Abstract
Although mutations in dozens of genes have been implicated in familial forms of amyotrophic lateral sclerosis (fALS) and frontotemporal degeneration (fFTD), most cases of these conditions are sporadic (sALS and sFTD), with no family history, and their etiology remains obscure. We tested the hypothesis that somatic mosaic mutations, present in some but not all cells, might contribute in these cases, by performing ultra-deep, targeted sequencing of 88 genes associated with neurodegenerative diseases in postmortem brain and spinal cord samples from 404 individuals with sALS or sFTD and 144 controls. Known pathogenic germline mutations were found in 20.6% of ALS, and 26.5% of FTD cases. Predicted pathogenic somatic mutations in ALS/FTD genes were observed in 2.7% of sALS and sFTD cases that did not carry known pathogenic or novel germline mutations. Somatic mutations showed low variant allele fraction (typically <2%) and were often restricted to the region of initial discovery, preventing detection through genetic screening in peripheral tissues. Damaging somatic mutations were preferentially enriched in primary motor cortex of sALS and prefrontal cortex of sFTD, mirroring regions most severely affected in each disease. Somatic mutation analysis of bulk RNA-seq data from brain and spinal cord from an additional 143 sALS cases and 23 controls confirmed an overall enrichment of somatic mutations in sALS. Two adult sALS cases were identified bearing pathogenic somatic mutations in DYNC1H1 and LMNA, two genes associated with pediatric motor neuron degeneration. Our study suggests that somatic mutations in fALS/fFTD genes, and in genes associated with more severe diseases in the germline state, contribute to sALS and sFTD, and that mosaic mutations in a small fraction of cells in focal regions of the nervous system can ultimately result in widespread degeneration.
Introduction
Amyotrophic lateral sclerosis (ALS), a disease in which premature loss of upper and lower motor neurons (UMNs and LMNs) leads to fatal paralysis, shows clinical, genetic, and pathological overlap with frontotemporal dementia (FTD), a neurodegenerative disorder characterized by behavioral, language, and memory dysfunction1. 5–22% of individuals with ALS develop FTD, and ≈ 15% of those with FTD eventually develop ALS2. ALS and FTD also share common pathology, with cytoplasmic inclusions of TAR DNA binding protein (TDP-43) found in almost all ALS brains and in half of FTD brains3,4. FTD brains lacking TDP-43 inclusions mainly show tau pathology. ALS typically begins focally and spreads regionally as the disease progresses5,6, although whether degeneration begins in UMNs, LMNs, or both simultaneously, has remained controversial7,8, with some studies suggesting that focality can manifest independently in UMNs and LMNs5,9. TDP-43 pathology also follows stereotypical patterns in ALS and FTD brains9–11, thought to reflect focal onset and intercellular transmission of TDP-43 inclusions in a prion-like manner, as shown in cell and animal models12–18.
Whereas over 30 genes are implicated in ALS and FTD19, most causative genes are linked to familial ALS (fALS) and FTD (fFTD), while 90–95% of cases are sporadic ALS (sALS) and FTD (sFTD) without a family history20. Prospective studies of ALS revealed a higher number of cases stemming from a genetic basis, regardless of whether a family history is documented21, with the underestimation of genetic cases probably reflecting multiple factors, including incomplete ascertainment, death from other causes before diagnosis, and incomplete disease penetrance. Therefore, genetic screening of ALS/FTD genes is needed to fully examine the contribution of germline mutations in sporadic cases.
The focal onset of ALS and FTD, their stereotypical spread, and the increased risk in smokers22, have raised interest in potential roles of somatic mosaic mutations in the pathogenesis of ALS and FTD23. Somatic mutations are increasingly recognized as prevalent in normal-appearing tissues, but somatic mutations responsible for neurological conditions are often limited to the central nervous system (CNS)24, and hence undetectable through DNA sequencing of non-CNS tissues. Recent studies have evaluated the contributions of somatic mutation to Alzheimer’s and Parkinson’s disease directly using postmortem brain tissues25.
In this study, we assessed potential contributions of germline and somatic mutations — distinguished by their variant allele frequencies (VAFs) — to sALS and sFTD using ultra-deep sequencing of a panel of neurodegeneration-associated genes on postmortem tissues of various brain regions and spinal cords from >400 unique sALS and sFTD cases. Our study revealed that pathogenic germline mutations are more common than previously appreciated in sALS and sFTD cases, supporting the underestimation of ALS and FTD cases with underlying genetic causes. In addition, we identified novel predicted pathogenic somatic mutations in 2.7% of the sALS and sFTD cases without known or novel pathogenic germline mutations. Protein-altering (missense/nonsense/frameshift) somatic mutations showed significant enrichment in sALS and sFTD cases and in disease-affected brain regions, supporting roles in disease pathogenesis. Regional analysis revealed focality of predicted pathogenic somatic mutations in primary motor cortex and spinal cord, supporting independent disease initiation in UMNs and LMNs, but also strongly supporting models of ALS and FTD in which the disease spreads beyond a relatively confined region containing a somatic mutation.
Results
Ultra-deep targeted sequencing of neurodegenerative genes in sALS and sFTD brains
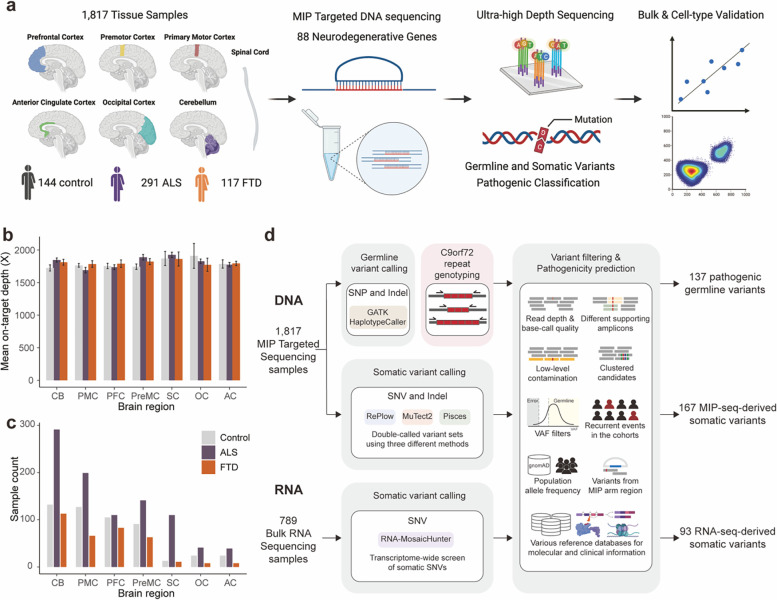
To directly detect somatic mutations in sALS and sFTD brains, we obtained post-mortem frozen tissues of several brain regions and spinal cords from individuals diagnosed with sALS or sFTD, as well as from age-matched controls through the Massachusetts Alzheimer’s Disease Research Center, Oxford Brain Bank, and Target ALS Foundation (Fig. 1a and Supplementary Table 1). Additional brain tissues from ALS, FTD and control cases, without a record of family history but with an age of death above 45 years old, were also obtained from the NIH NeuroBioBank. We designed a molecular inversion probe (MIP) panel targeting the exons and exon-intron junctions of 88 neurodegeneration-related genes26, which included 34 ALS/FTD genes, 10 Alzheimer’s disease genes, 28 Parkinson’s disease genes, and 16 genes associated with other rare neurodegenerative disorders (Supplementary Table 2). We performed MIP panel sequencing at ~1,800X average sequencing depth (Fig. 1b and Extended Data Fig. 1), with a similar distribution of sequencing depth across batches, disease conditions, and tissue regions (Extended Data Fig. 1). The variance of depth, along with the batch and sample information, were considered as factors in the mutation burden test. A total of 937, 364, and 516 samples from 291 ALS, 117 FTD, and 144 neurotypical control individuals respectively were sequenced (Fig. 1a, 1c and Supplementary Table 1). Of the ALS and FTD cases, 8 were diagnosed with both ALS and FTD, and were therefore counted for each condition.
Fig: 1. Experimental and analysis strategies.
(a) Overall scheme of the experiments. Genomic DNA isolated from 1,817 postmortem tissue samples of multiple brain regions and spinal cords of 144 control, 291 ALS, and 117 FTD cases were used for molecular inversion probe (MIP) capture sequencing with ultra-high depth. (b, c) Mean sequencing depth and number of tissue samples in different brain regions and spinal cords of control, ALS, and FTD cases. Control, n=516; ALS, n=937; FTD, n=364. CB: cerebellum; PMC: primary motor cortex; PFC: prefrontal cortex; PreMC: premotor cortex; SC: spinal cord; OC: occipital cortex; AC: anterior cingulate cortex. Error bars, 95% CI (d) Methodological pipelines to identify germline and somatic variants. Germline variants were called by GATK HaplotypeCaller. C9ORF72 genotype of ALS and FTD cases were determined by repeat-primed PCR. Somatic variants were called by RePlow, MuTect2, and Pisces. Additional somatic variants were called from 789 bulk RNA-seq profiles of multiple brain regions and spinal cords of ALS cases generated by the New York Genome Center ALS Consortium using RNA-MosaicHunter.
Pathogenic germline mutations in sALS and sFTD cases
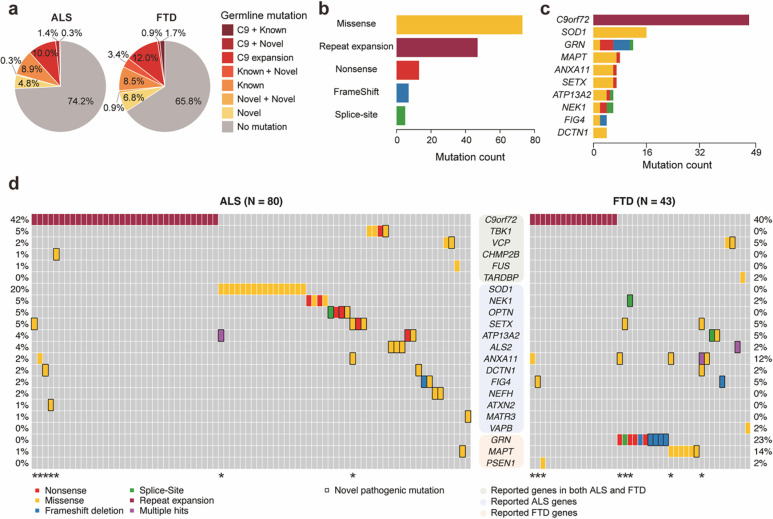
We first identified pathogenic germline single-nucleotide variants (SNVs) and short insertions and deletions (indels) using GATK followed by multiple variant filters (Fig. 1d). The functional impact and predicted pathogenicity of identified germline mutations were annotated using ANNOVAR27 and multiple clinical databases. In addition, the most common inherited cause of ALS and FTD, a hexanucleotide repeat expansion in the C9ORF72 gene28,29, was genotyped by a repeat-primed PCR assay (Extended Data Fig. 2). Overall, 20.6% (60/291) of ALS, 26.5% (31/117) of FTD and 0.7% (1/144) of control cases showed C9ORF72 repeat expansions or pathogenic germline mutations in ALS and FTD genes that have been previously reported (Fig. 2a, Supplementary Table 3, 4). Known and novel missense mutations in ALS/FTD genes represented the most prevalent mutation type (Fig. 2b). C9ORF72 repeat expansion was the most frequently mutated gene followed by known and novel pathogenic germline mutations in SOD1 for ALS, and GRN and MAPT mutations for FTD cases (Fig. 2c and 2d). The overall fractions of C9ORF72 repeat expansion carriers — 10.6% for ALS-only cases and 12.0% for FTD-only cases — slightly exceeded those reported in previous studies, yet they remained notably lower than the rates observed in fALS and fFTD cases30–32. Three carriers of the C9ORF72 repeat expansion also had known pathogenic mutations in other genes associated with ALS/FTD (Fig. 2d and Supplementary Table 3), aligning with previous studies that have demonstrated instances of oligogenic inheritance involving C9ORF72 repeat expansions and other pathogenic mutations in certain sALS and sFTD cases33,34.
Fig. 2: C9ORF72 repeat expansion and pathogenic germline variants in ALS/FTD genes are prevalent in ALS and FTD.
(a) Proportions of ALS and FTD cases with C9ORF72 repeat expansion, known, and novel pathogenic germline variants of ALS/FTD genes. Cases with multiple pathogenic mutations are indicated with ‘+’ sign. (b) Distribution of C9ORF72 repeat expansion and known and novel pathogenic germline variants in ALS/FTD genes classified by mutation types. (c) Ranking of the top 10 mutated ALS/FTD genes.(d) Visualization of ALS and FTD cases (vertical columns) with known and novel pathogenic germline variants (horizontal rows) in ALS/FTD genes. Color codes indicate the types of mutations. Rectangular outline represents novel variants. Genes are grouped by their known involvement in the diseases. * indicates cases with multiple pathogenic mutations.
Our pathogenicity prediction found pathogenic germline mutations in dominant ALS/FTD genes besides C9ORF72 repeat expansions in 14.1% of ALS, 19.7% of FTD, and 5.6% of control cases (Fig. 2a, Supplementary Table 3, 4). The odds ratios for the presence of pathogenic mutations in ALS and FTD cases, compared to control cases, were 2.78 (95% CI: 1.24–7.07, p=9.3×10−3) and 4.14 (95% CI: 1.70–11.17, p=8.2×10−4) respectively, suggesting pathogenic mutations are enriched in both ALS and FTD cases. Not surprisingly, all previously reported pathogenic mutations were predicted to be pathogenic. Most novel pathogenic mutations were nonsynonymous SNVs that would require experimental validation to confirm their functional impact. However, two novel GRN frameshift mutations (p.L46Rfs*18 and p.D250Tfs*6) identified in FTD cases are probably disease-causing (Supplementary Table 3), since loss-of-function GRN mutations are known to cause FTD in a dominant manner35,36.
When we considered previously unreported but likely pathogenic germline mutations, another 12 disease cases exhibited potential instances of oligogenic inheritance (Fig. 2d). Of these, five individuals carried C9ORF72 repeat expansions alongside novel pathogenic germline mutations in other ALS/FTD genes, while another five cases had known pathogenic germline mutations in GRN, SOD1, and MAPT genes, in combination with novel predicted pathogenic germline mutations in other ALS/FTD genes. Two patients carried multiple novel pathogenic germline mutations. These findings provide additional evidence for oligogenic inheritance of ALS and FTD33,34,37,38 (Fig. 2d). We also found 13 FTD cases to have germline mutations in genes previously linked to ALS only (NEK1, SETX, ATP13A2, ALS2, ANXA11, DCTN1, FIG4 and VAPB) and one ALS case to have a predicted pathogenic missense mutation in the FTD-associated MAPT gene (Fig. 2d). These crossover mutations between ALS and FTD reinforce the overlap between both diseases from shared underlying mechanisms.
Identification of somatic SNVs and indels from MIP sequencing data
We developed a custom pipeline integrating RePlow39, Mutect240, and Pisces41 for calling somatic SNVs and indels in our MIP sequencing data (Fig. 1d). We selected somatic mutations identified by at least two of the three callers (double-called mutations) followed by multi-step variant filters to remove false positive candidates. Unlike heterozygous germline mutations with variant allele fractions (VAFs) around 50%, heterozygous somatic mutations have VAFs less than 50%, and we only called somatic mutations with VAFs below 40%. To benchmark our pipeline, we performed a spike-in experiment by mixing two human samples from the Genome in a Bottle Consortium (GIAB) at variant allele fractions (VAFs) of 10%, 5%, 2.5%, 1%, and 0.5% (Extended Data Fig. 3a). Double-called mutations identified by Mutect2 and Pisces were excluded from the final call set due to high false positive and low validation rates (Extended Data Fig. 3b, c). High sensitivity and precision were achieved for the remaining Replow-based double-called mutations (Replow-Mutect2 and Replow-Pisces) while maintaining a low false positive rate across the low VAFs compared to the somatic mutations called by each caller. The MIP sequencing and our custom pipeline together allowed us to confidently identify somatic mutations with a low false positive rate at VAF as low as 0.5%. The observed VAFs of somatic mutations were well in line with the target VAFs at all five VAF levels (Extended Data Fig. 3).
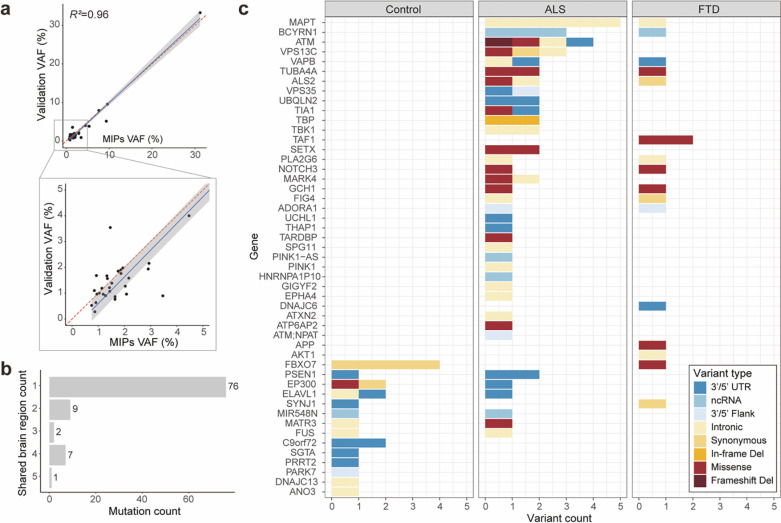
The custom pipeline identified 167 somatic SNVs and indels from our MIP sequencing data (Supplementary Table 5). The VAF distribution of identified somatic mutations was similar between disease and control cases at high VAF levels (>5%), but low-VAF mutations were more common in disease cases (Extended Data Fig. 4). Forty-one somatic candidates were selected for validation and 87.8% of them were confirmed by deep amplicon sequencing (Supplementary Table 6). The VAFs of validated candidates in amplicon sequencing showed a strong correlation with their original VAFs in the MIP sequencing data (Fig. 3a).
Fig. 3: Somatic variants in MIP sequencing data tend to be focal, protein-altering and are almost exclusively restricted to disease cases.
(a) The observed VAFs of somatic variants in amplicon sequencing validation were consistent with the VAFs in original MIP sequencing. Forty somatic variants were validated and included in the plot. (b) Total somatic variant counts classified by the number of brain regions in which a given variant was identified. (c) Distribution of somatic variants in all neurodegenerative genes. Color codes indicate variant types. Note that somatic variants identified in controls are unlikely to alter function, with just one missense mutation (red) and the remaining being synonymous or noncoding substitutions.
Somatic mutations in disease-relevant genes are enriched in ALS and FTD cases lacking pathogenic germline mutations
To examine the burden and potential roles of somatic mutations in ALS and FTD, we focused on cases that lacked known or novel pathogenic germline mutations (referred to as germline-free cases). Ninety-five unique somatic mutations in neurodegeneration-related genes were identified in 696, 243, and 516 samples from 216 ALS germline-free cases, 76 FTD germline-free cases, and 144 neurotypical controls, respectively. Most somatic mutations (80%, 76 out of 95 unique mutations) were focal, identified only in one tissue region of an individual (Fig. 3b), and at very low VAFs (Extended Data Fig. 4), suggesting that they likely arose after gastrulation42, and are likely to have been confined to nervous tissue. Mutational signature analysis using Mutalisk43 demonstrated that clock-like signatures (SBS5 and SBS1) were the predominant mutational signatures (Extended Data Fig. 5). Recent work has identified their presence in brain development44,45, and SBS1 reflects deamination of methylated cytosine during cell division and mitosis.
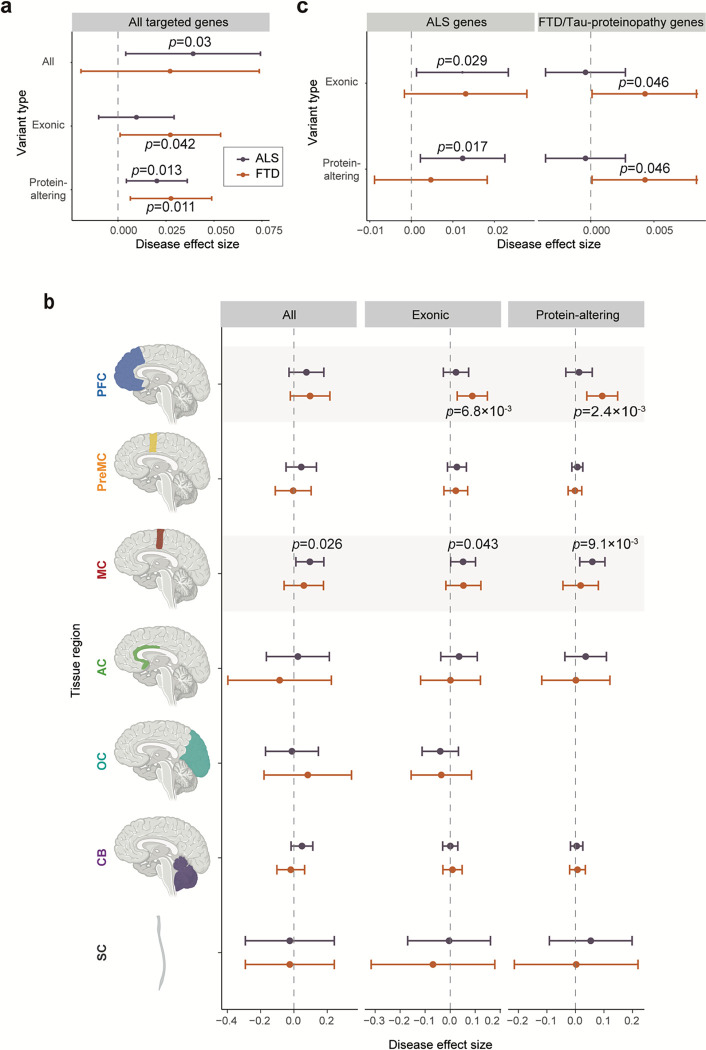
Our MIP panel contained not only ALS/FTD genes but also genes involved in other dementia. We first focused on somatic mutations in all the neurodegenerative genes. For the somatically mutated genes, there was a clear separation between the disease and control groups (Fig. 3c). Indeed, just one protein-altering somatic mutation was observed among all controls, while 15 and 7 were observed in ALS and FTD cases, respectively. These protein-altering somatic mutations were significantly enriched in ALS and FTD cases (Fig. 4a; p=0.013 and p=0.011) when tested using a linear mixed-effect regression model, which considers multiple potential confounding factors, suggesting that some or all of them were potentially disease-causing.
Fig. 4: Somatic variants are enriched in ALS and FTD cases and disease-related tissue regions.
(a) Enrichment of somatic variants in different genomic regions of germline-free ALS and FTD cases compared to normal controls. (b) Enrichment of somatic variants in different brain regions of germline-free ALS and FTD cases compared to normal controls. Significance of enrichment and 95% CI was estimated while controlling for potential confounding factors including average read-depth, sequencing batch, sampled individual using a linear mixed model. (c) Enrichment of exonic and protein-altering somatic variants in two different groups of disease-related genes (ALS genes and FTD/Tau-proteinopathy genes) compared to normal controls
The enrichments of somatic mutations in neurodegenerative genes showed striking topographic patterns, with exonic somatic mutations showing enrichment exclusively in disease-affected tissue regions for both FTD and ALS germline-free cases. The prefrontal cortex showed enrichment for somatic FTD mutations, and the primary motor cortex for ALS (Fig. 4b), while the premotor cortex—located immediately in between these two regions—showed no enrichment for either condition, as was the case for other tested cerebral cortical regions as well. The spinal cord in ALS had only a modest increase in protein-altering somatic mutations, although this analysis is limited by a small number of control spinal cord samples and resultant wide confidence intervals (Fig. 4b). For the prefrontal cortex of FTD and the primary motor cortex of ALS, enrichments of protein-altering somatic mutations in germline-free cases were even more significant than the overall enrichments of exonic somatic mutations (Fig. 4b; p=0.043 and p=9.1×10−3, p=6.8×10−3 and p=2.4×10−3 for exonic and protein-altering mutations in ALS and FTD germline-free cases, respectively; linear mixed model), further supporting the pathogenic roles of the identified somatic mutations.
We further assessed somatic mutations in genes specifically related to ALS and FTD and found that somatic mutations in each were enriched in genes relevant to that corresponding condition. Exonic and protein-altering mutations were specifically enriched in ALS-related genes in germline-free ALS samples (Fig. 4c; p=0.029 and p=0.017 for exonic and protein-altering mutations, linear mixed model). Moderate enrichments were observed for exonic and protein-altering mutations in FTD-related genes in germline-free FTD samples. In fact, less than half of FTD cases have pathological TDP-43 protein aggregates, while the other half have Tau protein aggregates4. We thus checked the contribution of Tau proteinopathy-related genes, including genes associated with Alzheimer’s disease (AD), together with FTD-related genes and found nominally significant enrichment of exonic and protein-altering somatic mutations only in germline-free FTD cases (Fig. 4c; p=0.046 for both exonic and protein-altering mutations, linear mixed model). Our FTD cases could not be categorized into those related to TDP-43 or Tau proteinopathies due to the lack of relevant pathological information, hindering our ability to examine the potential enrichment of somatic mutations within these distinct categories. On the other hand, no protein-altering mutation was observed in any of the ALS/FTD genes in control samples (Fig. 3c). The exclusive and diagnosis-specific enrichments of functional somatic mutations suggest that most or all somatic mutations contribute to the pathogenesis of sALS and sFTD.
Pathogenic somatic mutations have restricted regional distributions and are enriched in hypodiploid cells
Pathogenicity prediction of somatic mutations resulted in 8 predicted pathogenic somatic SNVs in previously known ALS and FTD/Tau-proteinopathy genes (Supplementary Table 7), which account for 3.2% and 2.6% of germline-free ALS and FTD cases, respectively (2.7% for all the germline-free sALS and sFTD cases). All mutations in ALS cases were observed in primary motor cortex or spinal cord, the most severely affected regions in ALS, emphasizing the remarkable topographic specificity of the mutations. In addition, a predicted pathogenic somatic SNV in APP (p.R328Q) was identified in primary motor cortex of a sporadic case that showed both ALS and FTD. All somatic mutations occurred in disease genes with dominant inheritance when found in the germline setting, except for one sALS case with a somatic ALS2 (p.T787R) mutation identified in spinal cord. ALS2 is an autosomal recessive disease gene46,47, and the same individual carried an ALS2 (p.Q24R) germline mutation in addition to the identified somatic mutation. Both ALS2 mutations were predicted to be pathogenic, suggesting that they initiate disease in a “second hit” autosomal recessive manner at the cellular level in a small proportion of cells in the spinal cord and again further supporting the likely contribution of somatic variants to pathogenesis.
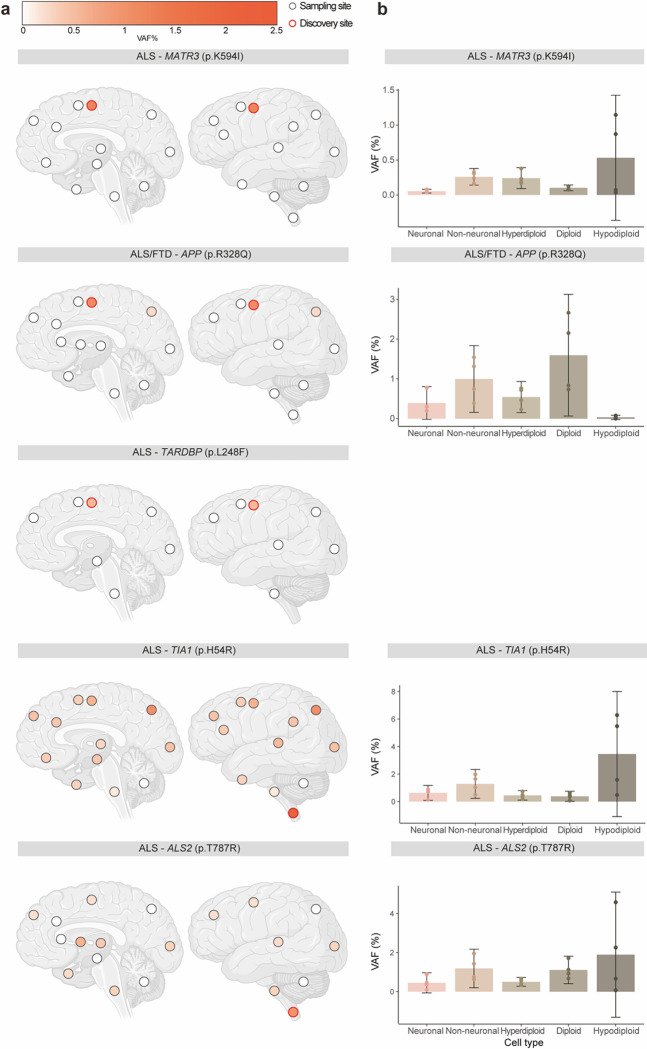
We selected four predicted pathogenic somatic SNVs in ALS/FTD genes-- TIA1 (p.H54R), MATR3 (p.K594I), ALS2 (p.T787R), and TARDBP (p.L248F), and the predicted pathogenic APP somatic SNV (p.R328Q)--to study in greater detail in terms of regional and cell-type distributions. Amplicon sequencing across multiple brain and spinal cord regions showed that three of the five somatic SNVs [MATR3 (p.K594I), APP (p.R328Q), TARDBP (p.L248F)] were restricted to the primary motor cortex (Fig. 5a and Supplementary Table 8). The other two somatic SNVs [TIA1 (p.H54R) and ALS2 (p.T787R)] had the highest VAFs in the spinal cord [2.16% for TIA1 (p. H54R) and 0.97% for ALS2 (p.T787R)], where they were originally identified, and were also present in other brain regions at very low VAFs [0.15–1.05% for TIA1 (p.H54R), 0.16% - 0.59% for ALS2 (p.T787R)] (Fig. 5a and Supplementary Table 8). All five somatic SNVs were absent in cerebellum. The ultra-low levels and limited distribution of these somatic SNVs suggest that they probably arose late in development and were thus likely excluded from non-CNS tissues. Together with the enrichment of exonic and protein-altering somatic mutations in disease-affected tissue regions, these findings also support the focal onset of ALS at the genetic level in these somatic cases. Cells carrying damaging somatic mutations could form initial lesions, likely TDP-43 inclusions, in UMNs and LMNs, but these must have ultimately spread to other regions of the motor system that lacked or carried exceedingly low levels of the mutation, but which nonetheless showed robust pathology post mortem otherwise indistinguishable from germline cases.
Fig. 5: Pathogenic somatic mutations have restricted regional distributions and are enriched in hypodiploid cells.
(a) Regional distribution of VAFs of somatic variants in individual brains and spinal cords. Brain cortex is annotated by Brodmann areas. The color spectrum indicates the VAFs of somatic variants in amplicon sequencing. Dots indicate unavailable regions and white indicates regions without the somatic variants. Red highlight indicates the region of initial detection by MIP sequencing. (b) VAFs of somatic variants in FANS sorted cell types. Five hundred neuronal (NeuN+), non-neuronal (NeuN−), diploid (DAPI), hyperdiploid (High DAPI) and hypodiploid (Low DAPI) cells were each sorted for amplicon sequencing with four replicates. Error bars, 95% CI.
We then determined the presence of these five somatic SNVs in different cell types by performing amplicon sequencing of DNA from neuronal (NeuN+), glial (NeuN−), diploid, polyploid, and hypodiploid nuclei isolated by fluorescence-activated nuclei sorting (FANS) from the tissue regions in which they were originally identified (Extended Data Fig. 6). Interestingly, TIA1 (p.H54R), MATR3 (p.K594I), and ALS2 (p.T787R) mutations were enriched in hypodiploid nuclei (Fig. 5b), which likely represent apoptotic cells with DNA fragmentation and cell death48,49. Enrichment of these three mutations in hypodiploid cells indicates a possible role in the pathogenic process, suggesting that they might be involved in inducing cell death. Surprisingly, these three mutations were identified in all cell fractions, but were more enriched in non-neuronal cells compared to neurons (Fig. 5b). This finding also implies that neurons may exhibit a cell-type specific vulnerability to damaging somatic mutations in ALS/FTD genes. In contrast, the depletion of the APP mutation from hypodiploid cells, and its relative enrichment in non-neuronal cells compared to neurons (Fig. 5b), align with models proposing important actions of AD risk genes in non-neuronal cells including microglia and astrocytes, potentially leading to secondary neuronal loss50. However, further research is needed to confirm and better understand these potential associations and mechanisms. The TARDBP (p.L248F) mutation was found in a primary motor cortex sample with a very low VAF (≈ 0.5% upon validation). However, when isolated cell fractions were tested, the mutation was not detected in any of them. This suggests that the mutation was only present in the specific area where it was initially discovered and did not extend to nearby regions. This conclusion was confirmed by amplicon sequencing of a second tissue sample from the primary motor cortex, where it was also absent.
RNA-MosaicHunter identifies additional pathogenic somatic mutations in bulk RNA-seq data of sALS cases
To complement our targeted sequencing of neurodegenerative genes, which identified pathogenic somatic mutations in a small proportion of sALS and sFTD cases in known genes, we performed a transcriptome-wide screen for somatic mutations using RNA-seq data to explore whether genes not normally associated with these conditions might cause them in the mosaic state. We profiled pathogenic somatic mutations in all expressed genes in bulk RNA-seq data generated from 789 postmortem brain and spinal cord tissue samples of 143 sALS cases and 23 age-matched controls by the New York Genome Center ALS Consortium (Supplementary Table 9; 81 and 11 of the sALS and control cases respectively were included in our MIP sequencing) using RNA-MosaicHunter, a tool capable of calling clonal somatic mutations from bulk RNA-seq data with a Bayesian probabilistic model. Because of the limited coverage of bulk RNA-seq data, RNA-MosaicHunter only has sensitivity to detect somatic mutations VAFs >≈5%, and discards somatic mutations at ultra-low levels. We found significant increases in total somatic mutations in sALS cases not carrying pathogenic germline mutations (Extended data Fig. 7; p=0.007). Additionally, there was a higher burden of somatic mutations predicted to be damaging in germline-free sALS cases; although this trend did not reach statistical significance (Extended data Fig. 7; p=0.058). Overall, these findings further confirmed that somatic mutations may contribute to the development of sALS.
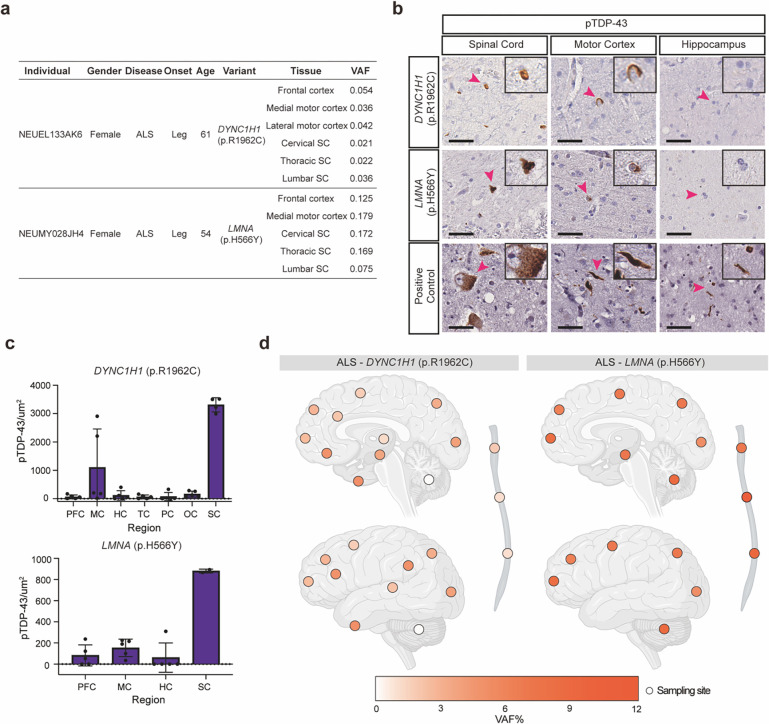
Interestingly, somatic SNVs in DYNC1H1 and LMNA were identified in multiple CNS regions of two sALS cases that did not harbor other pathogenic germline or somatic mutations (Fig. 6a and Supplementary Table 10, both cases were included in the MIP sequencing). Heterozygously acting, generally de novo, mutations in DYNC1H1 and LMNA have been found in patients with phenotypes resembling spinal muscular atrophy (SMA)51–54, a motor neuron disease genetically distinct but sharing some pathological overlap with ALS, including loss of lower motor neurons, denervation of neuromuscular junction, and muscle atrophy55. Analysis of whole-genome sequence data of the two cases for germline mutations in SMN1, the most commonly mutated genes in SMA, did not identify pathogenic germline mutations. Both individuals carrying these somatic mutations had leg-onset ALS with TDP43 pathology predominantly in spinal cord and to a lesser extent in motor cortex (Fig. 6a–c). We further investigated their regional mutation distribution using amplicon sequencing. The LMNA (p.H566Y) somatic mutation was detected in all the tested brain and spinal cord regions with VAFs ranging from 5.3 to 12.3% (Fig. 6d and Supplementary Table 8). The DYNC1H1 (p.R1962C) somatic mutation was also detected in all the tested CNS regions with VAFs ranging from 0.1% to 5.2%, but the VAFs of the mutation were extremely low in the cerebellum (0.1%), thoracic spinal cord (0.8%) and lumbar spinal cord (0.8%) (Fig. 6d and Supplementary Table 8). Notably, the DYNC1H1 (p.R1962C) mutation was undetectable in cultured fibroblasts from the patient (Supplementary Table 8), indicating that the mutation arose late in development and was likely limited to the CNS. The broad distribution of these two somatic mutations aligns with our previous finding that somatic mutations with more than 5% VAFs are typically detected throughout the CNS56, with the low levels of the mutation in lumbar spine potentially reflecting death of the motor neurons carrying this mutation. The DYNC1H1 p.R1962C mutation is known to be highly pathogenic, as it completely abolishes the motor function of the dynein complex in vitro57, and germline DYNC1H1 p.R1962C mutations have been found in patients with malformations of cortical development and delayed psychomotor development58,59. Although the LMNA (p.H566Y) mutation was not previously reported, LMNA mutations cause autosomal dominant laminopathies including Hutchinson-Gilford progeria and congenital muscular dystrophy, which are characterized by congenital defects and increased early lethality60,61. Thus, both genes can cause lethal diseases with pediatric age of onset, which may ordinarily preclude the appearance of ALS, but the mosaic state could allow for a normal early life and the onset of a degenerative disorder later in life. These data suggest that further genome-wide exploration of brain tissue for somatic mutations could reveal additional ALS genes that cause early lethality in the germline state.
Fig. 6: Somatic variants in DYNC1H1 and LMNA in sALS.
(a) Two pathogenic somatic SNVs that were shared by multiple tissue regions of the ALS cases. (b) Sections of the lumbar spinal cord, motor cortex, and hippocampus of the two sALS cases stained with a phospho-TDP43 antibody. Scale bar = 40 um. Arrowheads indicate the cells shown in the insets, which are magnified to twice their original size. (c) Quantification of phospho-TDP43 staining of CNS tissue sections of the two sALS cases with DYNC1H1 and LMNA somatic mutations. Error bars indicate SD (n = 5). PFC: prefrontal cortex. MC: primary motor cortex. HC: hippocampus. TC: temporal cortex. PC: parietal cortex. OC: Occipital cortex. SC: spinal cord. (d) Regional distribution of VAFs of somatic variants in individual brains and spinal cords. Brain cortex is annotated by Brodmann areas. The color spectrum indicates the VAFs of somatic variants in amplicon sequencing. Dots indicate unavailable regions and white indicates regions without the somatic variants.
Discussion
Our data provide several important insights into sALS and sFTD. First, we found that about 30% of both conditions show known or novel, likely pathogenic germline mutations in ALS or FTD genes, which advocates for a shift from family history-based to genetic testing-based classification of ALS and FTD cases. Second, we find that a small but important fraction (~2.7%) of germline-free sporadic cases harbor predicted pathogenic somatic variants in known ALS or FTD genes, with the distribution of these mutations being disease and brain region-specific, providing proof of concept of a potentially important contribution of somatic mutations to pathogenesis. Finally, we find examples of genes associated with severe pediatric degenerative diseases that can be present in ALS in the somatic state, potentially broadening the spectrum of causative genes for these conditions.
While the case-control enrichment of somatic variants suggests a role in pathogenesis, these somatic variants are present at surprisingly low VAFs and with patterns of topographic restriction that match disease onset. It is very likely that these pathogenic somatic mutations arose at a late stage of development and were not shared by other tissue regions. In the most extreme case, the TARDBP (p. L248F) somatic SNV was even undetectable in tissue adjacent to the original sampling site. The nature of these focal somatic events would prevent them from being identified through routine genetic testing with blood or other peripheral samples. The focality of these mutations in the nervous system also suggests a mechanism by which degeneration may spread from a site containing mutant cells to eventually cause loss of neurons in regions that do not carry the mutation. This process is thought to involve the TDP-43 proteinopathy as supported by recent studies in cell and animal models12–18. Identification of predicted pathogenic somatic mutations in the primary motor cortex and in spinal cord from individuals with ALS suggests potential onset of disease in either UMNs or LMNs but eventual involvement of both. Our cell-type analysis revealed that several predicted pathogenic somatic mutations were more enriched in glia than neurons. However, the reduced abundance in neurons might also reflect the loss of neurons carrying these somatic mutations. This was reinforced by our observations that three out of the four tested somatic mutations were more prevalent in hypodiploid cells, which likely represent apoptotic cells. The potential harm inflicted on neurons by these mutations once again bolsters the concept of a focal onset of ALS. Neurons carrying these mutations constitute the initial lesion and subsequently undergo cell death. The demise of these neurons could further reduce their presence, leading to a reduction in the VAFs of the mutations compared to their levels at the time of initial emergence.
Although only about 2.7% of germline-free ALS and FTD cases had predicted pathogenic somatic mutations in our MIP sequencing data, this is likely greatly underestimated because of the limited sensitivity of even our deep panel sequencing approach to detect somatic mutations at ultra-low levels (Extended data Fig. 4). The detection of somatic mutations with low VAFs remains a technical challenge45, but improved duplex sequencing approaches promise the ability in the future to systematically sample somatic mutations at virtually all allele frequency levels. Given that somatic mutations at very low levels and in focal regions appear capable of creating a spreading disease, it will require very deep analysis to determine the lower allele frequency range of variants that is capable of initiating this process. Variant detection is also limited by availability of samples from regions across the CNS.
Our identification of candidate somatic SNVs in DYNC1H1 (p. R1962C) and LMNA (p. H566Y) using RNA-seq analysis of sALS cases suggests that genes that predispose their carriers to ALS and FTD by somatic mutations may include genes distinct from those discovered in germline cases. Certain alleles in both DYNC1H1 and LMNA are associated with motor neuron degeneration in the form of SMA, so they are capable of predisposing to neuronal degeneration, but also in both cases, other alleles (including the DYNC1H1 p. R1962C allele58,59) cause severe pediatric disease that would normally mask the possibility of late-life ALS. This result suggests that a wider range of ALS genes and alleles could exist in the somatic state that cannot be observable in the germline state due to their association with early-onset severe disease. This raises an exciting prospect that future genome-wide approaches, such as deep whole-genome or exome sequencing of a cohort of ALS cases, could shed light not only on additional somatic genetic mechanisms and their contributions to ALS, but also on the topographic patterns of spread of pathology from focal sites.
Methods
Tissue sources and sample preparation
Fresh frozen postmortem human brain and spinal cord tissues were collected by the Massachusetts Alzheimer’s Disease Research Center, Oxford Brain Bank, Target ALS Foundation, and NIH NeuroBioBank (Supplementary Table 1) according to their respective institutional protocols, written authorization and informed consent; they were subsequently obtained for this study with the approval of the Boston Children’s Hospital Institutional Review Board. Research on these deidentified specimens and data was performed at Boston Children’s Hospital with approval from the Committee on Clinical Investigation. Sporadic ALS and FTD cases were selected based on available clinical records. ALS and FTD cases without clear recording of family histories were also selected if the age of death was above 45 years old. gDNA of these tissue samples was extracted using the EZ1 Advanced XL (Qiagen) system followed by an additional purification using AMPure XP beads (Beckman Coulter).
MIP panel design
A double-stranded DNA MIP panel targeting 1.4Mb across exons and exon-intron junctions of 88 neurodegenerative genes was designed using custom scripts incorporating MIPgen62 using the human reference genome, hg19, with Mly1 restriction sites masked with ‘N’ using bedtools. The final panel of 26,439 MIPs captures an average fragment length of 209bp, including the extension and ligation arms to ensure overlapping of the forward and reverse sequencing read. The panel successfully targets 92.7% of bases including flanking intronic regions, with >98% of exonic bases covered with an average of at least 2 unique MIPs. All MIPs were designed to include a custom backbone consisting of primer binding sites and dual 5nt unique molecular indexes (UMI). MIPs were rebalanced in the pool based on the percent of GC content within the regions. Common primer binding and Mly1 restriction enzyme sites were added to both ends of the oligo sequences to enable blunt-end removal of the primer binding sites. The forward and reverse compliment sequences were printed into a single ssDNA pool by CustomArray (Bothell, WA). The resulting panel was amplified at a low cycle number (12X), digested with Mly1 enzyme for 12 hrs at 37C, and purified using Qiagen Nucleotide removal kit.
MIP capture and library construction
Two hundred fifty ng of gDNA was first hybridized in a 15 ul reaction with 1.5 ul of Ampligase® 10X Reaction Buffer (VWR), 1.5 ul of the reverse blocking oligo (5’-NNNNGAAGTCGAAGGGCTATAGGCTGCCATCACANNNN-3’) and the MIP pool at 63 nM for 10 min at 95 °C and 24 hrs at 60 °C. Gap-fill/ligation was then performed by adding 1 unit of Phusion™ High-Fidelity DNA Polymerase (Thermo Fisher), 4 units of Ampligase® DNA Ligase (Epicentre), 0.2 ul of Ampligase® 10X Reaction Buffer, 0.6 ul of dNTPs (10 mM) and 1 ul of nuclease-free water to the MIP capture product and incubated at 60 °C for 1 hr. For exonuclease digestion, 50 units of Exonuclease III (Thermo Fisher), 10 units of Exonuclease I (Thermo Fisher), 0.2 ul of Ampligase® 10X Reaction Buffer (VWR), and 2.05 ul of nuclease-free water was added to the Gap-fill/ligation product, which was incubated for 40 min at 37 °C and 5 min at 95 °C. Ten ul of the captured library is amplified in a 50 ul final reaction by adding 1 unit of Phusion Hot Start II DNA Polymerase (Thermo Fisher), 10 ul of 5X HF buffer, 1 ul of dNTPs (10mM), 1 ul of the universal MIP barcode forward primer (10 uM), 1 ul of the individual barcode reserve primer (10 uM) and 26.5 ul of nuclease-free water. MIP library amplification was then performed under the following conditions: 98 °C for 30 s; 16 cycles of 98 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s; 72 °C for 2 min. MIP library was then purified using 2X AMPure XP Beads (Beckman Coulter,) and quantified by the Quant-iT™ dsDNA Assay HS Kit (Thermo Fisher). Ninety-six MIP libraries were pooled together and sequenced on one lane of Illumina Hiseq X.
Pre-processing and read mapping of MIP sequencing data
MIP sequencing primers were removed first from the raw FASTQ files using Cutadapt63 (v2.4, 5’ adapter of the first read: CATACGAGATCCGTAATCGGGAAGCTGAAG, 3’ adapter of the first read: ACACTACCGTCGGATCGTGCGTGT, 5’ adapter of the second read: GCTAAGGGCCTAACTGGCCGCTTCACTG, 3’ adapter of the second read: CTTCAGCTTCCCGATTACGGATCTCGTATG). Trimmed reads were aligned to the human reference genome (GRCh37) using BWA-mem64 (v0.7.15) and sorting and indexing were performed using samtools65 (v1.3.1). From the aligned BAM file, off-target reads were removed by checking the overlaps with the target regions using bedtools66 (v2.26.0). MIP arm regions were masked by soft-clipping for each read using BAMClipper67 (v1.0.1). Unique molecular identifier (UMI) information was extracted, and then mapped reads were deduplicated based on the mapping coordinate and the shared UMI using UMI-tools68 (v1.0.0). Base quality score recalibration and local realignment were performed using the Genome Analysis Toolkit (GATK, v3.7)69, generating final analysis-ready BAMs.
Variant calling for pathogenic germline mutations
Initial candidates of germline SNVs and indels were identified using GATK HaplotypeCaller with default parameter settings. Low-quality candidates were filtered out if any of the following conditions is not satisfied: 1) ≥ 10 variant-supporting reads, 2) ≥ 20 total read-depth at the variant site, 3) VAF ≥ 0.3, 4) GATK QUAL ≥ 50, and 5) identified in all brain regions from the same individual except for the samples failed to cover the variant site (<10 reads). Possible pathogenic germline variants were further selected by satisfying all the following conditions: 1) present in less than 0.1% of the population in any ethnic group of public databases including dbSNP70, the 1000 Genomes Project71, the Exome Aggregation Consortium (ExAC)72, the Genome Aggregation Database (gnomAD)73, the NHLBI Exome Sequencing Project (ESP6500)74, the Greater Middle East variome project (GME)75, and Kaviar database76, 2) candidates observed only in disease or control groups but not in both, 3) possible protein-altering candidates (missense, nonsense, frame-shift, or splicing variants), and 4) affecting 30 ALS- and FTD-related genes. Pathogenicity prediction module (see computational prediction of variant pathogenicity section below) was then applied to the remaining candidates, and predicted pathogenic variants were reported as final pathogenic germline mutations. ANNOVAR27 was used to annotate the genomic region, affected genes, population allele frequency, and exonic variant functions. SpliceAI77 was additionally utilized to identify more splice-altering variants. Candidates with delta score > 0.5 were considered to be possible splicing variants.
C9ORF72 repeat expansion genotyping
Repeat-primed PCR (RP-PCR) of the C9ORF72 repeat expansion was performed in a 30 ul PCR reaction with 150 ng of gDNA, 15 ul of 2X FastStart™ PCR Master (Roche), 2 ul of DMSO, 5 ul of 5X Q-solution (Qiagen), 1 ul of 5 mM 7-deaza-dGTP (NEB), 1 ul of 25 mM MgCl2 (Qiagen) and 1 ul of the primer mix (40 uM of the Forward primer: 5’-/56-FAM/AGTCGCTAGAGGCGAAAGC-3’, 20 uM of the Reverse primer: 5’-TACGCATCCCAGTTTGAGACGGGGGCCGGGGCCGGGGCCGGGG-3’ and 40 uM of the Anchor/tail primer: 5’-TACGCATCCCAGTTTGAGACG-3’. The reaction was performed with touchdown PCR cycling conditions consisting of 15 minutes at 95°C, followed by cycles of 94°C for 1 minute, annealing starting at 70°C for 1 minute, and extension at 72°C for 3 minutes, ending with a final extension step of 10 minutes at 72°C. The annealing temperature was decreased in 2°C steps as follows: 70°C for two cycles, 68°C for three cycles, 66°C for four cycles, 64°C for five cycles, 62°C for six cycles, 60°C for seven cycles, 58°C for eight cycles, and 56°C for five cycles. The RP-PCR products were separated by the SeqStudio Genetic Analyzer (Thermo Fisher) with the GeneScan™ 600 LIZ™ Dye Size Standard (Thermo Fisher). Results of fragment sizes were analyzed by Peak Scanner™ Software v1.0 (Thermo Fisher).
Somatic variant calling from MIP sequencing data
Three different callers RePlow (v1.1.0)39, Mutect2 (v4.1.5)40, and Pisces (v5.2.11)41 were used to generate initial candidate sets. Each sample was analyzed by all three callers with the single-sample mode. Default parameter settings were used except for the adjustments for disabling the coverage limit. Variants that passed all the filters from each caller were used to make three different initial sets. Candidates identified by only one caller were discarded, and those called at least two callers were retained as a double-call set. For indels, double-calls between Mutect2 and Pisces were used as somatic indel candidates since RePlow does not support indel detection. For SNVs, among double-calls Mutect2-Pisces pairs were additionally filtered out due to high false positive rates and low validation rates in the benchmarking data set (Supplementary Fig. 3). Remaining RePlow-based SNV double-calls and indel candidates were subject to multi-step variant filters to further remove false positive candidates.
Unlike germline variant calling, somatic variant calling aims to reliably detect low-VAF mutations up to ~0.5%, which requires enough supporting evidence to control the false positive rate. Calling thresholds such as variant-supporting read count, read-depth at the variant site, and average base-call quality were determined based on the benchmarking data. Somatic variants were selected satisfying all the following conditions: 1) ≥ 50 total read-depth at the variant site, 2) ≥ 15 variant-supporting reads excluding the reads with the variant allele on their probe-arm regions, 3) > 30 average base-call quality of variant allele, 4) ≥ 2 different types of variant-supporting amplicons, 5) 0.001 ≤ VAF ≤ 0.4, 6) ≤ 3 variant candidates within 20 bp window from the same sample, 7) present in less than 0.1% of the population in any ethnic group of public databases and 8) observed in < 5 different individuals.
We additionally found that low-level contamination of DNA from another sample occurred in a few samples. Germline variants from the contaminant mimicked low-VAF somatic mutations and generated false positive calls. We therefore implemented a module to identify low-level contamination and filter out candidates that originated from the contaminant. By comparing a somatic candidate set from a given sample with the germline call set of every individual, sample contamination was determined if the given sample has ≥ 40 low-VAF somatic candidates that are also observed in a specific individual as germline variants. In this case, germline variants of the matched individual are considered to be possible sources of false positive calls and all somatic candidates that are matched with these germline variants from the individual were filtered out. The remaining candidates were reported as final somatic variants.
Pathogenic somatic variants were further annotated with similar criteria for selecting pathogenic germline variants. Among final candidates, variants that are 1) observed only in disease or control groups but not in both, 2) possible protein-altering variants, and 3) affecting ALS- and FTD-related genes were selected and applied for the pathogenicity prediction module. ANNOVAR and SpliceAI were utilized to annotate variants with various genomic information and detect additional splice-altering variants, respectively.
Computational prediction of variant pathogenicity
Pathogenicity prediction module was applied to filtered germline and somatic variants to refine the pathogenic candidate sets. Variants that were previously reported as benign/likely benign in the clinical databases (ClinVar78 and Human Gene Mutation Database79) were excluded from the pathogenic candidate set. Nonsense, frameshift, and canonical splicing variants (±1–2 splice sites) were assumed to disrupt gene function and were included in the pathogenic set. For missense variants, the dbNSFP database80 was utilized to adopt multiple computational algorithms (SIFT81, PolyPhen282, LRT83, MutationTaster84, MutationAssessor85, FATHMM86, FATHMM-MKL87, PROVEAN88, MetaSVM89, MetaLR89), considering damaging effects at different levels such as biochemical property, protein structure, and evolutionary conservation. Categorical prediction results of each algorithm were delivered by ANNOVAR. A missense variant was selected to be pathogenic if at least three different algorithms predicted damaging effects (deleterious for SIFT, LRT, FATHMM, PROVEAN, MetaSVM and MetaLR; probably damaging for PolyPhen2; disease_causing for MutationTaster), while excluding possibly/likely damaging predictions from the counts for more conservative selection. For ALS/FTD-related genes, previously reported inheritance patterns (dominant/recessive) were carefully checked. For recessive genes, two independent mutations in the same gene were required to determine whether a given individual was affected by pathogenic mutations.
Benchmarking with spike-in datasets
Two Coriell cell lines (GM12878 and GM24695) were used to generate a spike-in data. Extracted DNA were mixed at five different levels to mimic low-level somatic mutations, targeting the VAFs of 0.5%, 1%, 2.5%, 5%, and 10%. Genomic DNA from GM12878 cells was spiked into DNA from GM24695, therefore unique germline SNPs in GM12878 were served as somatic mutations. Genomic position and genotype information for germline SNPs of Coriell samples were obtained from NIST high-confidence call sets90. A total of 165 SNPs (57 homozygous and 108 heterozygous SNPs) covered by our designed MIP panel were used as the benchmark variant set. RePlow, Mutect2, Pisces, and their combinations were tested. Detected mutations not in the benchmark set were considered to be false positives, except for GM24695 germline SNPs.
Somatic variant calling from RNA-seq data
Raw bam files of RNA-seq and matched WGS data for sALS and control cases of the New York Genome Center ALS Consortium were obtained from the New York Genome Center. RNA-seq reads extracted from raw bam files were aligned to the GRCh38 human reference genome by STAR (v2.5.0a)91 in the two-pass mode with the reference gene annotation (Gencode version 39). The aligned bam files were processed by Picard (v1.138) to remove duplicates, and then by GATK (v3.6)92 for SplitNCigarReads, indel realignment, and base quality recalibration. We further excluded reads that were improperly paired or with ambiguous alignment. Somatic SNVs were called by RNA-MosaicHunter (v1.0) with default parameters (https://gitlab.aleelab.net/august/rna-mosaichunter; manuscript in submission). Derived from MosaicHunter93, which was designed for somatic mutation calling in DNA sequencing, RNA-MosaicHunter incorporates a Bayesian genotyper and a series of empirical filters to systematically distinguish somatic mutations from technical artifacts and germline mutations, with 59% sensitivity and 94% precision benchmarked using cancer datasets. Specifically, germline mutations identified from the matched WGS data from the same individual were excluded. We excluded A-to-G candidates because they are most likely led by the widespread A-to-I(G) RNA editing events in the human genome. To remove recurrent artifacts, we only considered exonic candidates that were called in one or two individuals. We further excluded candidates present in human polymorphism databases including dbSNP70, the 1000 Genomes Project94, the Exome Sequencing Project95, and the Exome Aggregation Consortium96.
Nuclei isolation and whole genome amplification
Isolation of total (DAPI+), neuronal (NeuN+), non-neuronal (NeuN−), and damaged (low DAPI) nuclei were achieved by FANS together with nuclear staining of NeuN (Millipore, MAB377) and DAPI following a previously published study97. Five hundred nuclei of each cell population were sorted into wells of 96-well plates.
Sorted nuclei were subjected to genome amplification using the Primary Template-directed Amplification kit (BioSkryb, 100136) following the manufacturer’s protocol.
Amplicon sequencing
Primer sets targeting each identified somatic SNV were designed using BatchPrimer3 (Supplementary Table 11). Amplicon was amplified for 25 cycles in a 50 ul PCR reaction with 50 ng of gDNA, 1 unit of Phusion Hot Start II DNA Polymerase (Thermo Fisher), 10 ul of 5X HF buffer, 1 ul of dNTPs (10mM) and 10 ul of each primer (10 uM). Amplicon PCR products were then purified by a 0.65X + 1.05X double size selection with AMPure XP Beads (Beckman Coulter, A63882). Purified amplicons were then pooled based on the concentrations measured by the Quant-iT™ dsDNA Assay HS Kit (Thermo Fisher) and sequenced using Amplicon-EZ (Genewiz).
Burden analysis of somatic mutations using linear mixed model
Linear mixed-effect regression model was used to compare somatic mutation burden between clinical conditions while accounting for other covariates that may affect the burden. Clinical conditions and covariates of interest (e.g. age, gender, sequencing depth) were modeled as fixed effects and the batch and individual (donor) information were modeled as random effects, considering the uncertainty caused by sample clusters from the same origin (donor or batch). Somatic mutation count in each sample was normalized per megabase pair and modeled as a dependent variable. A covariate with a p-value < 0.05 was considered to be significant, based on a t-test using the Satterthwaite approximation of degrees of freedom. To test the burden of somatic mutations in different genomic regions, a linear mixed model was fitted to the mutation counts of specific type (e.g. exonic). To test the burden of somatic mutations in different brain regions, samples were first divided by the sequenced region and then a linear mixed model was fitted for each region group.
Immunohistochemistry
Immunohistochemistry was performed using DAB (3,3’-Diaminobenzidine) detection as previously described98. Briefly, 7μm formalin-fixed, paraffin-embedded (FFPE) sections were dewaxed using citrisolve, before being rehydrated through decreasing concentrations of ethanol. Antigen retrieval was performed using sodium citrate buffer pH 6.0 at 121°C for 15 mins. Endogenous peroxidases were blocked using 3% hydrogen peroxide solution, and non-specific binding was blocked using 10% normal goat serum. Sections were then incubated overnight at 4°C with primary antibody (pTDP-43 mouse monoclonal, CosmoBio CAC-TIP-PTD-P03, 1:10,000). After washing with TBS-Triton, sections were incubated with a Horseradish peroxidase (HRP)-conjugated Goat anti-mouse secondary (Dako) for one hour at room temperature. HRP signal was detected using DAB substrate (Dako) applied for 15 minutes. Counterstaining was performed using Coles hematoxylin for 1 minute. Sections were then dehydrated, cleared using citrisolve, and mounted using glass coverslips. All sections were viewed using a Leica upright light microscope and assessed for section quality prior to whole-slide digital scanning.
Quantification of p-TDP43 burden by immunohistochemistry
Stained sections were scanned using a NanoZoomer whole-slide digital imager at 40X magnification. Images were then visualized and quantified using QuPath image analysis software and algorithms described previously98. Briefly, for cortical/cerebellar sections 5 ROI measuring 3mm2 (1000 × 3000μm) were placed equidistantly around a single gyrus with the short end of the ROI placed at the pial surface. Pathology was then quantified using a positive pixel count within each ROI and measurements were averaged to provide an output of positive pixels/mm2. For spinal cord sections, square ROI (2.25mm2) was placed on each side of the central canal within the anterior horn and measurements were averaged.
Supplementary Material
Acknowledgements
We thank the Massachusetts Alzheimer’s Disease Research Center, Oxford Brain Bank, Target ALS Foundation (Biobank Core Facility at St. Joseph’s Hospital and Barrow Neurological Institute, Georgetown Brain Bank, Eleanor and Lou Gehrig ALS Center at Columbia University and UCSD ALS bank) and NIH NeuroBioBank (Harvard Brain Tissue Resource Center, Mount Sinai/JJ Peters VA Medical Center NIH Brain and Tissue Repository, Brain Endowment Bank of University of Miami, University of Pittsburgh Neuropathology Brain Bank, University of Maryland Brian and Tissue Bank and UCLA Human Brain and Spinal Fluid Resource Center) for providing fresh frozen human tissues. We thank the Target ALS Human Postmortem Tissue Core, New York Genome Center for Genomics of Neurodegenerative Disease, Amyotrophic Lateral Sclerosis Association and TOW Foundation for providing the bulk RNA-seq data. We thank the donors and families for their contributions, and J. E. Neil and J. Gonzalez for assistance with tissue procurement. We thank the Research Computing group at Harvard Medical School and Boston Children’s Hospital. The brains in Fig. 5 and Fig. 6 were illustrated by A. Lai with input from the authors. This work was supported by the PRMRP Discovery Award W81XWH2010028 (Z.Z.); the Edward R. and Anne G. Lefler Center postdoctoral fellowship (Z.Z.); the American Heart Association Career Development Award 23CDA1046074 (Z.Z.); the National Research Foundation of Korea (NRF) 2022R1C1C1010430 (J.K.); the Alzheimer’s Association research fellowship (A.Y.H.); R56 AG079857 (A.Y.H., C.A.W. and E.A.L.); A Cullen Education and Research Foundation Young Investigator Award from the Healey Center (M.N.); a Holloway Postdoctoral Fellowship from the Association for Frontotemporal Degeneration (M.N.); K08 AG065502 (M.B.M.); donors of the Alzheimer’s Disease Research program of the BrightFocus Foundation A20201292F (M.B.M.); the Doris Duke Charitable Foundation Clinical Scientist Development Award 2021183 (M.B.M.); K01 AG051791 (E.A.L.); the Suh Kyungbae Foundation (E.A.L.), DP2 AG072437 (E.A.L.); R01 NS032457 (C.A.W.); R01 AG070921 (C.A.W. and E.A.L.); a Massachusetts Alzheimer’s Disease Research Center pilot grant (C.L.-T. and C.A.W.); and the Allen Discovery Center program, a Paul G. Allen Frontiers Group advised program of the Paul G. Allen Family Foundation (C.A.W. and E.A.L.). C.L.-T. is supported by the Araminta Broch-Healey Endowed Chair in ALS. C.A.W. is an Investigator of the Howard Hughes Medical Institute. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Code availability
The source code and default configuration file of RNA-MosaicHunter are available at https://gitlab.aleelab.net/august/rna-mosaichunter.git. The implemented codes for preprocessing of MIP sequencing data, statistical test, and visualization will be available before publication.
Data availability
The bulk RNA-seq data for the NYGC ALS Consortium samples can be obtained upon request through the NYGC. The MIP targeted gene panel sequencing data generated in this study will be deposited to dbGaP with controlled use conditions set by human privacy regulations. Germline and somatic mutations identified and validated in this study are listed in the supplementary tables.
References
- 1.Ferrari R., Kapogiannis D., Huey E.D. & Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res 8, 273–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxon J.A. et al. Examining the language and behavioural profile in FTD and ALS-FTD. J Neurol Neurosurg Psychiatry 88, 675–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier-Tourenne C., Polymenidou M. & Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19, R46–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling S.C., Polymenidou M. & Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravits J.M. & La Spada A.R. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology 73, 805–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanouchi T., Ohkubo T. & Yokota T. Can regional spreading of amyotrophic lateral sclerosis motor symptoms be explained by prion-like propagation? J Neurol Neurosurg Psychiatry 83, 739–45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen A., Kim S. & Pant B. Amyotrophic lateral sclerosis (ALS): a phylogenetic disease of the corticomotoneuron? Muscle Nerve 15, 219–24 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Chou S.M. & Norris F.H. Amyotrophic lateral sclerosis: lower motor neuron disease spreading to upper motor neurons. Muscle Nerve 16, 864–9 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Gromicho M. et al. Spreading in ALS: The relative impact of upper and lower motor neuron involvement. Ann Clin Transl Neurol 7, 1181–1192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brettschneider J. et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol 74, 20–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brettschneider J. et al. Sequential distribution of pTDP-43 pathology in behavioral variant frontotemporal dementia (bvFTD). Acta Neuropathol 127, 423–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polymenidou M. & Cleveland D.W. Biological Spectrum of Amyotrophic Lateral Sclerosis Prions. Cold Spring Harb Perspect Med 7(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porta S. et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun 9, 4220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laferriere F. et al. TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates. Nat Neurosci 22, 65–77 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Peng C., Trojanowski J.Q. & Lee V.M. Protein transmission in neurodegenerative disease. Nat Rev Neurol 16, 199–212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rossi P. et al. FTLD-TDP assemblies seed neoaggregates with subtype-specific features via a prion-like cascade. EMBO Rep 22, e53877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamaki Y. et al. Spinal cord extracts of amyotrophic lateral sclerosis spread TDP-43 pathology in cerebral organoids. PLoS Genet 19, e1010606 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S.T. et al. Seeding the aggregation of TDP-43 requires post-fibrillization proteolytic cleavage. Nat Neurosci 26, 983–996 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosen D.R. et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Turner M.R. et al. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol 12, 310–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen P.M. & Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol 7, 603–15 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Wang H. et al. Smoking and risk of amyotrophic lateral sclerosis: a pooled analysis of 5 prospective cohorts. Arch Neurol 68, 207–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armon C. Acquired nucleic acid changes may trigger sporadic amyotrophic lateral sclerosis. Muscle Nerve 32, 373–7 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Jamuar S.S. et al. Somatic mutations in cerebral cortical malformations. N Engl J Med 371, 733–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proukakis C. Somatic mutations in neurodegeneration: An update. Neurobiol Dis 144, 105021 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Hardenbol P. et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol 21, 673–8 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renton A.E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeJesus-Hernandez M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majounie E. et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol 11, 323–30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrne S. et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol 11, 232–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahoney C.J. et al. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain 135, 736–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Blitterswijk M. et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet 21, 3776–84 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Testi S., Tamburin S., Zanette G. & Fabrizi G.M. Co-occurrence of the C9ORF72 expansion and a novel GRN mutation in a family with alternative expression of frontotemporal dementia and amyotrophic lateral sclerosis. J Alzheimers Dis 44, 49–56 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Baker M. et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–9 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Cruts M. et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–4 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Kuuluvainen L. et al. Oligogenic basis of sporadic ALS: The example of SOD1 p.Ala90Val mutation. Neurol Genet 5, e335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goutman S.A. et al. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol 21, 465–479 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J. et al. The use of technical replication for detection of low-level somatic mutations in next-generation sequencing. Nat Commun 10, 1047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamin D. et al. Calling Somatic SNVs and Indels with Mutect2. bioRxiv, 861054 (2019). [Google Scholar]
- 41.Dunn T. et al. Pisces: an accurate and versatile variant caller for somatic and germline next-generation sequencing data. Bioinformatics 35, 1579–1581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bizzotto S. et al. Landmarks of human embryonic development inscribed in somatic mutations. Science 371, 1249–1253 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J. et al. Mutalisk: a web-based somatic MUTation AnaLyIS toolKit for genomic, transcriptional and epigenomic signatures. Nucleic Acids Res 46, W102–W108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung C. et al. Comprehensive multi-omic profiling of somatic mutations in malformations of cortical development. Nat Genet 55, 209–220 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang A.Y. & Lee E.A. Identification of Somatic Mutations From Bulk and Single-Cell Sequencing Data. Front Aging 2, 800380 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hadano S. et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29, 166–73 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Yang Y. et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29, 160–5 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Ferlini C., Biselli R., Scambia G. & Fattorossi A. Probing chromatin structure in the early phases of apoptosis. Cell Prolif 29, 427–36 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Young N.A. et al. Use of flow cytometry for high-throughput cell population estimates in brain tissue. Front Neuroanat 6, 27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen D.V., Hanson J.E. & Sheng M. Microglia in Alzheimer’s disease. J Cell Biol 217, 459–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudnik-Schoneborn S. et al. Mutations of the LMNA gene can mimic autosomal dominant proximal spinal muscular atrophy. Neurogenetics 8, 137–42 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Harms M.B. et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology 78, 1714–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsurusaki Y. et al. A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics 13, 327–32 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Iwahara N., Hisahara S., Hayashi T., Kawamata J. & Shimohama S. A novel lamin A/C gene mutation causing spinal muscular atrophy phenotype with cardiac involvement: report of one case. BMC Neurol 15, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowerman M. et al. Pathogenic commonalities between spinal muscular atrophy and amyotrophic lateral sclerosis: Converging roads to therapeutic development. Eur J Med Genet 61, 685–698 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Lodato M.A. et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoang H.T., Schlager M.A., Carter A.P. & Bullock S.L. DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein-dynactin-cargo adaptor complexes. Proc Natl Acad Sci U S A 114, E1597–E1606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poirier K. et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet 45, 639–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H. et al. De Novo Variants in the DYNC1H1 Gene Associated With Infantile Spasms. Front Neurol 12, 733178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eriksson M. et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423, 293–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quijano-Roy S. et al. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann Neurol 64, 177–86 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Boyle E.A., O’Roak B.J., Martin B.K., Kumar A. & Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics 30, 2670–2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3 (2011). [Google Scholar]
- 64.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 (2013). [Google Scholar]
- 65.Danecek P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan A.R. & Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Au C.H., Ho D.N., Kwong A., Chan T.L. & Ma E.S.K. BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing. Sci Rep 7, 1567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith T., Heger A. & Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van der Auwera G.A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11 10 1-11 10 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherry S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29, 308–11 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Genomes Project C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karczewski K.J. et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 45, D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karczewski K.J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu W. et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott E.M. et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 48, 1071–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glusman G., Caballero J., Mauldin D.E., Hood L. & Roach J.C. Kaviar: an accessible system for testing SNV novelty. Bioinformatics 27, 3216–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jaganathan K. et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 176, 535–548 e24 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Landrum M.J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stenson P.D. et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 21, 577–81 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Liu X., Wu C., Li C. & Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat 37, 235–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P., Henikoff S. & Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4, 1073–81 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Adzhubei I.A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chun S. & Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res 19, 1553–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwarz J.M., Rodelsperger C., Schuelke M. & Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7, 575–6 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Reva B., Antipin Y. & Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39, e118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shihab H.A. et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 34, 57–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shihab H.A. et al. An integrative approach to predicting the functional effects of noncoding and coding sequence variation. Bioinformatics 31, 1536–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi Y., Sims G.E., Murphy S., Miller J.R. & Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong C. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 24, 2125–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zook J.M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol 32, 246–51 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DePristo M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang A.Y. et al. MosaicHunter: accurate detection of postzygotic single-nucleotide mosaicism through next-generation sequencing of unpaired, trio, and paired samples. Nucleic Acids Res 45, e76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Genomes Project C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tennessen J.A. et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lek M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Evrony G.D. et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nolan M. et al. Quantitative patterns of motor cortex proteinopathy across ALS genotypes. Acta Neuropathol Commun 8, 98 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq data for the NYGC ALS Consortium samples can be obtained upon request through the NYGC. The MIP targeted gene panel sequencing data generated in this study will be deposited to dbGaP with controlled use conditions set by human privacy regulations. Germline and somatic mutations identified and validated in this study are listed in the supplementary tables.