Abstract
Lambda Xis, which is required for site-specific excision of phage lambda from the bacterial chromosome, has a much shorter functional half-life than Int, which is required for both integration and excision (R. A. Weisberg and M. E. Gottesman, p. 489–500, in A. D. Hershey, ed., The Bacteriophage Lambda, 1971). We found that Xis is degraded in vivo by two ATP-dependent proteases, Lon and FtsH (HflB). Xis was stabilized two- to threefold more than in the wild type in a lon mutant and as much as sixfold more in a lon ftsH double mutant at the nonpermissive temperature for the ftsH mutation. Integration of lambda into the bacterial chromosome was delayed in the lon ftsH background, suggesting that accumulation of Xis in vivo interferes with integration. Overexpression of Xis in wild-type cells from a multicopy plasmid inhibited integration of lambda and promoted curing of established lysogens, confirming that accumulation of Xis interferes with the ability of Int to establish and maintain an integrated prophage.
Bacteriophage lambda employs integrated regulatory mechanisms to ensure the appropriate equilibrium between lysogeny and lytic growth. In addition to well-characterized controls for transcription initiation and termination, lambda also utilizes rapid and specific degradation of key regulatory proteins to influence the direction of its development. RecA-dependent degradation of the repressor cI as part of the SOS response returns the dormant prophage to the lytic cycle (22–24). Once the lytic decision is made and the level of cII expression has decreased, rapid degradation of cII by FtsH (HflB) ensures commitment to the lytic cycle (3). On the other hand, stabilization of cII results primarily in a lysogenic response (16, 19). In addition, the lytic N (antitermination) and O (replication) proteins are subject to rapid degradation (12, 13, 31). For each of these phage-encoded proteins, as in most instances of cytoplasmic degradation of proteins in Escherichia coli, degradation has been found to be mediated by an ATP-dependent protease (9).
One of the first suggestions for a role for protein degradation in the lambda life cycle came from studies of site-specific recombination between lambda and the bacterial chromosome. Site-specific integration of phage lambda into the bacterial chromosome requires the activity of the int gene product, while efficient excision of the phage lysogen requires Xis, the product of the xis gene, as well as Int (for a review, see reference 29). Robert Weisberg and Max Gottesman demonstrated in 1971 that Int and Xis have markedly different functional stabilities in vivo (half-life [t1/2], ∼60 min for Int versus ∼7 min for Xis) and observed that instability of Xis was at least partially due to an energy-dependent mechanism (30). They proposed that the efficient integration of phage lambda into the bacterial chromosome is enhanced by the instability of the excisionase activity. The idea that the relative quantities of the two proteins determine the efficiency and direction of the recombination reactions was eventually supported by in vitro data (6, 11, 18). Addition of Xis at levels that maximally stimulated excisive recombination in vitro completely inhibited integrative recombination in vitro (11).
Our objectives in this study were to determine if the Xis protein is degraded in vivo and, if so, if any of the known ATP-dependent proteases are responsible for its degradation. We also wanted to test whether the instability of Xis has significant effects on the ability of lambda to establish and maintain lysogeny.
Xis is degraded in vivo by Lon and FtsH.
Since the work by Weisberg and Gottesman, it has been found that most cytoplasmic protein degradation is energy dependent and that E. coli has at least five different energy-dependent proteases with different substrate specificities (for a review, see reference 9). In order to determine if Xis is specifically degraded by any of the known ATP-dependent proteases, we examined an isogenic set of protease mutant strains, each possessing a chromosomal copy of lacIq. We expressed Xis in the different genetic backgrounds from pRK5 (1), a multicopy plasmid with xis under plac control, and inhibited protein synthesis by addition of the translational inhibitor spectinomycin, and the remaining Xis was measured as a function of time by immunoblotting (Fig. 1).
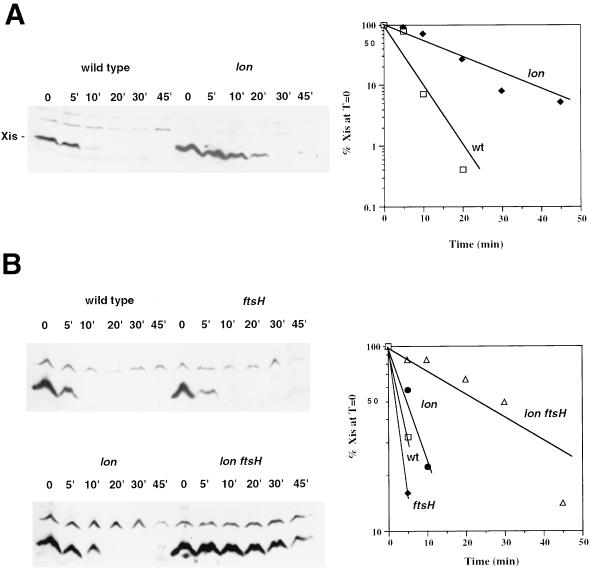
FIG. 1.
Stability of λ Xis. Cells containing the plasmid pRK5 (1) were grown to early log phase in LB medium with 50 μg of ampicillin per ml at 32°C, induced with IPTG (1 mM) for 20 to 30 min to express Xis, and then treated with spectinomycin (100 μg/ml) to inhibit protein synthesis. Aliquots were removed following addition of spectinomycin at different time points and precipitated with trichloroacetic acid and the pellets were resuspended in gel loading buffer. Samples were run on sodium dodecyl sulfate–16% polyacrylamide gel electrophoresis-Tricine gels, electroblotted (XCell System, Novex) to 0.1-μm-pore-size Protran nitrocellulose (Schleicher and Schuell), and immunoblotted with a rabbit polyclonal antiserum against the carboxy-terminal 15 amino acids of Xis (a gift from Carol Robertson and Howard Nash). Washes and detection were done with the ECL chemiluminescence system (Amersham Life Sciences). The relative intensities of the Xis band for the different time points on the developed film were determined with the Eagle Eye II video system (Stratagene). (A) SG22163 (lacIq) and SG22185 (lacIq Δlon-510) at 32°C; (B) SG22163, SG22166 (lacIq ftsH1), SG22185, and GL008 (lacIq Δlon-510 ftsH1) at 42°C. For spectinomycin chase experiments done at 42°C, the cultures were shifted to the higher temperature for 15 min prior to induction of xis with IPTG. Strain SG22163 is a malP::lacIq derivative of MC4100 (5). The protease mutant generated by P1 transduction of SG22163 by the appropriate P1 lysates (26).
Xis is in fact physically unstable in vivo, with a t1/2 of approximately 4 min at 32°C in wild-type cells (Fig. 1A). The only protease mutant that exhibited significant stabilization of Xis relative to that seen for the wild-type strain was the lon mutant. The t1/2 of Xis was extended to 10 to 12 min at 32°C in the lon background (Fig. 1A). None of the clp protease mutants stabilized Xis, either alone or in combination with a lon mutant (data not shown).
Because the FtsH (HflB) protease is essential to E. coli (2, 14), it was necessary to utilize a conditional lethal mutant (ftsH1 [25]) to assess the effects of FtsH activity on Xis stability. The temperature-sensitive ftsH mutation did not by itself appreciably stabilize Xis at 32 or 42°C but, in combination with the lon mutation, extended the t1/2 for Xis from approximately 4 min to about 25 min at the nonpermissive temperature (Fig. 1B). These results indicate that Xis is recognized and actively degraded in vivo by both the Lon and the FtsH proteases. The observations that Xis stabilization in the lon mutant background was not dramatic (two- to threefold) and that the ftsH genetic background by itself did not significantly stabilize Xis indicate that each protease degrades Xis rapidly, with Lon perhaps capable of degrading it more rapidly and thus playing the primary role in Xis degradation.
Integration of lambda is delayed in the lon ftsH double mutant.
Based on the observed inhibition of lambda integration by Xis in vitro (6, 11, 18), the prediction is that accumulation of Xis in the cell due to its stabilization would lead to abortive lysogeny by preventing Int from mediating integration of repressed phage and/or by working with Int to excise phage that did integrate. Analysis of the effects of Xis stabilization on the establishment and maintenance of lysogeny is complicated by the fact that both the lon and the ftsH mutations have other significant effects on the biology of phage lambda. ftsH mutations stabilize the cII and cIII proteins (3, 14, 15), resulting in greater levels of the cI repressor and thus shifting the lytic-lysogeny decision in favor of lysogeny. lon mutants stabilize λ N protein and in some unknown way destabilize the phage cII protein (13). Lambda infection of lon mutants results in a rate of lysogeny lower than that in wild-type cells, and lon lysogens commit rapidly and irreversibly to lysis following transient derepression.
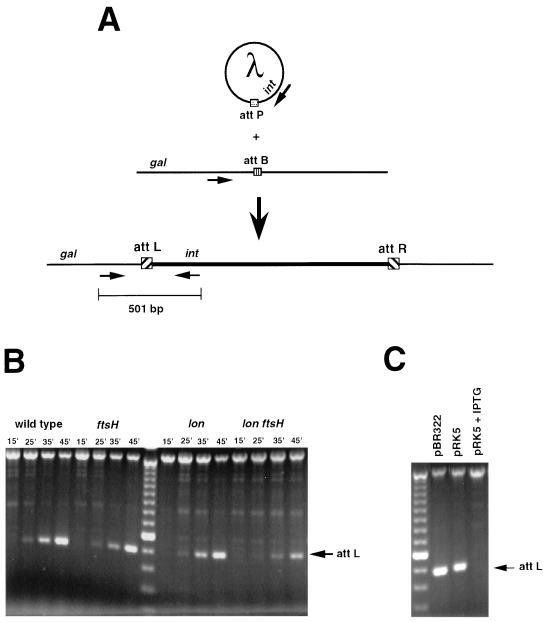
To have an assay that was independent of the pleiotropic effects of the ftsH and lon mutations on the life cycle of lambda, we utilized a PCR-based approach to directly monitor the kinetics of integration following infection with λcI857. Primers from within the int region of the phage and from the region between gal and attB of the bacterial chromosome were used to amplify the unique attL sequence of the integrated prophage (schematically shown in Fig. 2A) at different times following infection (20). The amount of attL detectable in each infection mixture at each time point following infection was approximately the same for the wild-type, ftsH, and lon cells (Fig. 2B). The accumulation of attL sequence in the ftsH lon infection mixture was delayed in comparison to that in the other strains by more than 10 min. Under these infection conditions, the final frequency of lysogeny among survivors (plated on Luria-Bertani [LB] agar at 32°C) approached 100% for the wild-type, ftsH, and lon ftsH strains, with the frequency in the lon strain being about 20% lower (data not shown), in agreement with the expected effects of these mutations on cII and therefore on lysogeny. These results are consistent with moderate stabilization of Xis in the double mutant, such that integration was delayed until the transiently expressed Xis (from pL) was degraded, and the more stable Int (expressed primarily from pI) could integrate the phage without interference. Although we did not see a consistent and large effect of the ftsH mutation in combination with lon on Xis degradation at permissive temperatures (data not shown), the delay in integration in the lon ftsH mutant suggests that temperature-sensitive FtsH is not fully functional at low temperatures.
FIG. 2.
Measurement of lambda integration by PCR amplification of attL. (A) Primers from within phage int and from the region between gal and attB of the bacterial chromosome were used to amplify a unique 501-bp attL sequence from lysogens following infection with λcI857 (20). (B) Kinetics of lambda integration in the different protease mutants. Cultures of SG22163 (lacIq), SG22166 (lacIq ftsH1), SG22185 (lacIq Δlon-510), and GL008 (lacIq Δlon-510 ftsH1) were grown to early log phase, their cell concentrations were normalized by measuring the optical density at 600 nm, and the cultures were then chilled on ice. Cells were mixed with λcI857 (multiplicity of infection, ∼17) and incubated on ice for 20 min in order to synchronize infection. Infection mixtures were warmed to 32°C, aliquots (∼3 × 107 cells) were taken at the time points indicated, and the cells were pelleted at 4,000 × g for 10 min at 4°C. Pelleted cells were washed twice with cold sterile water and resuspended in a final volume of 30 μl. A portion of each suspension was plated on LB-citrate agar at 32°C to determine titers for survivors, and 20 μl was used for PCR amplification as described elsewhere (20). Reconstruction experiments with a mixture of ∼2 × 103 lysogens and ∼2 × 107 wild-type cells demonstrated a strong attL amplification, suggesting that the absence of a signal in these samples represents an integration frequency of <10−4 (data not shown). In multiple experiments, cell killing under these infection conditions ranged from 50 to 90%. Lysogeny frequencies among survivors were determined by testing for λ immunity in cross-streaks and by plating at 39°C for SG22163 and SG22185. Lysogeny frequencies under these conditions were determined to be nearly 100% for wild-type, ftsH, and lon ftsH infection mixtures, with the lon survivors having ∼20% fewer lysogens. (C) Overexpression of Xis inhibits integration. SG22163/pBR322 and SG22163/pRK5 were grown to early log phase in LB medium with 50 μg of ampicillin per ml at 32°C and then treated either with no IPTG or with 1 mM IPTG for an additional 30 min at 32°C. The cells were then chilled on ice, infected as described above with lambda λcI857 (multiplicity of infection, ∼14) and harvested as described above for use in the attL amplification reaction. The DNA standard for the gels in panels B and C was the 100-bp ladder (Gibco-BRL).
Excess Xis inhibits integration of phage lambda into the bacterial chromosome.
If moderate stabilization of Xis delays integration, complete stabilization and, therefore, higher levels of Xis would be expected to inhibit integration more fully. We assumed that high-level expression from pRK5 (plac-xis+) would mimic to some degree the effects of stabilization by increasing the accumulation of Xis in the cell. We utilized the PCR approach described above to assess the degree of integration of λcI857 following infection of wild-type cells containing pBR322 or pRK5. Cells containing the control pBR322 plasmid (Xis−) and uninduced cells containing pRK5 had similar quantities of attL 25 min after infection (Fig. 2C). No attL joint sequence was detectable for the induced pRK5 (plus isopropyl-β-d-thiogalactopyranoside [IPTG]) culture, indicating that under these conditions an excess of Xis completely inhibited integration of lambda phage into the bacterial chromosome. Thus, both the kinetics of integration of lambda in a lon ftsH mutant and the inhibition of integration when Xis is overexpressed support a role for Xis instability in ensuring rapid integration of repressed phage.
Excess Xis promotes spontaneous curing of lambda lysogens.
Would accumulation of Xis in vivo promote excision of integrated and repressed prophage? The effects of excess Xis on the maintenance of established lysogens was assessed by measuring the curing frequency of λcI857 lysogens with and without overexpression of Xis from pRK5. Cultures of wild-type and lon lysogens (λcI857 Cmr) containing either the control pBR322 vector or the pRK5 plasmid were grown to early log phase and treated with different concentrations of IPTG for 1 h at 32°C, and then titers on LB agar plates were determined at 32 and 39°C. Survivors at 39°C represented cells that had been cured of the temperature-inducible lysogen due to transient derepression of xis followed by excision of the phage chromosome (all 39°C survivors tested were chloramphenicol sensitive). Without induction of the phage lytic cycle, the circularized phage chromosome is not replicated and consequently is lost during subsequent cell divisions.
The rate of curing (survivors at 39°C/total number of colonies at 32°C) for both wild-type and lon lysogens was dramatically increased by expression of Xis from pRK5, indicating that Xis, and not Int, was limiting for curing (Table 1). Curing of the lon lysogens increased from approximately 10−6 to approximately 10−2, while curing in the wild-type strain increased from 10−4 to 10−2. The difference in basal levels of curing for the two strains (10−4 for wild type versus 10−6 for lon) is not consistent with stabilization of Xis and must represent some other effect of the lon mutation on phage lambda curing. In agreement with the role of excess Xis in promoting curing, we saw a moderate 15-fold increase in curing for the lon mutant carrying the uninduced pRK5 plasmid (∼3 × 10−5) over that seen in the lon/pBR322 strain (2 × 10−6), but not in the equivalent wild-type strains. It is likely that the basal levels of expression of Xis from the pRK5 plasmid, combined with stabilization of Xis in the lon strain, were enough to partially overcome the inhibitory effects of the lon mutation on curing. Although these experiments do not directly address the level of Xis induced in a lysogen, these results suggest that if Xis were stable, the ability of phage lambda to maintain lysogeny would be diminished.
TABLE 1.
Excess Xis promotes curing of λcI857 lysogensa
| Concn of IPTG (mM) | Curing frequencyb
|
|||
|---|---|---|---|---|
| Wild type
|
lon
|
|||
| pBR322 | pRK5 | pBR322 | pRK5 | |
| None | 1.1 × 10−4 | 1.8 × 10−4 | 2.1 × 10−6 | 3.2 × 10−5 |
| 0.1 | 1.7 × 10−4 | 3.8 × 10−3 | 3.5 × 10−6 | 1.2 × 10−2 |
| 1.0 | 7.9 × 10−5 | 2.5 × 10−2 | 1.4 × 10−6 | 6.0 × 10−2 |
Cultures of GL076 (lacIq λcI857 Cmr) and GL078 (lacIq Δlon-510 λcI857 Cmr) containing either pBR322 or pRK5 were grown to early log phase at 30°C in LB with 50 μg of ampicillin per ml (to simplify the screen for lysogens, a derivative of λcI857 carrying a minitransposon that encoded chloramphenicol resistance [Cmr] was isolated). The cultures were then divided into three subcultures and treated with IPTG as indicated for 1 additional h at 30°C. Titers for each subculture were then determined on LB plates at 32°C to determine the total cell concentration and at 39°C on LB-citrate plates to determine the numbers of cells cured of the temperature-inducible prophage (all survivors tested were Cms).
Curing frequencies are expressed as CFU per ml at 39°C/CFU per ml at 32°C. Data presented are for a single experiment but are indicative of curing patterns seen in multiple experiments.
It seems unlikely that the high level of curing here is dependent on Int made after the temperature is increased, since we would expect lytic growth and cell killing under those conditions. Rather, we expect that the basal level of Int, combined with the high levels of Xis, leads to efficient curing and subsequent segregation of the repressed prophage and that longer times for segregation after Xis was expressed might have led to even higher curing rates. The increase in curing when Xis is overproduced from a plasmid demonstrates that the constitutive level of Int in these lysogens is sufficient for a significant level of excisive recombination, a point that emphasizes the usefulness of a specific and well-regulated excisionase factor in ensuring stable maintenance of the lysogen. In measurements of curing for a prophage that is dependent on attP × attB recombination, and thus not on Xis, curing was on the order of 10−3, consistent with the presence of a biologically active level of Int (4, 7). However, as pointed out by Campbell (4), this basal level of Int is not significant enough to reinsert greater than 1% of phage that are spontaneously cured, even in the absence of a stabilized Xis.
Conclusions.
We have determined that lambda Xis is rapidly degraded in vivo by two different ATP-dependent proteases, Lon and FtsH. The observations that large quantities of Xis in vivo, such as might be seen if Xis were stable, inhibit integration and promote excision are consistent with the proposal first made by Weisberg and Gottesman (30) that rapid degradation of Xis ensures rapid integration of lambda into the bacterial chromosome. Given the multiple strategies used by the phage to maintain the appropriate ratio of Int to Xis at different points in its life cycle (i.e., sib regulation of int from pL, the cII-dependent expression of int from pI, etc. [8]), it is likely that the difference in stability between the two proteins does in fact represent another important layer of regulation.
This is the first case in which we have seen overlap in substrate specificity for Lon and FtsH in E. coli, although an overlap of chaperone activity has been reported for the mitochondrial analogs of these proteins in Saccharomyces cerevisiae (21). These two proteases, while both ATP dependent, differ in many respects (for reviews, see references 9 and 10). They have different proteolytic mechanisms (a serine-active site for Lon and a Zn metalloprotease site for FtsH) and do not share any noticeable sequence similarity beyond the appearance of a Walker ATPase consensus sequence in each. FtsH has a transmembrane domain, while Lon is a fully cytoplasmic protein. The existence of a shared substrate suggests that the membrane association of FtsH does not preclude full access to cytoplasmic substrate proteins. As noted above, FtsH is responsible for degradation of λ cII; lon mutations actually lead to increased cII degradation (3, 13). The involvement of FtsH in degradation of other known Lon substrates (λ N protein and the bacterial proteins SulA and RcsA, for instance) has not been tested, but it is known that a lon mutant alone significantly stabilizes these proteins (13, 17, 27, 28). Thus, if FtsH contributes to their degradation, it plays a much more minor role than it does for Xis. It will be interesting to determine if FtsH and Lon recognize similar sites or structures on this small (72-amino-acid) protein.
Acknowledgments
We are grateful to Robert Weisberg for providing materials and useful suggestions during the course of this work and to Carol Robertson and Howard Nash for providing the anti-Xis antiserum. We also thank Robert Weisberg, Howard Nash, Nadim Majdalani, Michael Maurizi, and Laurence Van Melderen for their comments on the manuscript.
REFERENCES
- 1.Abremski K, Hoess R. Escherichia coli plasmid vectors for high-level regulated expression of the bacteriophage λ xis gene product. Gene. 1983;25:49–58. doi: 10.1016/0378-1119(83)90166-x. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama Y, Ogura T, Ito K. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention efficiency of a cytoplasmic reporter. J Biol Chem. 1994;269:5218–5224. [PubMed] [Google Scholar]
- 3.Banuett F, Hoyt M A, McFarlane L, Echols H, Herskowitz I. hflB, a new Escherichia coli locus regulating lysogeny and the level of bacteriophage lambda CII protein. J Mol Biol. 1986;187:213–224. doi: 10.1016/0022-2836(86)90229-9. [DOI] [PubMed] [Google Scholar]
- 4.Campbell A. Significance of constitutive integrase synthesis. Proc Natl Acad Sci USA. 1976;73:887–890. doi: 10.1073/pnas.73.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban M J, Cohen S. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vargas L M, Landy A. A switch in the formation of alternative DNA loops modulates λ site-specific recombination. Proc Natl Acad Sci USA. 1991;88:588–592. doi: 10.1073/pnas.88.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echols H. Constitutive integrative recombination by bacteriophage λ. Virology. 1975;64:557–559. doi: 10.1016/0042-6822(75)90133-6. [DOI] [PubMed] [Google Scholar]
- 8.Echols H, Guarneros G. Control of integration and excision. In: Hendrix R, Roberts J, Stahl F, Weisberg R, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 75–93. [Google Scholar]
- 9.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S. Roles for energy-dependent proteases in regulatory cascades. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 503–519. [Google Scholar]
- 11.Gottesman S, Abremski K. Purification of the bacteriophage λ xis gene product required for λ excisive recombination. J Biol Chem. 1982;257:9658–9662. [PubMed] [Google Scholar]
- 12.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 13.Gottesman S, Gottesman M E, Shaw J E, Pearson M L. Protein degradation in E. coli: the lon mutation and bacteriophage lambda N and cII protein stability. Cell. 1981;24:225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- 14.Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D’Ari R, Bouloc P. Cell growth and λ phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci USA. 1993;90:10861–10865. doi: 10.1073/pnas.90.22.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman C, Thevenet D, D’Ari R, Bouloc P. The HflB protease of Escherichia coli degrades its inhibitor λcIII. J Bacteriol. 1997;179:358–363. doi: 10.1128/jb.179.2.358-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyt M A, Knight D M, Das A, Miller H I, Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982;31:565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- 17.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of the SulA protein. Proc Natl Acad Sci USA. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nash H. Integrative recombination of bacteriophage lambda DNA in vitro. Proc Natl Acad Sci USA. 1975;72:1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obuchowski M, Shotland Y, Koby S, Giladi H, Gabig M, Wegrzyn G, Oppenheim O B. Stability of cII is a key element in the cold stress response of bacteriophage λ infection. J Bacteriol. 1997;179:5987–5991. doi: 10.1128/jb.179.19.5987-5991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell B S, Court D L, Nakamura Y, Rivas M P, Turnbough C L. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rep M, van Dijl J M, Suda K, Schatz G, Grivell L A, Suzuki C K. Promotion of mitochondrial membrane complex assembly by a proteolytically inactive yeast Lon. Science. 1996;274:103–106. doi: 10.1126/science.274.5284.103. [DOI] [PubMed] [Google Scholar]
- 22.Roberts J W, Roberts C W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci USA. 1975;72:147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts J W, Roberts C W, Craig N L. Escherichia coli recA gene product inactivates phage λ repressor. Proc Natl Acad Sci USA. 1978;75:4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts J W, Roberts C W, Mount D W. Inactivation and proteolytic cleavage of phage λ repressor in vitro in an ATP-dependent reaction. Proc Natl Acad Sci USA. 1977;74:2283–2287. doi: 10.1073/pnas.74.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos D, De Almeida D. Isolation and characterization of a new temperature-sensitive cell division mutant of Escherichia coli K-12. J Bacteriol. 1975;124:1502–1507. doi: 10.1128/jb.124.3.1502-1507.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 27.Stout V, Torres-Cabassa A, Maurizi M R, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres-Cabassa A S, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisberg R, Landy A. Site-specific recombination in phage lambda. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 211–250. [Google Scholar]
- 30.Weisberg R A, Gottesman M E. The stability of Int and Xis functions. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1971. pp. 489–500. [Google Scholar]
- 31.Wojtkowiak D, Georgopoulos C, Zylicz M. ClpX, a new specificity component of the ATP-dependent Escherichia coli Clp protease, is potentially involved in λ DNA replication. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]