Abstract
The Bacillus subtilis skin element confers resistance to arsenate and arsenite. The ars operon in the skin element contains four genes in the order arsR, ORF2, arsB, and arsC. Three of these genes are homologous to the arsR, arsB, and arsC genes from the staphylococcal plasmid pI258, while no homologs of ORF2 have been found. Inactivation of arsR, arsB, or arsC results in either constitutive expression of ars, an arsenite- and arsenate-sensitive phenotype, or an arsenate-sensitive phenotype, respectively. These results suggest that ArsR, ArsB, and ArsC function as a negative regulator, a membrane-associated protein need for extrusion of arsenite, and arsenate reductase, respectively. Expression of the ars operon was induced by arsenate, arsenite, and antimonite. Northern hybridization and primer extension analysis showed that synthesis of a full-length ars transcript of about 2.4 kb was induced by arsenate and that the ars promoter contains sequences that resemble the −10 and −35 regions of promoters that are recognized by EςA.
Sporulation in Bacillus subtilis is initiated in response to nutrient deprivation. During an early stage of sporulation, cells are divided into two compartments, the mother cell and the forespore. Later in sporulation, a DNA rearrangement occurs only in the mother cell; this rearrangement is catalyzed by a site-specific recombinase, SpoIVCA (28). The excision of the 48-kb skin (for sigK intervening) element (33) from the chromosome (13) creates a new composite gene, sigK, by fusion of the spoIIIC and spoIVCB genes (7, 14, 32).
Some putative proteins encoded by open reading frames (ORFs) in the skin element resemble those of the B. subtilis temperate phage φ105 and the defective phage PBSX, an observation that suggests that the skin element might be a cryptic remnant of an ancestral B. subtilis phage (16, 33). In addition, the skin element encodes homologs of GsiA (a protein-aspartyl phosphatase [18, 21]) and the Ars (arsenical resistance) proteins (3–5, 10, 19, 23, 31). The homolog of the ars operon in the skin element consists of four ORFs (arsR, ORF2, arsB [ORF1], and arsC) (33). The products of arsR and arsC are homologous to products of ars genes in Staphylococcus and Escherichia coli, ArsR and ArsC, respectively (33). The B. subtilis arsB product is also homologous to the Staphylococcus arsB product. However, the ArsB protein exhibits only limited sequence identity (24%) to the Staphylococcus ArsB. In addition, there is no homolog of the ORF2 product in Staphylococcus.
The ars operon in the staphylococcal plasmids pI258 and pSX258 endows the cell with resistance to arsenate (As5+), arsenite (As3+), and antimonite (Sb3+) (for reviews, see references 22 and 30). The arsR, arsB, and arsC genes encode an inducer-dependent repressor of transcription, an inner membrane protein (containing 12 membrane-spanning regions) of the arsenite extrusion pump, and a reductase that converts arsenate to arsenite, respectively (22, 30).
In this report we show that the ars operon in the skin element is inducible by arsenate, arsenite, and antimonite and endows the cells with resistance to arsenic (as both arsenate and arsenite).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of B. subtilis used in this study are listed in Table 1. Integration plasmid pMUTinT3 (1a) allows (i) selection of erythromycin resistance in B. subtilis, (ii) disruption of the region cloned into the plasmid and generation of a fusion transcript with a gene for β-galactosidase, and (iii) placement of genes downstream of Pspac-1 (38), which allows induction of transcription in the presence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG).
TABLE 1.
Strains of B. subtilis used
| Strain | Genotype | Source or reference |
|---|---|---|
| JH642 | pheA1 trpC2 | BGSCa |
| 4C12 | leuA8 metB4 nonB1 spoIVCA::cat | 27 |
| 4CJH | pheA1 trpC2 spoIVCA::cat | 4C12→JH642b |
| QUA2 | asa-2 leuA2 trpC2 | BGSC |
| ars-C | arsC::cat pheA1 trpC2 | 27 |
| sup1-3 | lys-1 purB6 Δskin | 27 |
| skin-1 | pheA1 trpC2 Δskin | This study |
| ars-R3 | pheA1 trpC2 arsR::pMUTinT3 | This study |
| ars-23 | pheA1 trpC2 ORF2::pMUTinT3 | This study |
| ars-B3 | pheA1 trpC2 arsB::pMUTinT3 | This study |
BGSC, Bacillus Genetic Stock Center, Ohio State University.
Transformation with 4C12 as donor and JH642 as recipient.
Growth inhibition.
Overnight cultures of JH642, skin-1, QUA2, ars-R3, ars-23, ars-B3, and ars-C were inoculated into fresh Luria-Bertani (LB) broth (5 Klett units; filter no. 66) with or without 1 mM IPTG and increasing amounts of arsenate or arsenite. Cells were grown for 8 h at 37°C, and then turbidity (Klett units) was measured. When required, chloramphenicol (5 μg/ml) or erythromycin (5 μg/ml) was added to the medium.
Measurement of β-galactosidase activity.
The specific activity of β-galactosidase was determined with o-nitrophenyl-β-d-galactoside as the substrate and expressed as units per absorbance unit of the culture at 600 nm as described elsewhere (15).
S1 nuclease mapping.
S1 nuclease mapping was performed as described previously (25). JH642 cells were grown in LB broth at 37°C to the exponential phase of growth (50 Klett units), and then 1 mM arsenate was added to the culture. Samples (10 ml each) were taken at various times after the addition of arsenate, and 50-μg aliquots of total RNA were extracted from the cells as described previously (9). These were allowed to hybridize with the 32P-labelled 573-bp PCR product (∼10 ng; 2 × 104 cpm) (33) that had been amplified with primer 3 (5′-CTTCTTGCGAGATGATG; nucleotides [nt] 457 to 441 upstream of the initiation codon of arsR) and 32P-labelled primer 4 (5′-AACACAGGTTTTTCCCC; complementary to the region from nt 117 to 101 downstream of the initiation codon of arsR). After S1 nuclease digestion (25), the nuclease-resistant DNA fragments were analyzed by gel electrophoresis on a denaturing 5% polyacrylamide gel and detected by autoradiography.
Primer extension.
Primer extension analysis was performed as described previously (25) with RNA isolated as described above. The site of initiation of transcription of ars mRNA was determined by the primer extension method with primer 4. Dideoxynucleotide sequencing reactions were performed with the 2,804-bp PCR product that had been amplified with primer 3 and 32P-labelled primer 5 (5′-CATATAAATATGCATCC; sequence complementary to the region from nt 37 to 21 downstream of the termination codon of arsC) as a template and the same primer as used to map the 5′ end of the ars mRNA.
Northern hybridization.
Northern hybridization was performed as described previously (25). Total RNA, extracted from JH642 cells that had been harvested 20 min after the addition of 1 mM arsenate, was hybridized with either the 573-bp PCR product or the 2,804-bp PCR product as a probe (20,000 cpm; approximately 10 ng) (33). Samples of 10 μg of RNA were loaded onto the lanes of a 1.0% agarose gel containing 2.2 M formaldehyde. After electrophoresis, the RNA was transfered to a nylon membrane (Hybond-N; Amersham).
RESULTS
Construction of ars mutants.
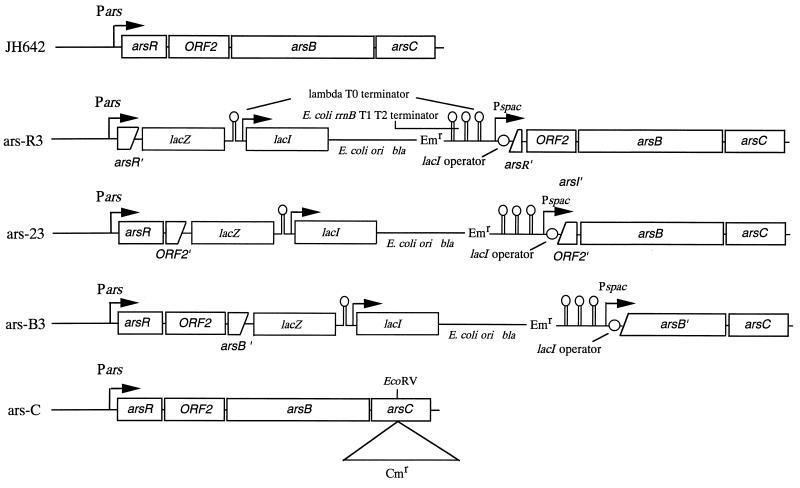
To determine whether the ars operon in the skin element is functional, we used an arsR mutant (ars-R3), an ORF2 mutant (ars-23), an arsB mutant (ars-B3), an arsC mutant (ars-C), and a skin-less mutant (skin-1) of B. subtilis JH642 (pheA1 trpC2). Details of the strains are shown in Table 1. PCR resulted in amplification of (i) a 210-bp internal segment of arsR with primers R1 (5′-GCCAAGCTTGCTACGGAAATATGAAC) and R2 (5′-CGCGGATCCATAATAACTCCATGTTC), (ii) a 325-bp internal segment of ORF2 with primers 21 (5′-GCCGAAGCTTGTAGGGGTTAACGTTGTC) and 22 (5′-CGCGGATCCCCCACTCGTTCCCATCTG), and (iii) a 337-bp internal segment of arsB with primers B1 (5′-GCCGAAGCTTCTTGCGATGGCATTGGGG) and B2 (5′-CGCGGATCCCATCGCAATACAGCGAGC). These primers contained either HindIII or BamHI sites (underlined residues). These PCR fragments were digested with BamHI and HindIII and cloned between the HindIII and BamHI sites (downstream of Pspac-1) of pMUTinT3 to create pMUTin-R3, pMUTin-23, and pMUTin-B3, respectively. The ars-R3 (pheA1 trpC2 arsR::pMUTinT3 [Emr]), ars-23 (pheA1 trpC2 ORF2::pMUTinT3 [Emr]), and ars-B3 (pheA1 trpC2 ORF1::pMUTinT3 [Emr]) strains were constructed by integration of pMUTin-R3, pMUTin-23, and pMUTin-B3 at the arsR, ORF2, and arsB loci, respectively. The organization of the ars operon in these strains is shown in Fig. 1. The skin-1 strain (pheA1 trpC2 Δskin) was constructed by transformation with a skin-less strain, sup1-3 (lys-1 purB6 Δskin) (27), as the donor and strain 4CJH (pheA1 trpC2 spoIVCA::cat) as the recipient. In addition, we used mutant strain QUA2 (asa-2 leuA2 trpC2), which had been isolated as an arsenate-sensitive mutant of B. subtilis 168 (1). Although the precise nature of the asa-2 mutation had not previously been determined, the asa-2 locus was mapped between pheA1 and aroD, which is also the location of the skin element.
FIG. 1.
Genetic construction of the ars mutants designated ars-R3, ars-23, ars-B3, and ars-C. The relevant portions of the genomes of the mutant strains are shown. Pars, ars promoter; Pspac, spac promoter; Emr, erythromycin resistance gene; Cmr, chloramphenicol resistance gene.
Growth of ars mutants in the presence of arsenate, arsenite, and antimonite.
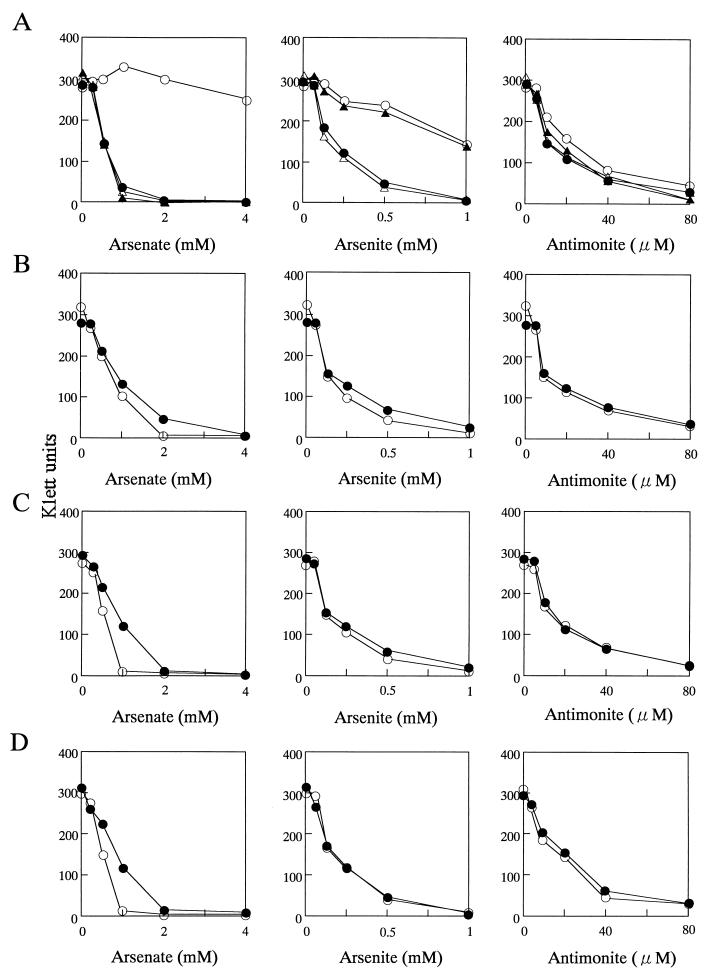
Wild-type (JH642) cells grew in the presence of 4 mM arsenate or 0.5 mM arsenite (Fig. 2). However, the skin-1 and QUA2 strains failed to grow in the presence of 1 mM arsenate or 0.5 mM arsenite (Fig. 2A). When the arsC gene was inactivated (strain ars-C), the cells became sensitive to arsenate but were still resistant to arsenite (Fig. 2A). These results indicate that the arsC gene was related to arsenate resistance. Similar results have been obtained with arsC mutants of E. coli R773 (6), Staphylococcus aureus pI258 (11), and Staphylococcus xylosus pSX267 (23). Resistance to arsenate is achieved by the reduction of arsenate to arsenite by the product of the arsC gene (8, 11, 12, 20). Inactivation of arsR (ars-R3) caused a decrease in resistance to arsenite and arsenate in the absence of IPTG. In the presence of IPTG, the cells became slightly resistant to arsenate and arsenite (Fig. 2B). This result suggests that ars expression from the spac promoter is not sufficient to induce arsenic resistance or that the ars mRNA produced from the spac promoter is unstable. The ars-23 and ars-B3 strains both had arsenate- and arsenite-sensitive phenotypes in the absence of IPTG (Fig. 2C and D). However, in the presence of 1 mM IPTG (for ars-23, arsB, and arsC were induced; for ars-B3, arsC was induced), these strains exhibited slightly reduced sensitivity to arsenate. This result suggested that arsB conferred resistance to arsenite by allowing cells to extrude this compound. However, the role of ORF2 is not established by these experiments. Overall, these results indicate that the ars operon in the skin element is functional and determines resistance to arsenicals in B. subtilis. The skin-1, QUA2, ars-23, and ars-B3 strains exhibited sensitivity to antimonite that was very similar to that of the wild-type strain.
FIG. 2.
Inhibition of growth of various B. subtilis strains by arsenate, arsenite, and antimonite. (A) Resistance of strains JH642 (○), skin-1 (•), ars-C (▴), and QUA2 (▵). (B) Resistance of strain ars-R3 with IPTG (•) (ORF2, arsB, and arsC are induced) or without IPTG (○) (ORF2, arsB, and arsC are not induced). (C) Resistance of strain ars-23 with IPTG (•) (arsB and arsC are induced) or without IPTG (○) (arsB and arsC are not induced). (D) Resistance of strain ars-B3 with IPTG (•) (arsC is induced) or without IPTG (○) (arsC is not induced). Overnight cultures of JH642, skin-1, ars-R3, ars-23, ars-B3, QUA2, and ars-C were inoculated into fresh LB broth (5 Klett units) with or without 1 mM IPTG (for panels B, C and D) and increasing amounts of arsenate, arsenite, or antimonite. Cultures were incubated for 8 h at 37°C, and then the turbidity (Klett units) was measured.
QUA2 is a skin-less mutant.
The asa-2 locus of the QUA2 strain mapped between pheA1 and aroD (1), where the skin element is also located. The pattern of resistance of QUA2 was similar to that of skin-1 (Fig. 2A). These results suggest that QUA2 might be a skin-less mutant carrying an uninterrupted sigK gene in vegetative cells. In an attempt to detect the intact sigK gene in vegetative QUA2 cells, we performed PCR with two primers, 5′-GCAGAGGACTTAATCTCC (nt 226 to 243 downstream of the initiation codon of sigK) and 5′-GTATCTATCACGTCTTC (complementary to the region from nt 476 to 460 downstream of the initiation codon of sigK). The sequences of these two primers are located in spoIVCB and spoIIIC, respectively, where they are separated by 48 kb in a wild-type strain (33). After PCR with chromosomal DNA of vegetative QUA2 cells, a 251-bp fragment corresponding in size to a fragment containing the spoIVCB-spoIIIC junction region was detected (data not shown). In contrast, this fragment was detected by PCR with DNA from wild-type cells only 6 h after initiation of sporulation (27). This result indicated that QUA2 was a skin-less mutant.
Induction of the ars operon by arsenate, arsenite, and antimonite.
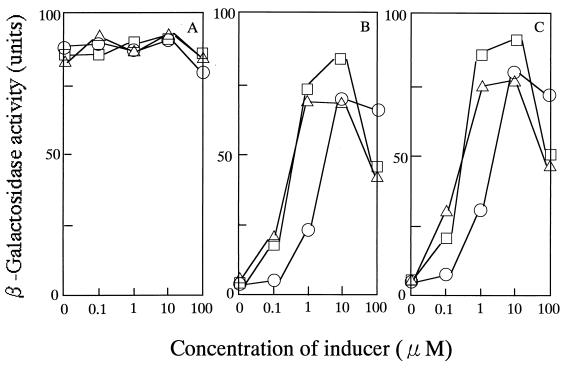
The expression of known ars operons is regulated by arsenic and antimony salts (29, 34). To characterize the regulation of expression of the ars operon in B. subtilis by arsenate, arsenite, and antimonite, we determined the β-galactosidase activity expressed by strains ars-R3 (an arsR-lacZ fusion strain), ars-23 (an ORF2-lacZ fusion strain), and ars-B3 (an arsB-lacZ fusion strain) (Fig. 3). Inactivation of arsR led to constitutive expression of ars, and the pattern of expression of the ORF2-lacZ fusion was similar to that of the arsB-lacZ fusion. Expression of the latter two lacZ fusions was repressed in the absence of an oxyanion inducer (arsenate, arsenite, or antimonite), but both were activated in the presence of such an inducer. Maximal induction was achieved with 1 to 10 μM arsenite or antimonite or with 10 to 100 μM arsenate. Similar results were obtained with an arsR-bla fusion in S. aureus pI258 (10).
FIG. 3.
Induction of the ars operon. Cells of strains ars-R3 (A), ars-23 (B), and ars-B3 (C) were grown in LB broth at 37°C to the exponential phase of growth (50 Klett units), and then the ars operon was induced by the addition of the indicated amount of arsenite (▵), antimonite (□), or arsenate (○). β-Galactosidase activity was measured 1 h after induction.
Size of the ars transcript.
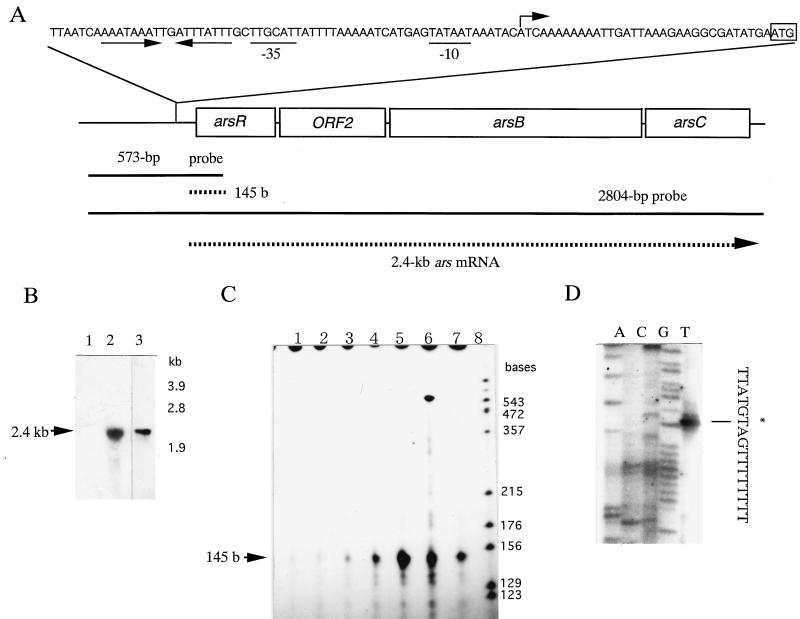
Northern hybridization analysis was performed to determine the size of the ars transcript (Fig. 4A). A transcript of approximately 2.4 kb, the size expected from the length of the ars operon, was found in the arsenate-induced wild-type strain, but no transcript was detected in the arsenate-induced skin-1 strain (Fig. 4B).
FIG. 4.
Analysis of the transcript of the ars operon. S1 nuclease mapping and Northern hybridization experiments were performed as described previously (25). (A) Structure of the ars operon and nucleotide sequence of the upstream region of the ars operon (24). Sizes of ORFs are shown to scale. Solid lines indicate the sizes and positions of the probes used. Dashed lines indicate the sizes and deduced positions of the S1 product and the observed mRNA. The site of initiation of transcription is indicated by an arrow. Putative −10 and −35 RNA polymerase-binding regions of the ars promoter are underlined, and the putative translation initiation codon of arsR is boxed. The converging arrows indicate the region of inverted repeats. b, bases. (B) Northern blotting analysis of the ars transcript. Lanes 1 and 2, transcript hybridized with the 2,804-bp probe. Lane 3, transcript hybridized with the 573-bp probe. Total RNA was isolated from strains skin-1 (lane 1) and JH642 (lanes 2 and 3) 20 min after induction by 1 mM arsenate. (C) S1 nuclease mapping of the ars transcript. The 32P-labelled 573-bp PCR product (20,000 cpm; approximately 10 ng) was hybridized with 50 μg of RNA that had been prepared as described previously (9). Lanes 1 through 7, S1 nuclease-protected fragments of the probe that had been hybridized with RNA isolated from JH642 cells which had been incubated for 0, 2, 5, 10, 20, 30, and 60 min, respectively, after the addition of 1 mM arsenate. Lane 8, 32P-labelled HpaII fragments of phage M13mp18 as size markers. (D) Primer extension mapping of the ars transcript. The primer extension reaction was performed as described previously (27). Dideoxynucleotide sequencing reactions, performed with the same primer as used to map the 5′ end of the ars transcript, were included in the analysis. The complementary base at the initiation site is indicated by an asterisk.
Time course of induction of expression of the ars operon by arsenate.
The transcription of the ars operon in B. subtilis probably starts in the region upstream of arsR, as shown by S1 nuclease mapping with the 573-bp probe that contained part of arsR and its upstream region. A single protected 145-nt band starting about 30 nt upstream from the initiation codon of arsR was detected (Fig. 4C). The rate of transcription of the ars operon reached a maximum ∼20 min after the start of arsenate induction. In the absence of arsenate, only a low basal level of expression of the ars operon was detected (data not shown).
Site for initiation of ars transcription.
The start site for ars mRNA was determined precisely by the primer extension method with primer 4. The 5′ terminus of ars mRNA was located 34 nt upstream from the initiation codon of arsR. The promoter region contains −10 and −35 regions similar to those of promoters recognized by EςA (−35, TTGACA; −10, TATAAT [17]) (Fig. 4A and D).
DISCUSSION
Kunkel et al. (14) reported that deletion of the entire skin element did not impair the growth and sporulation of B. subtilis. Our results suggest that an important physiological role of the skin element is to endow cells with resistance to arsenicals.
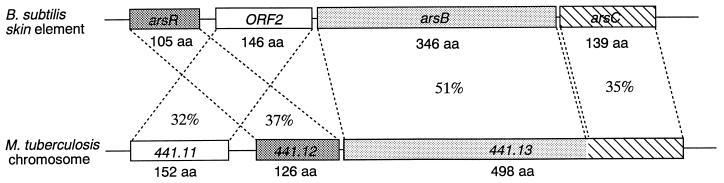
The ars operon of the E. coli plasmids R773 (5, 6) and R46 (3) consists of five genes (arsR, arsD, arsA, arsB, and arsC). The ArsR protein is a trans-acting repressor. The ArsD protein is a secondary regulator of transcription of the ars operon, and its absence has little effect on the level of resistance (35). The arsA gene encodes an arsenite- and antimonite-stimulated ATPase. The ArsB protein is an inner membrane protein (26, 36) which participates in the pumping of arsenite across the inner membrane. ArsC is an enzyme that reduces the less toxic arsenate ion to the more toxic arsenite ion (11). arsD and arsA are missing from staphylococcal plasmids and the E. coli chromosome. The product of the arsR gene of B. subtilis is 31 to 33% identical to ArsR proteins, while the arsC product is 17 to 18% identical to the ArsC proteins of gram-negative strains and 62 to 64% identical to the ArsC proteins of gram-positive strains. The newly reported homolog of the ars operon in the Mycobacterium tuberculosis chromosome consists of three ORFs (441.11, 441.12, and 441.13; the accession number for the nucleotide sequence containing these proteins is Z80225) (Fig. 5). The products of B. subtilis arsR and ORF2 are 37 and 32% identical to the hypothetical proteins 441.12 (126 amino acids) and 441.11 (152 amino acids), respectively. We also found that the amino acid sequence of the arsB product is strongly homologous (51%) to that of a hypothetical polypeptide, the amino-terminal region of protein 441.13 of M. tuberculosis (Fig. 5). It is of interest that the carboxy-terminal region of protein 441.13 is also similar to the product of arsC of B. subtilis. No functional analysis of proteins 441.11 and 441.13 has been reported to our knowledge.
FIG. 5.
Comparison of the B. subtilis ars operon and the probable ars operon of M. tuberculosis. aa, amino acid residues.
The arsB gene endows cells with resistance to arsenite. The ArsB protein is a hydrophobic protein, and it seems likely to be a membrane-associated protein. The B. subtilis ArsB protein shows very limited identity to the ArsB proteins of pI258, pSX267, R773, and E. coli (36). However, the hydropathy profile indicates that the B. subtilis ArsB protein has 10 membrane-spanning regions (data not shown), suggesting that the B. subtilis ArsB protein might have a function similar to that of other ArsB proteins (which have 12 membrane-spanning regions [36]). Recently, Bobrowicz et al. (2) revealed that three contiguous genes, ACR1, ACR2, and ACR3, are involved in arsenical resistance in Saccharomyces cerevisiae. Interestingly, they showed that the ACR3 gene has high similarity to B. subtilis arsB (ORF1), suggesting that B. subtilis and S. cerevisiae possess similar proteins for extrusion of arsenite.
The transcription of the ars operon starts just upstream of the initiation codon of arsR in staphylococcal plasmids (10, 24), in R773 of E. coli (34), and on the E. coli chromosome (37). Wu and Rosen (34) found that the ars operon of the E. coli plasmid R773 is repressed by an ArsR dimer bound to a region of imperfect 7-bp dyad symmetry that is located just upstream of the −35 region of the site of initiation of transcription of arsR. However, when the inducer (arsenite) is present, the ArsR dimer is released from the operator (34). In plasmid pI258 of S. aureus, ars transcription starts 6 bp downstream of a 6-bp inverted repeat (10), and Rosenstein et al. (24) showed that ArsR of S. xylosus (pSX267) also binds to a sequence with dyad symmetry (IR1, IR2, and IR3) within the ars promoter. Inactivation of B. subtilis arsR resulted in constitutive expression of ars, indicating a function for the ArsR protein as a negative regulator of the ars operon. In the promoter region of the ars operon of B. subtilis, there is an 8-bp inverted repeat that might function as a regulatory site (Fig. 4A), but the inverted repeat exhibits no significant similarity to those in the ars operons of R773, pI258, and pSX267.
More detailed analysis is necessary to define the similarities in and differences between the modes of regulation of the ars operons in B. subtilis, Staphylococcus, and E. coli and the probable ars operon of M. tuberculosis.
ACKNOWLEDGMENTS
We thank K. Asai and the Bacillus Genetic Stock Center (Ohio State University) for providing plasmid pMUTinT3 and strain QUA2, respectively.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Adams A, Oishi M. Genetic properties of arsenate-sensitive mutants of Bacillus subtilis 168. Mol Gen Genet. 1972;118:295–310. doi: 10.1007/BF00333565. [DOI] [PubMed] [Google Scholar]
- 1a.Asai, K. Personal communication.
- 2.Bobrowicz P, Wysocki R, Owsianik G, Goffeau A, Ulaszewski S. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces crevisiae. Yeast. 1997;13:819–828. doi: 10.1002/(SICI)1097-0061(199707)13:9<819::AID-YEA142>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 3.Bruhn D F, Li J, Silver S, Roberto F, Rosen B P. The arsenical resistance operon of IncN plasmid R46. FEMS Microbiol Lett. 1996;139:149–153. doi: 10.1111/j.1574-6968.1996.tb08195.x. [DOI] [PubMed] [Google Scholar]
- 4.Carlin A, Shi W, Dey S, Rosen B P. The ars operon of Escherichia coli confers arsenical and antimonical resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C-M, Misra T K, Silver S, Rosen B P. Nucleotide sequence of the structural genes for an anion pump: the plasmid-encoded arsenical resistance operon. J Biol Chem. 1986;261:15030–15038. [PubMed] [Google Scholar]
- 6.Chen C-M, Mobley H L T, Rosen B P. Separate resistances to arsenate and arsenite (antimonite) encoded by the arsenical resistance operon of R factor R773. J Bacteriol. 1985;161:758–763. doi: 10.1128/jb.161.2.758-763.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington J, Rong S, Rosenkrantz M S, Sonenshein A L. Transcriptional regulation and structure of the Bacillus subtilis sporulation locus spoIIIC. J Bacteriol. 1988;170:1162–1167. doi: 10.1128/jb.170.3.1162-1167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladysheva T B, Oden K L, Rosen B P. Properties of the arsenate reductase of plasmid R773. Biochemistry. 1994;33:7288–7293. doi: 10.1021/bi00189a033. [DOI] [PubMed] [Google Scholar]
- 9.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 10.Ji G, Silver S. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J Bacteriol. 1992;174:3684–3694. doi: 10.1128/jb.174.11.3684-3694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji G, Silver S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci USA. 1992;89:9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji G, Garber E A, Armes L G, Chen C-M, Fuchs J A, Silver S. Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry. 1994;33:7294–7299. doi: 10.1021/bi00189a034. [DOI] [PubMed] [Google Scholar]
- 13.Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the developmental transcription factor ςK is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–535. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel B, Sandman K, Panzer S, Youngman P, Losick R. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J Bacteriol. 1988;170:3513–3522. doi: 10.1128/jb.170.8.3513-3522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 16.Mizuno M, Masuda S, Takemaru K, Hosono S, Sato T, Takeuchi M, Kobayashi Y. Systematic sequencing of the 283-kb region (210° ∼232°) of the Bacillus subtilis genome containing the skin element and many sporulation genes. Microbiology. 1996;142:3103–3111. doi: 10.1099/13500872-142-11-3103. [DOI] [PubMed] [Google Scholar]
- 17.Moran C P, Jr, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 18.Mueller J P, Bukusoglu G, Sonenshein A L. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neyt C, Iriarte M, Thi V H, Cornelis G R. Virulence and arsenic resistance in yersiniae. J Bacteriol. 1997;179:612–619. doi: 10.1128/jb.179.3.612-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oden K L, Gladysheva T B, Rosen B P. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol. 1994;12:301–306. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- 21.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 22.Rosen B P, Silver S, Gladysheva T B, Ji G, Oden K L, Jagannathan S, Shi W, Chen Y, Wu J. The arsenite oxyanion-translocating ATPase: bioenergetics, functions, and regulation. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms. Washington, D.C: American Society for Microbiology; 1995. pp. 97–108. [Google Scholar]
- 23.Rosenstein R, Peschel A, Wieland B, Götz F. Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J Bacteriol. 1992;174:3676–3683. doi: 10.1128/jb.174.11.3676-3683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenstein R, Nikoleit K, Götz F. Binding of ArsR, the repressor of the Staphylococcus xylosus (pSX267) arsenic resistance operon to a sequence with dyad symmetry within the ars promoter. Mol Gen Genet. 1994;242:566–572. doi: 10.1007/BF00285280. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.San Francisco M J D, Tisa L S, Rosen B P. Identification of the membrane component of the anion pump encoded by the arsenical resistance operon of R-factor R773. Mol Microbiol. 1989;3:15–21. doi: 10.1111/j.1365-2958.1989.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 27.Sato T, Harada K, Kobayashi Y. Analysis of suppressor mutations of spoIVCA mutations: occurrence of DNA rearrangement in the absence of a site-specific DNA recombinase SpoIVCA in Bacillus subtilis. J Bacteriol. 1996;178:3380–3383. doi: 10.1128/jb.178.11.3380-3383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T, Samori Y, Kobayashi Y. The cisA cistron of Bacillus subtilis sporulation gene spoIVCA encodes a protein homologous to a site-specific recombinase. J Bacteriol. 1990;172:1092–1098. doi: 10.1128/jb.172.2.1092-1098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silver S, Budd K, Leahy K M, Shaw W V, Hammond D, Novick R P, Willsky G R, Malamy M H, Rosenberg H. Inducible plasmid-determined resistance to arsenate, arsenite, and antimony(III) in Escherichia coli and Staphylococcus aureus. J Bacteriol. 1981;146:983–996. doi: 10.1128/jb.146.3.983-996.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Ann Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 31.Sofia H J, Burland V, Daniels D L, Plunkett III G, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 33.Takemaru K, Mizuno M, Sato T, Takeuchi M, Kobayashi Y. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology. 1995;141:323–327. doi: 10.1099/13500872-141-2-323. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Rosen B P. Metalloregulated expression of the ars operon. J Biol Chem. 1993;268:52–58. [PubMed] [Google Scholar]
- 35.Wu J, Rosen B P. The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol Microbiol. 1993;8:615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 36.Wu J, Tisa L S, Rosen B P. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. J Biol Chem. 1992;267:12570–12576. [PubMed] [Google Scholar]
- 37.Xu C, Shi W, Rosen B P. The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem. 1996;271:2427–2432. doi: 10.1074/jbc.271.5.2427. [DOI] [PubMed] [Google Scholar]
- 38.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]