
The extent of protection of the FIS sites at each time point was determined by scanning autoradiographs with a Hoefer densitometer. Band intensities of the specific signals for each FIS site (−67, −77, and −78 for site I; −109 for site II; and −139 and −150 for site III) were normalized to a control band (G at position −58) that was determined previously from in vitro experiments to be unaffected by FIS binding. Positions −77 and −78 in site I were quantified together as one signal. Normalized values for band intensities were used to calculate the percent protection at each time point. It was determined that cells that remained in stationary phase for less than about 24 h retained some residual FIS. Therefore, in a few experiments it was necessary to correct for protection at early time points resulting from residual FIS in the inoculum. In the figures, maximal protection is defined as 100% and no protection is defined as 0%, comparable to the signal observed in a strain lacking the fis gene, or in vitro in the absence of FIS, or in cells remaining in stationary phase more than about 24 h. Eighty-five percent protection of signals in site I represents maximal occupancy observed in vivo and is interpreted to reflect complete site occupancy since it is equivalent to the maximal protection of site I observed with saturating amounts of FIS in vitro. The residual cleavage probably reflects partial accessibility of these positions to DMS in the presence of bound protein.
Western blotting.
Cultures of RLG911 (fis wild type) and RLG921 (fis::kan767) grown for 24 h at 30°C were diluted into fresh LB and grown under the same conditions as in the in vivo footprinting experiments. Aliquots of 0.25 to 1 ml of culture were removed at various times, pelleted in a microcentrifuge, resuspended in sample buffer (37), boiled for 5 min, sonicated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (37), blotted onto nitrocellulose (Bio-Rad), and probed with a polyclonal antibody to FIS (a gift from R. Johnson and C. Ball, University of California at Los Angeles). The FIS-antibody complexes were visualized by enhanced chemiluminescence (ECL kit; Amersham Life Science). No signal was observed from extracts made from RLG921 (data not shown).
In vitro transcription.
In vitro transcription was carried out essentially as described previously (48). Briefly, 25-μl reaction mixtures containing 0.2 nM supercoiled plasmid DNA were preincubated for 10 min at 22°C with 0, 45, or 90 nM purified FIS in a mixture containing 10 mM Tris-Cl (pH 8.0), 10 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 500 μM ATP, 100 μM CTP and GTP, and 10 μM UTP with [α-32P]UTP (Dupont NEN) at a specific activity of ca. 30 ci/mmol. Transcription was initiated by addition of purified RNAP (4 nM) and terminated after 15 min at 22°C by addition of 25 μl of 7 M urea–10 mM EDTA–1% sodium dodecyl sulfate–2× TBE–0.05% bromophenol blue–0.025% xylene cyanol. Equivalent aliquots of these samples were electrophoresed on 8% acrylamide–7 M urea gels containing 1× TBE for 2 h at 250 V.
Determination of promoter activities in vivo.
β-Galactosidase activities were determined from promoter-lacZ fusions in λ lysogens grown in LB with 2 mM isopropylthiogalactopyranoside (IPTG) at 30°C for four generations (to an OD600 of about 0.4 [36]).
mRNAs produced in vivo from chromosomal rrnBp1-lacZ fusions were quantified by primer extension. RLG1350 and RLG1831 were grown for 48 h in morpholinopropanesulfonic acid with 0.4% glucose and 0.8% Casamino Acids and diluted to an OD600 of 0.1 with LB prewarmed to 30°C, conditions in which cells were completely depleted for FIS and commenced growth with a minimal lag time. Cultures were grown with aeration, 6-ml samples were removed at different times, and RNA was isolated as described previously (31), except that approximately 5 μg of recovery marker RNA was added to each sample prior to the first phenol extraction. Recovery marker RNA was prepared from cells carrying an rrnBp1-lacZ fusion in the same λ cloning system as the test promoter but with a different promoter fragment endpoint (+50 instead of +1), resulting in a primer extension product of different length than that from the test promoter. Approximately 15 μg of RNA (including recovery marker) was mixed with 0.5 pmol of a 21-nucleotide γ-32P-end-labeled primer (sequence 5′TGGTGTTCGTCCCGGCTGTAA3′) and Sequenase reaction buffer (U.S. Biochemical). Following annealing, extension was performed at 45°C as described previously (31). After electrophoresis (10), bands were visualized and quantified with a PhosphorImager (Molecular Dynamics). Extension products were corrected for differences in culture density by normalization to OD600 at the time of sampling and for differences in RNA recovery, primer annealing, primer extension, and gel loading by normalization to the recovery marker.
Measurement of RNA half-lives.
Half-lives of the RNAs used in the quantitative primer extension experiments were determined after 5 and 60 min of growth in LB. Rifampin was added to aliquots of RLG1829 to a final concentration of 200 μg/ml, samples were removed for RNA extraction every 60 s, and RNAs were extracted, processed, and analyzed by primer extension as described above.
RESULTS
High levels of FIS are required for complete occupancy of rrnBp1 FIS sites in vivo.
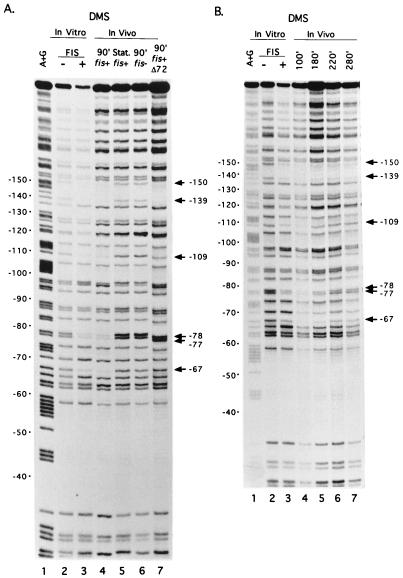
Intracellular levels of FIS vary widely with growth rate and growth phase. However, since FIS has so many cellular roles, it was possible that the variation in FIS levels was not related to its role in rRNA transcription. Therefore, we compared occupancy of the three rrnBp1 FIS binding sites with the intracellular concentration of FIS. Occupancy was assayed on plasmid-borne rrnBp1 throughout a growth cycle in rich medium by in vivo footprinting with DMS. Certain G residues in a protein binding site may be protected against methylation or show enhanced methylation when the protein is bound (29). We previously identified G residues protected by FIS in each of the three rrnBp1 binding sites, using DMS footprinting in vitro (8). These included top-strand positions −67, −77, and −78 in site I, −109 in site II, and −139 and −150 in site III (Fig. 1B; Fig. 2, lanes 3). The same G residues were protected in vivo during logarithmic growth (Fig. 2, lanes 4) but not in stationary-phase cells, where FIS is not detectable (3) (Fig. 3B), or in fis::kan cells (Fig. 2A, lanes 5 and 6). To further confirm that the in vivo protection signals reflected FIS binding, in vivo footprinting was carried out with rrnBp1 containing a mutation (Δ72) that abolishes FIS binding to site I in vitro (48). As expected, site I in the mutant construct was not protected under conditions where sites II and III were occupied by FIS (Fig. 2A, lane 7).
FIG. 2.
Binding of FIS to rrnBp1 in vitro and in vivo by DMS footprinting assays. (A) G residues protected by FIS in vitro and in vivo. Lane 1, A+G sequence marker; lanes 2 and 3, promoter fragment modified by DMS in vitro in the absence or presence of purified FIS (8); lane 4, wild-type promoter modified by DMS in vivo in a strain containing the wild-type fis gene (RLG911), 90 min (90′) after dilution of cells in fresh LB (exponential growth); lane 5, same as lane 4 except that cells were treated with DMS in stationary (Stat.) phase; lane 6, same as lane 4 except that cells (RLG921) contained the fis::kan767 allele; lane 7, same as lane 4 except that the rrnBp1 on the plasmid (pAJ4) contained the FIS site I mutation (Δ72; RLG912). The positions of G residues protected against DMS methylation by FIS in rrnB promoter sites I (−67, −77, and −78), II (−109), or III (−139, −150) are indicated. The degree of protection of particular positions is most easily evaluated by comparison to a reference band within the same lane (e.g., position −58, which is not protected by either FIS [8] or RNAP [38]). (B) Protection of FIS site residues in DMS footprints at different time points in a growth cycle. Plasmid DNA containing the wild-type rrnBp1 was modified in a wild-type (RLG911) strain at the indicated time point (100, 180, 220, or 280 min after dilution of a stationary-phase culture into fresh LB). Lanes 1 to 3, same as lanes 1 to 3 in panel A; lanes 4 to 7, in vivo footprints at the indicated times.
The DMS footprinting assay was used to determine the degree of FIS site occupancy in vivo at various times after dilution of stationary-phase cells into fresh medium (Fig. 2B, lanes 4 to 7, and data not shown; Fig. 3A). Occupancy of site I, as determined by protection of positions −67, −77, and −78, was 20% at 0.25 h after dilution, was at its maximal observed level (100% occupancy) at 1.5 to 1.6 h, and remained above 80% of maximum during mid-exponential growth phase, up to 3.5 h. Occupancy declined to <60% of maximum after 4.5 h as cells approached stationary phase. Complete loss of protection by FIS was observed only after cells had been in culture for at least 24 h. Occupancy of FIS sites II and III followed a similar pattern, although protection of FIS site II was slightly less complete (data not shown). The length of time during which high levels of protection were observed depended on the density of the starting culture and thus on the time the cells remained in exponential phase growth before the transition to stationary phase began (data not shown and reference 1). We also observed differences in the extent of methylation of two G residues in the core promoter (−8G and −34G) between stationary-phase and logarithmic growth (data not shown). These are positions where methylation by DMS is affected by bound RNAP in vitro (38). No evidence was found for interactions of other proteins with rrnBp1.
To determine whether the level of FIS site occupancy correlated temporally with FIS concentration, we performed Western blotting on extracts made from a culture grown under conditions similar to those used in the in vivo footprinting assays (Fig. 3B and C). FIS was barely detectable in stationary-phase cells. The concentration of FIS increased to 10% of its highest level within 0.5 h of dilution, was at its highest levels at 1.5 to 3 h, and then declined to 10% of its maximum level by 6 h, consistent with previous reports (3, 43). The maximum FIS site occupancy correlated with the highest concentration of FIS observed, and occupancy was partial at intermediate concentrations of FIS. However, the resolution of these experiments was insufficient to determine whether maximal FIS site occupancy requires maximal, rather than slightly lower, levels of FIS. We conclude that occupancy of the rrnBp1 FIS sites in vivo correlates qualitatively with cellular FIS concentration and is complete only at very high concentrations of FIS.
High cellular levels of FIS are required for maximal transcription activation.
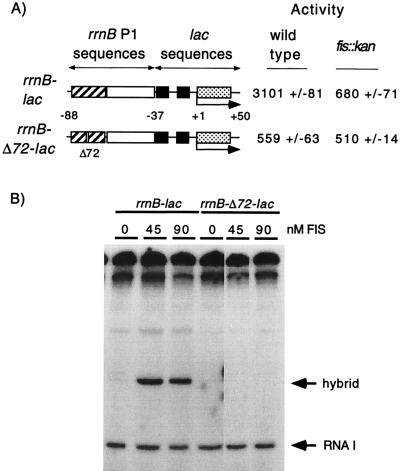
Since close to maximal occupancy of the FIS sites in the rrnBp1 promoter in vivo correlated with high concentrations of FIS, we predicted that maximal activation of rRNA transcription would also correlate with high concentrations of FIS. To determine the extent of FIS-dependent activation of transcription at different stages of growth, we used inducible hybrid promoters in which either a wild-type (rrnB-lac) or a Δ72 mutant (rrnB-Δ72-lac) rrnBp1 FIS site I was fused to the lac core promoter (Fig. 4A and reference 47). We used constructs with only FIS site I, since this FIS site is sufficient for a majority of activation, occupancy of sites II and III parallels that of site I, and use of site I allowed comparison with a non-FIS binding construct differing by only 1 bp. The position of the FIS site relative to the −35 consensus hexamer is the same in these promoters as in rrnBp1, but the lac core promoter region is not subject to the growth rate-dependent and stringent control systems that regulate the rrnBp1 core promoter (5, 31). The hybrid promoters are regulated by lac repressor binding to the lac operator site, allowing induction by IPTG and estimation of synthesis rates during a brief 15-min period of induction. The level of activation by FIS is calculated as the ratio of activities of the wild-type and Δ72 promoters.
FIG. 4.
Transcription of the rrnB-lac and rrnB-Δ72-lac hybrid promoters in vivo and in vitro. (A) Activation by FIS in vivo; residues −88 to −38 are from rrnBp1. Residues −37 to +59 are from lac and include the lac operator. Wild-type or mutant (Δ72) FIS site I (striped rectangle), the UP element (open rectangle), the −10 and −35 hexamers (filled squares), lac operator (stippled rectangle), and the transcription start site are indicated. β-Galactosidase activities at the right are from lysogens carrying the hybrid promoter-lacZ fusions (RLG1816, RLG1817, RLG1823, and RLG1824). (B) Activation by FIS in vitro. Supercoiled plasmids (pRLG1819 and pRLG1820) carrying the hybrid promoters pictured in panel A were transcribed in vitro in the presence of the indicated concentrations of purified FIS. Transcripts (about 220 nucleotides in length) from the hybrid promoter (containing the wild-type or mutant [Δ72] FIS site) and the RNA I promoter are indicated.
We first confirmed that FIS activates the rrnB-lac hybrid promoter similarly to the rrnBp1 promoter by measuring accumulated β-galactosidase activities from wild-type and mutant promoters after several generations of growth under inducing conditions (2 mM IPTG). The activity of the hybrid promoter with the wild-type FIS site was five- to sixfold higher than the activity of the promoter with the mutant FIS site (3,101 versus 559 U [Fig. 4A]), a difference comparable to that seen with rrnBp1 constructs containing only FIS site I (8, 48). FIS-dependent activation of the hybrid promoter constructs was also observed in vitro with purified FIS protein (Fig. 4B), confirming that the hybrid promoter is activated directly by FIS. The wild-type and Δ72 mutant hybrid promoters had similar activities in a fis::kan strain (680 versus 510 U [Fig. 4A]), and these activities were comparable to the activity of the Δ72 promoter in the wild-type strain, indicating that the difference in activity seen in the wild-type strain is due to FIS. These results contrast with experiments with the rrnBp1 promoter, where compensating regulatory mechanisms acting on the core promoter increased promoter activity in a fis::kan strain (48).
Synthesis of β-galactosidase from mutant and wild-type hybrid promoter-lacZ fusions during 15-min pulses of induction with 2 mM IPTG was determined at different times during the growth of a culture (Fig. 5). No activation was detected in the first 15 min of growth for cultures grown in LB (Fig. 5A), but activation was observed after 45 min and was at its maximal level of five- to sixfold approximately 1.5 to 3 h after dilution. Activation declined as cells approached stationary phase. Thus, activation by FIS followed the time course of site I occupancy observed in DMS footprints (Fig. 3A) and correlated with increased FIS concentrations observed in Western blots (Fig. 3B and C).
FIG. 5.
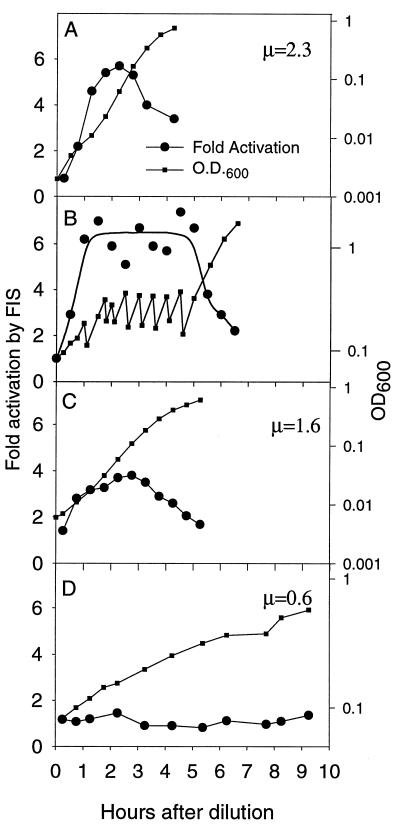
Activation of the rrnB-lac hybrid promoter by FIS as a function of growth phase in batch cultures grown in LB (A and B), M9 with glucose (C), and M9 with glycerol (D). In panel B, the cultures were kept at low density for several hours by dilution with prewarmed LB medium at 30-min intervals. Fold activation by FIS is the ratio of the β-galactosidase activities of strains RLG1816 and RLG1817 (Table 1) carrying the FIS-activated rrnB-lac and the non-FIS-activated rrnB-Δ72-lac promoter-lacZ fusions, respectively. Activity was measured after 15 min of induction by IPTG at various times after dilution of cells from stationary phase into fresh medium. Points on the graphs correspond to the end of the 15-min induction period. Peak levels of activation varied less than 15% in different experiments. Strains were isogenic and grew identically within the experiments illustrated in each panel; only one growth curve is presented for the sake of clarity. The growth rates (μ; doublings/hour) for both strains in each medium are indicated.
We wished to determine whether activation of transcription by FIS was limited to a fixed time interval after dilution into fresh medium, or whether activation by FIS could be maintained as long as cells remained in logarithmic growth. We used the hybrid promoters to measure activation of transcription by FIS in rapidly growing cultures maintained at a low density by repeated dilution with fresh prewarmed LB. Activation by FIS was maintained at a high level as long as cells were in logarithmic growth at low culture density; activation was not limited to a short, defined period of time following dilution (Fig. 5B). When the culture was allowed to increase without further dilution, the level of activation by FIS began to decline about one generation before cell growth slowed.
Activation of transcription by FIS was also measured in media supporting lower growth rates (Fig. 5C and D). The maximal extent of activation was lower in these media, corresponding with the reduced concentrations of FIS in cells grown in defined rather than in complex media (3, 43). Activation by FIS was observed only in logarithmic growth and was three- to fourfold in a defined medium (1.6 doublings/h), compared to five- to sixfold in LB (2.3 doublings/h). No activation was detected in a medium supporting a growth rate of 0.6 doublings/h. We conclude that the high levels of FIS observed during logarithmic growth in rich medium are necessary for maximal FIS-dependent activation of transcription.
rRNA synthesis and growth increase before FIS-dependent activation.
We and others have suggested previously that FIS may be important for the rapid increase in rRNA transcription upon outgrowth from stationary phase (17, 41, 42, 45, 48). In the experiments with hybrid promoters described above, FIS did not contribute measurably to promoter activity during the first 15 min of growth, and activation of transcription by FIS did not reach maximal levels until more than an hour after dilution into rich medium (Fig. 5A). However, the 15-min pulse times in these experiments were too long to provide an accurate estimate of the kinetics of this response. To directly and more precisely measure the contribution of FIS to the increase in rrnBp1 promoter activity during the first hour following dilution of the culture, we used quantitative primer extension (31) to measure the synthesis of an unstable mRNA expressed from the rrnBp1 promoter (as opposed to the hybrid promoter).
We constructed rrnBp1 promoter-lacZ fusions containing either the wild-type or the Δ72 mutant FIS site I. These promoters made identical mRNAs with a half-life that did not vary in a growth phase-dependent manner over the time course of the experiment (approximately 60 s when measured either 5 or 60 min after dilution of cells into fresh medium [see Materials and Methods; data not shown]). This short half-life allowed estimation of the rate of RNA synthesis by measuring the amount of RNA present. A recovery marker RNA was introduced during the RNA extraction process to correct for variable recoveries and for variable efficiency of primer annealing or extension. The marker RNA annealed to the same primer as was used for extension of the transcripts from the rrnBp1 promoters but yielded an extension product of different length.
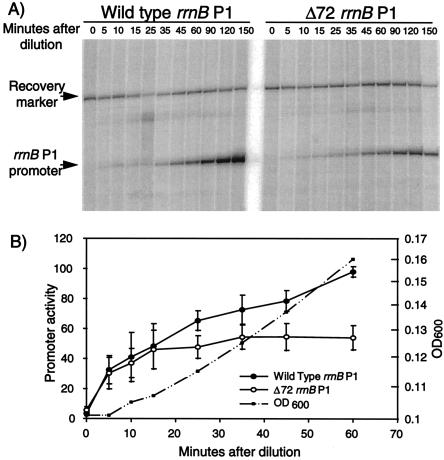
Cultures carrying either a wild-type or a Δ72 mutant rrnBp1-lacZ fusion were sampled at 5, 10, 15, 25, 35, 45, and 60 min following dilution of stationary-phase cells into fresh LB. After RNA extraction, primer extension and gel electrophoretic separation (Fig. 6A), the products were quantified by phosphorimaging and normalized to the recovery marker in the same sample (Fig. 6B). A large increase in RNA synthesis rate was observed for both the wild-type and mutant promoters during the first 20 min after dilution. This increase was the same for both promoters, indicating that a mechanism other than activation by FIS was responsible. A statistically significant difference in the activities of the two promoters was observed approximately 25 min following dilution, indicating the onset of activation by FIS. At 60 min, the wild-type promoter was approximately twofold more active than the non-FIS-activated Δ72 mutant promoter. Maximal FIS-dependent activation of rrnBp1 was achieved at a later time (data not shown), consistent with the kinetics of activation of the rrn-lac hybrid promoter (compare Fig. 5A and 6B). Thus, FIS contributed to rRNA transcription during exponential growth, but only after other system(s) had increased the rRNA synthesis rate following the upshift. The timing of this activation is consistent with the kinetics of occupancy of rrnBp1 FIS site I (Fig. 2), FIS concentration (Fig. 3), and activation of the hybrid promoter (Fig. 5).
FIG. 6.
FIS-dependent activation of rrnBp1 during outgrowth from stationary phase. (A) Primer extension products from wild-type and Δ72 mutant rrnBp1 promoters (from RLG1350 and RLG1831, respectively) were measured at the indicated times during the first 60 min following dilution of cells from stationary phase into fresh LB. Equal volumes of culture were used for all time points, and the data were subsequently normalized for culture density (B). Primer extension products from rrnBp1 promoters, extending to +1, and recovery marker products (from RLG1829 cells containing an rrnBp1 promoter, extending to +50) are indicated. (B) Activities of wild-type and Δ72 rrnBp1 promoters, normalized for recovery and increasing culture density (arbitrary units). Results from four experiments similar to that shown in panel A were quantified and averaged. Error bars represent standard deviations. The growth curve (OD600) is shown from one strain in one experiment for clarity, but the growth curves from both strains in all four experiments were very similar.
DISCUSSION
We investigated FIS binding to the rrnBp1 FIS sites and the contribution of FIS to rRNA synthesis during different phases of growth. The multiple regulatory systems acting on the rrnBp1 promoter and the pleiotropy of fis mutations required novel techniques to study the contribution of FIS to rrnp1 transcription without interference from other systems. By comparing activities of promoters with wild-type and mutant FIS binding sites, we were able to avoid the complications of working with strains with altered growth phenotypes caused by fis mutations. By using a hybrid promoter activated by FIS, we were able to monitor effects of FIS independently of regulatory systems acting on the rrnBp1 core promoter. Similar approaches should be applicable to the study of other systems involving pleiotropic activators or promoters subject to multiple regulatory inputs.
We observed that FIS was present at very low levels in stationary-phase cells, increased dramatically after the cells were diluted into fresh media, and was at its maximal concentration by 1.5 h after the start of growth, consistent with previous reports (3, 43). Maximal occupancy of rrnBp1 FIS site I in vivo, determined by footprinting with DMS, and maximal activation of transcription by FIS, determined by analysis of the hybrid promoter and by primer extension, correlated with high cellular FIS levels. The duration of the period of maximal activation varied as a function of the initial density of the batch culture and thus with the number of generations of logarithmic growth before the onset of stationary phase. Activation by FIS was not limited to a brief time interval following upshift as has been implied in previous studies but rather persisted until the culture was about one doubling from entering stationary phase (Fig. 5). The mechanism regulating fis to produce such a pattern of expression is not yet understood. Transcription of fis is known to be affected, either directly or indirectly, by FIS itself, integration host factor and ppGpp. However, these factors appear to affect the level, but not the timing, of FIS expression (3, 44–46).
It was previously suggested that FIS may be responsible for mediating the initial increase in rRNA synthesis upon dilution of stationary-phase cells into fresh medium (41, 43, 48). However, we observed a large increase in transcription from both the wild-type and FIS site I mutant rrnBp1 promoters immediately following dilution. Activation of the wild-type rrnBp1 by FIS was not detected until the cells had achieved their new growth rate, about 20 min following dilution. The rapid increase in transcription from rrnp1 promoters is consistent with previous observations that rRNA production increases too rapidly to result from de novo protein synthesis (19, 35). The thrU (tufB) promoter is also activated by FIS, and its transcription also increases upon upshift independently of fis (43). We suspect that this FIS-independent mechanism is likely to be general to many stable RNA promoters.
The rapid FIS-independent increase in rrnp1 transcription may be attributable to one or more of several regulatory systems that affect rrn expression. rrnp1 transcription is sensitive to the level of the initiating nucleotide (ATP in six rrnp1 promoters and GTP in the seventh). This mechanism contributes to growth rate-dependent regulation of rRNA synthesis (4, 18) and could potentially affect rRNA production upon dilution of stationary-phase cells. However, previous reports suggest that purine nucleoside triphosphate (NTP) concentrations do not increase immediately after upshift (6). The rapid decrease in ppGpp concentration upon upshift (26) may be a more likely contributor to the initial increase in rrnp1 activity following dilution.
The rapid increase in total rRNA production previously reported upon upshift (19) most likely reflects an increase in transcription from both rrnp1 and rrnp2 promoters. Transcripts from rrnp2 promoters increase more rapidly than transcripts from rrnp1 promoters during the first few minutes of upshift (1, 50). The rrnp2 promoters may therefore contribute substantially to the rapid increase in rRNA synthesis during outgrowth. Thus, changes in NTP and/or ppGpp levels (4, 18, 26) and transcription from the rrnp2 promoters could explain the increase in rRNA synthesis during upshift, and these mechanisms could also account for the nearly normal growth and rRNA transcription in strains with fis null mutations.
FIS is an important contributor to the activity of the rrnp1 promoters during logarithmic growth at low densities, acting in conjunction with other mechanisms to ensure production of sufficient rRNA for rapid growth. While having redundant mechanisms for rRNA transcriptional control would be a reasonable strategy for such an important biosynthetic pathway, the different systems may not overlap completely. Understanding the relative contributions of the FIS-dependent, NTP-dependent, and ppGpp-dependent mechanisms to rrnp1 activity, and the relationship between transcription of the p1 and p2 promoters, presents a challenge for the future and will provide a perspective on the interplay of control mechanisms in systems with multiple overlapping regulators.
ACKNOWLEDGMENTS
This work was supported by grant GM37048 from the National Institutes of Health to R.L.G. J.A.A. was supported in part by an NIH predoctoral training grant.
We thank Reid Johnson for antibodies to FIS and members of our lab for discussion and comments on the manuscript.
REFERENCES
- 1.Appleman, J. A., and R. L. Gourse. Unpublished results.
- 2.Bachmann B J. Linkage map of Escherichia coli K-12, edition 7. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 807–876. [Google Scholar]
- 3.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in FIS levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett, M., T. Gaal, W. Ross, and R. L. Gourse. RNA polymerase mutants defective in the NTP-sensing mechanism for growth rate-dependent control of rRNA transcription. Submitted for publication.
- 5.Bartlett M, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck C, Ingraham J, Maaloe O, Neuhard J. Relationship between the concentration of nucleoside triphosphates and the rate of synthesis of RNA. J Mol Biol. 1973;78:117–121. doi: 10.1016/0022-2836(73)90431-2. [DOI] [PubMed] [Google Scholar]
- 7.Blatter E E, Ross W, Tang H, Gourse R L, Ebright R H. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994;78:889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 8.Bokal A J, IV, Ross W, Gourse R L. The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J Mol Biol. 1995;245:197–207. doi: 10.1006/jmbi.1994.0016. [DOI] [PubMed] [Google Scholar]
- 9.Bokal A J, IV, Ross W, Gaal T, Johnson R C, Gourse R L. Molecular anatomy of a transcription activation patch: FIS-RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J. 1997;16:154–162. doi: 10.1093/emboj/16.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borukhov S, Sagitov V, Josaitis C A, Gourse R L, Goldfarb A. Two modes of transcription initiation in vitro at the rrnB P1 promoter of Escherichia coli. J Biol Chem. 1993;268:23477–23482. [PubMed] [Google Scholar]
- 11.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deBoer H A, Nomura M. In vivo transcription of rRNA operons in Escherichia coli initiates with purine nucleotide triphosphates at the first promoter and with CTP at the second promoter. J Biol Chem. 1979;254:5609–5612. [PubMed] [Google Scholar]
- 13.Dickson R R, Gaal T, deBoer H A, De Haseth P L, Gourse R L. Identification of promoter mutants defective in growth-rate-dependent regulation of rRNA transcription in Escherichia coli. J Bacteriol. 1989;171:4862–4870. doi: 10.1128/jb.171.9.4862-4870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emilsson V, Nilsson L. Factor for inversion stimulation-dependent growth rate regulation of serine and threonine tRNA species. J Biol Chem. 1995;270:16610–16614. doi: 10.1074/jbc.270.28.16610. [DOI] [PubMed] [Google Scholar]
- 15.Estrem, S., and R. L. Gourse. Unpublished data.
- 16.Filutowicz M, Ross W, Wild J, Gourse R L. Involvement of FIS protein in replication of the Escherichia coli chromosome. J Bacteriol. 1992;174:398–407. doi: 10.1128/jb.174.2.398-407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkel S, Johnson R. The FIS protein: it’s not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaal T, Bartlett M, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 19.Gausing K. Regulation of ribosome biosynthesis in E. coli. In: Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M, editors. Ribosomes: structure, function, and genetics. Baltimore, Md: University Park Press; 1980. pp. 693–718. [Google Scholar]
- 20.Gonzalez-Gil G, Bringmann P, Kahmann R. FIS is a regulator of metabolism in Escherichia coli. Mol Microbiol. 1996;22:21–29. doi: 10.1111/j.1365-2958.1996.tb02652.x. [DOI] [PubMed] [Google Scholar]
- 21.Gosink K K, Ross W, Leirmo S, Osuna R, Finkel S, Johnson R, Gourse R L. DNA binding and bending are necessary but not sufficient for Fis-dependent activation of rrnB P1. J Bacteriol. 1993;175:1580–1589. doi: 10.1128/jb.175.6.1580-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosink K K, Gaal T, Bokal A J, Gourse R L. A positive control mutant of the transcription activator protein FIS. J Bacteriol. 1996;178:5182–5187. doi: 10.1128/jb.178.17.5182-5187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gourse R L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gourse R L, de Boer H A, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 25.Gourse R L, Gaal T, Bartlett M S, Appleman J A, Ross W. rRNA transcription and growth rate dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez V J, Bremer H. Guanosine tetraphosphate (ppGpp) dependence of the growth rate control of rrnB P1 promoter activity in Escherichia coli. J Biol Chem. 1990;265:11605–11614. [PubMed] [Google Scholar]
- 27.Jinks-Robertson S, Gourse R L, Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983;33:865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- 28.Johnson R C, Ball C A, Pfeffer D, Simon M I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci USA. 1978;75:5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josaitis C, Gaal T, Gourse R L. Sequences upstream of the −35 hexamer of rrnB P1 affect promoter strength and upstream activation. Biochim Biophys Acta. 1990;1050:307–311. doi: 10.1016/0167-4781(90)90186-6. [DOI] [PubMed] [Google Scholar]
- 31.Josaitis C, Gaal T, Gourse R L. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keener J, Nomura M. Regulation of ribosome biosynthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: Cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1417–1431. [Google Scholar]
- 33.Koch C, Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986;261:15673–15678. [PubMed] [Google Scholar]
- 34.Lund E, Dahlberg J E. Initiation of Escherichia coli ribosomal RNA synthesis in vivo. Proc Natl Acad Sci USA. 1979;76:5480–5484. doi: 10.1073/pnas.76.11.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maaloe O, Kjeldgaard N O. Control of macromolecular synthesis. New York, N.Y: Benjamin Press; 1966. [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 37.Neville D M., Jr Molecular weight determination of protein-dodecyl sulfate complexes by gel electrophoresis in a discontinuous buffer system. J Biol Chem. 1971;246:6328–6334. [PubMed] [Google Scholar]
- 38.Newlands J T, Ross W, Gosink K K, Gourse R L. Factor-independent activation of Escherichia coli rRNA transcription. II. Characterization of complexes of rrnB P1 promoters containing or lacking the upstream activator region with Escherichia coli RNA polymerase. J Mol Biol. 1991;220:560–583. doi: 10.1016/0022-2836(91)90101-b. [DOI] [PubMed] [Google Scholar]
- 39.Newlands J, Josaitis C, Ross W, Gourse R L. Both FIS-dependent and factor-independent upstream activation of the rrnB P1 promoter are face of the helix dependent. Nucleic Acids Res. 1992;20:719–726. doi: 10.1093/nar/20.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsson L, Emilsson V. Factor for inversion stimulation-dependent growth rate regulation of individual tRNA species in Escherichia coli. J Biol Chem. 1994;269:9460–9465. [PubMed] [Google Scholar]
- 41.Nilsson L, Vanet A, Vijgenboom E, Bosch L. The role of FIS in trans-activation of stable RNA operons of E. coli. EMBO J. 1990;9:727–734. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson L, Verbeek H, Hoffmann U, Haupt M, Bosch L. Inactivation of the fis gene leads to reduced growth rate. FEMS Lett Microbiol. 1992;99:85–88. doi: 10.1016/0378-1097(92)90292-v. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson L, Verbeek H, Vijgenboom E, van Drunen C, Vanet A, Bosch L. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol. 1992;174:921–929. doi: 10.1128/jb.174.3.921-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ninnemann O, Koch C, Kahmann R. The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J. 1992;11:1075–1083. doi: 10.1002/j.1460-2075.1992.tb05146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osuna R, Lienau D, Hughes K, Johnson R C. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol. 1995;177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pratt T S, Steiner T, Feldman L S, Walker K A, Osuna R. Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and FIS. J Bacteriol. 1997;179:6367–6377. doi: 10.1128/jb.179.20.6367-6377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P, Record M T, Gourse R L. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–2435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 48.Ross W, Thompson F, Newlands J T, Gourse R L. E. coli FIS protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 50.Sarmientos P, Contente S, Chinali G, Cashel M. Ribosomal RNA operon promoters P1 and P2 show different regulatory responses. In: Hamer D H, Rosenberg M, editors. Gene expression. New York, N.Y: Alan R. Liss; 1983. pp. 65–74. [Google Scholar]
- 51.Thompson J F, de Vargas L M, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the λ site-specific recombination pathway. Cell. 1987;50:901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 52.Weinreich M, Reznikoff W. FIS plays a role in Tn5 and IS50 transposition. J Bacteriol. 1992;174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Johnson R C. Identification of genes negatively regulated by FIS: FIS and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1995;177:938–947. doi: 10.1128/jb.177.4.938-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J, Johnson R C. FIS activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J Bacteriol. 1995;177:5222–5231. doi: 10.1128/jb.177.18.5222-5231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]