Abstract
A model of the 66-kDa outer membrane protein (P66) of Lyme disease Borrelia spp. predicts a surface-exposed loop near the C terminus. This region contains an antigen commonly recognized by sera from Lyme disease patients. In the present study, this region of P66 and homologous proteins of other Borrelia spp. were further investigated by using monoclonal antibodies, epitope mapping of P66 of Borrelia burgdorferi, and DNA sequencing. A monoclonal antibody specific for B. burgdorferi bound to the portion of P66 that was accessible to proteolysis in situ. The linear epitope for the antibody was mapped within a variable segment of the surface-exposed region. To further study this protein, the complete gene of Borrelia hermsii for a protein homologous to P66 was cloned. The deduced protein was 589 amino acids in length and 58% identical to P66 of B. burgdorferi. The B. hermsii P66 protein was predicted to have a surface-exposed region in the same location as that of B. burgdorferi’s P66 protein. With primers designed on the basis of conserved sequences and PCR, we identified and cloned the same regions of P66 proteins of Borrelia turicatae, Borrelia parkeri, Borrelia coriaceae, and Borrelia anserina. The deduced protein sequences from all species demonstrated two conserved hydrophobic regions flanking a surface-exposed loop. The loop sequences were highly variable between different Borrelia spp. in both sequence and size, varying between 35 and 45 amino acids. Although the actual function of P66 of Borrelia spp. is unknown, the results suggest that its surface-exposed region is subject to selective pressure.
Members of the genus Borrelia, like other spirochetes, have two membranes (6, 21). The outer membrane of Borrelia spp. is more fluid and contains fewer integral membrane proteins than the outer membranes of many gram-negative bacteria, such as Escherichia coli (6, 31). Most of the spirochetal outer membrane proteins that have been identified to date have been lipoproteins. Presumably, these are anchored in the membrane by their lipid moieties and not by membrane-spanning regions of the protein. A 66-kDa protein is one of the few integral membrane proteins that have been identified in Borrelia burgdorferi, which is a cause of Lyme disease and the most commonly studied Borrelia species (7, 30). Based on its apparent size, this protein was originally identified as P66 and was shown to be commonly recognized by antibodies of patients with Lyme disease (4, 11, 16).
The genes for P66 of B. burgdorferi as well as of two other Lyme disease agents, Borrelia afzelii and Borrelia garinii, have been cloned and sequenced (10). The genes encode proteins of about 620 amino acids with predicted signal peptides of about 20 residues. A signal peptidase I cleavage site was confirmed by N-terminal sequencing of a native processed protein (10). The overall identity in amino acid sequences of P66 of these three species is about 90%.
The P66 proteins of the three Lyme disease Borrelia spp. are shortened by about 15 kDa when intact cells are treated with proteinase K (7, 10). Within the part that is lost from the cell’s surface is a region that is hydrophilic and predicted to be a flexible segment (10). Flanking this surface loop are more hydrophobic regions that could span membranes. Skare et al. demonstrated that native P66 had porin activity in liposomes and called the protein Oms66 for its outer membrane-spanning characteristics (38). However, the actual function of this protein remains unknown; the sequence was unlike that of any other protein in the database.
Studies of this novel membrane protein may contribute to the understanding of spirochetal outer membrane structure, provide further information about the role of this protein in the pathogenesis of Lyme disease, and identify another candidate antigen for diagnosis and immunoprophylaxis. As a step toward these goals, we further characterized a surface-exposed portion of P66 and sequences that flank it. We did this by producing monoclonal antibodies to P66, by using the antibodies to identify epitopes in the protein, and by comparing equivalent regions of homologous proteins of more distantly related Borrelia spp. We found that these surface-exposed regions of the P66 proteins of Borrelia spp. are highly variable in both size and sequence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Borrelia species and strains used were the following: B31 (ATCC 35210), Sh.2 (36), N40 (18), and PKa (41) of B. burgdorferi sensu stricto; ACAI (1) of B. afzelii; and Ip90 (23), NBS23A, NBS16 (9), Mal02, and Far02 (12) of B. garinii. The origins of strains of Borrelia hermsii HS1 (3), Borrelia turicatae Oz1 (14), Borrelia coriaceae (24), and B. anserina (17) have been described previously. The OspA− OspB− OspC− OspD− mutant B313 of B. burgdorferi was from the strain B31 lineage (35). Borrelia parkeri was originally provided to A.G.B. by H. Stoenner, Rocky Mountain Laboratories. Spirochetes were grown in BSK II medium and harvested as previously described (5, 8). Cells were counted in a Petroff-Hausser chamber under phase-contrast microscopy. E. coli TOP 10F′ (Invitrogen, Carlsbad, Calif.), BL21, and NovaBlue (DE3) (Novagen, Madison, Wis.) were grown in Luria-Bertani medium supplemented with carbenicillin (50 μg/ml) or kanamycin (50 μg/ml) when required.
Polyacrylamide gel electrophoresis (PAGE) and Western blot analysis.
Cell lysates were subjected to PAGE with 12.5% acrylamide as described previously (13). For Western blot analysis, proteins were transferred to nitrocellulose membranes (Bio-Rad Laboratories, Richmond, Calif.), which were then blocked with 3% dried nonfat milk in 10 mM Tris (pH 7.4)–150 mM NaCl (milk-TS) for 2 h (13). Membranes were incubated with human serum or hybridoma supernatants diluted 1:300 and 1:10, respectively, in 0.3% milk-TS. Alkaline phosphatase-conjugated recombinant protein A/G (Pierce Chemical Co., Rockford, Ill.) served as the second ligand. The blots were developed with nitroblue tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt as substrates (Pierce).
Production of recombinant P66 (rP66).
After cleavage of the signal peptide of 21 residues, the mature P66 protein of B. burgdorferi B31 begins with an alanine (10). DNA encoding P66 from this residue to the C terminus (nucleotides 290 to 2080 of the sequence with GenBank accession no. X87725) was cloned downstream of a thrombin recognition site in the pRSET expression vector (Invitrogen). This fusion polypeptide was expressed in E. coli BL21 and was recovered from lysed cells as inclusion bodies (2). After cleavage of the fusion protein with thrombin, the preparation was subjected to PAGE, and rP66 was electroeluted from the gel as described elsewhere (2). The identity of rP66 was verified by Western blotting with the monospecific polyclonal rabbit P66-specific antiserum (10) and by N-terminal amino acid sequencing with an Applied Biosystems model 477A Sequenator (Foster City, Calif.).
Antibodies.
Monoclonal antibodies to P66 were produced by first subcutaneously injecting adult C3H/HeN mice (Harlan Laboratories, Indianapolis, Ind.) with 20 μg of purified rP66 in Freund’s complete adjuvant on day 1. Thereafter, different immunization protocols were used. In one group, each mouse received 20 μg of rP66 in Freund’s incomplete adjuvant on day 21 and 60 μg on day 42. On days 70 and 84, each of these mice in this first group were injected intravenously and intraperitoneally with 5 × 108 B313 cells by each route. In a second group, each mouse was boosted intraperitoneally, intramuscularly, subcutaneously, and in the footpad with an equally divided dose of 70 μg of rP66 in Freund’s incomplete adjuvant on day 28. This second group then received 2 × 108 B31 cells intravenously on days 35, 42, and 49 and were boosted intravenously with 70 μg of rP66 in phospate-buffered saline on day 56. For both immunization protocols, spleens of the mice were collected 4 days after the last immunization. Spleen cells were fused to NS1 myeloma cells by a modification of the method of Oi and Herzenberg (27). Hybridoma supernatant fluids were screened by Western blot analysis. Purified monoclonal antibodies were obtained from hybridoma supernatants by using a protein A column (Pierce).
Serum specimens from three patients and five controls were from a reference panel provided by the Centers for Disease Control and Prevention, Fort Collins, Colo., and described elsewhere (34). Sera from these patients had previously been shown to contain antibodies that reacted to P66 of B. burgdorferi B31 by Western blot analysis (11).
Protease treatment of spirochetes.
Whole cells were treated with proteases by a modification of a method described previously (7). Briefly, harvested and washed spirochetes were resuspended in phosphate-buffered saline–Mg at a concentration of 109 cells/ml. To 450 μl of the cell suspension was added 50 μl of one of the following: distilled water, proteinase K (4 mg/ml in water; Boehringer Mannheim, Indianapolis, Ind.), 10−3 M HCl, or trypsin (1 mg/ml in 10−3 M HCl; Sigma). After incubation for 60 min at 20°C, proteolysis was inhibited by adding 10 μl of phenylmethylsulfonyl fluoride (Sigma; 50 mg/ml in isopropanol). The cells were centrifuged and washed twice with phosphate-buffered saline–Mg. The pellet was resuspended in a buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, and 5 mM MgCl2 and subjected to whole-cell protein extraction by being boiled in sodium dodecyl sulfate-PAGE sample buffer. The cell lysates were subjected to Western blot analysis to identify proteolytic products of P66 and the binding of monoclonal antibodies to these peptides.
Epitope mapping.
Overlapping recombinant peptides were produced to determine the epitopes for monoclonal antibodies. Recombinant plasmid pJB102 (10) was a source of DNA for a library of partial sequences of the P66 gene of B. burgdorferi B31. pJB102 was cleaved with BglII and MunI endonucleases to obtain a 570-bp segment encoding the C-terminal third of P66. This segment was processed in the NovaTope epitope mapping system (Novagen) to prepare an expression library as described by the manufacturer. Briefly, purified DNA was subjected to double-strand cleavage by DNase I in the presence of Mn2+ to generate fragments averaging 50 to 150 bp. These fragments were repaired at the 3′ ends by single deoxyribosyladenine tailing and ligated into an expression vector bearing single deoxyribosylthymine overhangs. The E. coli transformants were transferred to nitrocellulose filters, lysed, and screened with antibody. Positive clones were verified by Western blotting of E. coli lysates.
Cloning of P66 genes of other Borrelia spp.
Database searching with the DNA sequence of the P66 gene of B. burgdorferi had revealed a similarity between part of its internal segment and a deposited sequence of B. hermsii with an open reading frame of unknown function (GenBank accession no. M58430) (32). This partial sequence provided the starting point for cloning the entire gene. BglII-digested total genomic DNA from B. hermsii was separated on an agarose gel and blotted on a nylon membrane. The blotted DNA was hybridized with randomly-primed digoxigenin-labeled probe (DIG DNA labeling kit; Boehringer Mannheim). The probe DNA was generated by PCR with a pair of primers directed at positions 1 to 24 (positive-strand primer; 5′TTAGAACAATACAGCTCAGATGTC3′) and 349 to 373 (negative-strand primer; 5′CAAATTGAAGTTTATTCTCTTTTGG3′) of M58430. The homologous sequence targeted by the probe contained an internal BglII restriction site (32). The BglII-digested DNA fraction corresponding to hybridizing bands was extracted from the gel, ligated into BamHI-precut pZErO-2.1 vector (Invitrogen), and transformed into E. coli TOP 10F′ cells. Recombinant colonies were lifted on nylon filters and screened by hybridization with the same probe. After direct cloning of the DNA fragment did not succeed, inverse PCR was used. The template DNA was prepared by self-ligating 1 μg of the BglII-digested DNA fragments in 100 μl of the ligation reaction mixture. A pair of diverging primers used in the PCR targeted positions 31 to 56 (negative-strand primer; 5′CCAAAGTTTAATTCAAATGGAGTTTC3′) and 60 to 83 (positive-strand primer; 5′CTCAGGAGCAATCGGAAATTCAAC3′) of the available sequence. The DNA was amplified with the Expand Long Template PCR system (Boehringer Mannheim) under these conditions: 94°C for 10 s, 49°C for 30 s, and 68°C for 2 min for 10 cycles, followed by 94°C for 10 s, 49°C for 30 s, and 68°C for 2 min with a 20-s increment each cycle for 15 cycles, and finally by one cycle of 68°C for 7 min. The PCR product was purified by gel extraction (QIAquick gel extraction kit; Qiagen Inc., Chatsworth, Calif.), ligated into pCR2.1 vector (TA cloning kit; Invitrogen), and transformed into E. coli.
Fragments of homologous sequences of other Borrelia spp. were produced by PCR with a pair of primers directed to conserved regions at the 3′ half of the gene (see Results). The DNA template for PCR was prepared by resuspending a small amount of harvested spirochetes in water and boiling the cell suspension for 5 min. The cell debris was removed by centrifugation at 7,000 × g for 10 min. DNA was amplified with a PCR Core kit (Boehringer Mannheim) for 35 cycles under the following conditions: 94°C for 1 min, 40°C for 2 min, and 72°C for 2 min. The PCR product was ligated into pCR2.1 vector and transformed into E. coli.
Sequence analysis.
Both strands of inserts of recombinant plasmids produced in epitope mapping and gene cloning experiments were sequenced by the dideoxy chain-termination method on double-stranded templates. Sequencing was performed on a Perkin-Elmer ABI377 automatic DNA sequencer at the Biotech Diagnostic Institute (Laguna Niguel, Calif.).
For comparison of protein sequences, the following Borrelia spp. sequences were used: B. burgdorferi sensu stricto B31 P66 (accession no. X87725), B. afzelii ACAI (X87726), and B. garinii Ip90 (X87727). Multiple sequences were aligned with the aid of the Higgins-Sharp routine with default values of the MacDNASIS Pro suite (version 3.6) of programs from Hitachi Software (San Bruno, Calif.). Predictions of transmembrane or membrane-associated regions of proteins were made with the TMpredict algorithm (Bioinformatics Group, Swiss Institute for Experimental Cancer Research).
Nucleotide sequence accession number.
The following GenBank accession numbers have been assigned for the sequences described here: B. hermsii (AF016408), B. turicatae (AF016540), B. parkeri (AF016650), B. coriaceae (AF016651), and B. anserina (AF016652).
RESULTS
Production of P66-specific monoclonal antibodies.
Our previous study had shown that a surface-exposed region of P66 was frequently bound by antibodies of patients with Lyme disease (11). This region demonstrated the greatest sequence variability among B. burgdorferi, B. afzelii, and B. garinii. To further characterize this hypervariable region of the membrane protein, we raised monoclonal antibodies to P66. Previously, the mutant B313, which lacks Osp proteins A to D, had been used to generate monoclonal antibodies to the p13 outer membrane protein of the spirochetes (33). These cells also expressed P66 (10). In this study, B313 cells were used as a final booster immunization in attempt to select for antibodies against the surface-exposed portion of the P66 molecule. This approach yielded a monoclonal antibody designated H1337. Another monoclonal antibody (H914) was generated from mice primarily immunized with purified, full-length recombinant P66 protein and boosted with wild-type B31 cells.
The specificities of H1337 and H914 for different strains of Lyme disease species and other Borrelia spp. were determined by Western blotting (Table 1). H914 reacted to P66 of all Lyme disease Borrelia spp.; H1337 bound to P66 of B. burgdorferi sensu stricto strains but not to P66 of B. afzelii and B. garinii. Neither monoclonal antibody bound to the relapsing fever agent B. hermsii or to B. coriaceae.
TABLE 1.
Summary of Western blot analyses of binding of P66-specific monoclonal antibodies with different Borrelia species
| Isolate | Reactivity with monoclonal antibody:
|
|
|---|---|---|
| H1337 | H914 | |
| B. burgdorferi | ||
| B31 | ||
| Untreated | + | + |
| Proteinase K treated | − | + |
| Trypsin treated | + | + |
| N40, Sh.2, PKa | + | + |
| B. afzelii ACAI | − | + |
| B. garinii Ip90, NBS23A, NBS16, Mal02, Far02 | − | + |
| B. hermsii | − | − |
| B. coriaceae | − | − |
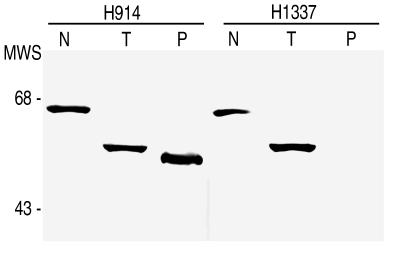
Reactivity of antibodies to untreated and protease-treated spirochetes has previously been used to identify and characterize proteins exposed on the Borrelia surface (7). To begin to localize the epitopes for H1337 and H914, we compared their Western blot reactivity with cells treated with trypsin or proteinase K and with untreated cells of B. burgdorferi B31 (Table 1; Fig. 1). Both antibodies bound to P66 in the cell lysates of untreated spirochetes. H914 also reacted with a 50-kDa polypeptide of proteinase K-treated cells. This polypeptide had previously been shown to be P66 truncated at its C-terminal end (10, 30). H1337 bound to the 52-kDa polypeptide of trypsin-treated cells but not to the 50-kDa polypeptide. These findings indicate that the epitope for H914 resides on that part of P66 not affected by proteases. In contrast, the epitope for H1337 is likely to be surface exposed and, consequently, accessible to proteases. Moreover, this epitope was retained after treatment of the cells with trypsin, a more site-specific protease than proteinase K. This suggested that the epitope for H1337 resided somewhere between a trypsin cleavage site (arginine at position 459 or lysine at position 461, 487, or 500) and the predicted transmembrane segment ending at position 458 (Fig. 2). Given the 2-kDa difference between trypsin and proteinase K fragments, we concluded that trypsin was most likely cleaving the loop at position 487.
FIG. 1.
Western blot analysis of reactivity of P66-specific monoclonal antibodies against protease-treated B. burgdorferi B31. Spirochetes were treated with buffer alone (N), trypsin (T), or proteinase K (P). After PAGE and transfer to membrane, the cell components were reacted with murine monoclonal antibody H914 or H1337. Molecular weight standards (MWS) in thousands are shown to the left.
FIG. 2.
Monoclonal antibody H1337 epitope mapping. Shown are sequences of overlapping recombinant peptides (fragments 1 to 5) representing amino acids 440 to 528 of P66 of B. burgdorferi. Origins of fragments (Frag.) 1 and 5 of P66 and their Western blot reactivities with serum specimens from patients with Lyme disease have been described elsewhere (11). The amino acids of predicted transmembrane regions flanking the putative surface-exposed loop of P66 are underlined. Lysine and arginine residues at predicted trypsin cleavage sites are indicated by double underlines. Amino acids are numbered according to the processed P66 sequence of B. burgdorferi B31 (10).
Mapping of a species-specific epitope.
The epitope for H1337 was further mapped by assessing reactivity with overlapping peptides. Secondary structure analysis of mature P66 by computer algorithm predicted that the two hydrophobic regions (positions 440 to 458 and 503 to 528) flanking the loop region (459 to 502) were predominantly alpha-helical and, thus, likely to span the outer membrane (Fig. 2). We next examined recombinant peptides that had been previously generated and included all or parts of the putative transmembrane regions (11). We also produced peptides representing different parts of the loop. The expressed peptides were assayed by Western blotting for reactivity with the species-specific monoclonal antibody H1337 or with serum from each of three North American patients with Lyme disease. These peptides were not bound by monoclonal antibody H914 or by antibodies of human controls (data not shown).
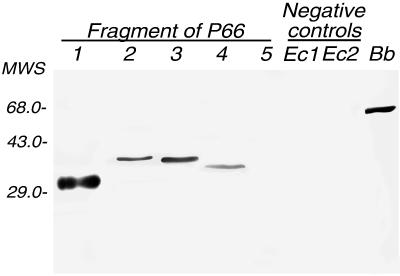
H1337 and the human serum antibodies bound, as expected, to full-length P66 but not to the 50-kDa polypeptide of proteinase K-treated cells or fragment 5 of Fig. 2 (residues 498 to 597); neither of these fragments includes the loop (reference 11 and this study). A smaller peptide, which was comprised of residues 449 to 496 (fragment 1) and contained almost the entire loop, was bound by the monoclonal antibody (Fig. 3) and by antibodies in each of the three patient serum samples (data not shown).
FIG. 3.
Western blot reactivity of monoclonal antibody H1337 to fragments 1 to 5. The E. coli lysates containing pGEX-encoded glutathione S-transferase or a product from pTOPE bearing an irrelevant insert were used for negative controls in lanes Ec1 and Ec2, respectively. Fragments 1 to 5 are described in the legend to Fig. 2. Reactivity of H1337 to P66 of B. burgdorferi B31 is shown in the rightmost lane (Bb). MWS, molecular weight standards (in thousands).
To further define H1337’s epitope, an expression library of 20- to 80-bp fragments of the last one-third of the P66 gene was created in E. coli, and the transformants were screened for reactivity with H1337. Three immunoreactive clones were recovered, and their inserts were sequenced. The three peptides encoded by these clones were amino acids 473 to 499, 474 to 502, and 479 to 490 of the mature P66; all were within the predicted surface-exposed loop of P66 (Fig. 2). They overlapped at positions 479 to 490, and thus, the epitope could be localized to these 12 amino acids. The probable trypsin cleavage site at position 487 is in this hydrophilic sequence. If it was cleaved thus, the H1337 epitope can be further restricted to TEQSSTSTK. Antibodies in serum specimens from three patients with Lyme disease bound to peptides overlapping by 23 amino acids (positions 474 to 496), which included the epitope for H1337, but the patient antibodies did not detectably bind to the smallest peptide (data not shown).
Cloning of the B. hermsii P66 gene.
Having previously demonstrated that the predicted loop of P66 varied in sequence between different Lyme disease Borrelia spp. and that species-specific antibodies were directed against this region (11), we next examined whether more distantly related Borrelia spp. also had this protein. Evidence of a homologous protein in relapsing fever Borrelia spp. was the following: (i) the finding of a 66-kDa protein that was shortened by protease treatment of intact cells (3), (ii) the reactivity of polyclonal antiserum to P66 of B. burgdorferi to a similarly sized protein in B. hermsii (30), and (iii) the presence in B. hermsii of a chromosomal sequence that was highly similar to part of the P66 gene of B. burgdorferi (32).
We first sought a homologous gene in B. hermsii. Southern blot analysis of B. hermsii genomic DNA with a 373-bp probe, which was for part of the suspected P66 gene of B. hermsii (32), identified hybridizing BglII fragments of 1.4 and 3.9 kb (data not shown). Sequence analysis of the cloned 1.4-kb fragment revealed that it included 937 nucleotides representing the 3′ half of the gene as well as several hundred nucleotides downstream. Direct cloning of the 3.9-kb fragment, which presumably included the 5′ part of the gene and its promoter, was unsuccessful.
To obtain the sequence of the gene’s 5′ end, inverse PCR with a pair of diverging primers was carried out. The expected PCR product of 3.9 kb was cloned, and that part of the insert containing the 5′ end of the gene and its flanking region was sequenced. The combined sequence of the B. hermsii gene has been deposited with accession no. AF016408 with GenBank. Positions 873 to 1250 of this sequence were identical to the fragment reported by Rosa et al. (32).
Sequence analysis revealed an open reading frame of 1,794 nucleotides that would encode a polypeptide of 598 amino acids. The open reading frame was preceded by a consensus ribosomal binding sequence (AGGAG). Upstream from the latter, the hexanucleotides TTGTTA and CAATAT were consistent in sequence and spacing with −35 and −10 elements of a ς70-type promoter and the promoter regions for the P66 genes of B. burgdorferi, B. afzelii, and B. garinii (10). The TAA stop codon was followed by a rho-independent terminator sequence. A possible signal peptidase I site was the alanine at position 21 of the deduced amino acid sequence (39). A protein so processed would be 578 amino acids and have a molecular mass of 63,744 Da. This compares to 597 amino acids and a mass of 65,808 Da for mature P66 of B. burgdorferi. The overall DNA sequence identity between the P66 genes of B. burgdorferi B31 and B. hermsii was 69%; the proteins, with 58% identity, were less similar.
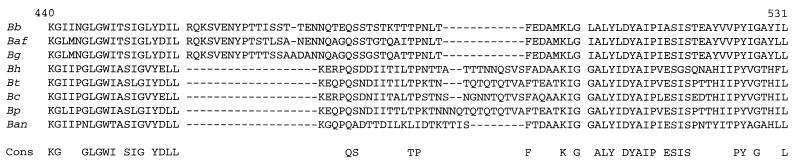
The majority of insertions or deletions accounting for the size difference between the proteins of B. burgdorferi and B. hermsii occurred in the C-terminal half of the protein and more specifically in the loop region (Fig. 4). In B. hermsii, as in B. burgdorferi, this region was flanked by two hydrophobic stretches. In the predicted loop of B. hermsii, only 9 (20%) positions were identical to the 44 amino acids comprising the loop of B. burgdorferi P66. The rest of the proteins were 61% identical.
FIG. 4.
Sequence comparison of predicted surface-exposed region and flanking transmembrane regions of P66 from Lyme disease Borrelia spp., relapsing fever Borrelia spp., B. coriaceae, and B. anserina. The surface-exposed segment is delimited from the transmembrane regions by spaces. Gaps are indicated by hyphens. Consensus (Cons) amino acids at each position were those that occurred in at least six of the eight sequences. Amino acids are numbered according to the processed P66 sequence of B. burgdorferi B31. Bb, B. burgdorferi B31; Baf, B. afzelii ACAI; Bg, B. garinii Ip90; Bh, B. hermsii; Bp, B. parkeri; Bt, B. turicatae; Bc, B. coriaceae; Ban, B. anserina.
Cloning and sequence analysis of partial P66 genes of other Borrelia spp.
The finding of a complete P66 gene in B. hermsii, an agent of relapsing fever, suggested that the other Borrelia spp. may have this gene as well. To determine if other species had P66 and, if they did, whether there was a region comparable to the surface-exposed loop, we used PCR to amplify DNA from B. turicatae, B. parkeri, B. coriaceae, and B. anserina. The primers were based on sequences that were nearly identical between the P66 genes of Lyme disease Borrelia spp. and B. hermsii: 5′CTGATTAWGGAATAGATCCWTTTGC3′ (positions 1130 to 1154) for the forward primer and 5′TTTGACTCCCATCCAAGWGAWATTGT3′ (positions 1855 to 1880 of AF016408) for the reverse primer. Products of approximately 750 bp were expected to include the loop region, hydrophobic flanking regions immediately flanking the loop, and several hundred more base pairs on either side.
The PCR products ranging in size from 730 to 755 bp were obtained, cloned in E. coli, and sequenced. The Lyme disease agents B. burgdorferi, B. afzelii, and B. garinii were 89 to 92% identical in DNA sequence for these fragments (Table 2). The relapsing fever species, B. hermsii, B. turicatae, and B. parkeri, together with B. coriaceae and B. anserina, were 76 to 84% identical to each other and 63 to 67% identical to the three members of the Lyme disease group.
TABLE 2.
Percent identity between partial DNA sequences of P66 genes of Borrelia spp.
| Sp. | B. burgdorferi B31 | B. afzelii ACAI | B. garinii Ip90 | B. hermsii | B. parkeri | B. turicatae | B. coriaceae | B. anserina |
|---|---|---|---|---|---|---|---|---|
| B. burgdorferi B31 | 100 | 90 | 89 | 65 | 67 | 66 | 66 | 65 |
| B. afzelii ACAI | 100 | 92 | 64 | 67 | 66 | 65 | 64 | |
| B. garinii Ip90 | 100 | 63 | 66 | 65 | 65 | 64 | ||
| B. hermsii | 100 | 82 | 84 | 84 | 81 | |||
| B. parkeri | 100 | 94 | 80 | 76 | ||||
| B. turicatae | 100 | 82 | 78 | |||||
| B. coriaceae | 100 | 78 | ||||||
| B. anserina | 100 |
Alignments of the partial amino acid sequences of P66 proteins of the eight Borrelia spp. are shown in Fig. 4. This figure displays the loop and the hydrophobic flanking sequences of B. burgdorferi, the corresponding sequences of the other seven species, and a consensus sequence below. The flanking sequences of each of the species were hydrophobic and alpha-helical by prediction. The predicted loops ranged in size from a low of 30 amino acids for B. anserina to a high of 45 amino acids for B. garinii. The 19 residues to the left of the loop and the 16 residues to the right of the loop were 74 to 75% identical or nearly identical across all species. In contrast, the more hydrophilic loops were identical or nearly identical in only 7 (12%) out of a possible 57 positions that included insertions or deletions. The least variable part of the loop was at its C-terminal end: three (38%) of eight positions were identical without gaps across species. In the remaining part of the loop, only QS{X}7TP was constant, with the exception of B. anserina. The epitope for H1337 partially overlapped this motif.
DISCUSSION
The present study started with the expectation but not the certainty that the P66 outer membrane protein previously identified in Lyme disease Borrelia spp. also occurs in other Borrelia spp. This was confirmed in the five other species we examined. In addition, we also further mapped what may be the only surface-exposed part of P66, an approximately 50-residue-long hydrophilic region in the C-terminal third of the protein. We have referred to this region as the surface loop of the protein.
A Borrelia species-specific antigenic determinant had previously been mapped to this loop region by using sera from patients with Lyme disease (11). In the present study, a species-specific monoclonal antibody, H1337, bound to the loop sequence, but a monoclonal antibody, H914, that was not species specific bound elsewhere. The epitope for the specific antibody was localized to 12 residues of the loop of B. burgdorferi’s P66. The experiments with trypsin indicated that one of the preferred trypsin sites in P66 in situ was contained within this 12-residue peptide (Fig. 2). From this finding, we concluded that the epitope was likely at most 9 amino acids. The study also indicated that trypsin sites predicted to be adjacent to the membrane were not accessible to this protease.
The loop region that was bound by the species-specific antibody and by patients’ antibodies is the most variable part of the protein. Across species there was considerable sequence and size heterogeneity in the predicted loops. This agrees with the observation that insertions or deletions in protein sequence commonly occur in loop regions (28). The flanking hydrophobic regions were more conserved in sequence within Borrelia spp., as would be expected for transmembrane regions (20). The loop also exhibits segmental mobility (10), a predicted secondary structure characteristic of antigenic determinants of proteins (40). Notably, TEQSSTS, by prediction the most mobile segment of B. burgdorferi P66 (10), is contained within the epitope for monoclonal antibody H1337.
The diversity among P66 sequences of different Borrelia spp. was most pronounced in the loop region. Overall, the amount of variation in P66 amino acid sequences falls between that of flagellin and the Vmp and OspC proteins of Borrelia spp. P66 proteins of Borrelia spp. are approximately 60% identical. Borrelia flagellin sequences are 94 to 95% identical across species (26). A large Vmp protein of B. hermsii and its closest relative in B. burgdorferi are about 40% identical (42). Small Vmp proteins and the homologous OspC proteins are 40 to 50% identical (13, 15, 25). Sequence polymorphism of the antigenic determinants exposed on the surface of borreliae may be a result of selection by the host’s immune system or for certain environments (14, 29). Unlike the entirely surface-exposed Vmp and OspC molecules, only the loop of P66 is likely to be subjected to such selective pressure. The remainder of the protein may be as inaccessible as periplasmic flagella to circulating antibodies.
The function of P66 is not known. Embedded in liposomes, this protein exhibits channel conductance, an activity consistent with bacterial porins (38). Yet P66 is not typical of a bacterial porin with respect to size, predicted secondary structure, or tendency to oligomerize (22). Other spirochetal outer membrane proteins with porin activity oligomerize and are predicted to span the membrane as beta-sheets, rather than the alpha-helix that P66 appears to use (19, 37). In this sense, P66 is novel.
If it is true that the loop of P66 is the only point of interaction of this protein with the environment, perhaps some clue to P66 function can be found in further study of this region. If the loop region is under selection by both the immune system and competition for niches in the host, then the loop would likely be variable but also demonstrate some conservation in structure. An example of the latter may be the QS{X}7TP motif present in the loop of all species studied here except B. anserina, a bird pathogen. If any function of the loop is associated with this motif, it may be specific for spirochete-mammalian interactions. The only other strains to date that differ from this motif sequence are bird-associated strains of B. garinii. Four of five B. garinii strains isolated from the seabird tick Ixodes uriae had QKS{X}7TP instead of QS{X}7TP (11).
ACKNOWLEDGMENTS
We thank Carol Carter and Jill Schurr for expert technical assistance and Ariadna Sadziene for advice and her early contributions to this study.
This work was supported by NIH grants AI32748 and AI24424 to A.G.B. and Medical Research Council grant 07922 to S.B.
REFERENCES
- 1.Åsbrink E, Hovmark A, Hederstedt B. The spirochetal etiology of acrodermatitis chronica atrophicans Herxheimer. Acta Dermato-Venereol. 1984;64:506–512. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1993. [Google Scholar]
- 3.Barbour A. Clonal polymorphisms of surface antigens in a relapsing fever Borrelia spp. In: Jackson G, editor. Pathogenesis of bacterial infection. Heidelberg, Germany: Springer-Verlag; 1985. pp. 235–245. [Google Scholar]
- 4.Barbour A G. Immunochemical analysis of Lyme disease spirochetes. Yale J Biol Med. 1984;57:581–586. [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour A G, Tessier S L, Hayes S F. Variation in a major surface protein of Lyme disease spirochetes. Infect Immun. 1984;4:94–100. doi: 10.1128/iai.45.1.94-100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbour A G, Tessier S L, Todd W J. Lyme disease spirochetes and Ixodes tick spirochetes share a common surface antigen determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergström S, Olsen B, Burman N, Gothefors L, Jaenson T G T, Jonsson M, Mejlon H A. Molecular characterization of Borrelia burgdorferi isolated from Ixodes ricinus in Northern Sweden. Scand J Infect Dis. 1992;24:181–188. doi: 10.3109/00365549209052610. [DOI] [PubMed] [Google Scholar]
- 10.Bunikis J, Noppa L, Bergström S. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol Lett. 1995;131:139–145. doi: 10.1111/j.1574-6968.1995.tb07768.x. [DOI] [PubMed] [Google Scholar]
- 11.Bunikis J, Noppa L, Ostberg Y, Barbour A G, Bergstrom S. Surface exposure and species specificity of an immunoreactive domain of a 66-kilodalton outer membrane protein (P66) of the Borrelia spp. that cause Lyme disease. Infect Immun. 1996;64:5111–5116. doi: 10.1128/iai.64.12.5111-5116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunikis J, Olsen B, Fingerle F, Bonnedahl J, Wilske B, Bergstrom S. Molecular polymorphism of the Lyme disease agent Borrelia garinii in northern Europe is influenced by a novel enzootic Borrelia focus in the North Atlantic. J Clin Microbiol. 1996;34:364–368. doi: 10.1128/jcm.34.2.364-368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadavid D, Pennington P M, Kerentseva T A, Bergström S, Barbour A G. Immunologic and genetic analyses of VmpA of a neurotropic strain of Borrelia turicatae. Infect Immun. 1997;65:3352–3360. doi: 10.1128/iai.65.8.3352-3360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadavid D, Thomas D D, Crawley R, Barbour A G. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J Exp Med. 1994;179:631–642. doi: 10.1084/jem.179.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter C J, Bergström S, Norris S J, Barbour A G. A family of surface-exposed proteins of 20 kilodaltons in the genus Borrelia. Infect Immun. 1994;62:2792–2799. doi: 10.1128/iai.62.7.2792-2799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dressler F, Whalen J A, Reinhardt B N, Steere A C. Western blotting in the serodiagnosis of Lyme disease. J Infect Dis. 1993;167:392–400. doi: 10.1093/infdis/167.2.392. [DOI] [PubMed] [Google Scholar]
- 17.Ferdows M S, Serwer P, Barbour A G. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J Bacteriol. 1995;178:793–800. doi: 10.1128/jb.178.3.793-800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fikrig E, Barthold S W, Kantor F S, Flavell R A. Protection of mice against the Lyme disease agent by immunizing with recombinant OspA. Science. 1990;250:553–556. doi: 10.1126/science.2237407. [DOI] [PubMed] [Google Scholar]
- 19.Haake D A, Champion C I, Martinich C, Shang E S, Blanco D R, Miller J N, Lovett M A. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J Bacteriol. 1993;175:4225–4234. doi: 10.1128/jb.175.13.4225-4234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haltia T, Freire E. Forces and factors that contribute to the structural stability of membrane proteins. Biochim Biophys Acta. 1995;1228:1–27. doi: 10.1016/0005-2728(94)00161-w. [DOI] [PubMed] [Google Scholar]
- 21.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;38:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jap B K, Walian P J. Structure and functional mechanism of porins. Physiol Rev. 1996;76:1073–1088. doi: 10.1152/physrev.1996.76.4.1073. [DOI] [PubMed] [Google Scholar]
- 23.Kryuchechnikov V N, Korenberg E I, Scherbakov S V, Kovalevsky Y V, Levin M L. Identification of Borrelia isolated in the USSR from Ixodes persulcatus schulze ticks. J Microbiol Epidemiol. 1988;12:41–44. [PubMed] [Google Scholar]
- 24.Lane R S, Burgdorfer W, Hayes S F, Barbour A G. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus: a possible agent of epizootic bovine abortion. Science. 1985;230:85–87. doi: 10.1126/science.3898367. [DOI] [PubMed] [Google Scholar]
- 25.Margolis N, Hogan D, Cieplak W, Schwan T, Rosa P. Homology between Borrelia burgdorferi OspC and members of the family of Borrelia hermsii variable major proteins. Gene. 1994;143:105–110. doi: 10.1016/0378-1119(94)90613-0. [DOI] [PubMed] [Google Scholar]
- 26.Noppa L, Barbour A G, Sadziene A, Bergström S. The expression of the flagellin gene in Borrelia is controlled by an alternate sigma factor. Microbiology. 1995;141:85–93. doi: 10.1099/00221287-141-1-85. [DOI] [PubMed] [Google Scholar]
- 27.Oi V T, Herzenberg L A. Immunoglobulin-producing hybrid cell lines. In: Mishell B B, Shiigi S M, editors. Selected methods in cellular immunology. W. H. San Francisco, Calif: Freeman and Co.; 1980. pp. 351–372. [Google Scholar]
- 28.Pascarella S, Argos P. Analysis of insertions/deletions in protein structures. J Mol Biol. 1992;224:461–471. doi: 10.1016/0022-2836(92)91008-d. [DOI] [PubMed] [Google Scholar]
- 29.Pennington P, Allred C D, Cadavid D, Norris S, Barbour A G. Arthritis severity and spirochete burden are determined by serotype in the Borrelia turicatae-mouse model of Lyme disease. Infect Immun. 1997;65:285–292. doi: 10.1128/iai.65.1.285-292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Probert W S, Allsup K M, LeFebre R B. Identification and characterization of a surface-exposed 66-kilodalton protein from Borrelia burgdorferi. Infect Immun. 1995;63:1933–1939. doi: 10.1128/iai.63.5.1933-1939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radolf J D. Role of outer membrane architecture in immune evasion by Treponema pallidum and Borrelia burgdorferi. Trends Microbiol. 1994;9:307–311. doi: 10.1016/0966-842x(94)90446-4. [DOI] [PubMed] [Google Scholar]
- 32.Rosa P A, Hogan D, Schwan T G. Polymerase chain reaction analyses identify two distinct classes of Borrelia burgdorferi. J Clin Microbiol. 1991;29:524–532. doi: 10.1128/jcm.29.3.524-532.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadziene A, Thomas D D, Barbour A G. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect Immun. 1995;63:1573–1580. doi: 10.1128/iai.63.4.1573-1580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadziene A, Thompson P A, Barbour A G. In vitro inhibition of Borrelia burgdorferi growth by antibodies. J Infect Dis. 1993;167:165–172. doi: 10.1093/infdis/167.1.165. [DOI] [PubMed] [Google Scholar]
- 35.Sadziene A, Wilske B, Ferdows M S, Barbour A G. The cryptic OspC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skare J T, Champion C I, Mirzabekov T A, Shang E S, Blanco D R, Erdjument-Bromage H, Tempst P, Kagan B L, Miller J N, Lovett M A. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J Bacteriol. 1996;178:4909–4918. doi: 10.1128/jb.178.16.4909-4918.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skare J T, Mirzabekov T A, Shang E S, Blanco D R, Erdjument-Bromage H, Bunikis J, Bergstrom S, Tempst P, Kagan B L, Miller J N, Lovett M A. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect Immun. 1997;65:3654–3661. doi: 10.1128/iai.65.9.3654-3661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Heijne G. Patterns of amino acids near signal sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 40.Westhof E, Altschuh D, Moras D, Bloomer A C, Mondragon A, Klug A, Van Regenmortel M H V. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature. 1984;311:123–126. doi: 10.1038/311123a0. [DOI] [PubMed] [Google Scholar]
- 41.Wilske B, Preac-Mursic V, Schierz G, Busch K V. Immunochemical and immunological analysis of European Borrelia burgdorferi strains. Zentralbl Bakteriol Hyg A. 1986;263:92–102. doi: 10.1016/s0176-6724(86)80108-0. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J-R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]