Abstract
Streptomyces reticuli produces a 35-kDa cellulose-binding protein (AbpS) which interacts strongly with crystalline forms of cellulose (Avicel, bacterial microcrystalline cellulose, and tunicin cellulose); other polysaccharides are recognized on weakly (chitin and Valonia cellulose) or not at all (xylan, starch, and agar). The protein could be purified to homogeneity due to its affinity to Avicel. After we sequenced internal peptides, the corresponding gene was identified by reverse genetics. In vivo labelling experiments with fluorescein isothiocyanate (FITC), FITC-labelled secondary antibodies, or proteinase K treatment revealed that the anchored AbpS protrudes from the surfaces of the hyphae. When we investigated the hydrophobicity of the deduced AbpS, one putative transmembrane segment was predicted at the C terminus. By analysis of the secondary structure, a large centrally located α-helix which has weak homology to the tropomyosin protein family was found. Physiological studies showed that AbpS is synthesized during the late logarithmic phase, independently of the carbon source.
Streptomycetes are gram-positive aerobic soil bacteria. Numbering about 106 to 108 germs per gram of soil, they make up one of the dominant groups, owing to their optimum adaptation to their natural environment (1). The streptomycetes exhibit a differentiated growth cycle, starting with the germination of spores and proceeding to the formation of substrate mycelia. The cycle closes with the formation of aerial hyphae, in which spores are present (17). These spores are resistant to heat, dryness, and cold, and their survival is guaranteed in periods when no nutrients are available. In addition, streptomycetes are able to produce a wide range of pigments, antibiotics, and fungicides to inhibit the growth of competing organisms (4). Biopolymers like chitin, starch, xylan, and cellulose, which constitute abundant carbon sources for organisms in the soil, are very efficiently hydrolyzed by streptomycetes, due to the action of corresponding catabolic enzymes (23). The synthesis of these extracellular proteins is strictly and specifically regulated. In order to survive, the organisms constantly have to monitor the external conditions and to adjust their structures, physiologies, and behaviors accordingly. To this end, bacteria have devised several different sophisticated signalling systems to elicit a variety of adaptive responses to their environment. Proteins with two-component systems make up the major group of such signal transduction proteins (21). They consist of a sensor (mostly membrane integrated) and a cytoplasmically located response regulator, the communication of which is mediated by de- and phosphorylation. These modules are widespread among bacteria; they handle a broad range of signalling tasks (e.g., host detection and invasion, leading to symbiosis or pathogenesis; metabolic adaptation to changes in carbon, nitrogen, and phosphate sources; responses to osmolarity and stress; and chemotaxis) (for a review, see reference 22). Another signal system was recently discovered in Escherichia coli. It regulates the ferric citrate uptake systems, with the ferric citrate functioning as an inducer. FecR, a protein located in the periplasm and integrated into the inner membrane, seems to transmit a signal from the outer-membrane-pore-forming protein FecA to the cytoplasmic transcriptional activator FecI in the presence of the energy-transmitting Ton system (for a review, see reference 5). Studying the virulence of bacteria, several research groups could show that the contact of bacteria with eukaryotic host cells may trigger the expression of corresponding bacterial genes. Contact may produce an additional signal. There are already precedents for this in environmental bacteriology. In Pseudomonas aeruginosa biofilms, transcription of algC, a gene required for the synthesis of alginate, is specifically activated upon attachment to a glass surface and Teflon (7). A lateral flagellum-encoding gene (laf) from Vibrio parahaemolyticus is activated only when the strain is grown on agar (3). Vandevivere and Kirchman (37) demonstrated surface activation of exopolysaccharide biosynthesis by a subsurface bacterium. The signal transduction cascade or the surface proteins necessary for these processes are not yet known, although some authors posit membrane-bound sensory proteins (37).
Streptomyces reticuli, the organism used for our studies, is able to utilize crystalline cellulose (Avicel), due to its production of an exoglucanase (Cell or Avicelase) (27). Physiological studies showed that Avicel, to which S. reticuli strongly adheres during cultivation (26), is the only known inducing carbon source (40). A low-molecular-weight inducer, such as the breakdown products of cellulose (glucose and cellobiose, -triose, -tetraose, and -pentaose), able to enter mycelia could be excluded (39). On the molecular level, the regulation of the S. reticuli cel1 gene is achieved by both transcriptional activation and repression.
In this article, we identify and characterize a novel surface-anchored cellulose binding protein (AbpS) from S. reticuli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation.
The wild-type S. reticuli strain Tü45 described by Wachinger et al. (38) was obtained from H. Zähner, Tübingen, Germany. The E. coli plasmids pUC18 and pUC19 (44) were used as cloning vectors for DNA sequence analysis. E. coli transformants were grown in Luria-Bertani medium containing 100 μg of ampicillin ml−1. Streptomycetes were cultivated in pH-stable medium (MM3) supplemented with a carbon source (1%, wt/vol), as outlined previously (39).
Isolation of DNA.
Genomic DNA from S. reticuli was isolated as described previously (13). Plasmids were isolated from E. coli with a Qiagen plasmid kit.
General DNA techniques.
Restriction enzyme digestions, ligation reactions, and analyses of DNA with nucleases and polymerases were carried out by standard procedures (25).
Hybridizations.
The NH2 terminus of an internal peptide was used to deduce and synthesize a 39-mer oligonucleotide (5′GARGARGCSGACGCSCTSTTCGARGARACSCGSGCSAAG3′). This oligonucleotide was labelled with digoxigenin and hybridized at 50, 55, 60, and 65°C with SalI DNA fragments transferred from an agarose gel onto a nylon membrane (25). After 20 h, the membrane was washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (5 min at room temperature) and twice in 0.1× SSC (15 min at the temperature corresponding to the hybridization conditions). Immunodetection was performed according to the specifications of the DNA labelling and detection kit supplied by Boehringer.
Preparation and screening of a subgenomic DNA library.
Total DNA (200 μg) from S. reticuli was cleaved with SalI, and resulting fragments were separated on an agarose gel. Fragments of about 2.8 to 3.4 kb were eluted and ligated to SalI-digested pUC18. The ligation mixture was transformed to E. coli XL1 Blue by the CaCl2 method (25). Ampicillin-resistant transformants were tested for the presence of the desired insert by colony hybridization with the synthesized oligonucleotide.
DNA sequencing.
A 1,912-bp SmaI fragment containing the complete reading frame was sequenced by dideoxy chain termination (T7 sequencing kit from Pharmacia) and digoxigenin-labelled oligonucleotides which corresponded to those of the lacZ system or were deduced from internal sequences of abp1.
Identification of AbpS.
S. reticuli mycelia were disrupted by sonification (41) at 4°C. After the cell debris was removed, the proteins were incubated with Avicel (15 mg/ml) in 50 mM potassium phosphate buffer, pH 7, for 30 min. Avicel recovered by centrifugation was washed three times with 50 mM potassium phosphate buffer containing 1 M NaCl. The mixture was treated with the same buffer without NaCl and then heated in sodium dodecyl sulfate (SDS) sample buffer for 5 min at 95°C (25) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). After the gels were stained with Coomassie brilliant blue, the AbpS was identified by its size or by immunodetection with antibodies (see below).
Enzyme assays.
AbpS bound to Avicel was released by adding 7 M urea, precipitated with NH4SO4 (90% saturation), and suspended in 50 mM potassium phosphate buffer, pH 7. Cellulolytic activity was studied with the help of pNPC (para-nitrophenylcellobioside) (40). Alternatively, the protein was incubated in SDS sample buffer at 30°C for 10 min. After electrophoretic separation on a 10% polyacrylamide gel containing 0.1% (wt/vol) SDS (19) and hydroxyethyl cellulose (HEC) (40), the proteins were renatured by washing the gel twice in 0.1% Triton X-100 at 30°C and subsequently in 20 mM Tris-HCl, pH 7.5, for 30 min. Cellulolytic activities were tested after incubation at 30°C for 5 h and staining of the gel in a 0.1% solution of Congo red in water, as described by Schwarz et al. (32).
Determination of amino acid sequences.
The protein was blotted onto a polyvinylidine difluoride membrane (Immobilon P; Millipore), as outlined previously (27). Cleavage of AbpS and sequencing of the internal peptides by Edman degradation were done by P. Jungblut, Wita, Berlin, Germany.
Generation of antibodies and immunological studies.
Protein (200 μg) was subjected to preparative SDS-polyacrylamide gels (see above) and, after its transfer to a polyvinylidine difluoride membrane, eluted in a solution containing 2% SDS, 1% Triton X-100, and 0.1 mM dithiothreitol. After precipitation with two volumes of acetone, the protein was used for immunization. Antiserum against AbpS was obtained by immunization of a rabbit with 200 μg of pure AbpS (Eurogentec).
For Western blot studies, proteins were transferred onto a nylon membrane and incubated in phosphate-buffered saline (PBS; 150 mM NaCl, 2.6 mM KCl, 1.4 mM KH2PO4, 8.1 mM Na2PO4 [pH 7.0]) containing a 1:105 dilution of the antiserum. After three washes, the blot was incubated with alkaline phosphatase-conjugated AffiniPure F(ab′)2 fragment goat anti-rabbit immunoglobulin G (IgG) (dianova, Hamburg, Germany) diluted 1:15,000. Color development was performed according to the method of West et al. (42).
Isolation of membranes and associated proteins.
Washed S. reticuli mycelia were disrupted by sonification (see above). After removal of cell debris (10,000 × g, 20 min), the crude extract was centrifuged at 100,000 × g for 30 min. The supernatant was used for analyses of soluble protein. The membrane-containing pellet was washed twice with 50 mM potassium phosphate buffer (pH 7.0) and suspended in 1% Tween 20 in 50 mM phosphate buffer (pH 7.0). Insolubilized particles were removed by an additional centrifugation step (100,000 × g).
FITC-labelling studies.
Washed S. reticuli mycelia were incubated with fluorescein isothiocyanate (FITC) labelling solution (8 mg of FITC in 1 ml of 0.5 M sodium carbonate buffer supplemented with 0.15 M NaCl [pH 9.5]) and kept for 1 h at room temperature. Unbound FITC was removed by washing the mycelia with 500 mM glycine in sodium carbonate buffer (pH 9.5). In order to minimize binding to proteins different from AbpS, the pH was decreased to 5. The mycelia were washed with 3 liters of sodium carbonate buffer (pH 5) and subsequently used for the extraction of AbpS.
Studies with proteinase K.
Washed mycelia from S. reticuli were incubated with proteinase K (50 mg ml−1) for 3, 5, and 7 h at 30°C. Then the mycelia were washed several times with at least 1 liter of 50 mM Tris-HCl (pH 7) containing 2 mM Pefabloc (a proteinase inhibitor from Boehringer). After cell disruption, the proteins were analyzed by immunodetection with different antibodies.
In vivo immunolabelling of AbpS.
Washed S. reticuli mycelia were incubated with PBS with 1% bovine serum albumin for 1 h and subsequently with PBS containing a 1:1,000 dilution of anti-AbpS antiserum for 4 h at room temperature. To remove unbound antibodies, the mycelia were washed three times with PBS and then treated with PBS containing 1% bovine serum albumin and FITC-labelled AffiniPure F(ab′)2 fragment goat anti-rabbit IgG (1:300) for 4 h at room temperature. After three washes, the mycelia were analyzed for fluorescence labelling under UV light with an Axiovert microscope (Zeiss). For visualization, a charge-coupled-device camera (SenSys; Photometrics) and the software IPLab, version 1.7, were used.
Nucleotide sequence accession number.
The nucleotide sequence reported in this article will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under the accession no. Z97071.
RESULTS
Identification and purification of Avicel-binding protein.
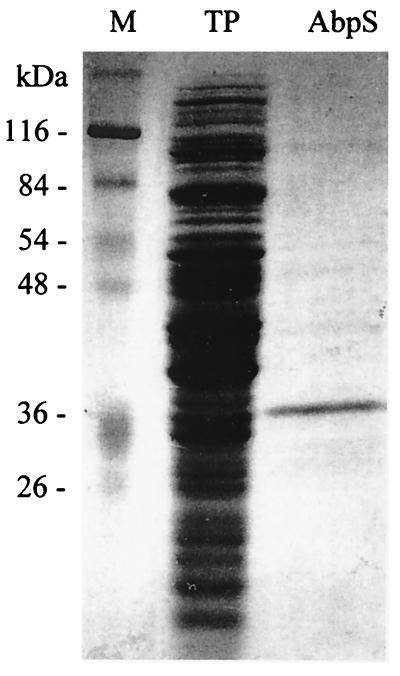
S. reticuli was grown with glucose as the sole carbon source. Proteins of the crude extract prepared from the mycelia were mixed with insoluble cellulose (Avicel). To inhibit nonspecific binding, the mixture was washed with 1 M NaCl. Under these conditions, only one protein with an apparent molecular mass of 36 kDa was found to adhere specifically to Avicel (Fig. 1). It was named AbpS (stands for Avicel-binding protein of Streptomyces).
FIG. 1.
Identification of AbpS. One milligram of total protein prepared from S. reticuli mycelia was incubated with Avicel. After being washed, the bound proteins were released from Avicel by heating them in SDS sample buffer and then subjected to SDS-PAGE (lane AbpS). As controls, 30 μg of the total protein was loaded onto lane TP and the molecular mass standard was loaded onto lane M. The gels had been stained with Coomassie brilliant blue.
Determination of the amino acid sequence and identification of the corresponding gene.
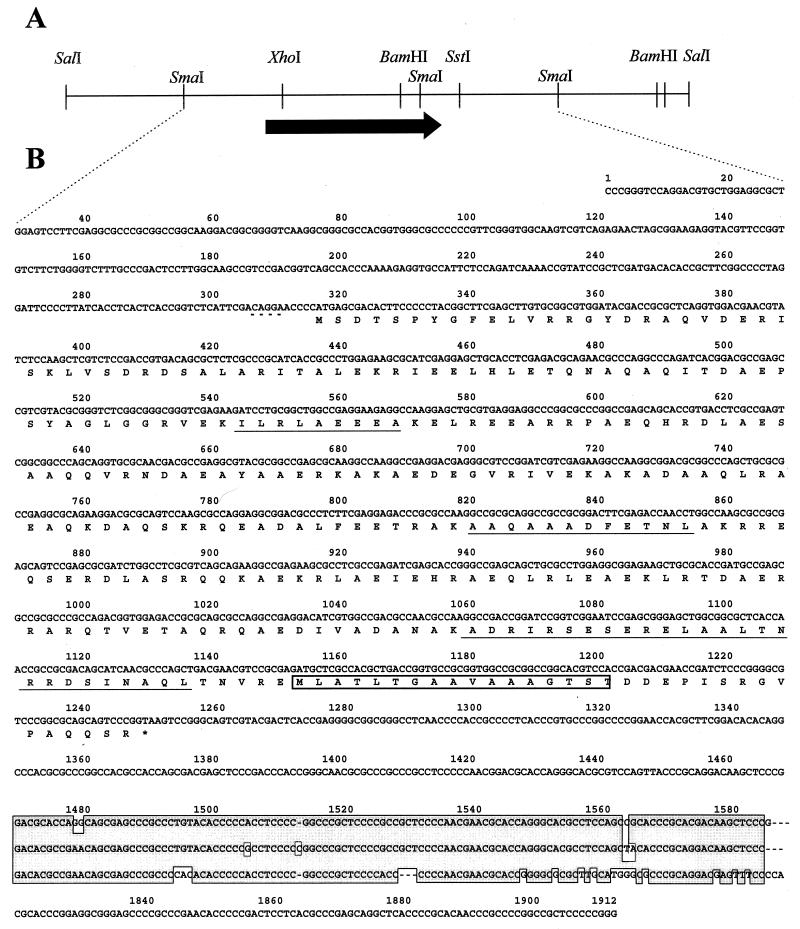
Larger quantities of the Avicel-bound protein were subjected to electrophoresis on preparative SDS-polyacrylamide gels, as described under Materials and Methods. After transfer to a nylon membrane, the AbpS was eluted and concentrated. As the NH2-terminal amino acids of AbpS could not be determined by Edman degradation, AbpS was cleaved with the protease LysC and the NH2 termini of several internal peptides were analyzed. The peptide from which the longest consecutive stretch of amino acids could be obtained was used to deduce and synthesize a 39-mer oligonucleotide, which was hybridized with total DNA from S. reticuli cleaved with SalI. At 60°C, only one fragment (3.2 kb) was found to hybridize (data not shown).
A subgenomic DNA library in E. coli XL1 Blue (containing 2.9- to 3.4-kb SalI fragments of S. reticuli DNA) was established in pUC18 and hybridized with the 39-mer oligonucleotide. Four transformants carried the expected constructs. After analyses of the inserted DNA with restriction enzymes (Fig. 2A), the DNA was subcloned. The sequences of overlapping fragments were determined from both strands. One complete reading frame (abpS) (Fig. 2B) has a G+C content of 73% (77% in the first, 47% in the second, and 96% in the third position of the codons), which is typical of streptomycetes. A start codon (ATG) with a putative Shine-Dalgarno sequence (AGGA), as well as a stop codon (TAA), could be identified. Downstream of the reading frame, we found three direct repeats, each of them consisting of 120 nucleotides, which have a high degree of homology (Fig. 2B).
FIG. 2.
Restriction map and sequence analysis. (A) Restriction map of the 3.2-kb SalI fragment. (B) The nucleotide sequence of the 1,912-bp SmaI fragment and the deduced amino acid sequence of AbpS are given. The NH2-terminal amino acids of the internal peptides (determined by Edman degradation) are underlined. The putative Shine-Dalgarno sequence is marked by a broken line. The three direct DNA repeats are aligned and marked by an outlined shaded area. The hydrophobic segment is marked by an open box.
The deduced amino acid sequence had a calculated molecular mass of 34.7 kDa, which is in good agreement with the estimated apparent molecular mass of AbpS (36 kDa). The NH2-terminal sequences from the sequenced internal peptides were refound, proving that the cloned fragment contains the corresponding abpS gene (Fig. 2B). By comparison with sequences from the SwissProt and EMBL data banks, a weak homology to tropomyosin proteins, streptococcal M proteins, or TolA from E. coli could be ascertained.
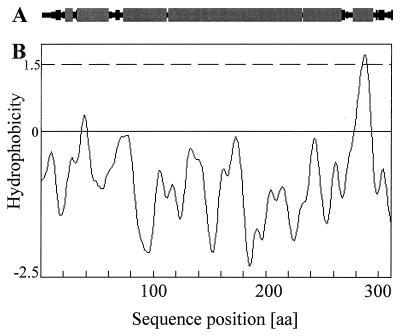
When we analyzed the hydrophobicity of the deduced protein, one putative transmembrane segment of 17 amino acids, which seems to have an α-helical structure (see below), was identified at the C-terminal part (Fig. 2B and 3B). An additional computer-supported analysis predicted that AbpS consists of dominant α-helical structures, organized in one large, centrally located unit and three smaller units (Fig. 3A).
FIG. 3.
Prediction of the hydrophobicity and secondary structure of AbpS. (A) The predicted secondary structure of AbpS. α-helical structures are indicated with shaded boxes, and β-sheets are indicated with filled boxes. (B) Hydrophobicity values were calculated according to the method of Kyte and Doolittle (18). Transmembrane segments correspond to the values above the dotted line. The predictions were made with the help of the computer program PSAAM. aa, amino acid.
Characterization of AbpS.
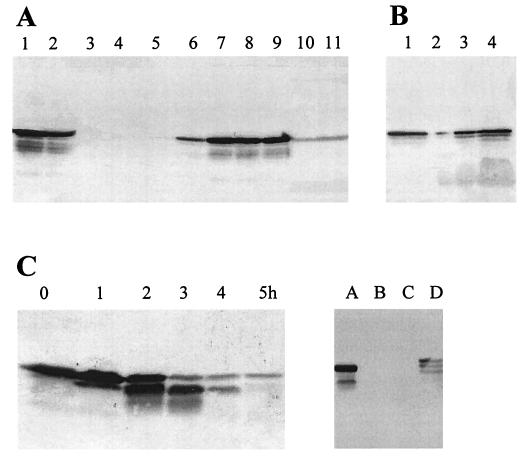
AbpS interacts with several forms of crystalline cellulose, like β-cellulose of tunicin, microcrystalline cellulose from bacteria (Acetobacter spp.), and commercial microcrystalline cellulose (Avicel), regardless of its grain size (with various diameters between 0.1 and 0.02 mm). Weaker binding of AbpS to α-chitin from crab shells, Whatman filters, or cellulose from Valonia, a eukaryotic alga, was observed. In contrast, AbpS did not bind to starch, xylan, agar-agar (Fig. 4A), soluble forms of cellulose (carboxymethyl cellulose [CMC] and HEC), or sugars. Neither glucose, cellobiose, a mixture of cellodextrins (cellobiose, -triose, -tetraose, and -pentaose), CMC, nor HEC competed in the interaction of AbpS with Avicel (Table 1). After the release of proteins bound to various types of crystalline cellulose, in addition to those bound to the full-length AbpS, low amounts of smaller proteins also cross-reacting with anti-AbpS antibodies were detected (Fig. 4A). As demonstrated in Fig. 4C, this process is due to proteolytic activities of endogenous S. reticuli proteases. The degradation of AbpS could not be inhibited by 1 M NaCl or 2 mM Pefabloc, a serine protease inhibitor, but by EDTA, typical of metalloproteases.
FIG. 4.
Characterization of AbpS. (A) One milligram of total protein was incubated with 25 mg of bacterial microcrystalline cellulose (lane 1); tunicin cellulose (lane 2); agar-agar (lane 3); xylan (lane 4); starch (lane 5); chitin (lane 6); Avicel with grain diameters of 0.02 mm (lane 7), 0.05 mm (lane 8), and 0.1 mm (lane 9); Valonia cellulose (lane 10); and a Whatman filter (lane 11) for 30 min at room temperature. Having been washed, the bound proteins were released by heating them in SDS sample buffer and then subjected to SDS-PAGE. After blotting and immunological detection, the amount of AbpS was determined. (B) Ten milligram of total protein (10 ml) was used to isolate the membranes and their associated proteins. The membranes were solubilized in 5 ml of phosphate buffer containing 1% Tween 20, and 5 μl (lane 2), 20 μl (lane 3), and 80 μl (lane 4) were transferred to an SDS-polyacrylamide gel. In order to determine the distribution of AbpS (membrane-associated and soluble forms), 10 μl (lane 1) of soluble proteins was additionally loaded onto the SDS-polyacrylamide gel. After immunological detection, the amount of AbpS was calculated densitometrically. (C) Thirty micrograms of total protein was incubated for 0, 1, 2, 3, 4, and 5 h at 37°C (left). Additionally, 30 μg of total protein (right) was incubated for 30 min in buffer (lane A) or for 4 h in the same buffer in the presence of a proteinase inhibitor (2 mM Pefabloc) (lane B), 1 M NaCl (lane C), or 50 mM EDTA (lane D). After SDS-PAGE and blotting, all samples were analyzed with anti-AbpS antibodies.
TABLE 1.
Characteristics of the AbpS-Avicel interaction
| Supplement or adjustment in pHa | % of AbpS bound to Avicel |
|---|---|
| Control | 100 |
| Carbohydrates | |
| 1% glucose | 100 |
| 1% cellobiose | 100 |
| 1% cellodextrins | 100 |
| 1% HEC | 100 |
| 1% CMC | 100 |
| Detergents | |
| 1% Tween 80 | 100 |
| 1% Triton X-100 | 100 |
| 1% Mega 9 | 100 |
| 1% Aminoxide | 100 |
| 1% Nonidet P-40 | 100 |
| 1% Tween 20 | 100 |
| 1% dodecylmaltoside | 100 |
| 0.1% SDS | 80 |
| Chaotropic agents or salts | |
| 1 M NaCl | 100 |
| 5 M NaCl | 75 |
| 4 M urea | 20 |
| 5 M urea | 0 |
| 7 M urea | 0 |
| 4 M Gdn-HCl | 35 |
| 5 M Gdn-HCl | 0 |
| 6 M Gdn-HCl | 0 |
| 90% (NH4)2SO4 | 0 |
| Other supplements | |
| 50 mM EDTA | 100 |
| 1% mercaptoethanol | 100 |
| Distilled water | 100 |
| Adjustment to pH: | |
| 10 | 100 |
| 7 | 100 |
| 4 | 0 |
| 3 | 0 |
One milligram of total protein was (i) incubated with 20 mg of Avicel in 1 ml of phosphate buffer containing the indicated supplements or (ii) adjusted to the indicated pH. After being washed with 3 ml of the supplemented buffers, the mixtures were subjected to SDS-PAGE and Coomassie brilliant blue staining and the amounts of bound proteins were denistometrically determined and compared to that bound by the control (100%). Gdn-HCl, guanidinium hydrochloride.
The AbpS-Avicel interaction occurred very rapidly; that is why the amount of AbpS which bound to Avicel after 1 min was identical with that bound in the course of 2 h (data not shown). Nonionic or ionic SDS detergents (1%, wt/vol), salts in high concentrations (1 M NaCl or 5 M KCl), and the chelating agent EDTA (50 mM) did not induce a release of Avicel-associated AbpS (Table 1). In contrast, the interaction of AbpS with Avicel could be completely inhibited by the addition of 5 M guanidium hydrochloride, 5 M urea, or NH4SO4 (90% saturation). pH values less than 5 entailed a release of the protein, whereas values of 5 to 10 had no effect (for details see Table 1).
Anti-AbpS antibodies, added to AbpS before the binding to Avicel occurred, prohibited the interaction completely, whereas the supplementation of Avicel-bound AbpS with anti-AbpS antibodies did not release the protein (data not shown).
No enzymatic activity of AbpS could be detected when HEC, para-nitrophenylcellobioside, para-nitrophenylglucoside, and methylum-belliferylcellobioside were used as substrates. These results are in agreement with the fact that the deduced protein does not have amino acids identical to those of catalytic domains of cellulases or glycosylhydrolases.
Localization of AbpS.
As outlined above, the deduced amino acid sequence of AbpS contains a putative, C-terminally located transmembrane segment. Therefore, the distribution of AbpS among membrane-associated and cytoplasmic proteins was investigated. Its relative amounts were determined after separation of all proteins, followed by immunodetection. Five to 10% of the total amount of AbpS was found in the membrane fraction (Fig. 4B).
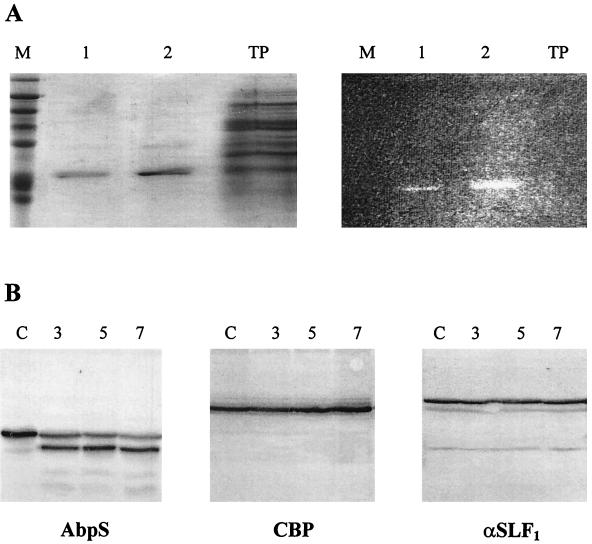
The C-terminally located stretch of 17 hydrophobic amino acids from AbpS is preceded by 15 randomly distributed lysine residues (Fig. 2B and 3). The NH2 terminus and lysine residues are known to interact with FITC. To test whether part of AbpS is surface exposed, S. reticuli hyphae were treated with FITC under conditions preventing nonspecific labelling (see Materials and Methods). Total hyphal proteins were gained, and those which bound to Avicel were separated on SDS-polyacrylamide gels. One prominent protein—isolated by its binding to Avicel and corresponding in size to AbpS—displayed high fluorescence (Fig. 5A). Within the total proteins of the mycelial extract which had not been treated with Avicel (control), hardly any labelled proteins could be detected. These results suggest that AbpS can be effectively labelled by FITC in native S. reticuli hyphae.
FIG. 5.
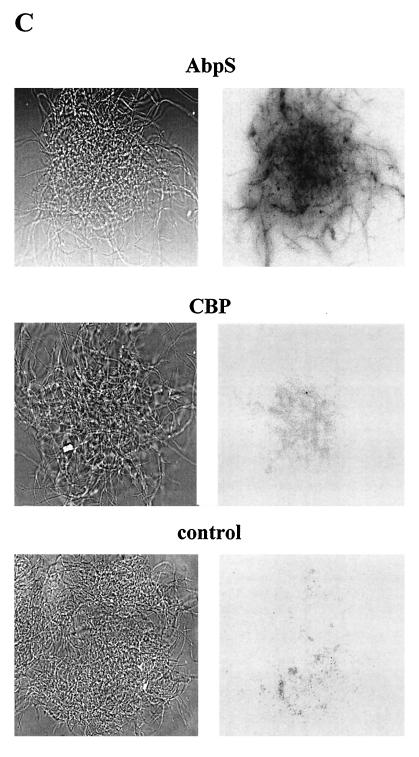
Localization of AbpS. (A) S. reticuli mycelia were incubated with FITC, and AbpS was subsequently isolated by its interaction with Avicel and separated by SDS-PAGE. Lanes M, molecular mass standards; AbpS isolated from 0.1 mg of total protein; lanes 2, AbpS isolated from 1 mg of total protein; lanes TP, 10 μg of total protein. The gel was analyzed under UV light (right gel) or after Coomassie brilliant blue staining (left gel). (B) Mycelia from S. reticuli grown with glucose as the sole carbon source were washed and incubated for 3 (lanes 3), 5 (lanes 5), and 7 (lanes 7) h in a proteinase K-containing buffer and for 7 h without proteinase K (lanes C). After being washed, crude cell extracts were prepared. Aliquots (10 μg) of each sample were subjected to SDS-PAGE. After being blotted, the proteins were incubated with antibodies raised against AbpS (left blot), CBP (middle blot) (28), or α-SLF1 (right blot) (8). (C) Washed S. reticuli mycelia were incubated with anti-AbpS antibodies (top section) and anti-CBP antibodies (middle section), or without primary antibodies (bottom section) for 4 h at room temperature. After unbound antibodies were removed, secondary FITC-labelled anti-rabbit IgG2ab was used to determine the presence of bound primary antibodies on the surfaces of S. reticuli hyphae, with the help of UV light and an Axiovert microscope.
To study the location in more detail, native S. reticuli mycelia were treated with or without (control) proteinase K for various times (3, 5, and 7 h) and the presence of AbpS was immunologically determined. Contrary to what occurred with the control, a major portion of AbpS was found to be truncated by approximately 3 kDa. However, isolated pure AbpS is completely degraded by proteinase K. The studies indicate that within native hyphae, only a small AbpS region is accessible to proteinase K and that the protection of the major portion of the protein is due to the surrounding envelope structure. This assumption is supported by the fact that the formerly described cellobiose-binding protein (CBP) from S. reticuli, which faces the outer side of the cell membrane and is fixed by a lipid anchor, is not degraded (28) by proteinase K treatment within S. reticuli hyphae (Fig. 5B). The S. reticuli α-subunit of the ATPase (12) (detected by antibodies previously raised against the α-subunit of the Streptomyces lividans F1 ATPase [α-SLF1]) was also found to be resistant to proteinase K.
Further investigations showed that anti-AbpS antibodies, unlike anti-CBP antibodies, cross-reacted with protein within native hyphae (Fig. 5C). These data clearly demonstrate that epitopes of AbpS are surface exposed.
Physiological studies.
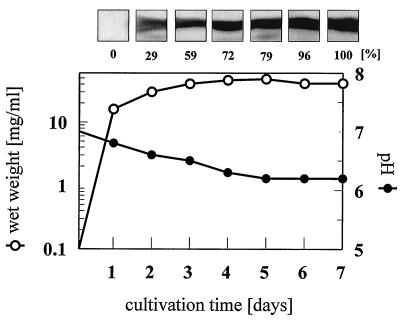
In the presence of the carbon sources chitin, xylan, CMC, cellobiose, glucose, and Avicel (1% in minimal medium), S. reticuli produced AbpS in increasing amounts from the early stationary to the late stationary growth phase, as shown exemplarily for glucose (Fig. 6).
FIG. 6.
Temporal course of AbpS synthesis. Minimal medium supplemented with 1% glucose was inoculated with S. reticuli spores and incubated at 30°C. After the indicated cultivation periods, aliquots were taken, and the wet weights of the mycelia, as well as their pHs, were determined. In addition, 30 μg of total protein prepared from each aliquot was separated by SDS-PAGE. After blotting, AbpS was detected immunologically, and after the blots were scanned, AbpS’s quantity was determined densitometrically with the Cybertech program CAM 2.0.
DISCUSSION
In the course of our studies, we found that S. reticuli synthesizes an up to now unique hyphal protein (AbpS) which binds tightly to crystalline forms of cellulose (Avicel, bacterial microcrystalline cellulose, and tunicin cellulose) but not to other polysaccharides (e.g., starch, agar, and xylan). The fact that AbpS binds very quickly to Avicel and can be released only by chaotropic or denaturing agents emphasizes its high specifity.
Cellulose-binding modules have been discovered in several cellulases from different microorganisms. They are functionally and structurally separate protein domains (cellulose-binding domains [CBDs] and are located on the COOH- or NH2-terminal part of the catalytic domain [for a review, see reference 10]). On the basis of their primary structures, 13 families of CBDs were defined (2). The primary structure of the deduced AbpS protein sequence shows no significant homology with that of members of the established families. Crystallographic analyses of several CBDs from the families I, II, and III revealed that their binding to cellulose is mediated by aromatic amino acid residues (tryptophane and tyrosine), exposed on one side of the CBD (15, 16, 36, 43). The distances between the aromatic amino acid side chains correlate with those between the repetitive glucose units within the cellulose molecule. At the NH2 terminus of AbpS, four tyrosines were found. Whether these aromatic amino acids participate in the interaction with highly crystalline cellulose remains to be investigated.
It was shown that CBDs are also present in cellulolytically inactive proteins, such as CipA from Clostridium thermocellum (9) and its homolog CbpA from Clostridium cellulovorans (34). Both organisms produce large cellulolytic complexes (cellulosomes), in which several cellulases are tightly bound to the scaffolding proteins CipA and CbpA, whose CBDs mediate contact with cellulose. The affinities of these binding domains are very high (11), which is also true for AbpS from S. reticuli.
A lectin-like chitin-binding protein (CHB1) is secreted by Streptomyces olivaceoviridis (31). This CHB1 (18.7 kDa) lacks any enzymatic and antifungal activities, has a high affinity to α-chitin, and seems to participate in the targeting of α-chitin-containing organisms (45). Unlike the secreted CHB1, AbpS was found to be associated with the surfaces of S. reticuli hyphae. The results of in vivo FITC-labelling and proteinase K experiments suggest that AbpS protrudes out of the polyglucane layer, which also occurs with certain proteins anchored to the cell wall of gram-positive pathogenic bacteria; these include protein A from Staphylococcus aureus and group A Ig-binding P and M proteins from several streptococci (6, 14, 33, 35). The function of these surface proteins appears to be either to conceal the bacterial surface from the host’s defense system or to promote adhesion of the pathogen to host tissues. These proteins are exposed on their cell surfaces and anchored to bacterial cell walls. Anchoring requires a 35-residue sorting signal, which is situated in the C terminus and consists of an LPXTG motif, followed by a hydrophobic domain and a tail of largely positively charged residues (20, 29, 30). This protein-sorting sequence, as well the NH2-terminal signal sequence, is not found in the deduced AbpS protein. Therefore, it remains to be elucidated if the small C-terminally located hydrophobic segment of AbpS participates in anchoring.
AbpS and M proteins also seem to be similar in their secondary structures. M molecules are highly α-helical coiled-coil dimers, the structures of which resemble those of tropomyosin and other filament-forming proteins belonging to the same family (24, 33). The deduced AbpS protein also shows weak homology to tropomyosin molecules and to the above-described M proteins. The predicted secondary structure of AbpS indicates the presence of a large central α-helix. Further studies, including spectroscopical and microscopical analyses, have to be performed to determine the structure of AbpS and its possible dimerization and interaction with other proteins.
Physiological studies showed that AbpS synthesis is independent of the carbon source and stimulated during late logarithmic growth of S. reticuli. Under these conditions, the depletion of nutrients is expected. In order to survive, the organism has to monitor its environment to develop new nutrient sources. In this context the possible function of AbpS is the mediation of the contact between the biopolymer and the hyphae. This assumption is substantiated by the fact that S. reticuli hyphae grow in tight connection with Avicel in the course of cultivation (26). This entails an effective hydrolysis of the biopolymer by secreted cellulases and an enhanced uptake of the breakdown products. To what extent the induction of the corresponding genes is dependent on the attachment of the mycelia to the insoluble substrate has be studied in the future. To this end, comparative investigations of an abpS-negative mutant of S. reticuli are required; however, genetic manipulations of the strain have proven to be difficult.
ACKNOWLEDGMENTS
We are grateful to A. Schlösser and E. Wolfsholz for having performed initial cloning experiments, to H. Chanzy (Grenoble, France) for cellulose substrates, to P. Jungblut (Wita) for protein sequencing, to D. Müller for photographical work, and to M. Lemme for her support in the writing of the manuscript. We thank G. Deckers-Hebestreit (University of Osnabrück) and A. Schlösser for the provision of antibodies raised against α-SLF1 and CBP, respectively.
The work was financed in part by the Sonderforschungsbereich (grant 171/C14).
REFERENCES
- 1.Alexander M. Introduction to soil microbiology. New York, N.Y: John Wiley & Sons; 1977. [Google Scholar]
- 2.Béguin P, Aubert J-P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 3.Belas R, Simson M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berdy J. Recent advances in prospects of antibiotic research. Process Biochem. 1980;15:15–30. [Google Scholar]
- 5.Braun V. Surface signalling: novel transcription initiation mechanism starting from the cell surface. Arch Microbiol. 1997;167:325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 6.Cleary P, Retnoningrum D. Group A streptococcal immunoglobulin-binding proteins: adhesins, molecular mimicry or sensory proteins? Trends Microbiol. 1994;2:131–136. doi: 10.1016/0966-842x(94)90600-9. [DOI] [PubMed] [Google Scholar]
- 7.Davies D G, Geesey G G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61:860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deckers-Hebestreit G, Altendorf K. The F0F1-type ATP synthases of bacteria: structure and function of the F0 complex. Annu Rev Microbiol. 1996;50:791–824. doi: 10.1146/annurev.micro.50.1.791. [DOI] [PubMed] [Google Scholar]
- 9.Gerngross U T, Romaniec M P M, Kobayashi T, Huskisson N S, Demain A L. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993;8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 10.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein M A, Takagi M, Hashida S, Shoseyov O, Doi R H, Segel I H. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J Bacteriol. 1993;175:5762–5768. doi: 10.1128/jb.175.18.5762-5768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hensel M, Lill H, Schmid R, Deckers-Hebestreit G, Altendorf K. The ATP synthase (F1F0) of Streptomyces lividans: sequencing of the atp operon and phylogenetic considerations with subunit beta. Gene. 1995;152:11–17. doi: 10.1016/0378-1119(95)00673-t. [DOI] [PubMed] [Google Scholar]
- 13.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 14.Jenkinson H F. Cell surface protein receptors in oral streptococci. FEMS Microbiol Lett. 1994;121:133–140. doi: 10.1111/j.1574-6968.1994.tb07089.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson P E, Joshi M D, Tomme P, Kilburn D G, McIntosh L P. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:14381–14394. doi: 10.1021/bi961612s. [DOI] [PubMed] [Google Scholar]
- 16.Kraulis P J, Clore G M, Nilgers M, Jones T A, Pettersson G, Knowles J, Gronenborn A M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 17.Kutzner H J. The family Streptomycetaceae. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H, editors. The prokaryotes: a handbook on habitats, isolation and identification of bacteria. Berlin, Germany: Springer-Verlag; 1981. pp. 2028–2090. [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 21.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 22.Parkinson J S, Kofoid E C. Communication modules in bacterial signalling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 23.Peczynska-Czoch W, Mordarski M. Actinomycete enzymes. In: Goodfellow M, Williams S T, Mordarski M, editors. Actinomycetes in biotechnology. London, United Kingdom: Academic Press; 1988. pp. 219–283. [Google Scholar]
- 24.Phillips G N, Jr, Flicker P F, Cohen C, Manjula B N, Fischetti V A. Streptococcal M protein: α-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci USA. 1981;78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schlochtermeier A, Niemeyer F, Schrempf H. Biochemical and electron microscopic studies of the Streptomyces reticuli cellulase (Avicelase) in its mycelium-associated and extracellular forms. Appl Environ Microbiol. 1992;58:3240–3248. doi: 10.1128/aem.58.10.3240-3248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlochtermeier A, Walter S, Schröder J, Moormann M, Schrempf H. The gene encoding the cellulase (Avicelase) Cel1 from Streptomyces reticuli and analysis of protein domains. Mol Microbiol. 1992;6:3611–3621. doi: 10.1111/j.1365-2958.1992.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 28.Schlösser A, Schrempf H. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/-triose transport system from the cellulose degrader Streptomyces reticuli. Eur J Biochem. 1996;242:332–338. doi: 10.1111/j.1432-1033.1996.0332r.x. [DOI] [PubMed] [Google Scholar]
- 29.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 30.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnellmann J, Zeltins A, Blaak H, Schrempf H. The novel lectin-like protein CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridis gene and binds specifically to α-chitin of fungi and other organisms. Mol Microbiol. 1994;13:807–819. doi: 10.1111/j.1365-2958.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz W H, Bronnenmeier K, Gräbnitz F, Staudenbauer W L. Activity staining of cellulases in polyacrylamide gels containing mixed linkage β-glucans. Anal Biochem. 1987;164:72–77. doi: 10.1016/0003-2697(87)90369-1. [DOI] [PubMed] [Google Scholar]
- 33.Scott J R, Caparon M G. Streptococcus. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 53–63. [Google Scholar]
- 34.Takagi M, Hashida S, Goldstein M A, Doi R H. The hydrophobic repeated domain of the Clostridium cellulovorans cellulose-binding protein (CbpA) has specific interactions with endoglucanases. J Bacteriol. 1993;175:7119–7122. doi: 10.1128/jb.175.21.7119-7122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talay S R, Grammel M P, Chhatwal G S. Structure of a group C streptococcal protein that binds to fibrinogen, albumin and immunoglobulin G via overlapping modules. Biochem J. 1996;315:577–582. doi: 10.1042/bj3150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 37.Vandevivere P, Kirchman D L. Attachment stimulates exopolysaccharide synthesis by a bacterium. Appl Environ Microbiol. 1993;59:3280–3286. doi: 10.1128/aem.59.10.3280-3286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachinger G, Bronnenmeier K, Staudenbauer W L, Schrempf H. Identification of mycelium-associated cellulase from Streptomyces reticuli. Appl Environ Microbiol. 1989;55:2653–2657. doi: 10.1128/aem.55.10.2653-2657.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter S, Schrempf H. Studies of Streptomyces reticuli cel-1 (cellulase) gene expression in Streptomyces strains, Escherichia coli, and Bacillus subtilis. Appl Environ Microbiol. 1995;61:487–494. doi: 10.1128/aem.61.2.487-494.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter S, Schrempf H. Physiological studies of cellulase (Avicelase) synthesis in Streptomyces reticuli. Appl Environ Microbiol. 1996;62:1065–1069. doi: 10.1128/aem.62.3.1065-1069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter S, Schrempf H. The synthesis of the Streptomyces reticuli cellulase (Avicelase) is regulated by both activation and repression. Mol Gen Genet. 1996;251:186–195. doi: 10.1007/BF02172917. [DOI] [PubMed] [Google Scholar]
- 42.West S, Schröder J, Kunz W. A multiple-staining procedure for the detection of different DNA fragments on a single blot. Anal Biochem. 1990;190:254–258. doi: 10.1016/0003-2697(90)90189-g. [DOI] [PubMed] [Google Scholar]
- 43.Xu G-Y, Ong E, Gilkes N R, Kilburn D G, Muhandiram D R, Harris-Brandts M, Carver J P, Kay L E, Harvey T S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 45.Zeltins A, Schrempf H. Visualization of α-chitin with a specific chitin-binding protein (CHB1) from Streptomyces olivaceoviridis. Anal Biochem. 1995;231:287–294. doi: 10.1006/abio.1995.0053. [DOI] [PubMed] [Google Scholar]