Abstract
The Saccharomyces cerevisiae SCS2 gene has been cloned as a suppressor of inositol auxotrophy of CSE1 and hac1/ire15 mutants (J. Nikawa, A. Murakami, E. Esumi, and K. Hosaka, J. Biochem. 118:39–45, 1995) and has homology with a synaptobrevin/VAMP-associated protein, VAP-33, cloned from Aplysia californica (P. A. Skehel, K. C. Martin, E. R. Kandel, and D. Bartsch, Science 269:1580–1583, 1995). In this study we have characterized an SCS2 gene product (Scs2p). The product has a molecular mass of 35 kDa and is C-terminally anchored to the endoplasmic reticulum, with the bulk of the protein located in the cytosol. The disruption of the SCS2 gene causes yeast cells to exhibit inositol auxotrophy at temperatures of above 34°C. Genetic studies reveal that the overexpression of the INO1 gene rescues the inositol auxotrophy of the SCS2 disruption strain. The significant primary structural feature of Scs2p is that the protein contains the 16-amino-acid sequence conserved in yeast and mammalian cells. The sequence is required for normal Scs2p function, because a mutant Scs2p that lacks the sequence does not complement the inositol auxotrophy of the SCS2 disruption strain. Therefore, the Scs2p function might be conserved among eukaryotic cells.
The Saccharomyces cerevisiae SCS2 gene was identified as a multicopy suppressor of inositol auxotrophy of CSE1 and ire15 mutants (25). CSE1 mutants show dominant inositol auxotrophy in the presence of choline in the growth medium (14). CSE1 mutants cannot activate the expression of INO1, which encodes inositol-1-phosphate synthase, an essential protein for inositol biosynthesis in yeast cells (8). In yeast, the expression of INO1 and other phospholipid biosynthetic genes is regulated in response to the amount of the soluble lipid precursors inositol and choline (3, 27). Genetic analysis of CSE1 mutants has revealed that CSE1 is a factor involved in the regulation of INO1 expression, although the gene has not been cloned yet (13).
ire15 mutants have defects in the expression of the inositol transporter gene (ITR1) in addition to that of INO1. Three human genes which can suppress the growth defect of ire15 mutants have been isolated. They encode transforming growth factor β receptor type IIB, protein phosphatase type 2A subunit A, and the 14-3-3 protein (23). These results suggest that yeast cells contain a signal transduction mechanism resembling the human transforming growth factor β receptor-mediated pathway to induce the expression of inositol biosynthetic genes (23). The gene responsible for the ire15 mutation is identical to HAC1, which encodes a transcriptional factor with a basic leucine zipper motif (24, 29).
A relationship between inositol metabolism and a signal transduction mechanism is also suggested by recent work on the IRE1 gene (5, 20). Although IRE1 was originally identified as a gene required for inositol prototrophy (26), it is also required for the transcription of KAR2, which encodes a protein chaperon, BiP, in response to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). The IRE1 gene product is a member of transmembrane serine/threonine kinases and lies in the ER/nuclear membrane. It is thought that Ire1p transmits a signal of unfolded-protein accumulation in the ER to the nucleus by a mechanism similar to those found in transmembrane kinases in the plasma membranes of higher eukaryotic cells (35).
Recently, a gene which has partial homology to SCS2 has been cloned from Aplysia californica (37). The gene encodes the synaptobrevin/VAMP-associated protein VAP-33, which was identified by using the yeast two-hybrid system (37). Synaptobrevin is localized to the surface of synaptic vesicles and associates with syntaxin and SNAP-25, which are localized to the presynaptic membranes (38). Through the interaction of these proteins, the synaptic vesicles fuse with the presynaptic membranes and neurotransmitters are released from the vesicles. Since presynaptic injection of antibodies to VAP-33 inhibited the synaptic transmission, VAP-33 is considered to be required for the exocytosis of neurotransmitters (37). As synaptobrevin homologs have been isolated from yeast and are involved in protein secretion pathways (10, 32), it is likely that VAP-33 homologs exist in yeast and participate in the regulation of yeast exocytic pathways.
The ability of SCS2 to suppress the inositol auxotrophy of CSE1 and hac1/ire15 mutants and its structural relationship to VAP-33 lead to the assumption that SCS2 is involved both in the regulation of membrane biogenesis through the activation of phospholipid biosynthetic gene expression and in intracellular membrane transport through the activation of fusion of transport vesicles. To investigate this assumption, we have characterized the SCS2 gene product. In this paper we show that the gene is involved in the activation of the INO1 expression and that the gene product (Scs2p) is a 35-kDa type II integral membrane protein. On the other hand, we failed to obtain any line of evidence that the gene is required for protein secretion, although Scs2p is localized to the ER, where protein and lipid biosynthesis and transport vesicle formation take place.
MATERIALS AND METHODS
Media and strains.
Yeast extract-peptone-dextrose (YPD) and yeast minimal media were described by Kaiser et al. (16) and Klig et al. (17), respectively. When added, inositol (myo-inositol; Sigma) was at a final concentration of 100 μM. The preparation of INO1 and INO2 genes was described by Kagiwada et al. (15). Strains used were CTY182 (MATa ura3-52 his3-Δ200 lys2-801 [2]), YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 [36]), YPH501 (MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 [36]), KY356 (CTY182 scs2Δ::URA3), and KY360 (YPH500 scs2Δ::TRP1).
Construction of SCS2 vectors.
The SCS2 gene was amplified by PCR from pSC2, which contains the HindIII-ClaI fragment of the SCS2 genome (25). The forward and reverse primers used were 5′-CCAAGCTTTGCATAGCGCACGC-3′ and 5′-CCGAATTCTAGTATTGTAAAGGC-3′, respectively. An EcoRI site was engineered at the 5′ region of the reverse primer. The PCR fragment was cut with HindIII and EcoRI, and the 1.3-kb fragment was inserted into the same sites of YEplac195 and YCplac33 (11) to generate pKY134 [YEp(SCS2)] and pKY151 [YCp(SCS2)], respectively.
SCS2 disruption.
To disrupt the SCS2 gene, the coding region corresponding to amino acids 4 through 219 was replaced with the URA3 or TRP1 gene. To this end, an additional PstI site, other than the endogenous one, was generated in the SCS2 gene by mutagenesis of the nucleotide at position +9 from T to A (position +1 refers to the A residue of the ATG start codon). The resultant plasmid was cut with PstI and self-ligated to generate pKY144, which lacks 648 nucleotides from position +11 to +658. The URA3 or TRP1 marker gene was incorporated into the PstI site of pKY144 to generate pKY145 or pKY159, respectively. The HindIII-EcoRI fragments of pKY145 and pKY159 were used for one-step gene replacement (16). YPH500, YPH501, and CTY182 were used for SCS2 disruption. The identities of disruption strains were verified by PCR analysis of genomic DNA prepared from transformed cells and Western blotting.
Polyclonal antibody.
A glutathione S-transferase (GST)–Scs2 fusion protein was constructed for preparation of an anti-Scs2p polyclonal antibody. PCR was performed with pKY134 as a template. The forward and reverse primers, 5′-GGATCCCCTGACGTGTTGGTG-3′ and 5′-GAATTCATTTTCTGCAGGTACG-3′, respectively, were constructed to place a BamHI site at the 5′ end and an EcoRI site at the 3′ end of the 645-bp segment of SCS2 which corresponds to residues 7 through 221. The PCR product was cut with BamHI and EcoRI and ligated into BamHI-EcoRI-digested pGEX4T-1 (Pharmacia Biotech) to yield pKY149. Proteins which were expressed in response to induction with isopropyl-β-d-thiogalactopyranoside were purified from DH5α cells harboring pKY149 by using Bulk GST Purification Modules (Pharmacia Biotech). Rabbit antiserum against the purified proteins was passed through GST-Sepharose 4B beads to remove antibodies against GST, and then the serum was affinity purified with antigen conjugated to Sepharose 4B beads.
HA epitope tagging.
A 9-amino-acid epitope recognized by antihemagglutinin (anti-HA) antibody was introduced into the SCS2 coding sequence by the following method. The SCS2 gene was modified by mutagenesis of the nucleotide at position +12 from T to A to introduce an AccI site. This modification does not change the SCS2 amino acid sequence. The AccI site was used for insertion of the PCR product amplified by using the forward primer 5′-GTATACCCATACGATGTTCCAGATTACGCTGAAATTTCCCCTGACGTG-3′ and the reverse primer 5′-CCGAATTCTAGTATTGTAAAGGC-3′. The forward primer was constructed to place the HA-coding sequence 5′ to nucleotide +13. The resulting fragment was subcloned into YCplac33 and YEplac195 to generate pKY166 [YCp(HA-SCS2)] and pKY167 [YEp(HA-SCS2)]. By this construction, the N terminus of Scs2p was changed from MSAVEI to MSAVYPYDVPDYAEI (the HA epitope residues are underlined).
SCS2(Δ36-53) construction.
To construct SCS2(Δ36-53), a DNA fragment (AA1-35) which contains 989 bp from nucleotide −442 to +547 was ligated with a DNA fragment (AA54-243) which contains 726 bp from nucleotide +602 to +1327. AA1-35 was amplified by PCR with the primer 5′-CCAAGCTTTGCATAGCGCACGC-3′ and 5′-CTGCAGTGGTTTGGTCTGAATTGTTGG-3′. AA54-243 was amplified with the primers 5′-CTGCAGTTGTTGCTCCAGGTG-3′ and 5′-CCGAATTCTAGTATTGTAAAGGC-3′. AA1-35 was digested with HindIII and PstI and subcloned into HindIII-PstI-digested pKY144 to create pKY168. AA54-243 was cut with PstI and subcloned into PstI-digested pKY168 to create YEp(SCS2Δ36-53). YEp(SCS2Δ36-53) was cut with HindIII and EcoRI and was inserted into the same site of YCplac33 to generate YCp(SCS2Δ36-53).
Cell fractionation.
Preparation of subcellular fractions was performed as described by Cleves et al. (4) with minor modifications. Appropriate yeast strains were grown to an optical density at 600 nm of 0.4 in 100 ml of YPD with shaking at 26°C. The cells were washed with 10 mM NaN3, converted to spheroplasts by resuspension in 10 ml of spheroplasting buffer (1.1 M sorbitol, 50 mM potassium phosphate [pH 7.4], 10 mM NaN3) containing 20 μg of Zymolyase 20T (Seikagaku Kogyo) per ml and 5 μg of β-mercaptoethanol per ml, and incubated at 30°C for 60 min. Spheroplasts were washed with the spheroplasting buffer and resuspended in 6 ml of ice-cold lysis buffer (0.3 M sorbitol, 10 mM potassium phosphate, pH 7.4). The cells were incubated on ice for 20 min with occasional gentle agitation. Lysates were adjusted to 1.1 M sorbitol and centrifuged at 800 × g for 5 min to remove unlysed cells. The low-speed supernatant (LSS) was centrifuged at 12,000 × g for 15 min to yield pellet (12P) and supernatant (12S) fractions. The 12S fraction was centrifuged at 100,000 × g for 1 h in a TLA45 rotor (Beckman Instruments) to yield pellet (100P) and supernatant (100S) fractions.
Treatment of membranes.
In order to extract peripheral proteins associated with the 12P fraction, 40 μl of the 12P fraction was incubated with 10 μl of either H2O, 5 M KCl, 10 M urea, 0.5 M Na2CO3, or 5% Triton X-100. The mixtures were incubated for 30 min on ice and centrifuged at 100,000 × g for 30 min. Pellets were resuspended in 50 μl of lysis buffer. For the protease protection assay, the 12P fraction of KY360 harboring YCp(HA-SCS2) was incubated with 0.1 mg of trypsin (type III; Sigma) per ml in the presence or absence of 0.1% Triton X-100. At the beginning or end of the incubation (4°C, 30 min), soybean trypsin inhibitor (Sigma) was added to a final concentration of 0.4 mg/ml.
Immunoblotting.
Protein samples were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and transferred to a nitrocellulose membrane (0.45-μm pore size; Toyo Roshi). Immunoblotting was performed by using the anti-Scs2p polyclonal antibody, a mouse anti-HA monoclonal antibody (clone 12CA5; Boehringer Mannheim), a mouse anti-Dpm1p monoclonal antibody (clone 5C5-A7; Molecular Probes), a rabbit anti-Kar2p polyclonal antibody (a gift from K. Kohno, Nara Institute of Science and Technology, Nara, Japan) (41), or a rabbit anti-Sec14p polyclonal antibody (a gift from V. A. Bankaitis, University of Alabama at Birmingham) (2). Secondary antibodies were alkaline phosphatase-conjugated anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG (Promega).
Immunofluorescence.
Appropriate yeast strains were grown to the early logarithmic stage in complete or uracil-deficient medium for plasmid maintenance. Cells were fixed by direct addition of formaldehyde (final concentration, 4%) and incubation with gentle agitation at 30°C for 30 min. Cells were centrifuged at 700 × g for 5 min, resuspended in 1 ml of spheroplasting buffer containing 4% formaldehyde, and incubated overnight at 4°C with gentle agitation. Fixed cells were centrifuged and resuspended in 0.5 ml of spheroplasting buffer containing 20 μg of Zymolyase 20T per ml and 5 μg of β-mercaptoethanol per ml and incubated for 1 h at 30°C. Spheroplasts were centrifuged, washed once, and applied to poly-l-lysine-coated coverslips. Cells were treated with ice-cold methanol for 5 min and blocked with 1 mg of bovine serum albumin per ml dissolved in phosphate-buffered saline (PBS). Primary antibodies used were the rabbit anti-Scs2p polyclonal antibody, a rabbit anti-HA polyclonal antibody (a gift from R. Hirata, The Institute of Physical and Chemical Research [RIKEN], Wako, Japan) (12), and the anti-Dpm1p monoclonal antibody. Secondary antibodies were fluorescein isothiocyanate-conjugated anti-mouse IgG and tetramethylrhodamine isothiocyanate-conjugated anti-rabbit IgG (Biomedical Technologies). Antibody incubations were for 1 h at room temperature, with four washes with PBS–0.1% Tween 20. Prior to a final rinse, cells were incubated with 5 μg of DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Sigma) per ml. Cells were mounted with 90% glycerol–10% PBS containing 1 mg of p-phenylenediamine (Sigma) per ml. Fluorescence images were recorded with a fluorescence microscope equipped with a cooled charge-coupled device camera (PROVIS AX-70; Olympus).
Secretion assay.
Secretion of invertase was analyzed by invertase activity staining (21). Cells were grown in YPD medium at 25°C and shifted to YP with 0.1% glucose. After incubation at 30°C for 2 h, the cells were washed with ice-cold 10 mM NaN3 and were converted to spheroplasts as described above. After centrifugation of the spheroplasts at 800 × g for 3 min, intracellular (pellet) and extracellular (supernatant) fractions were obtained. The intracellular fraction was resuspended in 0.5 ml of 10% glycerol containing 2% Triton X-100. Samples were resolved on 6.75% nondenaturing polyacrylamide gels. After electrophoresis, gels were incubated in 0.2 M sodium acetate (pH 4.8) containing 0.2 M sucrose for 1 h at 30°C and were stained with 0.1% 2,3,5-triphenyltetrazolium chloride in 0.1 M NaOH. A halo assay was carried out according to the method of Sprague (39) with RC687 (MATa sst2) as a tester strain.
RESULTS
SCS2 disruption mutants show inositol auxotrophy.
In order to study the physiological role of the SCS2 gene, we have constructed SCS2 disruption (scs2Δ) strains. A diploid strain (YPH501) was transformed with the scs2::URA3 gene, in which the SCS2 coding region for residues 7 through 221 was replaced with URA3, and Ura+ transformants were selected and purified. Transformants which had both intact SCS2 and scs2::URA3 genes were selected and subjected to sporulation and tetrad analysis. All four viable spores from 20 tetrads grew on a YPD plate. The ability to generate haploid yeast strains with the scs2 null allele as the sole copy of this gene demonstrated that the SCS2 gene was not essential for yeast viability. This finding made it possible to construct SCS2 disruption strains by transforming haploid cells directly. To this end, two independent yeast strains, CTY182 and YPH500, were transformed with scs2::URA3 and scs2::TRP1, respectively, and transformants were purified. Since isogenic strains were available, we studied the nature of scs2Δ by using the disruption strains (KY356 and KY360) derived from CTY182 and YPH500 for further studies.
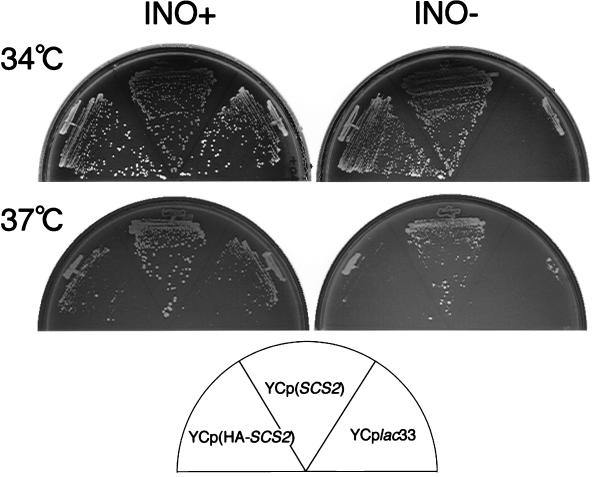
As the SCS2 gene was originally isolated as a suppressor of the inositol auxotrophy of CSE1 and hac1/ire15 mutants (25), we examined the viability of scs2Δ mutants on inositol-free medium. As expected, an scs2Δ strain (KY360) could not grow well on inositol-free medium compared to the parental strain (YPH500). The growth defect was marked when cells were incubated at temperatures of above 34°C (Fig. 1). Another scs2Δ strain (KY356) derived from CTY182 also showed a similar growth deficiency. Therefore, the inositol auxotrophy was independent of genetic background and marker genes. To prove that the inositol auxotrophy was caused by the SCS2 gene disruption, we examined whether incorporation of the SCS2 gene into the scs2Δ strain rescues the auxotrophy. As shown in Fig. 1, scs2Δ strains harboring SCS2 on a centromere-based (CEN) plasmid [YCp(SCS2)] could grow on inositol-free medium at 37°C. Interestingly overproduction of SCS2 from a multicopy 2μm plasmid [YEp(SCS2)] could not rescue the defect efficiently (see Fig. 5B). Since even wild-type cells (CTY182) did not grow well at 37°C when the SCS2 gene was overexpressed by the 2μm plasmid (data not shown), overproduction of the SCS2 gene would be toxic to yeast at 37°C.
FIG. 1.
Inositol auxotrophy of scs2Δ strains. KY360 (scs2Δ::TRP1) cells transformed with YCplac33 (control), YCp(SCS2), or YCp(HA-SCS2) were streaked for isolation on either inositol-containing (INO+) or inositol-free (INO−) minimal medium and incubated at 34°C for 72 h or at 37°C for 96 h.
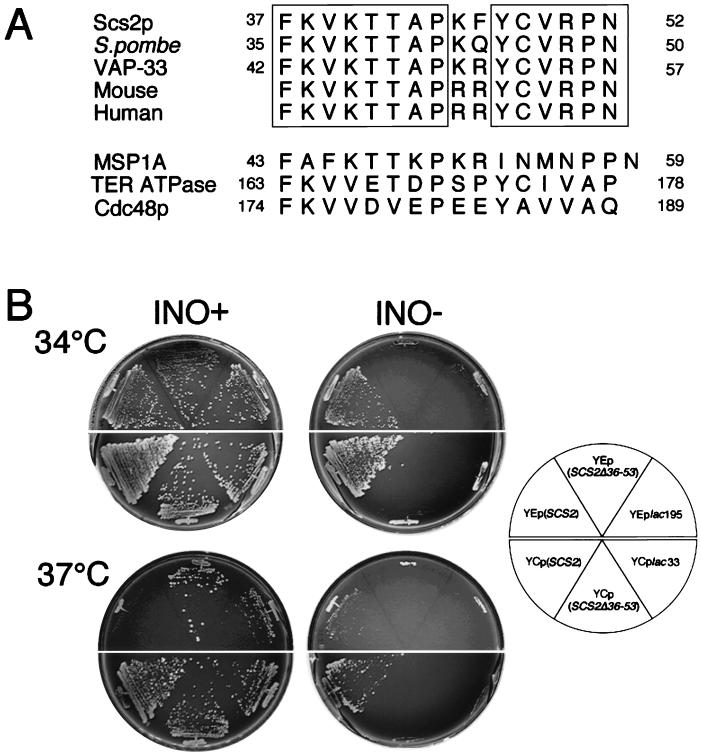
FIG. 5.
The 16-amino-acid conserved region is required for Scs2p activity. (A) Alignment of a 16-amino-acid sequence of Scs2p with those of S. pombe, A. californica (VAP-33), and mouse and human homologs. The sequences of the mouse and human homologs are deduced from nucleotide sequences of expressed sequence tags. Residues identical in all five sequences are boxed. Homologous sequences of MSP1A, TER ATPase, and Cdc48p are also shown. For Scs2p, the S. pombe homolog, VAP-33, MSP1A, TER ATPase, and Cdc48p, the numbers refer to amino acid positions. GenBank accession numbers are D44493 (Scs2p), Z73099 (S. pombe), U36779 (VAP-33), W54842 (mouse), N34715 (human), P53021 (MSP1A), U11760 (TER ATPase), and X56956 (Cdc48p). (B) Overproduction of Scs2pΔ36-53 failed to suppress the inositol auxotrophy of the scs2Δ strain. KY360 (scs2Δ::TRP1) cells transformed with YEplac195, YEp(SCS2), YEp(SCS2Δ36-53), YCplac33, YCp(SCS2), or YCp(SCS2Δ36-53) were streaked for isolation on either inositol-containing (INO+) or inositol-free (INO−) minimal medium and incubated at 34°C for 72 h or at 37°C for 96 h.
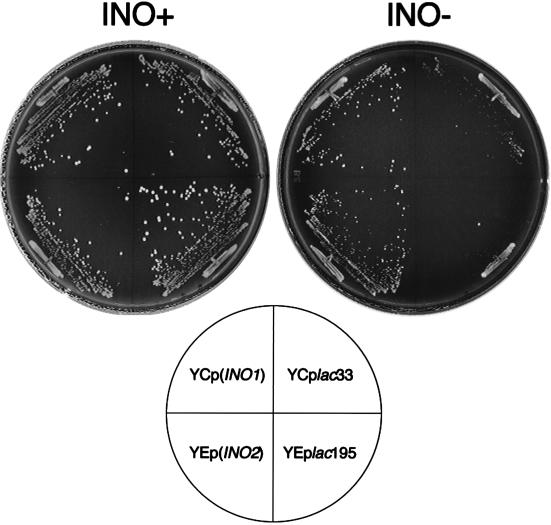
In yeast an essential step of inositol biosynthesis is the conversion of glucose-6-phosphate to inositol-1-phosphate, which is catalyzed by the INO1 gene product. The expression of the INO1 gene is controlled by the positive regulators INO2 and INO4, which encode basic helix-loop-helix proteins. The INO2 and INO4 gene products form a heterodimer that interacts with the upstream activating sequence of the INO1 gene and activates INO1 expression (1). To investigate the cause for the scs2Δ inositol auxotrophy, the INO1 or INO2 gene was incorporated into an scs2Δ strain (KY360). As shown in Fig. 2, overproduction of INO1 from the CEN plasmid [YCp(INO1)] and of INO2 from the 2μm plasmid [YEp(INO2)] could rescue the growth defect. The results suggest that an increase in INO1 expression levels can rescue the scs2Δ inositol auxotrophy.
FIG. 2.
Inositol auxotrophy of scs2Δ strains transformed with INO1 or INO2. KY360 (scs2Δ::TRP1) cells transformed with YCplac33 [vector control for YCp(INO1)], YEplac195 [vector control for YEp(INO2)], YCp(INO1), or YEp(INO2) were streaked for isolation on either inositol-containing (INO+), or inositol-free (INO−) minimal medium and incubated at 34°C for 72 h.
Scs2p is a 35-kDa type II integral membrane protein.
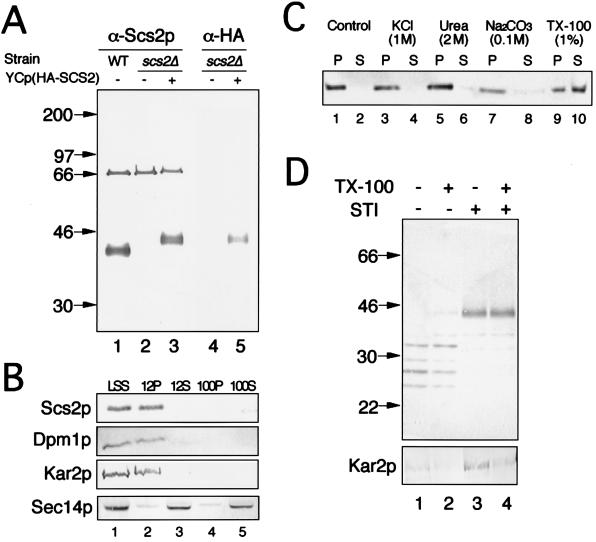
To gain insight into the physiological role of SCS2, we investigated the nature of the SCS2 gene product (Scs2p). An affinity-purified polyclonal antibody raised against a GST-SCS2 fusion protein recognized a 35-kDa band in the LSS of wild-type cells (YPH500) (Fig. 3A, lane 1), and the signal intensity of the band was increased in the lysate of cells carrying YEp(SCS2) (data not shown). The 35-kDa band was not observed in the fraction prepared from an scs2Δ strain (KY360) (Fig. 3A, lane 2), suggesting that structurally homologous proteins with similar molecular masses were not expressed to the extent that they could be visualized by Western blotting. Other than the 35-kDa band, a 66-kDa band, which was not reacted with a preimmune serum, was detected in lysates of scs2Δ cells (Fig. 3A, lane 2).
FIG. 3.
Characterization of Scs2p by Western blotting. (A) Strains were grown in minimal medium, converted to spheroplasts, and lysed osmotically. The resultant lysate was centrifuged at 800 × g to yield LSS. The LSS was separated by SDS–10% PAGE and immunoblotted with the anti-Scs2p polyclonal antibody (α-Scs2p) (lanes 1 to 3) or the anti-HA monoclonal antibody (α-HA) (lanes 4 and 5). The strains employed were YPH500 (wild type [WT]) (lane 1) and KY360 (scs2Δ::TRP1) harboring either YCplac33 (lanes 2 and 4) or YCp(HA-SCS2) (lanes 3 and 5). (B) The LSS fraction of wild-type cells (CTY182) was subjected to two rounds of differential centrifugation to yield 12,000 × g pellet and supernatant fractions (12P and 12S, respectively) and 100,000 × g pellet and supernatant fractions (100P and 100S, respectively). These fractions were resolved by SDS–10% PAGE and immunoblotted for the presence of Scs2p and for markers of the ER membrane (Dpm1p), ER lumen (Kar2p), and cytoplasm/Golgi membrane (Sec14p). (C) The 12P fraction of wild-type cells (CTY182) was incubated with 0.2 volume of one of the following solutions: H2O (control), 5 M KCl, 10 M urea, 0.5 M Na2CO3 (pH 11), or 5% Triton X-100 (TX-100). After incubation at 4°C for 30 min, samples were centrifuged at 100,000 × g for 30 min to separate supernatant (S) and pellet (P) fractions. These fractions were resolved by SDS–10% PAGE and immunoblotted with the anti-Scs2p polyclonal antibody. (D) The 12P fraction of KY360 (scs2Δ::TRP1) harboring YCp(HA-SCS2) was incubated with 0.1 mg of trypsin per ml in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 0.1% Triton X-100 (TX-100). Soybean trypsin inhibitor (STI) was added at the beginning (lanes 3 and 4) or end (lanes 1 and 2) of the incubation. Samples were resolved by SDS–10% PAGE and immunoblotted with the anti-HA monoclonal antibody (top panel) or the anti-Kar2p polyclonal antibody (bottom panel). In panels A and C, the numbers on the left are molecular masses in kilodaltons.
The nucleotide sequence of SCS2 predicts a protein of 26.9 kDa. The discrepancy between the estimated and the observed molecular masses indicates the existence of posttranslational modifications. In fact, the SCS2-encoded sequence contains three potential N-linked glycosylation sites (25). However, since Scs2p does not appear to be sensitive to digestion with endoglycosidase Hf (data not shown), it is unlikely that these sites are utilized. In addition, phosphorylation of Scs2p was not detected by immunoprecipitation of cell lysates labeled with [32P]orthophosphate (data not shown).
A hydrophobic stretch of 16 amino acids at the carboxy terminus of Scs2p (25) suggests that it is bound to membranes by insertion of this region into the membranes. Cell lysates of a wild-type strain (CTY182) were subjected to a series of centrifugation steps at 800, 12,000, and 100,000 × g. The supernatant fractions at 800, 12,000, and 100,000 × g are referred to as LSS, 12S, and 100S, respectively, and the pellet fractions at 12,000 and 100,000 × g are called 12P and 100P, respectively. As shown in Fig. 3B, Scs2p detected by Western blotting was found exclusively in the LSS and 12P fractions, in which the ER and the nuclear membranes are enriched (4). In fact, the ER membrane protein marker, dolichol phosphate mannose synthase (Dpm1p) (31), was found exclusively in the LSS and 12P fractions. The ER luminal protein marker, Kar2p (34), was also enriched in those fractions, indicating that the integrity of the membrane fractions remained intact during the centrifugation. As expected, the cytosolic/Golgi protein marker, a phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) (2, 4), was associated mainly with the 12S and 100S fractions. These results indicate that Scs2p is associated with membranes. This was also confirmed by differential solubilization of the 12P fraction. Scs2p was not released into the supernatant by treatment with a high salt concentration (1 M KCl), sodium carbonate (pH 11), or 2 M urea, all of which extract peripheral membrane proteins (Fig. 3C, lanes 1 to 8). On the other hand, about 60% of Scs2p was solubilized in the presence of 1% Triton X-100 (Fig. 3C, lanes 9 and 10). These results suggest that Scs2p is an integral membrane protein.
The topology of Scs2p with respect to the cytosol was examined by a protease protection assay. For this assay, the N-terminal region of the protein should be recognized specifically by a monoclonal antibody. To this end, we constructed an HA-SCS2 gene, which encodes an SCS2 gene product tagged with nine amino acids from the influenza virus HA protein (the HA tag) at its amino terminus. The fusion protein (HA-Scs2p) encoded by the HA-SCS2 gene retained the normal Scs2p activity because it suppressed the inositol auxotrophy of the scs2Δ strain at 34°C, although the suppression was not sufficient at 37°C (Fig. 1). On Western blots, an anti-HA monoclonal antibody (clone 12CA5) recognized a 40-kDa band (Fig. 3A, lane 5). This band was not present in extracts made from strains lacking the HA-SCS2 gene (Fig. 3A, lanes 1, 2, and 4) and was more abundant in strains containing HA-SCS2 on the 2μm plasmid (data not shown). Treatment of the 12P fraction from yeast cells harboring HA-SCS2 with trypsin (0.1 mg/ml) for 30 min at 4°C digested the HA-Scs2p protein into several peptides, irrespective of whether the treatment was carried out in the absence or presence of Triton X-100 (Fig. 3D, top panel). On the other hand, Kar2p was not digested in the absence of Triton X-100 (Fig. 3D, bottom panel), indicating that membranes in the 12P fraction were not destroyed during the treatment. Taken together, all of these data are consistent with the idea that Scs2p has the topology of a type II integral membrane protein, with the N-terminal domain located in the cytoplasm and the C-terminal hydrophobic domain located inside membranes.
Scs2p is localized to the ER membrane.
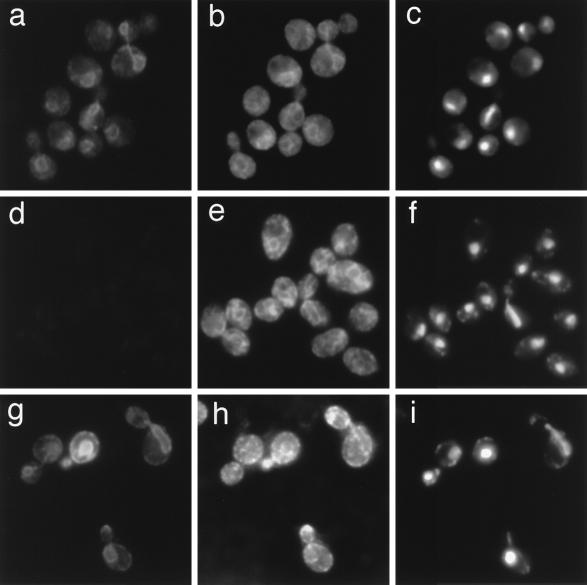
In order to study the localization of Scs2p, we used indirect immunofluorescence. Fixed and permeabilized cells (YPH500) were double labeled with the affinity-purified anti-Scs2p polyclonal antibody and the anti-Dpm1p monoclonal antibody (Fig. 4a to c). The anti-Scs2p antibody stained cells in a pattern that includes the nuclear membrane, projections of membrane from the nucleus, and membranes just beneath the plasma membrane (Fig. 4a). The staining pattern is very similar to that with the anti-Dpm1p antibody (Fig. 4b), indicating that Scs2p is colocalized with Dpm1p, the ER membrane protein. On the other hand, only faint staining of the cytoplasm was observed in scs2Δ cells with the anti-Scs2p antibody (Fig. 4d), while localization of Dpm1p was similar to that in wild-type cells (Fig. 4b and e). The anti-HA polyclonal antibody showed HA-Scs2p localization much more clearly, probably because of the high specificity of the antibody (Fig. 4g). These results exclude the possibility that the staining pattern in Fig. 4a shows the localization of the 66-kDa protein observed in Fig. 3A. Thus, cumulatively, these results indicate that Scs2p is an integral ER membrane protein.
FIG. 4.
Localization of Scs2p. Indirect immunofluorescence was carried out with YPH500 cells (wild type) (a, b, and c), KY360 cells (scs2Δ::TRP1) (d, e, and f), and KY360 cells harboring YCp(HA-SCS2) (g, h, and i). The cells in panels a to f were incubated with the rabbit anti-Scs2p polyclonal antibody and the mouse anti-Dpm1p monoclonal antibody, and cells in panels g to i were incubated with the rabbit anti-HA polyclonal antibody and the mouse anti-Dpm1p monoclonal antibody. Samples were stained with tetramethylrhodamine isothiocyanate-conjugated anti-rabbit IgG to detect Scs2p (a, d, and g), fluorescein isothiocyanate-conjugated anti-mouse IgG to detect Dpm1p (b, e, and h), and DAPI to indicate the position of the nuclei (c, f, and i).
Scs2p has a 16-amino-acid conserved sequence.
Protein and nucleotide database searches by using FASTA and BLAST protocols have revealed that a 16-amino-acid sequence which corresponds to residues 37 through 53 of Scs2p is well conserved between yeast and mammalian gene products (Fig. 5A). In the sequences shown in Fig. 5A, mouse and human homologs were deduced from cDNA sequences of expressed sequence tags. The Schizosaccharomyces pombe homolog function is unknown. The Aplysia homolog (VAP-33) was studied at the protein level (see below). Two other ER membrane proteins of yeast and rat liver, Cdc48p (an ER membrane protein required for fusion of ER membranes [9, 18]) and TER ATPase (a protein associated with transition vesicles between the ER and the Golgi complex [44]), have the consensus sequence, although similarities are not significant. Nematode MSP1A, which also has a similar sequence, is a member of the major sperm protein family expressed specifically in crawling sperm (33, 40).
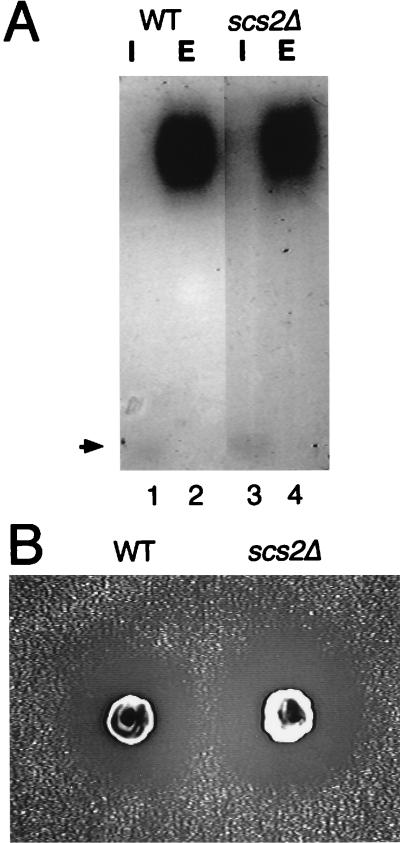
The Aplysia homolog is a synaptobrevin/VAMP binding protein, VAP-33, required for neurotransmitter release. Since Scs2p has 26.8% identity and 66.3% similarity in the N-terminal 190 residues with VAP-33, we examined whether Scs2p is involved in protein secretion pathways. However, scs2Δ cells secreted invertase, a major yeast secretory protein, as well as did wild-type cells in YPD medium at 30°C. Moreover, there was no difference between wild-type and scs2Δ cells in the electrophoretic mobility of the secreted invertase (Fig. 6A), suggesting that the sugar modification of invertase in scs2Δ cells was normal under these conditions. The secretion of another marker, α-factor, was also examined by the halo assay (39). As shown in Fig. 6B, scs2Δ cells could secrete the protein as efficiently as wild-type cells.
FIG. 6.
Disruption of SCS2 has no effect on protein secretion. (A) Sugar modification of invertase. Cells were grown in YPD medium at 25°C and shifted to YP medium with 0.1% glucose. After 2 h of incubation at 30°C, cells were converted to spheroplasts and separated into the intracellular (I) and extracellular (E) fractions. These fractions were subjected to native gel activity staining for invertase. The position of nonglycosylated (cytosolic) invertase is indicated by an arrow. Strains used were CTY182 (wild type [WT]) (lanes 1 and 2) and KY356 (scs2Δ::URA3) (lanes 3 and 4). (B) Halo assay for α-factor production. YPH500 (WT) and KY360 (scs2Δ::TRP1) cells were spotted on a lawn of a-mating-type cells (MATa sst2) on a plate and incubated at 30°C for 48 h to allow halos to develop.
To investigate whether the conserved region of Scs2p is crucial for its function, we constructed a mutant Scs2p protein which lacks the region by removing residues 36 through 53. As shown in Fig. 5B, overproduction of the protein (Scs2pΔ36-53) from the 2μm plasmid failed to suppress inositol auxotrophy of the scs2Δ strain, although Scs2pΔ36-53 expression was not lessened significantly (data not shown). Since Scs2p overproduction was toxic, as described above, there is a possibility that the failure to rescue the inositol auxotrophy is due to the toxic effect. However, overproduction of Scs2pΔ36-53 from the CEN plasmid also failed to suppress the auxotrophy (Fig. 6B). The results suggest that the region is required for normal Scs2p function.
DISCUSSION
In this study we found that scs2Δ strains showed inositol auxotrophy. They could form isolated colonies in the absence of inositol at 30°C, even though the growth rate was not as high as that of wild-type cells. Significant growth defects in inositol-free medium were observed when cells were incubated at temperatures of above 34°C (Fig. 1). The observed inositol auxotrophy was relatively weak compared to those of other inositol-auxotrophic mutants (ino1 and ino2 mutants) (7, 14, 19, 28). The finding that overproduction of INO1 or INO2 rescued the inositol auxotrophy (Fig. 2) suggests that Scs2p is a transcriptional factor like Ino2p or Ino4p (1). In fact, both SCS2 and INO2 are multicopy suppressors of CSE1 inositol auxotrophy, and an increase in INO1 mRNA levels is observed when SCS2 is overproduced in CSE1 mutants (25). However, since we have found that Scs2p is an integral membrane protein of the ER (Fig. 4), it is unlikely that Scs2p is a conventional transcriptional factor which directly binds to the upstream activation site of the INO1 gene.
Studies on sterol-regulatory element binding protein 1 (SREBP-1), found in mammalian cells, provide a possibility for the Scs2p function. SREBP-1 is a transcriptional factor which is associated with ER membranes and has a molecular mass of 125 kDa. Surprisingly, in cells deprived of cholesterol, the protein is cleaved to release a 68-kDa N-terminal fragment. The 68-kDa fragment is then targeted to the nucleus, where it binds to the sterol-regulatory element of the low-density lipoprotein receptor (42, 43). By analogy, it seems likely that Scs2p is a novel membrane-bound transcriptional factor which moves to the nucleus from the ER in response to inositol starvation and induces the expression of INO1. Unfortunately, we have failed to find migration of Scs2p to the nucleus in response to inositol starvation by immunofluorescence analysis (unpublished data). More detailed studies using cell-free systems might reveal the localization change.
Ire1p is one of the ER membrane proteins whose disruption causes inositol auxotrophy (5, 20, 26). Ire1p is an ER transmembrane kinase and transmits a signal of unfolded-protein accumulation in the ER to the nucleus. A basic leucine zipper transcription factor, Hac1p/Ire15p, is required for the unfolded-protein response (UPR) and binds to the UPR element in the promoters of UPR-regulated genes (6). As inositol-containing lipids are major phospholipid components of yeast membranes, Ire1p is postulated to regulate the coordinated biogenesis of both the protein and lipid components of the ER (30). Since overproduction of Scs2p suppresses inositol auxotrophy of ire15/hac1 mutants and scs2Δ strains are sensitive to tunicamycin treatment that induces the UPR (26a), it seems likely that Scs2p is involved in a signal transduction pathway similar to the Ire1p pathway. While Ire1p is activated by the UPR, Scs2p may be activated by heat shock, because the scs2 inositol auxotrophy become significant at high temperatures (Fig. 1 and 5B).
VAP-33, which is required for neurotransmitter release, is the only characterized protein which has an overall similarity with Scs2p. Since we have failed to obtain any line of evidence that Scs2p is involved in protein secretion, function may not be conserved between Scs2p and VAP-33, although there remains a possibility that a functionally redundant protein(s) may substitute for Scs2p function. The localization of Scs2p on the ER membrane (Fig. 4) does not favor the assumption that Scs2p is directly associated with yeast synaptobrevin homologs (Snc1p and Snc2p [10, 32]), because synaptobrevin is a membrane protein of secretory vesicles which are derived from the Golgi complex. However, it might be possible that Scs2p is associated with the other yeast synaptobrevin homologs found on vesicular carriers responsible for protein transport from the ER to the Golgi complex (22). Identification of a protein(s) which binds to Scs2p would give insight into this question.
Although the biochemical activity of Scs2p is still unknown, the existence of the conserved 16-amino-acid sequence (Fig. 5A) suggests that the Scs2p function is conserved between yeast and mammalian cells. Interestingly, Scs2p, VAP-33, Cdc48p, and TER ATPase, all of which contain the conserved sequence, are associated with the cytoplasmic face of biomembranes (Fig. 3) (9, 18, 37, 44). This fact implies that the sequence serves as a targeting or anchoring signal for those proteins to associate with this specific membrane region. Therefore, more detailed studies of the sequence are expected to reveal a novel protein motif which is required for the association with membranes in various eukaryotic cells.
ACKNOWLEDGMENTS
We thank Vytas Bankaitis and Ryogo Hirata for providing strains and antibodies, Kenji Kohno for the anti-Kar2p antibody, and Makoto Muroi and members of Animal and Cellular Systems Laboratory of RIKEN for helpful discussions. We also thank the Division of Laboratory Animal Research of RIKEN for production of the anti-Scs2p polyclonal antibody.
S.K. was supported by the special postdoctoral scientist program of RIKEN.
REFERENCES
- 1.Ambroziak J, Henry S A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 2.Bankaitis V A, Malehorn D E, Emr S D, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman G M, Henry S A. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 4.Cleves A E, McGee T P, Whitters E A, Champion K M, Aitken J R, Dowhan W, Goebl M, Bankaitis V A. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox J S, Shamu C E, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 6.Cox J S, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 7.Culbertson M R, Henry S A. Inositol requiring mutants of Saccharomyces cerevisiae. Genetics. 1975;80:23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean J M, Henry S A. Biosynthesis of inositol in yeast. Primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J Biol Chem. 1989;264:1274–1283. [PubMed] [Google Scholar]
- 9.Fröhlich K U, Fries H W, Rudiger M, Erdmann R, Botstein D, Mecke D. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J Cell Biol. 1991;114:443–453. doi: 10.1083/jcb.114.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerst J E, Rodgers L, Riggs M, Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc Natl Acad Sci USA. 1992;89:4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 12.Hirata R, Graham L A, Takatsuki A, Stevens T H, Anraku Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 13.Hosaka K, Nikawa J, Kodaki T, Yamashita S. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J Biochem. 1992;111:352–358. doi: 10.1093/oxfordjournals.jbchem.a123761. [DOI] [PubMed] [Google Scholar]
- 14.Hosaka K, Nikawa J, Kodaki T, Yamashita S. Cloning and characterization of the SCS1 gene required for the expression of genes in yeast phospholipid synthesis. J Biochem. 1994;115:131–136. doi: 10.1093/oxfordjournals.jbchem.a124287. [DOI] [PubMed] [Google Scholar]
- 15.Kagiwada S, Kearns B G, McGee T P, Fang M, Hosaka K, Bankaitis V A. The yeast BSD2-1 mutation influences both the requirement for phosphatidylinositol transfer protein function and derepression of phospholipid biosynthetic gene expression in yeast. Genetics. 1996;143:685–697. doi: 10.1093/genetics/143.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 17.Klig L S, Homann M J, Carman G M, Henry S A. Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: pleiotropically constitutive opi1 mutant. J Bacteriol. 1985;162:1135–1141. doi: 10.1128/jb.162.3.1135-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latterich M, Fröhlich K U, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- 19.Loewy B S, Henry S A. The INO2 and INO4 loci of Saccharomyces cerevisiae are pleiotropic regulatory genes. Mol Cell Biol. 1984;4:2479–2485. doi: 10.1128/mcb.4.11.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori K, Ma W, Gething M J, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 21.Muroi M, Shiragami N, Nagao K, Yamasaki M, Takatsuki A. Folimycin (concanamycin A) and bafilomycin A(1), inhibitors specific for V-ATPase, exert similar but distinct effects on intracellular translocation and processing of glycoproteins. Biosci Biotech Biochem. 1994;58:425–427. [Google Scholar]
- 22.Newman A P, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikawa J. A cDNA encoding the human transforming growth factor β receptor suppresses the growth defect of a yeast mutant. Gene. 1994;149:367–372. doi: 10.1016/0378-1119(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 24.Nikawa J, Sugiyama M, Hayashi K, Nakashima A. Suppression of the Saccharomyces cerevisiae hac1/ire15 mutation by yeast genes and human cDNAs. Gene. 1997;201:5–10. doi: 10.1016/s0378-1119(97)00418-6. [DOI] [PubMed] [Google Scholar]
- 25.Nikawa J, Murakami A, Esumi E, Hosaka K. Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae. J Biochem. 1995;118:39–45. doi: 10.1093/oxfordjournals.jbchem.a124889. [DOI] [PubMed] [Google Scholar]
- 26.Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol prototrophy in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 26a.Nikawa, J. Unpublished data.
- 27.Nikoloff D M, Henry S A. Genetic analysis of yeast phospholipid biosynthesis. Annu Rev Genet. 1991;25:559–583. doi: 10.1146/annurev.ge.25.120191.003015. [DOI] [PubMed] [Google Scholar]
- 28.Nikoloff D M, Henry S A. Functional characterization of the INO2 gene of Saccharomyces cerevisiae: a positive regulator of phospholipid biosynthesis. J Biol Chem. 1994;269:7402–7411. [PubMed] [Google Scholar]
- 29.Nojima H, Leem S-H, Araki H, Sakai A, Nakashima N, Kanaoka Y, Ono Y. Hac1: a novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5279–5288. doi: 10.1093/nar/22.24.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunnari J, Walter P. Regulation of organelle biogenesis. Cell. 1996;84:389–394. doi: 10.1016/s0092-8674(00)81283-0. [DOI] [PubMed] [Google Scholar]
- 31.Piper R C, Whitters E A, Stevens T H. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur J Cell Biol. 1994;65:308–318. [PubMed] [Google Scholar]
- 32.Protopopov V, Govindan B, Novick P, Gerst J E. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- 33.Roberts T M, Stewart M. Nematode sperm locomotion. Curr Opin Cell Biol. 1995;7:13–17. doi: 10.1016/0955-0674(95)80039-5. [DOI] [PubMed] [Google Scholar]
- 34.Rose M D, Mistra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian Bip/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 35.Shamu C E, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 36.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skehel P A, Martin K C, Kandel E R, Bartsch D. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science. 1995;269:1580–1583. doi: 10.1126/science.7667638. [DOI] [PubMed] [Google Scholar]
- 38.Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 39.Sprague G F., Jr Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- 40.Theriot J A. Worm sperm and advances in cell locomotion. Cell. 1996;84:1–4. doi: 10.1016/s0092-8674(00)80068-9. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga M, Kawamura A, Kohno K. Purification and characterization of Bip/Kar2 protein from Saccharomyces cerevisiae. J Biol Chem. 1992;267:17553–17559. [PubMed] [Google Scholar]
- 42.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. SREBP-1, a membrane-bound transcriptional factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 43.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 44.Zhang L, Ashendel C L, Becker G W, Moore D J. Isolation and characterization of the principal ATPase associated with transitional endoplasmic reticulum of rat liver. J Cell Biol. 1994;127:1871–1883. doi: 10.1083/jcb.127.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]