Abstract
Signal transduction in the archaeon Halobacterium salinarum is mediated by three distinct subfamilies of transducer proteins. Here we report the complete htrVIII gene sequence and present analysis of the encoded primary structure and its functional features. HtrVIII is a 642-amino-acid protein and belongs to halobacterial transducer subfamily B. At the N terminus, the protein contains six transmembrane segments that exhibit homology to the heme-binding sites of the eukaryotic cytochrome c oxidase. The C-terminal domain has high homology with the eubacterial methyl-accepting chemotaxis protein. The HtrVIII protein mediates aerotaxis: a strain with a deletion of the htrVIII gene loses aerotaxis, while an overproducing strain exhibits stronger aerotaxis. We also demonstrate that HtrVIII is a methyl-accepting protein and demethylates during the aerotaxis response.
An aerotactic response in the archaeon Halobacterium salinarum has previously been characterized as an accumulation of motile cells around an air bubble (4, 30). In H. salinarum, the aerotactic response was shown to be methylation dependent (21). Inhibition of methylation in H. salinarum by depletion of the methyl donor S-adenosylmethionine resulted in defective aerotaxis, providing further evidence that methylation is involved in aerotaxis in H. salinarum (21). Recently, a transducer for aerotaxis and other energy-dependent responses was identified in Escherichia coli (3, 24). Membranes with overexpressed Aer protein contained high levels of noncovalently associated flavin adenine dinucleotide (FAD) (3).
Signal transduction in the archaeon H. salinarum is mediated by a family of 13 putative transducers (28, 35). On the basis of hydropathy plot analysis and protein fractionation by ultracentrifugation, it was shown that this family contains both soluble and membrane-bound putative transducer proteins. There are three distinct subfamilies of these proteins: subfamily A consists of eubacterial chemotaxis-type transducers, containing periplasmic and cytoplasmic domains connected by two transmembrane segments; subfamily B contains transducers with two or more transmembrane segments and lacking a periplasmic domain, such as the SRI transducer HtrI (34); and subfamily C contains soluble transducer proteins (7, 35). Here, we report the full primary structure of HtrVIII, a member of subfamily B, and present experimental evidence that it is an aerotaxis transducer in H. salinarum.
MATERIALS AND METHODS
Strains and plasmids.
Halobacterial strain Flx15 (29), which lacks bacteriorhodopsin and halorhodopsin, was used for the identification of the htrVIII gene. After appropriate restriction enzyme digestion, halobacterial DNA fragments were cloned into the commercial vector pDELTA1 (Gibco BRL, Gaithersburg, Md.). H. salinarum Pho81, which lacks bacteriorhodopsin, halorhodopsin, SRI, and SRII (31), was used for the deletion mutant construction. E. coli JM109 was used in routine cloning experiments. Halobacterial shuttle vector pMDS20 was a generous gift of M. Dyall-Smith (University of Melbourne, Melbourne, Australia).
Media and growth conditions.
H. salinarum Flx15 and Pho81 were grown aerobically in tryptone medium at 37°C in the dark. E. coli JM109 cells, containing recombinant plasmids, were grown overnight in Luria-Bertani medium supplemented with the appropriate antibiotic.
Chemicals and electrophoresis reagents.
All chemicals were reagent grade. Novobiocin was purchased from Sigma, St. Louis, Mo.
DNA isolation, restriction analysis, and cloning.
The 1.8-kbp PstI and 6.4-kbp BamHI genomic fragments were used for the identification and cloning of the htrVIII gene. Southern hybridization and Western blotting analysis with antibody HC23 were performed as described previously (35). Standard molecular biological methods were followed, if not otherwise indicated, as described in reference 35.
DNA sequencing and data analysis.
Double-stranded DNA was sequenced by the chain termination method with a Sequenase kit (United States Biochemicals). We used four sequencing strategies: the bidirectional deletion factory method with the pDELTA system (Gibco BRL), primer walking, the Erase-a-Base deletion system (Promega, Madison, Wis.), and automatic DNA sequencing (Applied Biosystems model 373; Perkin-Elmer).
Secondary-structure analysis.
A prediction of the secondary structures of the halobacterial transducers was generated by a consensus among computer prediction algorithms PHD sec (25–27), Predator (9, 10), and four other prediction methods courtesy of SOPMA (8, 12–14, 20) on the Internet. The Kyte-Doolittle hydrophobicity plot was used to determine the approximate borders of the transmembrane regions. Generation of the prediction consensus was done with Microsoft Excel and was based on the individual predictions with the highest frequencies. All diagrams illustrating the predicted secondary structures were created with Microsoft PowerPoint.
Isolation of the deletion mutant htrVIII gene.
The 6.4-kbp BamHI-BamHI fragment containing the full-length htrVIII gene was subcloned into vector pGEM-7Zf+ (Promega) to yield recombinant plasmid phtrVIIIG. The halobacterial shuttle vector pMDS20 (17) was digested with PstI and SmaI restriction endonucleases to produce a 2.9-kbp fragment containing the gyrB gene, conferring resistance to novobiocin. The resultant 2.9-kbp fragment was cloned into PstI sites of phtrVIIIG, replacing most of the coding sequence of the htrVIII gene (from bp 56 to 1864) with a SmaI-PstI adapter. The final plasmid (phtrVIIIGnov) was transformed into halobacterial strain Pho81 according to a standard polyethylene glycol-mediated protocol (19). Transformants were grown on 2% agar plates containing novobiocin (2 μg/ml). Primary screening of the deletion mutants was performed by two independent methods: Southern hybridization with a 27-mer conserved oligonucleotide probe and immunoblot detection with the antibody HC23 (35). This polyclonal antibody was raised against a 23-amino-acid peptide representing the most highly conserved region of all the transducers. We used a synergy system to synthesize a 23-amino-acid multiply antigenic peptide as an antigen to generate polyclonal antibody HC23, described in detail by Zhang et al. (35). For additional control of the deletion construction, we performed PCR with a set of specific primers to detect the presence of the gyrB cassette in the genome.
Construction of an HtrVIII overexpression strain.
The 6.4-kbp BamHI-BamHI fragment containing the full-length htrVIII gene was subcloned into the multicopy halobacterial shuttle vector pMDS20. The resultant expression vector, phtrVIIIGwt2, which contained the htrVIII gene under the control of its own promoter, was transformed into strain Pho81. Transformants were grown on 2% agar plates containing novobiocin (2 μg/ml). The htrVIII++/htrVIII+ overexpression strains were screened by immunoblotting with our HC23 polyclonal antibody. The htrVIII++/htrVIII-1 strain was chosen for the high-level expression of htrVIII.
Radiolabeling with [methyl-3H]methionine, electrophoresis, immunoblotting, and fluorography.
Radiolabeling experiments were performed according to the method of Alam and Hazelbauer (1). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed essentially by the procedure of Laemmli (18), with modifications described by Randall and Hardy (23). Immunoblotting and fluorography were performed according to the method of Alam and Hazelbauer (1). The demethylation in vivo flow assay was performed according to the method of Lindbeck et al. (21). We used the scintillation cocktail Scinti-Verse BD and polyethylene scintillation vials (Fisher Scientific, Santa Clara, Calif.).
Aerotaxis assay.
Highly motile halobacterial cells were grown to an optical density at 660 nm of 0.6 to 0.7. Cells were washed and resuspended in basal salt medium (25). Microslide capillaries (VitroCom, Mountain Lakes, N.J.) were filled halfway with washed cells. Both ends of the capillary were sealed and the capillary was placed for 5 to 6 h on a microscope stage that had been prewarmed to 37°C. The distribution of halobacterial cells close to the surface of an air bubble was recorded by time-lapse dark-field microscopy.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF031641).
RESULTS
Cloning, sequencing of the htrVIII gene, and analysis of the deduced amino acid sequence.
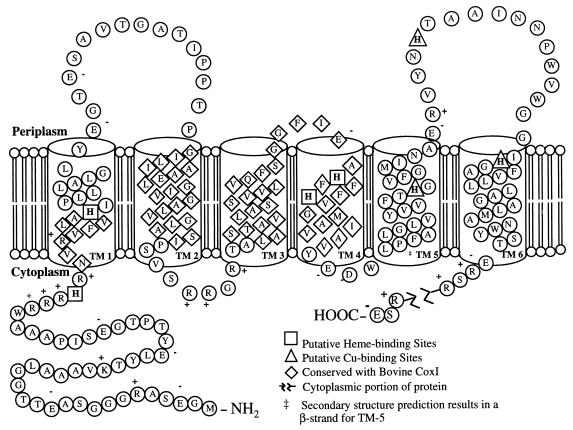
The htrVIII gene encodes a 642-amino-acid protein. A portion of the gene was identified in the 1.8-kbp PstI fragment, and the full-length gene was identified in the 6.4-kbp BamHI genomic fragment. The calculated molecular mass of the HtrVIII protein is 67.1 kDa, and it has a pI of 4.1. The newly identified transducer is a member of subfamily B (6, 35). It has six putative transmembrane segments (residues 1 to 209) (Fig. 1) that have homology with eukaryotic mitochondrial cytochrome c oxidase subunit I (COXI). Transmembrane 4 (TM4) of HtrVIII has 50% identity with helix X of bovine COXI, and TM5 and TM6 have 36% identity with helices VII and VIII (33). HtrVIII residues 386 to 550 are about 31% identical and residues 459 to 492 are 71% identical to the cytoplasmic signaling domains of eubacterial methyl-accepting chemotaxis proteins (12).
FIG. 1.
Schematic representation of the transmembrane portion of HtrVIII. The putative transmembrane segments predicted as α-helices lie within the cylinders. Diamond-shaped residues have high homology with bovine COXI. The extensive cytoplasmic portion of the protein has been left out and is designated by the jagged break in the chain.
Secondary-structure prediction for the HtrVIII protein.
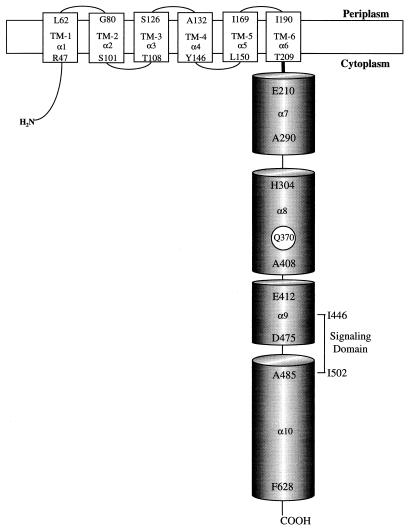
Generation of the secondary structure from the primary sequence was accomplished with alignments and computer prediction algorithms as described in the Materials and Methods section. The algorithms predicted the presence of four α-helices in the cytoplasmic domain of HtrVIII (Fig. 2) instead of the predicted six helices in the eubacterial chemotransducers (16).
FIG. 2.
Schematic representation of the secondary-structure prediction of the cytoplasmic domain of HtrVIII. The six transmembrane portions are represented as boxes, the cytoplasmic α-helices are represented as cylinders, and the linker regions are represented as broad lines.
Isolation of the htrVIII deletion strain gene.
The htrVIII deletion mutant was isolated by a standard gene knockout technique using the gyrB cassette. Southern hybridization with a 27-mer oligonucleotide probe showed that the 1.8-kbp PstI htrVIII-specific fragment is missing in the ΔhtrVIII-24 strain (data not shown). An immunoblot with HC23 antibody of total cell lysate of Pho81 and the ΔhtrVIII-24 strain indicated that one of the cross-reacting bands is absent in the deletion strain (Fig. 3A). In sodium dodecyl sulfate-polyacrylamide gel electrophoresis, recombinant HtrVIII expressed in E. coli showed a mobility similar to that of the native protein from the halobacterial strain Pho81 (5).
FIG. 3.
Immunoblot and fluorograph of Pho81, the htrVIII++/htrVIII+-1 strain, and the ΔhtrVIII-24 strain. (A) Western blotting analysis with HC23 antibody. Lanes: 1, Pho81; 2, the ΔhtrVIII-24 strain; 3, the htrVIII++/htrVIII+-1 strain. (B) Electrophoretic analysis of methyl-3H-labeled transducers. Lanes: 1, Pho81; 2, the ΔhtrVIII-24 strain; 3, the htrVIII++/htrVIII+-1 strain. The molecular mass is in kilodaltons. Arrows indicate the position of HtrVIII.
HtrVIII is a methyl-accepting protein.
The expression level of HtrVIII in strain Pho81 is relatively low compared to that in the other halobacterial transducers (Fig. 3A). To demonstrate that HtrVIII is indeed a methyl-accepting protein, we needed to express enough of HtrVIII for it to be distinctly visible in fluorography. Thus, we constructed an overexpression htrVIII+/htrVIII++-1 strain based on the multicopy shuttle vector pMDS20 (13). The immunoblot clearly shows that the expression level of HtrVIII in the htrVIII+/htrVIII++-1 strain is higher than in the Pho81 strain (Fig. 3A). Fluorography of radiolabeled htrVIII+ (strain Pho81), ΔhtrVIII-24, and overexpression strains demonstrates that the specific radiolabeled band is missing in the deletion strain. The same band is present in wild-type and overexpression strains, and its intensity corresponds to the expression levels of the HtrVIII protein in those strains (Fig. 3B).
HtrVIII is involved in aerotaxis.
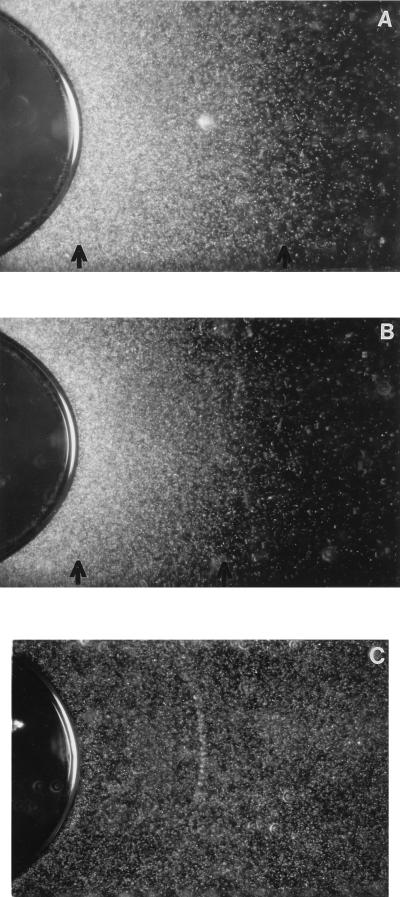
The ability of the cells to sense an oxygen gradient and thus to concentrate around the trapped air in the flat microcapillary was analyzed by time-lapse dark-field microscopy. Wild-type halobacterial cells congregated around the interface between air and cell suspensions (Fig. 4A). In contrast, cells of the ΔhtrVIII-24 strain failed to gather around the air boundary (Fig. 4C). The aerotaxis band of the overexpression strain after 5 to 6 h at 37°C is much broader than that of the wild-type strain (Fig. 4A and B). We postulate that this difference in aerotaxis response is due to the multicopy plasmid bearing the wild-type htrVIII gene. The swimming speeds of the deletion and overexpression strains are comparable with that of the wild-type strain.
FIG. 4.
Aerotaxis response (seen as white ring designated by two arrows in the dark background) in the air bubble assay. (A) htrVIII++/htrVIII+-1 overexpression strain; (B) wild-type Pho81; (C) ΔhtrVIII-24 deletion strain.
HtrVIII is involved in demethylation during the adaptation to the aerotaxis response.
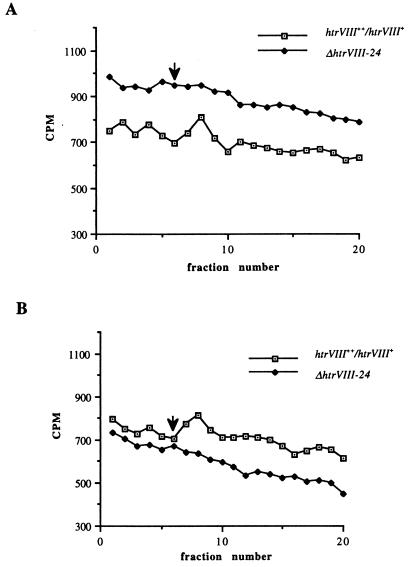
Halobacterial cells transiently release methanol, which is an indication of carboxymethyl group turnover by chemostimuli and photostimuli (2, 30). The effects of sensory stimuli on the rate of release of methanol in H. salinarum do not exhibit the same symmetry as in E. coli; both positive and negative stimuli result in an increased rate of methanol release. Lindbeck et al. showed transient increases in the methanol production by H. salinarum cells in response to a step up or down in the oxygen concentration (21). Because fluorography showed that HtrVIII is indeed a methyl-accepting protein, transient increases in methanol release should not be seen in the deletion strain (the ΔhtrVIII-24 strain) in response to the addition or removal of oxygen in a flow assay. To test our hypothesis, we studied the methylesterase activity in the deletion and overexpression strains. Indeed, unlike the overexpression strain, the ΔhtrVIII-24 strain showed no transient changes in the methanol production following a step up or down in the oxygen concentration (Fig. 5). These results further confirmed that HtrVIII is a methyl-accepting protein and that covalent modifications of putative methylation residues are involved in aerotaxis.
FIG. 5.
Aerotaxis-induced changes in the rate of release of [3H]methyl groups under conditions of nonradioactive chase of the htrVIII++/htrVIII+-1 overexpression strain and the ΔhtrVIII-24 deletion mutant. Arrows indicate switch from N2-equilibrated buffer to O2-equilibrated buffer (A) and from O2 to N2 buffer (B).
DISCUSSION
HtrVIII was originally cloned as a putative transducer protein based on its homology in the highly conserved signaling domain with other methyl-accepting proteins (35). The position −22 to −28 bp from the start codon of htrVIII has a strong homology with the putative archaeal promoter element (32).
The BLAST sequence homology search of the six transmembrane segments in HtrVIII revealed that this region has high homology with eukaryotic COXI (Fig. 1). The C-terminal domain has high homology with the eubacterial methyl-accepting chemotaxis protein. Cytochrome c oxidase is a heme/copper protein that catalyzes the four-electron reduction of dioxygen to water. Therefore, we suspected that HtrVIII functions as an oxygen-sensing transducer protein.
All three methylation-dependent taxis responses (chemo-, photo-, and aerotaxis) described so far for H. salinarum exhibit similar profiles of methanol release. Methylation is not involved in E. coli aerotaxis (22). The putative oxygen sensor DcrH from Desulfovibrio vulgaris was shown to be methylated, but no physiological data regarding its involvement in aerotaxis has been reported so far (11).
These experimental results provide the answer to the first key question, i.e., whether HtrVIII is an aerotaxis transducer in H. salinarum. The next logical question is: what is the active site(s) that acts as the oxygen-sensing center? Several types of oxygen-sensing proteins are now known, including FixL (Rhizobium meliloti) and the recently discovered aerotaxis protein Aer in E. coli. A functional distinction between these two proteins is the nature of their oxygen-sensing sites. In the case of FixL, the active site is a heme chromophore, while in Aer it is a putative FAD. In the case of Aer, the oxygen sensing is believed to be indirect, with electron transfer to the FAD occurring via the respiratory chain, while FixL directly binds dioxygen to the heme iron (15).
Given the primary sequence of HtrVIII, we cannot rule out the possibility of any sort of prosthetic group, but the presence of histidine residues in HtrVIII (Fig. 1) is consistent with metal binding. Cytochrome c oxidase binds oxygen via a five-coordinate heme a chromophore located in subunit I. The conserved histidine residues are diagnostic for the oxidase family. The corresponding residues His-376 and His-378 of bovine COXI are conserved in HtrVIII; they are His-133 and His-135. The corresponding position of His-290 of COXI is His-163 in HtrVIII, and His-291 is replaced by Gly-164 in HtrVIII. It is tempting to speculate that HtrVIII is a heme protein (based upon the homology between HtrVIII TM4 and subunit I helix X of COXI). An alternative hypothesis is that HtrVIII contains a multinuclear copper center similar to the binuclear copper centers of hemocyanins (also oxygen-binding proteins) or a FAD as in the aerotaxis transducer of E. coli (3, 24). In the case of the hemocyanin binuclear copper sites, six histidines are required.
This report describes the first example of a methyl-accepting aerotaxis protein in the archaea. Further studies are under way to determine the nature of the oxygen-binding chromophore and the molecular mechanism of signal transduction by HtrVIII in the archaeon H. salinarum.
ACKNOWLEDGMENTS
We thank R. Berger and P. Patek for critical reading and discussion of the manuscript. We also thank Weisheng Zhang for excellent technical support in the initial phase of this work and M. Dyall-Smith, University of Melbourne, who kindly provided us with shuttle vector pMDS20.
This work was supported by National Science Foundation grant MCB-9600860 and National Institutes of Health Shannon Award R55 GM53149-01A1 to M.A. T.F. is the recipient of a Minority Access to Research Careers Honors Program (MARC) award.
REFERENCES
- 1.Alam M, Hazelbauer G L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991;173:5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam M, Lebert M, Oesterhelt D, Hazelbauer G L. Methyl accepting taxis proteins in Halobacterium halobium. EMBO J. 1989;8:631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikov S I, Skulachev V P. Mechanism of phototaxis and aerotaxis in Halobacterium halobium. FEBS Lett. 1989;243:303–306. [Google Scholar]
- 5.Brooun, A., and M. Alam. Unpublished data.
- 6.Brooun, A., F. Villablanca, T. Freitas, and M. Alam. Unpublished data.
- 7.Brooun A, Zhang W, Alam M. Primary structure and functional analysis of the soluble transducer protein HtrXI from the archaeon Halobacterium salinarium. J Bacteriol. 1997;179:2963–2968. doi: 10.1128/jb.179.9.2963-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 9.Frishman D, Argos P. Incorporation of long-distance interactions into a secondary structure prediction algorithm. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 10.Frishman D, Argos P. 75% accuracy with a new approach in utilizing related sequences for protein secondary structure prediction. Proteins Struct Funct Genet. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Fu R, Wall J D, Voordouw G. DerA, a c-type heme-containing methyl-accepting protein from Desulfovibrio vulgaris Hildenborough, senses the oxygen concentration or redox potential of the environment. J Bacteriol. 1994;176:344–350. doi: 10.1128/jb.176.2.344-350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 13.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 14.Gibrat J F, Garnier J, Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 15.Gilles G M, Gonzales G, Perutz M F, Kriger L, Marden M C, Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autooxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 16.Hazelbauer G L. Bacterial chemoreceptors. Curr Opin Struct Biol. 1992;2:505–510. [Google Scholar]
- 17.Holmes M, Pfiefer F, Dyall-Smith M. Improved shuttle vectors for Haloferax volcanii including a dual resistance plasmid. Gene. 1994;146:117–121. doi: 10.1016/0378-1119(94)90844-3. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lam W L, Doolittle W F. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc Natl Acad Sci USA. 1989;86:5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin J M, Robson B, Garnier J. An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 21.Lindbeck J C, Goulbourne E A, Johnson M S, Taylor B L. Aerotaxis in Halobacterium salinarium is methylation-dependent. Microbiology. 1995;141:2945–2953. doi: 10.1099/13500872-141-11-2945. [DOI] [PubMed] [Google Scholar]
- 22.Niwano M, Taylor B L. Novel sensory adaptation mechanism in bacterial chemotaxis to oxygen and phosphotransferase substrates. Proc Natl Acad Sci USA. 1982;79:11–15. doi: 10.1073/pnas.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randall L L, Hardy S J S. Synthesis of exported proteins by membrane-bound polysomes from E. coli. Eur J Biochem. 1977;75:43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- 24.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer and the serine chemoreceptor Tsr independently sense intracellular energy levels and transducer oxygen, redox, and energy signal for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins Struct Funct Genet. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 26.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 27.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 28.Rudolph J, Nordmann B, Storch K-F, Grunberg H, Oesterhelt D. A family of halobacterial transducer proteins. FEMS Lett. 1996;139:161–168. doi: 10.1111/j.1574-6968.1996.tb08197.x. [DOI] [PubMed] [Google Scholar]
- 29.Spudich E N, Spudich J L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other signal transduction mutants of Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79:4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoeckenius W, Wolff E K, Hess B. A rapid population method for action spectra applied to Halobacterium halobium. J Bacteriol. 1988;170:2790–2795. doi: 10.1128/jb.170.6.2790-2795.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundberg S A, Bogomolni R A, Spudich J L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985;164:282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomm M, Which G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of the higher eukaryotes. Nucleic Acids Res. 1988;102:117–122. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of 13-subunit oxidized cytochrome c oxidase of 2.8 Å. Science. 1995;269:1069–1074. [Google Scholar]
- 34.Yao V J, Spudich J L. Primary structure of an archaebacterial transducer, a methyl-accepting taxis protein associated with sensory rhodopsin I. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Brooun A, MacCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]