Abstract
BACKGROUND & AIMS:
Metabolic dysfunction-associated steatotic liver disease (MASLD) affects over 25% of the population and currently has no effective treatments. Plasma proteins with causal evidence may represent promising drug targets. We aimed to identify plasma proteins in the causal pathway of MASLD and explore their interaction with obesity.
METHODS:
We analysed 2,941 plasma proteins in 43,978 European participants from UK Biobank. We performed genome-wide association study (GWAS) for all MASLD-associated proteins and created the largest MASLD GWAS (109,885 cases/1,014,923 controls). We performed Mendelian Randomization (MR) and integrated proteins and their encoding genes in MASLD ranges to identify candidate causal proteins. We then validated them through independent replication, exome sequencing, liver imaging, bulk and single-cell gene expression, liver biopsies, pathway, and phenome-wide data. We explored the role of obesity by MR and multivariable MR across proteins, body mass index, and MASLD.
RESULTS:
We found 929 proteins associated with MASLD, reported five novel genetic loci associated with MASLD, and identified 17 candidate MASLD protein targets. We identified four novel targets for MASLD (CD33, GRHPR, HMOX2, and SCG3), provided protein evidence supporting roles of AHCY, FCGR2B, ORM1, and RBKS in MASLD, and validated nine previously known targets. We found that CD33, FCGR2B, ORM1, RBKS, and SCG3 mediated the association of obesity and MASLD, and HMOX2, ORM1, and RBKS had effect on MASLD independent of obesity.
CONCLUSIONS:
This study identified new protein targets in the causal pathway of MASLD, providing new insights into the multi-omics architecture and pathophysiology of MASLD. These findings advise further therapeutic interventions for MASLD.
Keywords: metabolic dysfunction-associated steatotic liver disease, non-alcoholic fatty liver disease, genomics, proteomics, drug target
Introduction
Metabolic dysfunction-associated steatotic liver disease (MASLD), formerly known as non-alcoholic fatty liver disease (NAFLD)1, is characterised by an excessive accumulation of fat in the liver (>5%) that is not caused by excessive alcohol consumption or other known liver disease aetiologies. Its estimated to affect more than 25% of the global population, making it one of the most common liver diseases worldwide2. Its prevalence is increasing rapidly, which is attributed to the increase in obesity. MASLD is a major risk factor for liver fibrosis, metabolic dysfunction associated steatohepatitis (MASH), formerly known as non-alcoholic steatohepatitis (NASH), cirrhosis, liver failure, and liver cancer in Western countries2. Currently, there are no specific medications for MASLD, and lifestyle modifications are the mainstay of treatment, which is not achievable by many patients. Thus, there is an urgent need to improve our understanding of MASLD and identify and evaluate therapeutic targets or interventions to treat it more effectively.
Over 95% of all currently known drugs target proteins3, highlighting the importance of understanding the role of proteins in the development of MASLD. Understanding how proteins relate to MASLD, especially distinguishing the proteins in the causal pathway of MASLD, could provide insights into the underlying mechanisms of the disease and identify potential therapeutic targets for drug development. Studying liver tissue for proteomic analysis in MASLD is challenging due to a paucity of liver tissue samples available. The liver is the central organ producing and metabolising plasma proteins. Circulating plasma proteins can be an informative read-out of liver function for the discovery phase, followed by a validation in tissue specific biological data such as liver imaging, transcriptomics and biopsy.
The present study aims to identify proteins in the causal pathway of MASLD by analyzing data on 2,941 plasma proteins measured by Olink® in 43,978 European participants in UK Biobank. We find that 929 proteins (31.6%) are observationally associated with MASLD and identify 17 proteins that may be on the causal pathway to MASLD based on Mendelian randomization (MR) and genetics data. These include CD33, GRHPR, HMOX2, and SCG3 which are reported as protein/genetic targets of MASLD or NAFLD for the first time, and AHCY, FCGR2B, ORM1, and RBKS which are identified on their protein levels with MASLD or NAFLD for the first time. We further confirmed their validity using a range of independent methods, including exome sequencing, liver imaging, bulk and single-cell gene expression, liver biopsies, pathway analysis, and phenome-wide data.
Materials and Methods
Study participants
We conducted our study within UK Biobank which comprised over 500,000 participants aged 37 to 73 years during the recruitment period (2006 to 2010). Participant data include genome-wide genotyping, exome sequencing, whole-body magnetic resonance imaging, electronic health record linkage, blood and urine biomarkers, and physical and anthropometric measurements. Further details are available online4. All participants provided electronically signed informed consent. UK Biobank has approval from the North West Multi-centre Research Ethics Committee, the Patient Information Advisory Group, and the Community Health Index Advisory Group. The current study is a part of UK Biobank project 53639 and 65851.
Human liver biopsies were obtained from female patients undergoing elective surgery, including bariatric procedures and gall bladder removal. All participants gave full, informed, written consent for the liver biopsy (NHS research ethics reference: 17/WM/0130). Clinical and biochemical data were collected on the day of surgery; 36 female European participants were included in the analysis (Supplementary Table 2). An experienced histopathologist assessed all liver biopsies for steatosis and MASLD activity score according to the Kleiner classification5.
Phenotype definitions
We defined MASLD cases based on magnetic resonance imaging-estimated proton density fat fraction (MRI-PDFF)6 ≥5% (~42K participants available) or from primary care records (~250K participants available), hospital admission or death registration (~500K participants available) using International Classification of Diseases (ICD)9 (code 5715 and 5718) and ICD10 (code K758, K760 and K746)7 until November 2022. We excluded participants with excessive alcohol consumption or any secondary causes of hepatic steatosis (Supplementary Table 3)8 from both cases and controls of MASLD analyses. The definition of other traits and covariates are descripted in Supplementary Table 4.
Genomic data processing
UK Biobank array genotyping was conducted using bespoke Affymetrix UK BiLEVE Axiom® Array or UK Biobank Axiom® array. All genetic data were quality controlled and imputed as described previously9. Participants who had gender mismatch, failed quality control, significant missing data, or had heterozygosity were excluded following UK Biobank’s recommendation10. Non-European participants were excluded from current analysis. Whole-exome sequencing was measured in 454,787 participants from UK Biobank using a previously described method11.
Proteome measurement
High-throughput proteomics measures were performed by Olink® in a randomly selected 46,673 participants, of which 43,978 Europeans were included in the current proteomics analyses. Details of the Olink proteomics assay, data processing and quality control were descripted previously12. We included 2,941 protein variables, of which 12 molecules mapped to two or more possible proteins were analysed as one variable, and 18 proteins duplicated among panels were analysed separately in our analysis. A general map of the proteomics is shown in Supplementary Table 1. The protein values were rank-based inverse normal transformed to ensure that the results were comparable across the entire dataset. Analysis of proteins in the liver biopsies was performed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), which is fully described in Supplemental Materials.
Statistical analysis
All analyses were performed in R statistical software (version 4.0.3) unless otherwise specified. Two-tailed tests were considered. Detailed data preparation and missing imputation is in Supplemental Materials.
Proteome associations
For the association of proteins and outcomes, we used logistic regression for binary outcomes and linear regression for continuous outcomes for each protein separately. We used two models for the regression analysis in UK Biobank: model 1 included age, age squared, sex, fasting time, and batches of proteomic measurement; model 2 included the covariates in model 1 and common lifestyle factors, including smoking status, number of pack-years of smoking, grams of alcohol consumption per week, education, and physical activity. Multiple testing was considered by Matrix Spectral Decomposition (MSD) method13. The explanation of each significance threshold in our study is descripted in Supplemental Materials. Linear regression analysis was used to examine the association between specific proteins and features of hepatic steatosis in liver biopsy tissue. The covariates adjusted for in the model included age, and study groups.
We further explored the full STRING protein-protein networks and enrichment facilities of the 929 proteins for MASLD through STRING database14 using the high confidence (≥ 0.90) based on experiments or databases source (false discovery rate, FDR<0.05). The network was clustered into ten clusters based on KMeans clustering. STRING database included 905 proteins available for MASLD.
Genome-wide association study (GWAS) and meta-analysis
We performed GWAS for all proteins (n=41,286), PDFF (n=38,174), and MASLD (11,947 cases and 313,042 controls) in UK Biobank using a whole-genome regression approach implemented in REGENIE15. It considers relatedness, population structure, polygenicity, and unbalanced binary traits by Firth logistic regression. We used participants with European ancestry based on both questionnaires and their genetic background. The relatedness included in the analysis was adjusted for using the genotype relatedness matrix. Covariates in the model included age, age squared, sex, the interaction of age and sex, the interaction of age squared and sex, batch and chip of genotyping process, and the first ten principal components. For MASLD GWAS sources, we also included three previous studies: (1) GWASGhodsian, conducted by N Ghodsian, et al16; (2) GWASFinngen, conducted using Finngen R917; and (3) GWASMVP, conducted by M Vujkovic, et al, using unexplained chronic alanine aminotransferase (ALT) elevation as a proxy for MASLD18. By METAL19, considering study specific weight, genomic control, and sample overlap, we incorporated a meta-analysis of GWASGhodsian, GWASFinngen, and GWASUKB, correcting for 16.3% overlapped cases and 64.4% overlapped controls(GWASmeta) and a meta-analysis of GWASGhodsian, GWASFinngen, GWASUKB, and GWASMVP, correcting for 2.9% overlapped cases and 56.3% overlapped controls (GWASmeta+MVP). GWAS summary statistics for Somalogic proteome were obtained from previous publication which included 35,559 Icelanders20. GWAS summary statistics for liver volume, liver iron content, liver fat percentage, and visceral adipose tissue volume were obtained from previous publication which considered over 38,000 participants in UK Biobank21.
MR, multivariable MR, phenome-wide MR (PheMR) and colocalization
We selected cis-protein quantitative trait loci (pQTLs) (p<5×10−8, minor allele frequency>1.0×10−3), cis-expression quantitative trait loci (eQTLs) (p<5×10−8, minor allele frequency>0.01) or genetic determinants of BMI (p<5×10−8, minor allele frequency>0.01) from the GWAS summary statistics22,23 based on +/−500kbs of the encoding gene and r2<0.01 using a reference panel with 10,000 random European individuals from UK Biobank24. We used R package TwoSampleMR, MendelianRandomization, and MRPRESSO to perform different MR methods, including inverse-variance weighted, weighted mode, weighted median, simple mode, and MR-Egger, Cochran’s Q statistic for heterogeneity effect, MR-Egger intercept test and regression25, and MR-PRESSO26 for pleiotropic effect, and contamination mixture model27 for valid instrumental variables, when allowed. Multivariable MR was performed using R package GRAPPLE with the function grappleRobustEst to estimate the causal effects of individual proteins and body mass index (BMI) under a random effect model of the pleiotropic effects28. For PheMR, we used the most completed GWAS summary statistics from Pan-UKB team29 for 916 selected diseases, which include European cases based on three-digit ICD10 codes. Colocalization analysis was based on HyPrColoc30. We used non-uniform priors, including genetic variants +/− 500kbs of the encoding genes, and investigated posterior alignment probabilities. We assumed a probability of ≥ 0.7 to indicate significant colocalization.
Whole exome sequencing analyses
Exome-wide association analysis was performed through REGENIE15 in the European population only to exclude the ancestry heterogeneity as previous publication11. Variants were annotated using Variant Effect Predictor (VEP) versions, and Loss-Of-Function Transcript Effect Estimator (LOFTEE) plug-ins, to select the most severe consequence of each variant among all protein-coding transcripts. We focused on variants that are highly or moderately like to influence the function of phenotypes defined through snpEff31, including transcript ablation/amplification, splice acceptor/donor variants, loss/gain-of-function mutations, missense variants, inframe insertion or deletion, and protein-altering variants.
Gene expression related analyses
Default FUMA32 pipeline based on GTEx (v8) dataset22 were used to demonstrate the expression levels of the encoding genes of target proteins. It included the gene expression levels through log2 transformed average expression values and enrichment analysis based on pre-calculated differentially expressed genes (DEG) sets across 54 different general tissue types. Genes with log2 transformed expression values greater than 2.84, which corresponds to an expression level approximately 6.5-fold higher than the median expression level across all genes, were considered to be highly expressed. The GWAS summary statistics of gene expression in the liver and adiposity tissues were obtained from GTEx (v8) dataset22 and STARNET dataset33 and used to identify the genetic variants/determinants of the gene expression (p<5×10−8).
The bulk RNA-seq data of liver biopsies were obtained from a previous transcriptomic study, which have snap-frozen biopsies from 206 MASLD patients and 10 healthy obese control cases without any biochemical or histological evidence of MASLD processed for RNA sequencing on the Illumina NextSeq 500 system34. Sixteen of the 17 candidate causal proteins were available, except for SCG3 which was not measured in the full set of control groups (n<10). GEO2R platform was used to perform the analysis35.
Analysis of single nucleus RNA sequence (snRNA-seq) and single cell RNA sequence (scRNA-seq)
We integrated six independent studies of liver cell snRNA-seq and scRNA-seq data, considering the differences in sample processing techniques and donor health conditions across the studies36–42. Detailed procedure is descripted in Supplemental Materials. For each the liver comprising cell types, we calculated the fraction of cells and the mean expression levels of the encoding genes of target proteins. For genes detected in more than 15% of the cells and a mean expression level greater than 75% of the genes, we compared the differential gene expression levels through the Mann-Whitney-Wilcoxon test across disease groups. We used data from Ramachandran et al.’s study38 for MASLD and non-MASLD groups. As there were no available MASLD data for hepatocytes and stellate cells, we used alternative data from NASH or alcoholic liver disease studies38–40.
Pathway enrichment analyses
Comprehensive enrichment analyses among all the potential causal targets of MASLD were performed through Metascape pipeline43. Functional enrichment analyses have been carried out by hypergeometric test and Benjamini-Hochberg p-value correction algorithm with the following ontology sources: GO Biological Processes, DisGeNET44, and PaGenBase45. All genes in the genome have been used as the enrichment background. Terms with a p < 0.01, a minimum count of three, and an enrichment factor > 1.5 (the enrichment factor is the ratio between the observed counts and the counts expected by chance) were collected and grouped into clusters based on their membership similarities.
Results
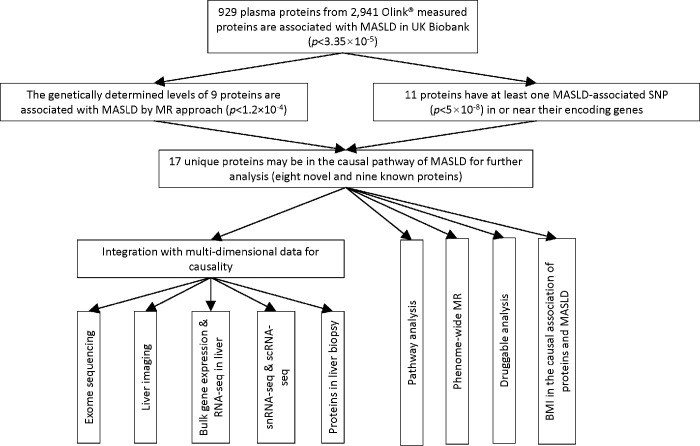
A flow chart of the study design is presented in Figure 1.
Figure 1. Flow chart of the study design.
Cohort description
We studied 43,978 randomly selected European participants from UK Biobank for proteomic analyses. After excluding participants with excessive alcohol consumption or secondary liver disease, 1,181 MASLD patients and 30,719 controls were identified. Individuals with MASLD had a higher prevalence of obesity, male gender, diabetes, hypertension, and various metabolic abnormalities compared to controls (p<1.0×10−3; Table 1). Only 8.4% of participants (3,676 out of 43,978) had liver imaging data, while a comparison between those with and without liver imaging data shows a high correlation in the association of MASLD with its common risk factors (r=0.89, p=2.4×10−11; Supplementary Figure 1; Supplementary Table 5), suggesting no evidence of major selection bias in the patients included.
Table 1.
Characteristics of the study population for proteomic analyses.
| Total (n = 30,719) | Controls (n = 29,538) | Cases (n = 1,181) | |
|---|---|---|---|
| Age (years), Mean ± SD | 56.98 ± 8.07 | 57 ± 8.09 | 56.63 ± 7.46 |
| Male, n (%) | 12,534 (40.82) | 11,943 (40.43) | 591 (50.04) |
| BMI (kg/m2), Mean ± SD | 27.35 ± 4.84 | 27.23 ± 4.79 | 30.30 ± 5.15 |
| Waist circumference (cm), Mean ± SD | 89.49 ± 13.52 | 89.15 ± 13.42 | 98.04 ± 13.20 |
| Smoking status, n (%) | |||
| Never | 18,354 (59.75) | 17,681 (59.86) | 673 (56.99) |
| Previous | 9,796 (31.89) | 9,385 (31.77) | 411 (34.80) |
| Current | 2,569 (8.36) | 2,472 (8.37) | 97 (8.21) |
| Smoking pack years, Median (IQR) | 9.0 (0.025, 23.4) | 8.75 (0.025,23.25) | 11.25 (0.025, 26.06) |
| Alcohol grams per week, Median (IQR) | 80.0 (48.0, 118.4) | 80.0 (48.0, 118.40) | 84.0 (56.20, 124.0) |
| Education, n (%) | |||
| College or University degree | 9,847 (32.06) | 9,492 (32.13) | 355 (30.06) |
| A levels AS levels or equivalent | 3,507 (11.42) | 3,361 (11.38) | 146 (12.36) |
| CSEs or equivalent | 1,661 (5.41) | 1,597 (5.41) | 64 (5.42) |
| NVQ orHND or HNC or equivalent | 1,956 (6.37) | 1,875 (6.35) | 81 (6.86) |
| 0 levels GCSEs or equivalent | 6,623 (21.56) | 6,367 (21.56) | 256 (21.68) |
| Other professional qualifications | 1,637 (5.33) | 1,563 (5.29) | 74 (6.27) |
| None | 5,488 (17.87) | 5,283 (17.89) | 205 (17.36) |
| Physical activity, n (%) | |||
| High | 12,328 (40.13) | 11,939 (40.42) | 389 (32.94) |
| Moderate | 12,576 (40.94) | 12,086 (40.92) | 490 (41.49) |
| Low | 5,815 (18.93) | 5,513 (18.66) | 302 (25.57) |
| Glucose (mmol/L), Mean ± SD | 5.07 ± 0.92 | 5.06 ± 0.91 | 5.29 ± 1.22 |
| HbAlc (mmol/mol), Mean ± SD | 36.01 ± 5.66 | 35.93 ± 5.55 | 37.90 ± 7.56 |
| Type 2 diabetes, n (%) | 1,589 (5.17) | 1,437 (4.86) | 152 (12.87) |
| Anti-diabetic medications, n (%) | 1,086 (3.54) | 988 (3.34) | 98 (8.3) |
| Hypertension, n (%) | 15,799 (53.8) | 15,085 (53.42) | 714 (63.19) |
| Systolic blood pressure (mmHg), Mean ± SD | 136.98 ± 18.51 | 136.91 ± 18.55 | 138.66 ± 17.45 |
| Diastolic blood pressure (mmHg), Mean ± SD | 81.56 ± 10.08 | 81.48 ± 10.07 | 83.7 ± 9.89 |
| Anti-hypertensive medications, n (%) | 6,949 (22.62) | 6,570 (22.24) | 379 (32.09) |
| Triglycerides (mmol/L), Median (IQR) | 1.48 (1.05, 2.12) | 1.46 (1.04, 2.1) | 1.9 (1.41, 2.73) |
| HDL-cholesterol (mmol/L), Mean ± SD | 1.43 ± 0.37 | 1.44 ± 0.37 | 1.25 ± 0.31 |
| Lipid-lowering medications, n (%) | 6,308 (20.53) | 5,991 (20.28) | 317 (26.84) |
| AST(U/L), Median (IQR) | 24.1 (20.8, 28.3) | 24 (20.8, 28.1) | 26.3 (22.2, 32.4) |
| ALT (U/L), Median (IQR) | 19.56 (15.12, 26.29) | 19.38 (15.01, 25.92) | 25.97 (19.18, 37.27) |
| GGT (U/L), Median (IQR) | 24.3 (17.6, 36.4) | 24 (17.5, 35.9) | 33.6 (23.8, 52.9) |
| CRP(mg/L), Median (IQR) | 1.32 (0.65, 2.75) | 1.29 (0.64, 2.71) | 1.93 (1.04, 3.83) |
| MRI-PDFF (%), Median (IQR) | 2.9 (2.1, 5) | 2.49 (1.99, 3.16) | 8.6 (6.1, 13.1) |
: Never smokers were not considered when calculating pack years.
: Never drinkers were not considered when calculating alcohol grams per week. SD: standard deviation; BMI: body mass index; IQR: interquartile range; AS: Advanced Subsidiary; CES: Certificate of Secondary Education; NVQ: National Vocational Qualification; HND: Higher National Diploma; HNC: Higher National Certificates; GCSE: General Certificate of Secondary Education; HbAlc: Haemoglobin A1c; HDL: high-density lipoprotein; AST: Aspartate aminotransferase; ALT: Alanine aminotransferase; GGT: γ-glutamyl transferase; CRP: c-reactive protein; MRI-PDFF: Proton Density Fat Fraction measured by magnetic resonance imaging (MRI).
Proteins associated with MASLD
After adjusted significance threshold for multiple testing (p<3.35×10−5), 1,022 (34.8%) of the 2,941 plasma proteins were found to be associated with MASLD based on the baseline model (Supplementary Table 6). Adjustment for common lifestyle factors additionally reduced the number of proteins associated with MASLD by 9.1% to 929 (Figure 2A, Supplementary Figure 2). BMI, the most important risk factor for MASLD, was found to be significantly associated with most of the proteins associated with MASLD (99.0%) apart from nine proteins (Supplementary Table 6; Figure 2B).
Figure 2. Association of proteins with MASLD and BMI.
A) Proteome-wide association of MASLD based on model 2. B) Comparison of the proteome-wide association between MASLD and BMI. T-statistics were obtained from logistic regression under model 2. Fitted linear regression model and Pearson’s correlation are shown. Axis labels show the number of cases and controls, or total samples used in the association analysis.
The 929 proteins associated with MASLD were further investigated for their co-regulation through network and enrichment analyses using the STRING database14. The proteins showed active interactions in the networks (Supplementary Figure 3) and were enriched in various biological pathways, diseases, and tissues, particular in immune system, lipoprotein, and visceral adipose and liver tissue-related pathways (Supplementary Table 7).
Identification of proteins may be in the causal pathways of MASLD
For causal inference, we conducted MR analyses to distinguish proteins that cause MASLD from those that are affected by the disease or other biases. We used cis-pQTL data from GWAS of individual protein in UK Biobank for the MR analyses. We identified 824 out of the 929 MASLD-associated proteins having at least one cis-pQTL.
To increase the power of GWAS summary statistics for MASLD, we utilized multiple sources including three previous studies (GWASGhodsian,16 GWASFinngen,17 and GWASMVP46) and three in-house sources: GWASUKB, GWASmeta, and GWASmeta+MVP (Methods; Supplementary Table 8; Supplementary Figure 4). GWASmeta included 19,477 MASLD cases and 886,736 controls, and GWASmeta+MVP included 109,885 MASLD or proxy (unexplained chronic ALT elevation) cases and 1,014,923 controls, as the largest MASLD GWAS up to date. Our in-house GWAS identified five unique loci which were reported to be genome-wide significantly associated with MASLD for the first time (p<5×10−8; Table 2; Supplementary Figure 5). They included four loci based on GWASmeta (APP, CYP7A1/UBXN2B, RPL7P6/CAPZA3, and ZNF737), and two loci based on GWASmeta+MVP (PEMT, and ZNF737). Table 2 demonstrates the directional consistency of the associations across the various GWASs.
Table 2.
Genetic loci (top SNPs) firstly reported to be genome-wide significantly associated with MASLD based on in-house GWAS of MASLD
| GWAS source | CHR | POS | RSID | Band | Mapped Gene | EA | OA | EAF | BETA | SE | P | Direction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GWAS-meta | 8 | 59398461 | rs10504255 | 8q12.1 | CYP7A1; UBXN2B | A | G | 0.663 | −0.029 | 0.005 | 1.05E-08 | |
| GWAS-meta | 12 | 19133136 | rs2130719 | 12p12.3 | RPL7P6; CAPZA3 | C | G | 0.192 | 0.033 | 0.006 | 3.02E-08 | +++ |
| GWAS-meta+MVP, GWAS-meta | 19 | 20732074 | rs2099124 | 19p12 | ZNF737 | T | G | 0.045 | 0.034 | 0.006 | 6.14E-09 | ++++ |
| GWAS-meta | 21 | 27581283 | rs458358 | 21q21.3 | APP | A | G | 0.236 | 0.031 | 0.006 | 2.01E-08 | +++ |
| GWAS-meta+MVP | 17 | 17456604 | rs4646380 | 17p11.2 | PEMT | A | G | 0.061 | 0.028 | 0.005 | 2.88E-08 | ++++ |
GWAS-UKB: GWAS based on latest UK Biobank data only. GWAS-meta (direction order): a meta-analysis of GWASChodsian, GWASFinngen, and GWASUKB. GWAS-meta+MVP (direction order): a meta-analysis of GWASMVP, GWASUKB, GWASChodsian, and GWASFinngen. CHR: chromosome; POS: position; RSID: SNP ID; EA: effect allele; OA: other allele; EAF: effect allele frequency; BETA: effect estimate; SE: standard error; P: p-value.
Our MR analysis of the 824 plasma proteins and MASLD GWAS sources identified the genetic predisposition of nine proteins associated with MASLD, directionally consistent with the findings in the observational study. It included plasma APOE, CD33, GRHPR, HMOX2, IL1RN, KRT18, and SORT1 with a higher risk of MASLD and plasma NCAN and SCG3 with a lower risk of MASLD (p<1.2×10−4; Figure 3A, Supplementary Table 9). Among them, CD33, GRHPR, HMOX2, and SCG3 are newly identified proteins associated with MASLD or NAFLD. The association was robust using various MR methods, including simple mode, weighted mode, weighted median, MR Egger, and/or MR-PRESSO25,26 (Figure 3B). Further MR-Egger intercept test, MR-Egger regression and contamination mixture method27 also revealed that pleiotropy, and heterogeneity did not influence the associations between the nine significant proteins and MASLD (Supplementary Table 9). Colocalization analysis suggested that the genetic associations with plasma APOE, KRT18, and IL1RN were due to the same genetic variants associated with MASLD30 (posterior probability ≥ 0.7; Supplementary Figure 6). Comparing MR results in independent MASLD GWAS sources (GWASMVP, GWASFinngen, and GWASUKB) revealed directionally consistent and significant associations with MASLD for APOE, IL1RN, NCAN, SORT1, and a novel target, SCG3 (p<0.05; Figure 3A). Further MR analyses using previous Somalogic proteome data in an independent population20 replicated the association of APOE, IL1RN, NCAN, and SCG3 with MASLD (p<8.3×10−3; Figure 3C; Supplementary Table 10).
Figure 3. Association of proteins and MASLD by MR analysis.
A) Association of proteins and MASLD by MR analysis through different MASLD GWAS sources. Proteins significantly associated with at least one MASLD GWAS source are shown (p<1.2×10−4). B) Association of proteins and MASLD by different MR methods, if applicable. The figure shows the MASLD GWAS source underlying the most significant result by inverse variance weighted MR. C) Association of proteins measured by Somalogic in decode and MASLD by MR analysis through different MASLD GWAS sources. Six proteins are available and shown. D) Association of proteins and liver imaging variables. Solid point indicates p-value less than 0.05. Hollow point indicates p-value not less than 0.05. Detailed data are presented in Supplementary Table 9, 10 and 13.
We hypothesized that proteins observationally associated with MASLD are more likely to be on the causal pathway of the disease if their encoding genes are also associated with MASLD. We found that 11 of the 929 MASLD-associated proteins with at least one MASLD-associated single-nucleotide polymorphism (SNP; p<5×10−8) in or near their encoding genes. In addition to the proteins also identified in the MR above (APOE, IL1RN, and NCAN) these included AHCY, ANPEP, APOC1, FCGR2B, KRT8, LPL, ORM1, and RBKS (Supplementary Table 11). Among them, AHCY, FCGR2B, ORM1, and RBKS were identified on their plasma protein levels with MASLD or NAFLD for the first time.
By combining observational and genetic analyses, we identified 17 unique proteins that may play a role in the causal pathway of MASLD, including eight novel MASLD-associated plasma proteins (AHCY, CD33, FCGR2B, GRHPR, HMOX2, ORM1, RBKS, and SCG3) and nine known MASLD-associated proteins (ANPEP, APOC1, APOE, IL1RN, KRT18, KRT8, LPL, NCAN, and SORT1).
Integration of the 17 candidate causal MASLD proteins with multi-dimensional data
To further understand the causal pathways implicated by the 17 MASLD-associated proteins, we conducted further cross-omics annotation, which are summarized in Figure 1 and Table 3.
Table 3.
Summary of the cross-omics annotation of the 17 candidate causal MASLD proteins
| AHCY | ANPEP | APOCI | APOE | CD33 | FCGR2B | GRHPR | HM0X2 | IL1RN | KRT18 | KRT8 | LPL | NCAN | 0RM1 | RBKS | SCG3 | SORTI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Direction of the association with MASLD | + | + | - | + | + | + | + | + | + | + | + | - | - | + | + | - | + | ||
| Discovery analysis | MR | MR using either MASLD GWAS sources | X | X | X | X | X | X | X | X | X | ||||||||
| MR using independent MASLD GWAS sources | X | X | X | X | X | ||||||||||||||
| Colocalization | X | X | X | ||||||||||||||||
| MR replicated by Somalogic | X | X | X | X | |||||||||||||||
| MASLD GWAS | Encoding gene genome widely associated with MASLD | X | X | X | X | X | X | X | X | X | X | X | |||||||
| Integrating with multi-dimensional data | Whole exome sequence data | Whole exome sequence data | X | X | |||||||||||||||
| Liver imaging data | Encoding gene genomewide significantly associated with liver MRI | X | X | X | X | ||||||||||||||
| MR with liver MRI | X | X | X | ||||||||||||||||
| Bulk gene expression data in liver tissues | Significant differential gene expression | X | X | X | X | X | X | X | X | X | X | X | |||||||
| Cis-eQTL in or near the encoding gene | X | X | X | X | |||||||||||||||
| MR of gene expression and MASLD | X | X | x | ||||||||||||||||
| Bulk gene expression data in visceral adipose tissues | Significant differential gene expression | X | X | X | X | X | X | X | X | X | X | X | X | ||||||
| Cis-eQTL in or near the encoding gene | X | X | X | X | X | ||||||||||||||
| MR of gene expression and MASLD | X | X | X | X | |||||||||||||||
| RNA-seq data in liver tissue | Significant differential gene expression between MASLD and controls | X | X | X | X | X | |||||||||||||
| Single-cell RNA-seq data in the liver tissue | Significant differential gene expression between MASLD and controls in hepatocytes | X | X | X | X | X | |||||||||||||
| Significant differential gene expression between MASLD and controls in other liver cells except for hepatocytes | X | X | X | X | X | X | X | X | X | X | |||||||||
| Protein data in liver biopsies | Associated with liver steatosis features | X | X | ||||||||||||||||
| Druggability | X | X | X | X | X | X | X | X | |||||||||||
| External exploration | Pathway analysis | Enriched in fatty liver disease | X | X | X | X | X | X | X | ||||||||||
| Enriched in cholesterol metabolism or serum total cholesterol measurement | X | X | X | X | X | X | X | ||||||||||||
| Enriched in liver tissue | X | X | X | X | |||||||||||||||
| MR-PheWAS | Beneficial effect | X | X | X | X | X | |||||||||||||
| Unbeneficial effect | X | X | |||||||||||||||||
| Associated with BMI | Observational association | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |
| MR with BMI (BMI as exposure) | X | X | X | X | X | X | X | X | X | X | X | X | |||||||
| MR with BMI (BMI as outcome) | X | X | |||||||||||||||||
| Associated with MASLD after adjusted for BMI by multivariable MR | X | X | X | X | X | X | X | X | X | X | X | X | |||||||
Proteins newly associated with MASLD in the current study are marked in bold.
Integrating with exome sequencing data
We tested whether genetic variants in the exome sequencing data from UK Biobank (n~470K, p<5×10−8; Methods) were associated with both MASLD and the candidate causal proteins in-cis and directionally consistent with the observational findings. We focused on variants with moderate to high impact on gene function31. We found that the missense variant at 19:19219115 (minor allele frequency, MAF = 0.075) was associated with plasma NCAN and MASLD, and the common missense variants at 19:44905910 (MAF = 0.36) and 19:44908684 (MAF=0.15) were associated with plasma APOE and MASLD (Supplementary Figure 7; Supplementary Table 12).
Integrating with liver imaging data
We then explored various evidence from liver imaging for the 17 candidate causal proteins. We studied MRI-PDFF, liver volume, liver iron content, liver fat percentage, and visceral adipose tissue volume, assessed by liver MRI in UK Biobank. We found that APOC1, APOE, NCAN, and RBKS have at least one SNP, in or near their encoding genes (+/−500kb), genome-wide significantly associated with PDFF and liver volume, and NCAN and RBKS have at least one SNP genome-wide significantly associated with liver fat percentage (p<5×10−8; Supplementary Figure 8). MR analyses showed that genetically determined plasma APOE and NCAN levels were associated with PDFF, liver fat percentage, and liver volume, whereas genetically determined IL1RN levels were associated with liver iron content (p<3.3×10−3, Figure 3D; Supplementary Table 13).
Integrating with gene expression data in the liver and visceral adipose tissue
We further investigated the 17 plasma proteins with gene expression data to understand whether these candidate causal proteins might originate from the liver. We found that the encoding genes of AHCY, ANPEP, APOC1, APOE, GRHPR, HMOX2, IL1RN, KRT18, KRT8, ORM1, and RBKS were highly expressed in liver, and the encoding genes of AHCY, ANPEP, APOC1, APOE, CD33, FCGR2B, GRHPR, HMOX2, KRT18, KRT8, LPL, SGSH, and SORT1 were highly expressed in visceral adipose tissue (Supplementary Figure 9A). The up-regulated differential expression of these encoding genes was enriched in liver and visceral adipose tissues than other tissues (p<0.05/54; Supplementary Figure 9B). We identified ANPEP, CD33, FCGR2B, and SORT1 having at least one cis-eQTL for liver tissue in or near their encoding genes, and ANPEP, CD33, FCGR2B, GRHPR, and HMOX2 having at least one cis-eQTL for visceral adipose tissue (p<5×10−8; Supplementary Table 14). MR analyses show that the genetically determined expression levels of ANPEP, FCGR2B and SORT1 in liver, and the genetically determined expression levels of CD33, FCGR2B, GRHPR, and HMOX2 in visceral adipose tissue associated with MASLD, directionally consistent with the association between plasma proteins and MASLD (Supplementary Table 15). Using bulk RNA-seq data of liver biopsies from 206 MASLD patients and 10 controls34, we found that expression levels of APOC1, APOE, HMOX2, KRT18 and KRT8 were significantly different in the liver tissues of MASLD patients and non-MASLD controls, and in the consistent direction as shown in our observational findings (p<3.3×10−3, Supplementary table 16).
Integrating with snRNA-seq and scRNA-seq data
A further exploration on liver tissue cell-type-based differential gene expression of the encoding genes of the 17 candidate causal MASLD proteins was performed, combining snRNA-seq and scRNA-seq data from human liver biopsies from six previous publications36–42 (Supplementary Figure 10). We found that ANPEP, APOC1, APOE, GRHPR, ORM1, and RBKS were differentially expressed in hepatocytes than other genes in hepatocytes. ANPEP, APOE, GRHPR, ORM1, and RBKS expression levels were significantly (p<1.0×10−3) up-regulated in non-alcoholic fatty liver (NAFL)/NASH patients39, directionally consistent with our observational study in plasma. APOE, KRT8, KRT18, ORM1, RBKS, and SORT1 were differentially expressed in cholangiocytes and up-regulated significantly in MASLD patients38. AHCY, CD33, and FCGR2B were differentially expressed in myeloid cells, and HMOX2 was differentially expressed in T-lymphocytes, and up-regulated significantly in MASLD patients38.
Integrating with protein data in liver biopsy
We next associated the proteins with histological hepatic steatosis features using human liver biopsies from 36 women undergoing elective surgery (Methods). Eight candidate causal proteins (AHCY, ANPEP, APOC1, APOE, GRHPR, KRT8, KRT18, and RBKS) were available amongst the proteins measured in the liver biopsies using mass spectrometry. We identified KRT18 and RBKS levels in these liver biopsies were positively significantly associated with percentage of steatosis (p<8.3×10−3; Supplementary Table 17).
Pathway enrichment analysis
We performed enrichment analyses of the 17 candidate causal proteins to understand biological mechanisms of MASLD (Supplementary Table 18)43,47. The top pathway enriched was cholesterol metabolism (APOC1, APOE, LPL, SORT1 and HMOX2), followed by extrinsic apoptotic signalling pathway (KRT8, KRT18, and SORT1) and regulation of interleukin-1 beta production (ANPEP, CD33, FCGR2B, HMOX2, LPL, and ORM1). The top three diseases enriched were alcoholic liver diseases (IL1RN, KRT8, KRT18, LPL, and NCAN), fatty liver disease (AHCY, ANPEP, APOE, FCGR2B, IL1RN, KRT18, and LPL), and serum total cholesterol measurement (APOC1, APOE, IL1RN, LPL, NCAN, and SORT1). APOC1, APOE, GRHPR and ORM1 were enriched in liver tissue and APOC1, IL1RN, and ORM1 were enriched in human liver cancer cell line (Hep-G2).
PheMR
We performed a PheMR to explore potential effects of the 17 candidate causal proteins on other diseases beyond MASLD (p<1.0×10−5, Supplementary Figure 11, Supplementary Table 19). The PheMR results show that for five proteins in the causal pathway, modifying their plasma levels for lowering MASLD risks also reduces the risk of other diseases, including obesity and arthritis with APOE, depressive episode and ulcerative colitis with FCGR2B, acute myocardial infarction with LPL, type 2 diabetes with NCAN, and tympanic membrane disorders with SCG3. However, modifying the plasma APOE levels for MASLD may increase the risk of neurological degenerative disorders, such as Alzheimer’s disease, vascular dementia, cognitive disorders, and delirium, whereas modifying plasma RBKS levels may increase the risk of iron deficiency anaemia.
Druggable analysis
To leverage the 17 potential causal proteins to therapeutic avenues for MASLD, we investigated their potential druggability through OpenTargets48 and Therapeutic Target Database49. APOC1, APOE, IL1RN, LPL, NCAN, ORM1, and SCG3 are secreted proteins which are easier accessible sources of therapeutics. We also identified nine unique drugs targeting five proteins, including ANPEP (Tosedostat/IP10C8), APOE (AEM-28), CD33 (Gemtuzumab ozogamicin/Lintuzumab/M195/AVE-9633), FCGR2B (Obexelimab), and LPL (Clofibrate).
BMI in the causal association of proteins and MASLD
We further explored the role of BMI, the most important risk factor of MASLD, in the association between the candidate causal proteins and MASLD. We found that genetically determined BMI was associated with AHCY, APOC1, CD33, FCGR2B, IL1RN, KRT18, KRT8, LPL, NCAN, ORM1, and SCG3 (Figure 4A); while genetically determined APOE and SCG3 were associated with BMI (p<3.3×10−3; Supplementary Table 20; Figure 4B). Multivariable MR analysis showed that after adjusting for the causal effect of BMI on MASLD, genetically determined plasma ANPEP, APOC1, APOE, GRHPR, HMOX2, IL1RN, KRT18, KRT8, LPL, ORM1, RBKS, and SORT1 were still significantly associated with MASLD (p<3.3×10−3; Supplementary Table 21; Figure 4C). We did not detect significance between genetic determined plasma AHCY, CD33, FCGR2B, NCAN, and SCG3 with MASLD after adjusting for the causal effect of BMI on MASLD (Figure 4D).
Figure 4. Role of BMI in the association of candidate causal proteins and MASLD.
A) Proteins as mediators in the causal association of BMI to MASLD. B) BMI as a mediator in the causal association of proteins to MASLD. C) Proteins directly associated with MASLD after adjusted for the causal effect of BMI by multivariable MR. D) Proteins not directly associated with MASLD after adjusted for the causal effect of BMI by multivariable MR. IVs: instrumental variables. Arrows indicate direction from MR analysis, or knowledge (i.e., IVs causes exposures, and BMI causes MASLD).
Discussion
Using large-scale genomic and proteomic data, we identified 929 (31.6%) plasma proteins associated with MASLD. Of these proteins, 17 were potentially involved in the causal pathway of MASLD (Table 3). By adopting a plasma proteome-first approach and integrating with human genetics, transcriptomics, liver imaging, and biopsy proteomics, we found four novel protein/genetic targets of MASLD, namely CD33, GRHPR, HMOX2, and SCG3. Additionally, our study provided evidence supporting AHCY, FCGR2B, ORM1, and RBKS on MASLD from proteins, and validated previously known MASLD targets by MR, including ANPEP, APOC1, APOE, IL1RN, KRT18, KRT8, LPL, NCAN, and SORT1.
We used the largest proteomics dataset currently available to discover proteins associated with MASLD. Comparing to a recent study by Sveinbjornsson et al.50, which found one protein associated with NAFLD by MR, we identified nine proteins potentially on the causal pathway to MASLD using MR. Our GWAS helped to identify five novel MASLD loci (APP, CYP7A1, PEMT, RPL7P6/CAPZA3, and ZNF737), of which the expression of CYP7A1 and PEMT were reported previously with NAFLD51,52. These novel loci were associated (p<5×10−8) with other traits such as Alzheimer’s disease (APP), height (APP, PEMT, and RPL7P6/CAPZA3), BMI and lung function (RPL7P6/CAPZA3), waist-hip index and coronary artery disease (PEMT), lipid levels (APP, CYP7A1, PEMT, and ZNF737) and gallstones, cholelithiasis, liver enzymes, Insulin-like growth factor 1 levels and bilirubin (CYP7A1) 53.
We have identified CD33, GRHPR, HMOX2, and SCG3 as novel targets of MASLD. Among them, GRHPR and HMOX2 had independent effects on MASLD beyond obesity/BMI. Glyoxylate and hydroxypyruvate reductase (GRHPR) reduces glyoxylate into less reactive glycolate and is mainly present in the liver. Previous studies have reported impaired glyoxylate detoxification in MASLD54, which may explain the causal association of plasma GRHPR and MASLD beyond obesity. Heme oxygenase 2 (HMOX2) is a genetically distinct isozymes of heme oxygenase 1 (HMOX1), and previous studies have reported that increased expression of HMOX1 in human liver biopsies reflects the severity of MASLD55. The colocalization of HMOX2 expression in tibial artery and muscle with blood insulin-like growth factor (IGF) 1 levels suggests potential pathways through artery and/or muscle to MASLD. Secretogranin III (SCG3) is a secreted protein which has been reported previously to regulate IGF transport and insulin secretion56 and is associated with BMI53,57. Our study provides robust causal evidence of the effect of plasma SCG3 on MASLD, which was disappeared when adjusted for the causal effect of BMI on MASLD, suggesting the importance of obesity in the association of SCG3 and MASLD. Few previous studies focused on the association of CD33 and MASLD, though approved drugs of CD33 are available.
Moreover, we deduced the causal association of plasma AHCY, FCGR2B, ORM1, and RBKS with MASLD involving MASLD GWAS studies. These targets were not previously highlighted in the GWAS, potentially because GWAS tend to map target genes based on the top genetic loci/SNP, ignoring secondary associations with neighbouring genes. Our study’s pipeline caught this missing information by looking up the full set of genes mapped in any genome-wide significant SNPs of MASLD, validated by their encoding proteins associated with MASLD. Ribokinase (RBKS) is one of the genes which was in a known MASLD region but over-shadowed by the primary association with GCKR. Our study found a consistent association of liver RBKS levels with steatosis features in liver biopsies, along with significant differential gene expression in liver tissue, mainly hepatocytes and cholangiocytes. This finding suggests that carbohydrate metabolism, the main pathway of RBKS, in the liver may alter the incidence of MASLD. Orosomucoid 1 (ORM1) is another target of MASLD, which was masked by AKNA. It is a secreted protein and transports proteins in the blood stream but is also highly expressed in human hepatocytes and cholangiocytes38,39. Its isoform ORM2 regulates de novo lipogenesis, one of the most important mechanisms of MASLD, in the mouse liver, which may indicate the potential causal association of ORM1 and MASLD58,59. A few studies have reported the potential association of Adenosylhomocysteinase (AHCY)60 and Fc gamma receptor IIb (FCGR2B) with MASLD61,62. Our study found that their causal association with MASLD disappeared after adjusting for the causal effect of BMI.
Our study provides causal evidence for previous MASLD targets, including ANPEP, APOC1, APOE, IL1RN, KRT18, KRT8, LPL, NCAN, and SORT163–69. Notably, there have been approved drugs targeting ANPEP, APOE, LPL and SORT148,49, though their effect on liver diseases is still not clear. We also provided various evidence for these targets, including gene expression in liver or single cells, liver imaging, liver biopsies, and potential beneficial or unbeneficial effects to other diseases. Our study examined the association of these targets with BMI, showing that BMI has causal mediating effect on the association of APOE to MASLD, and APOC1, IL1RN, KRT18, KRT8, and LPL have causal mediating effects on the association of BMI to MASLD. Additionally, we found that the causal association between NCAN and MASLD may be fully explained by the causal effect of BMI; while ANPEP, APOC1, IL1RN, KRT18, KRT8, LPL, and SORT1 have causal effects on MASLD beyond obesity. This suggests that, besides obesity, these proteins may be involved in other pathways relevant for MASLD that requires further exploration.
Despite being a comprehensive study identified many innovative findings of the targets for MASLD, this study has some limitations. Firstly, liver imaging was not available for all participants, which may have led to an underestimation of MASLD cases and increased the rate of false-negative results in MASLD associations. Secondly, the study relied on plasma proteins to identify potential therapeutic targets for MASLD, which may not necessarily reflect protein levels in liver tissue. However, this approach is informative, and partially validated by tissue data. Thirdly, the MASLD GWAS by Vujkovic M, et al used chronic ALT elevation as a proxy for MASLD, but their conclusion was well validated70 and our findings were generally consistent across cohorts (Figure 3). Fourthly, the shared genetic variation between MASLD and proteins was limited, which may be due to violations of the single causal variant assumption or the known high false-negative rate for colocalization71. Fifthly, when comparing snRNA-seq and scRNA-seq data between MASLD/NASH and non-MASLD groups, selection bias from limited participants may drive the conclusions.
Overall, our study underscores the immense potential of large-scale multi-omics in enhancing our understanding of complex diseases, such as MASLD in our study, and identifying potential targets for translational research. By leveraging these innovative technologies, we were able to shed new light on the pathophysiology of MASLD and uncover promising new avenues for therapeutic intervention.
Supplementary Material
Acknowledgements:
This research was conducted using data from UK Biobank, a major biomedical database (https://www.ukbiobank.ac.uk/) via application no. 53639 and 65851. We thank the participants, contributors, clinicians, and researchers for making data available for this study. We are grateful to the research & development leadership teams at the thirteen participating UKB-PPP member companies (Alnylam Pharmaceuticals, Amgen, AstraZeneca, Biogen, Bristol-Myers Squibb, Calico, Genentech, Glaxo Smith Klein, Janssen Pharmaceuticals, Novo Nordisk, Pfizer, Regeneron, and Takeda) for jointly funding the proteomics study in UK Biobank. We thank the team at Olink Proteomics for their consistent logistic support throughout the project. We thank Dipender PS Gill for his help in facilitating MVP collaboration. This research is based on data from the MVP, Office of Research and Development, Veterans Health Administration and was supported by award MVP000 (mvp003/028b). MVP MASLD data was generated with Department of Veterans Affairs (VA) funding support (I01-BX003362: K.M.C., P.S.T., and M.V.) with additional support from resources and facilities of the VA Informatics and Computing Infrastructure (VINCI), VA HSR RES 13-457. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. M.V. acknowledges support for this work from the National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases, grant R01 DK134575. J.L. is supported by a Novo Nordisk Postdoctoral Fellowship and N.D. through a Novo Nordisk clinical research training fellowship run in partnership with the University of Oxford. The work has been supported by the Medical Research Council (JWT) and by the Oxford NIHR Biomedical Research Centre (JWT).
ABBREVIATIONS:
- MASLD
metabolic dysfunction-associated steatotic liver disease
- NAFLD
nonalcoholic fatty liver disease
- MASH
metabolic dysfunction-associated steatohepatitis
- NASH
non-alcoholic steatohepatitis
- MR
Mendelian randomization
- MRI-PDFF
magnetic resonance imaging - estimated proton density fat fraction
- ICD
International Classification of Diseases
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MSD
Matrix Spectral Decomposition
- GWAS
genome-wide association study
- ALT
alanine aminotransferase
- PheMR
phenome-wide Mendelian randomization study
- pQTL
protein quantitative trait locus
- eQTL
expression quantitative trait locus
- BMI
body mass index
- VEP
variant effect predictor
- LOFTEE
loss-of-function transcript effect estimator
- DEG
differentially expressed genes
- snRNA-seq
single nucleus RNA sequence
- scRNA-seq
single cell RNA sequence
- age2
age squared
- SNP
single-nucleotide polymorphism
- NAFL
non-alcoholic fatty liver
- GRHPR
Glyoxylate and hydroxypyruvate reductase
- HMOX2
heme oxygenase 2
- HMOX1
heme oxygenase 1
- IGF
insulin-like growth factor
- SCG3
secretogranin III
- RBKS
ribokinase
- ORM1
Orosomucoid 1
- AHCY
adenosylhomocysteinase
- FCGR2B
Fc gamma receptor IIb
Footnotes
Conflict of interest statement: The authors declare the following competing interests: S.H., L.C., C.D., C.A.P.M., M.T., R.M., T.M. and J.M.M.H. are full-time employees of Novo Nordisk. T.G.R. was part-time employee of Novo Nordisk during the project running. J.L. is supported by a University of Oxford Novo Nordisk Research Fellowship. N.D. was supported by a University of Oxford Novo Nordisk Research Training Fellowship. The remaining authors declare no competing interests.
Data availability:
UK Biobank data are publicly available to bona fide researchers upon application at http://www.ukbiobank.ac.uk/using-the-resource/. Publicly available summary statistics are obtained from https://gwas.mrcieu.ac.uk/, https://www.ebi.ac.uk/gwas/, https://www.gtexportal.org/home/datasets, and https://pan.ukbb.broadinstitute.org. Bulk liver RNA-seq data are available in Gene Expression Omnibus (GEO) under accession GSE135251. SnRNA-seq and scRNA-seq data are available in GEO under accession GSE189175, GSE185477, GSE136103, GSE212837, GSE189600, and GSE192740. Other source of data or web source were clarified in the methods. Relevant detailed results generated in this study were presented in supplementary tables. Clinical data with respect to the liver biopsy samples were assessable through contacting J.W.T.
References
- 1.Rinella M. E. et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 79, 1542–1556 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Loomba R., Friedman S. L. & Shulman G. I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UK Biobank. Available at: https://biobank.ndph.ox.ac.uk/showcase/. Accessed Jun 7, 2022. [Google Scholar]
- 5.Kleiner D. E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatol. Baltim. Md 41, 1313–1321 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Linge J. et al. Body Composition Profiling in the UK Biobank Imaging Study. Obes. Silver Spring Md 26, 1785–1795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayward K. L. et al. Detecting non-alcoholic fatty liver disease and risk factors in health databases: accuracy and limitations of the ICD-10-AM. BMJ Open Gastroenterol. 8, e000572 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairfield C. J. et al. Genome-Wide Association Study of NAFLD Using Electronic Health Records. Hepatol. Commun. 6, 297–308 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UK Biobank Genotyping of 500,000 UK Biobank participants. version 2.0. https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/ukb_dna_processing.pdf (2017). 20 Jun, 2022. [Google Scholar]
- 11.Backman J. D. et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature 599, 628–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J. & Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Szklarczyk D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mbatchou J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Ghodsian N. et al. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep. Med. 2, 100437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finngen Freeze R9; Available at: https://www.finngen.fi/en/access_results; Accessed Sep 22, 2022. [Google Scholar]
- 18.Vujkovic M. et al. A trans-ancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for nonalcoholic fatty liver disease with histological and radiological validation. 10.1101/2020.12.26.20248491 (2021) doi: 10.1101/2020.12.26.20248491. [DOI] [PMC free article] [PubMed]
- 19.Willer C. J., Li Y. & Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferkingstad E. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 53, 1712–1721 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Liu Y. et al. Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. eLife 10, e65554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GTEx Consortium. Genotype-Tissue Expression (GTEx). Available at https://www.gtexportal.org/, Accessed Jul 7, 2022. [Google Scholar]
- 23.Yengo L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kibinge N. K., Relton C. L., Gaunt T. R. & Richardson T. G. Characterizing the Causal Pathway for Genetic Variants Associated with Neurological Phenotypes Using Human Brain-Derived Proteome Data. Am. J. Hum. Genet. 106, 885–892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemani G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verbanck M., Chen C.-Y., Neale B. & Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S., Foley C. N., Allara E., Staley J. R. & Howson J. M. M. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat. Commun. 11, 376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J. et al. Causal inference for heritable phenotypic risk factors using heterogeneous genetic instruments. PLoS Genet. 17, e1009575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan-UKB team. Pan-UKB 2020. Available at: https://pan.ukbb.broadinstitute.org. Accessed July 7, 2023.
- 30.Foley C. N. et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun. 12, 764 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cingolani P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe K., Taskesen E., van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koplev S. et al. A mechanistic framework for cardiometabolic and coronary artery diseases. Nat. Cardiovasc. Res. 1, 85–100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Govaere O. et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 12, eaba4448 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez M. et al. Human liver single nucleus and single cell RNA sequencing identify a hepatocellular carcinoma-associated cell-type affecting survival. Genome Med. 14, 50 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews T. S. et al. Single-Cell, Single-Nucleus, and Spatial RNA Sequencing of the Human Liver Identifies Cholangiocyte and Mesenchymal Heterogeneity. Hepatol. Commun. 6, 821–840 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandran P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S. et al. An autocrine signaling circuit in hepatic stellate cells underlies advanced fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 15, eadd3949 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao Y. et al. Hepatocytes demarcated by EphB2 contribute to the progression of nonalcoholic steatohepatitis. Sci. Transl. Med. 15, eadc9653 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilliams M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aizarani N. et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piñero J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan J.-B. et al. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PloS One 8, e80747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vujkovic M. et al. A multiancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for nonalcoholic fatty liver disease with histological and radiological validation. Nat. Genet. 54, 761–771 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe K., Taskesen E., van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ochoa D. et al. The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 51, D1353–D1359 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y. et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 50, D1398–D1407 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sveinbjornsson G. et al. Multiomics study of nonalcoholic fatty liver disease. Nat. Genet. 54, 1652–1663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piras I. S. et al. Hepatic PEMT Expression Decreases with Increasing NAFLD Severity. Int. J. Mol. Sci. 23, 9296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X. & Deng R. Dysregulation of Bile Acids in Patients with NAFLD. in Nonalcoholic Fatty Liver Disease - An Update (ed. Hamdy Gad E.) (IntechOpen, 2019). doi: 10.5772/intechopen.81474. [DOI] [Google Scholar]
- 53.GWAS Catalog EMBL-EBI. Available at: https://www.ebi.ac.uk/gwas/. Accessed Sep 22, 2023. [Google Scholar]
- 54.Gianmoena K. et al. Epigenomic and transcriptional profiling identifies impaired glyoxylate detoxification in NAFLD as a risk factor for hyperoxaluria. Cell Rep. 36, 109526 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Malaguarnera L., Madeddu R., Palio E., Arena N. & Malaguarnera M. Heme oxygenase-1 levels and oxidative stress-related parameters in non-alcoholic fatty liver disease patients. J. Hepatol. 42, 585–591 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Lin C.-C. et al. Serum Secretogranin III Concentrations Were Increased in Subjects with Metabolic Syndrome and Independently Associated with Fasting Plasma Glucose Levels. J. Clin. Med. 8, 1436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J. et al. Genomics and phenomics of body mass index reveals a complex disease network. Nat. Commun. 13, 7973 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L. et al. Mitigation of non-alcoholic steatohepatitis via recombinant Orosomucoid 2, an acute phase protein modulating the Erk1/2-PPARγ-Cd36 pathway. Cell Rep. 42, 112697 (2023). [DOI] [PubMed] [Google Scholar]
- 59.Zhou B., Luo Y., Ji N., Hu C. & Lu Y. Orosomucoid 2 maintains hepatic lipid homeostasis through suppression of de novo lipogenesis. Nat. Metab. 4, 1185–1201 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Murphy S. K. et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 145, 1076–1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishikawa T., Yokoyama H., Matsuura T. & Fujiwara Y. Fc gamma RIIb expression levels in human liver sinusoidal endothelial cells during progression of non-alcoholic fatty liver disease. PloS One 14, e0211543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shu T. et al. Fc Gamma Receptor IIb Expressed in Hepatocytes Promotes Lipid Accumulation and Gluconeogenesis. Int. J. Mol. Sci. 19, 2932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao A. et al. Implications of Sortilin in Lipid Metabolism and Lipid Disorder Diseases. DNA Cell Biol. 36, 1050–1061 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Wu M.-J. et al. Role of NCAN rs2228603 polymorphism in the incidence of nonalcoholic fatty liver disease: a case-control study. Lipids Health Dis. 15, 207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Speliotes E. K. et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 7, e1001324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pihlajamäki J. et al. Serum interleukin 1 receptor antagonist as an independent marker of non-alcoholic steatohepatitis in humans. J. Hepatol. 56, 663–670 (2012). [DOI] [PubMed] [Google Scholar]
- 67.van den Berg E. H., Corsetti J. P., Bakker S. J. L. & Dullaart R. P. F. Plasma ApoE elevations are associated with NAFLD: The PREVEND Study. PloS One 14, e0220659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feldstein A. E. et al. Cytokeratin-18 fragment levels as noninvasive biomarkers for nonalcoholic steatohepatitis: a multicenter validation study. Hepatol. Baltim. Md 50, 1072–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawanaka M. et al. Correlation between serum cytokeratin-18 and the progression or regression of non-alcoholic fatty liver disease. Ann. Hepatol. 14, 837–844 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Serper M. et al. Validating a non-invasive, ALT-based non-alcoholic fatty liver phenotype in the million veteran program. PloS One 15, e0237430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuber V. et al. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am. J. Hum. Genet. 109, 767–782 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are publicly available to bona fide researchers upon application at http://www.ukbiobank.ac.uk/using-the-resource/. Publicly available summary statistics are obtained from https://gwas.mrcieu.ac.uk/, https://www.ebi.ac.uk/gwas/, https://www.gtexportal.org/home/datasets, and https://pan.ukbb.broadinstitute.org. Bulk liver RNA-seq data are available in Gene Expression Omnibus (GEO) under accession GSE135251. SnRNA-seq and scRNA-seq data are available in GEO under accession GSE189175, GSE185477, GSE136103, GSE212837, GSE189600, and GSE192740. Other source of data or web source were clarified in the methods. Relevant detailed results generated in this study were presented in supplementary tables. Clinical data with respect to the liver biopsy samples were assessable through contacting J.W.T.