Abstract
Thermomonospora fusca E4 is an unusual 90.4-kDa endocellulase comprised of a catalytic domain (CD), an internal family IIIc cellulose binding domain (CBD), a fibronectinlike domain, and a family II CBD. Constructs containing the CD alone (E4-51), the CD plus the family IIIc CBD (E4-68), and the CD plus the fibronectinlike domain plus the family II CBD (E4-74) were made by using recombinant DNA techniques. The activities of each purified protein on bacterial microcrystalline cellulose (BMCC), filter paper, swollen cellulose, and carboxymethyl cellulose were measured. Only the whole enzyme, E4-90, could reach the target digestion of 4.5% on filter paper. Removal of the internal family IIIc CBD (E4-51 and E4-74) decreased activity markedly on every substrate. E4-74 did bind to BMCC but had almost no hydrolytic activity, while E4-68 retained 32% of the activity on BMCC even though it did not bind. A low-activity mutant of one of the catalytic bases, E4-68 (Asp55Cys), did bind to BMCC, although E4-51 (Asp55Cys) did not. The ratios of soluble to insoluble reducing sugar produced after filter paper hydrolysis by E4-90, E4-68, E4-74, and E4-51 were 6.9, 3.5, 1.3, and 0.6, respectively, indicating that the family IIIc CBD is important for E4 processivity.
Thermomonospora fusca is a filamentous soil bacterium that degrades most plant cell wall polymers, including cellulose. T. fusca secretes six different cellulases, including two exocellulases (E3 and E6), three endocellulases (E1, E2, and E5), and one unusual endocellulase (E4). E4 is a 90.4-kDa protein and consists of four domains: an N-terminal 51.4-kDa family 9 catalytic domain, a family IIIc cellulose binding domain (CBD), a fibronectin III-like domain, and a C-terminal family II CBD (Fig. 1). The gene for E4 has been sequenced (17), and the corrected sequence is in GenBank (accession no. L20093). E4 is unique in that it has relatively high activity on bacterial microcrystalline cellulose (BMCC) and synergizes with both classes of exocellulases and with endocellulases E2 and E5 (16). The other endocellulases from T. fusca do not synergize with each other (16). E4 retains more than 70% of its activity from pH 4.7 to pH 10.1.
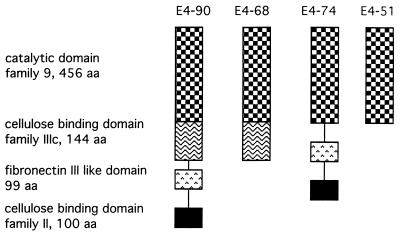
FIG. 1.
Schematic diagrams of the E4 domain combinations used in this study. aa, amino acids.
The crystal structure of a 68-kDa fragment of E4 (E4-68) has been solved (22) and consists of an (α/α)6 barrel catalytic domain joined with only limited flexibility to a family IIIc CBD, which has an antiparallel β-sandwich fold. These domains interact in two loop regions so that the shallow catalytic cleft is aligned with the flat face of the family IIIc CBD in such a way that a cellulose strand could bind along both domains (22). Tormo et al. (24) have determined the crystal structure of the Clostridium thermocellum family IIIa CBD from the cellulosomal scaffoldin subunit and proposed that it is this flat surface which interacts with cellulose. The structure of the catalytic domain of a homologous family 9 protein, C. thermocellum CelD, has been solved and includes a small N-terminal domain that is not present in E4 (18). The role of this domain is unknown, and it is not aligned with the active cleft of the catalytic domain.
The enzyme-product complex structure observed after the soaking of cellopentaose (CP) into E4-68 crystals was modeled as two overlapping binding modes of cleaved CP, each at 50% occupancy (22). These complexes identified the active center of the enzyme and showed the locations of six glucosyl binding sites numbered, from the nonreducing end, −4 to +2. Family 9 enzymes cleave cellulose with inversion at the anomeric carbon (11). Glu424 was identified as the active acidic residue, while Asp55 and Asp58 could both act as a base to deprotonate the nucleophilic water. All of these residues are conserved in family 9 proteins (17). The observed cleavage product clearly showed the α configuration of the anomeric carbon at the point of hydrolysis (22). The crystal structure also showed an induced fit, a closing of amino acid residues over the −3, −2, and −1 glucosyl binding sites, which was dependent on the occupancy of the +1 site (22). The open conformation appears to be complementary to a linear (unkinked) substrate, while the closed conformation shifts Glu424 within hydrogen bonding distance of the scissile oxygen and forces the substrate [especially Glc(−2) and Glc(−1)] toward the nucleophilic water (22).
In the work described here, we have studied the activities and properties of four combinations of the E4 domains, as well as the Asp55Cys mutants of two of these constructs. These data are discussed, along with previous data, in an effort to understand the mechanism of cellulose degradation by E4.
MATERIALS AND METHODS
Cloning of E4-68, E4-68 Asp55Cys, E4-51, E4-51 Asp55Cys, and E4-74.
Standard nucleic acid techniques were used as described by Sambrook et al. (23). DNA sequence analysis was performed by the Cornell Biotechnology Resource Center using a Perkin Elmer/Applied Biosystems automated sequencer. The strains used were Escherichia coli DH5α (13) and Streptomyces lividans TKM31 (19).
The molecular mass of the E4-68 protein created by limited digestion of E4-90 with papain (22) was determined by mass spectrometry to be 68.3 kDa, and the N-terminal sequence of the C-terminal digestion fragment was GlyGlnGlnProGlyGly. A plasmid was then constructed to place a stop codon after Gly613 in the E4 sequence to produce the 68-kDa protein without papain digestion. A 2.1-kb DNA fragment containing the E4-68 gene was obtained by PCR using pEJ3 (17) as the DNA template, a reverse primer (5′gcctcccggtactagttagccgccgggctc3′ [mismatched bases are underlined]) which created a stop codon plus a SpeI site after Gly613, and a forward primer (5′gactacttaagcttcacccattgaac3′) which created a HindIII site 130 bases upstream from the start codon. This fragment was ligated into S. lividans-E. coli shuttle vector pSES1 (19) at the HindIII and SpeI sites to produce pSZ46.
Mutant E4-68 Asp55Cys was created by the PCR overlap extension method (14) as previously described (27). The internal reverse primer sequence was 3′ccgccgaccatgacgcgtccgctggtgc5′, and the internal forward primer sequence was 5′gcggctggtactgcgcaggcgaccacg3′. The resulting PCR fragment was cloned to produce pSZ47 by the same methods used for E4-68.
Plasmid pD767, coding for E4-51, was constructed by digestion of pSZ46 with HindIII and BglII to obtain the 1.65-kb DNA fragment. This was ligated to HindIII- and XbaI-digested pUC19 by using linkers which created BglII and SpeI sticky ends, as well as adding a stop codon and a PstI site (linker G [5′gatctaataactgcaga3′] and linker H [3′attattgacgtctgatc5′]). This places the stop codon after Ile 463, the second amino acid in the first beta strand of the family IIIc CBD.
Plasmid pD768, coding for E4-74, was constructed by ligating three DNA fragments: (i) The 1.65-kb HindIII-BglII fragment used for pE4-51, which contains the catalytic domain; (ii) an 870-bp PCR product containing the fibronectin III-like region and the type II CBD, made by using forward primer 5′gaggaaggggaagagatctctggcggagaaggac3′ and reverse primer 5′gtgagcggataactagttcacacaggaaacagc3′, digested with BglII and SpeI; and (iii) pUC19 digested with HindIII and XbaI. This construction joins Ile 463 via a Ser residue to Gly613 and deletes the region from Phe464 to Gly612. The E4-51 Asp55Cys plasmid was created by gel purifying the 1.65-kb HindIII-BglII fragment from pSZ47 and ligating it into the gel-purified BglII-HindIII vector fragment of pD767.
The S. lividans plasmids were constructed by ligating pD768 or pD767 cut with HindIII and EcoRI, pSES1 cut with EcoRI and SspI, and SspI-HindIII linkers (L2A [5′attgcgtacgtagcga3′] and L2B [3′taacgcatgcatcgcttcga5′]). These ligation mixtures were transformed directly into S. lividans TKM31 (15). All PCR-produced clones were sequenced to check for accidental mutations, and none were found.
Production of E4 proteins from S. lividans.
Each S. lividans transformant was grown and harvested as previously described (26), except that cultures were harvested after only 41 to 48 h. The supernatant, containing 1.2 M (NH4)2SO4, was loaded onto a CL-4B phenyl-Sepharose column and eluted as previously described (26). Only the fractions containing the desired protein (as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and activity assays) were combined and applied to a Q-Sepharose column. E4 contamination by endogenous S. lividans endocellulase (26) was avoided by using a short fermentation time and carefully selecting the phenyl-Sepharose fractions to be combined. Elution from Q Sepharose was with 0.01 M bis-Tris (pH 5.4)–10% glycerol buffer containing NaCl gradients (0.18 to 0.4 M for E4-68, 0.18 to 0.5 M for E4-74, 0 to 0.4 M for E4-51, and 0.2 to 0.6 M for E4-90). The purest fractions for each protein were combined and concentrated as previously described (22) and stored in buffer containing 10% glycerol.
Protein concentration and cellulase activity assays.
Protein concentrations were determined by measuring A280 by using extinction coefficients calculated from the predicted amino acid compositions. Filter paper (FP; Whatman no. 1; 8.7 mg/ml), BMCC (Cellulon; Weyerhaeuser; 2.5 mg/ml), acid-swollen cellulose (SW; 2 mg/ml) (8), and carboxymethyl cellulose (CMC; Sigma Low Viscosity; 10 mg/ml) assays were performed as previously described (16), in 0.4-ml volumes at 50°C for 16, 16, 2, and 2 h, respectively. The reducing sugar produced was determined by the dinitrosalicylic acid method (12, 20). The buffer was 0.05 M Na acetate (pH 5.5) with 15 mM CaCl2 added for filter paper reactions. All assays were done in triplicate by using increasing amounts of the E4 proteins. Cellulase synergism and the distribution of reducing ends between filter paper and supernatant were measured as described by Irwin et al. (16).
Calculation of cellulase activity.
Cellulase activity assays are nonlinear, at least partly due to substrate heterogeneity. To compare the activities of the different E4 proteins, we assayed them for a fixed amount of time by using various amounts of protein and calculated the activities at a fixed amount of reducing sugar produced. The assays were set up to produce between 0.1 and 0.6 μmol of reducing sugar per reaction. The dinitrosalicylic acid standard curve was made by using cellobiose as the reducing sugar. The nanomoles of protein used (X) versus the micromoles of reducing sugar (Y) were plotted and fitted to the equation Y = m1 · X/(m2 + X) by using the program KaleidaGraph (Synergy Software) (m1 and m2 are constants determined from the curve fit). This curve was used to determine the amount of enzyme required to produce the arbitrary value of 0.424 μmol of reducing sugar in the reaction (or 1.06 μmol/ml). If this target amount of product could not be produced, then the activity was calculated from the micromoles of cellobiose produced by 0.6 nmol of enzyme (1.5 nmol/ml).
Binding assays.
Binding assays were done in siliconized 1.5-ml Eppendorf tubes by using 0.05 M Na acetate buffer (pH 5.5)–10% glycerol. Binding assays contained 0 to 3 mg of BMCC per ml and 1 nmol of the desired E4 protein per ml. Tubes were rotated end over end at room temperature for 1 h and centrifuged for 5 min. The supernatants were filtered through 0.45-μm-pore-size CA micro-spin filters (Lida) which had been pretreated with 300 μl of bovine serum albumin at 1 mg/ml and then rinsed with buffer. The amount of E4 remaining unbound was determined by CMC assays or by protein concentration (A280).
Viscosity assays.
Viscosity assays were done by using a size 100 Ostwald-Fenske viscometer at 50°C in 0.05 M Na acetate buffer (pH 5.5). Enzyme (100 μl) was added to 10 ml of 0.3% Hercules CMC 4H1F. The time of outflow was measured at intervals. Samples for reducing sugar assays were assayed by the bicinchoninate method (7, 10).
Identification of hydrolysis products by TLC.
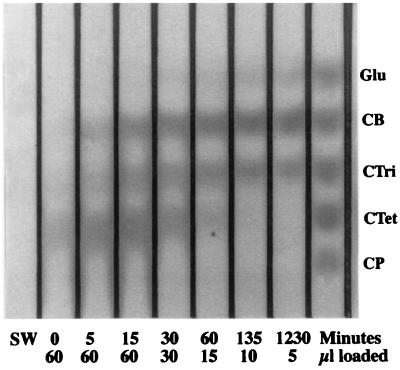
E4 proteins were reacted with SW (6.3 mg/ml) in 12.5 mM Na acetate (pH 5.5) at 50°C in a total volume of 1.2 ml. After preincubation for 5 min, enzyme was added and mixed and an aliquot was withdrawn immediately and frozen on dry ice; further aliquots were withdrawn at the time points indicated (see Fig. 3) and frozen. The aliquots were analyzed by thin-layer chromatography (TLC) as previously described (5, 17).
FIG. 3.
Time course of products released by E4-90 digestion of SW. CB, cellobiose; Ctri, cellotriose.
RESULTS
Effect of each CBD on activity and thermostability.
The different E4 domain constructs are diagrammed in Fig. 1, and their specific activities on different cellulose substrates are given in Table 1. E4-90 has the highest activity of any T. fusca cellulase on BMCC, while the two constructs which lack the family IIIc CBD, E4-74, and E4-51, have almost no activity on BMCC. Only E4-90 could reach the target of 4.5% digestion on FP, illustrating that both the family II and IIIc CBDs are necessary for degradation of this crystalline substrate. The other constructs had only minimal FP activity, which did not increase with the addition of further enzyme. The CMC activities show that the family II CBD is not required but there is a fourfold decrease in the activity of the proteins lacking the family IIIc CBD. E4-51, which has no binding domain at all, has only 3% of the activity of E4-90 on SW. In comparison, the catalytic domains of two other T. fusca endocellulases, E2 and E5, retain 62 and 95% of their SW activity, respectively. The E4-68 Asp55Cys mutant has low, but not zero, activity, which agrees well with its proposed role as one of the two catalytic bases. The activities of E4-90 reported here are substantially higher than those reported earlier (16); this may be due to the slightly different purification methods and the use of 10% glycerol in the final purification buffers.
TABLE 1.
Properties of E4 domain combinations
| Protein | Molecular mass (kDa) | Sp act (μmol of cellobiose/min-μmol)
|
FP hydrolysis
|
||||
|---|---|---|---|---|---|---|---|
| CMC | SW | FP | BMCC | % Insoluble reducing sugar produced | Soluble/insoluble ratio | ||
| E4-90 | 90.4 | 475.0 | 202.0 | 1.03 | 19.1 | 13 | 6.9 |
| E4-68 | 68.0 | 488.0 | 54.0 | 0.24a | 6.1 | 22 | 3.5 |
| E4-74 | 74.0 | 121.0 | 23.2 | 0.27a | 0.3a | 44 | 1.3 |
| E4-51 | 51.4 | 108.0 | 6.3 | 0.13a | 0.2a | 63 | 0.6 |
| E4-68 D55C | 68.0 | 3.7a | 2.1a | NDb | ND | ND | ND |
| E4-51 D55C | 51.4 | 0.8a | <0.1a | ND | ND | ND | ND |
Target digestion was not achieved; activity was calculated at 0.6 nmol of protein. The value is an overestimate of the true activity.
ND, not determined.
The results of synergism assays with the different E4 constructs are shown in Table 2. At the lower percent digestion, removal of the various domains does not have much effect on activity. However, if activity is calculated at 7.3% digestion, the three component mixtures with E4-90 and E4-68 show higher activity than the E3-E5 mixture alone, while the mixtures with E4-51 and E4-74 show decreased activity. (The activity of these mixtures per micromole of protein is less than that of the E3-E5 mixture because there is less E3 and E5 in a three-way mixture and the E4-51 and E4-74 species do not compensate for this.) Assuming that the more easily hydrolyzed, less crystalline filter paper is hydrolyzed first, this shows that the family IIIc CBD contributes to the hydrolysis of the more difficult to hydrolyze, more crystalline substrate.
TABLE 2.
Synergism between cellulases during FP hydrolysis
| Cellulase(s) | Activity (μmol of cellobiose/min-μmol) at:
|
|
|---|---|---|
| 5.2% digestion | 7.3% digestion | |
| E4 | 1.0 | |
| E3 + E5 | 4.1 | 2.7 |
| E4 + E3 + E5 | 5.4 | 4.3 |
| E4-68 + E3 + E5 | 5.2 | 3.9 |
| E4-74 + E3 + E5 | 3.9 | 1.6 |
| E4-51 + E3 + E5 | 4.6 | 2.0 |
To test the thermostability of the different constructions, the proteins were incubated at temperatures of 0 to 65°C for 16 h and then assayed for CMC activity at 50°C as usual. A plot of activity versus temperature showed that 50% of the activity was retained after incubation at 63°C for the constructions with the type IIIc CBD and after incubation at 56°C for those lacking this domain. The two loop regions in the crystal structure where the two domains interact (22) could provide the extra thermostability.
Processivity.
Although we do not have an assay to measure processivity directly, one indication of the degree of processivity is to compare the ratio of the reducing sugar ends in the supernatant (soluble) to the reducing sugar ends left in a filter paper circle (insoluble). We have previously shown that there is a clear distinction between endo- and exocellulases (16) in that the soluble/insoluble ratio is about 2.2 for endocellulases E2 and E5 and 12 to 23 for exocellulases (16). E4-90 is unusual in that its ratio falls between values for exo- and endocellulases (Table 1). The soluble/insoluble ratio of the E4 constructs decreases with the removal of the family II CBD and decreases much more with the removal of the family IIIc CBD. The effect is most dramatic when both CBDs are removed, giving 63% insoluble products.
Reduction of the viscosity of a CMC solution.
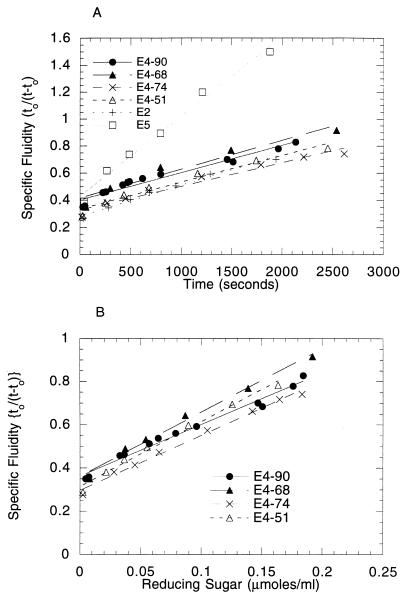
E4-51 and E4-74 required a fivefold higher enzyme concentration than did E4-90 or E4-68 to reduce the viscosity of a CMC solution (Fig. 2A). E4 is a much less active endocellulase than E5 in viscosity experiments but is as active as E2. The specific fluidity as a function of the amount of reducing sugar produced showed no significant difference between the different E4 constructions (Fig. 2B).
FIG. 2.
Viscosity reduction of CMC. Time of flow of buffer is t0; time of flow of CMC solution is t. The concentration of E4-90, E4-68, E2, and E5 was 0.005 nmol/ml, and the concentration of E4-74 and E4-51 was 0.025 nmol/ml.
Substrate binding in the catalytic cleft and analysis of products.
TLC analysis of the products from a time course of hydrolysis of SW by E4-90 is shown in Fig. 3. This experiment clearly shows that the initial cleavage of SW produces cellotetraose (CTet) almost exclusively. This same TLC pattern was seen for E4-68, while for E4-51, the activity was so low that only CTet could be seen on the TLC plate (data not shown). Over time, CTet is cleaved to cellobiose or to cellotriose and glucose in roughly equal amounts and does not accumulate (as previously shown [17]). Barr et al. (1) showed that CP labeled with 18O at the reducing-end anomeric carbon was cleaved by E4 with the 18O distributed as follows: 11% cellotriose and 89% cellobiose or 20% CTet and 80% glucose. This fits well with the crystallographic data showing that CP binds in the E4 cleft a large majority of the time in one of two modes: in −3 to +2 or in −4 to +1. To form the low-frequency products, about 5% of the time CP binds with its nonreducing end in the −2 position and 10% of the time with its nonreducing end in the −1 position. Since the catalytic domain only has sites −4 to +2, this suggests there are some binding interactions on the flat face of the family IIIc CBD which is aligned with the catalytic cleft.
Cellulose binding properties and effect of an Asp55Cys mutation.
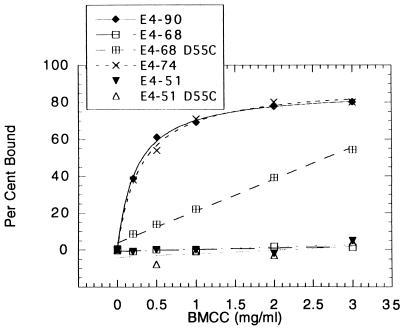
The binding of the different constructs to BMCC is shown in Fig. 4. E4-90 and E4-74 bind equally well to BMCC, and yet E4-74 has very little activity on FP or BMCC. Surprisingly, E4-68 does not bind to BMCC, even though it retains 32% of the E4-90 BMCC activity. In contrast, the E4-68 Asp55Cys mutant demonstrates significant binding to BMCC but E4-51 Asp55Cys does not bind at all. The inability of E4-68 to bind to BMCC suggests that once a strand has been hydrolyzed, its binding becomes very weak. A rationale for this result could be that tight binding is not desirable if the role of the family IIIc CBD is to guide the cellulose chain toward the catalytic cleft to aid in processivity.
FIG. 4.
Binding of the E4 constructs to BMCC.
DISCUSSION
Gal et al. (9) have studied C. cellulolyticum CelG, which contains a family 9 catalytic domain and a family IIIc CBD, as well as a dockerin domain specific to cellulosomal hydrolases. The amino acids of the catalytic (CelGcat) and CBD (GST-CBDCelG) domains show 51 and 39% similarity to E4. Constructs of these two domains alone and also of CelGS, which is missing only the dockerin domain, were made. Neither CBDCelG nor CelGcat was able to bind to SW, Avicel, or BMCC, which is in agreement with the results presented here. CelG and CelGS were able to bind to Avicel but had weaker binding than the family IIIa CBD of miniCipC1 (21).
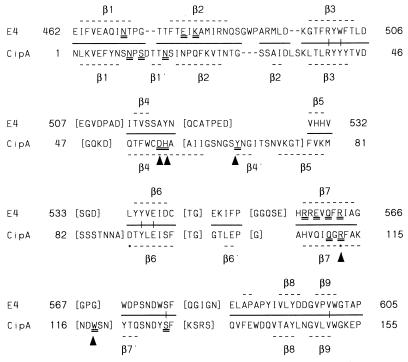
The C. thermocellum scaffoldin family IIIa Cip-CBD (24) serves to bind the cellulosome to cellulose, and only one CBD has been found in any scaffoldin. An alignment based on the three-dimensional crystal structures of the CipA CBD and the E4 family IIIc CBD is shown in Fig. 5. Yaron et al. (25) have mutated to Ala 10 residues of CipA considered likely to affect cellulose binding. Mutation of any one of the residues participating in the planar aromatic strip (Asp56, His57, Tyr67, Arg112, and Trp118) decreased binding significantly (3- to 10-fold). On the other hand, mutation of the “anchoring” residues (Asn10, Asn16, Gln110, Ser12, and Ser133), which are proposed to participate in hydrogen bonding to cellulose, did not reduce the affinity significantly. The Cip CBD aromatic strip residues are not conserved in the E4 family IIIc CBD. The E4 family IIIc CBD conserved residues suggested by Sakon et al. (22) to be well aligned to interact with a cellulose chain leading into the E4 active cleft are also noted in Fig. 5, and only three of the seven are somewhat conserved in CipA. While the CipA scaffoldin CBD has a relatively lengthy 11-residue beta 4 strand (Gln51-Ile61) terminating in a hairpin turn including a three-residue beta 4′ strand (24), E4 has only a three-residue beta 4 strand (Thr514-Ser516) (22). Although these two molecules have similar overall topologies, the family IIIc CBDs clearly play a role very different from that of the family IIIa CBDs.
FIG. 5.
Alignment of the E4 family IIIc CBD with the CipA scaffoldin CBD based on superposition of the three-dimensional structures (22, 24) by the strategy of Chothia and Lesk (6) using a 3Å cutoff to define structurally equivalent residues (denoted by black lines between the sequences). Residues where the structures diverge in their Cα placement are bracketed to emphasize that the alignment in these regions is not meaningful. Gaps are indicated by hyphens. The β strands are labeled above or below each sequence. The E4 residues which are double underlined, Asn470, Glu478, Lys480, Arg557, Glu559, Gln561, and Arg563, are conserved in family IIIc and lie in a region (colored red in Fig. 5a of reference 22) that could interact well with an extended cellulose chain from the active site. CipA residues which were mutated to Ala by Yaron et al. (25) are double underlined, and those which resulted in 3- to 10-fold lower binding to cellulose are marked by arrows. Residues marked by asterisks have the same Cα orientation, but the directions of their side chains are markedly different; the CipA Arg112 side chain is involved in a salt bridge with an Asp, while the corresponding E4 Arg563 is not. Bayer et al. (2) have identified six well-conserved residues of family III CBDs that form a shallow groove on the opposite side of the molecule from the “binding” residues; these are represented by vertical lines between the two sequences.
Based on the results presented in this paper and the crystal structure, we hypothesize that after E4 makes an initial cleavage (either endo or exo), the cellulose strand is processed, from the nonreducing end, with the help of the family IIIc binding domain to refill the six glucosyl sites of the catalytic cleft and then CTet is cleaved from the nonreducing end. E4 is probably able to give synergy with exocellulases because it can bind and cleave internal sites, while it is able to give synergy with other endocellulases because of its processive activity. Analysis of the activities of the various E4 constructs on different substrates provides clues to the roles of the different domains. The constructs without the family IIIc CBD are only 25% as efficient in CMC hydrolysis. There is no evidence from the viscosity experiments that CMC hydrolysis is processive. Probably, the carboxymethyl groups of CMC prevent processivity. This suggests that one role of this domain is simply to help attract a CMC chain to the vicinity of the catalytic cleft.
Pairwise comparisons of SW hydrolysis by proteins with a family IIIc CBD versus those without (E4-90 versus E4-74 and E4-68 versus E4-51) show 8.8- and 8.6-fold differences in activity, respectively. Similar comparisons of proteins with and without the family II CBD (E4-90 versus E4-68 and E4-74 versus E4-51) show a 3.7-fold decrease in both cases. Loss of both CBDs (E4-90 versus E4-51) results in a 32-fold difference, illustrating that the effects of the two domains are multiplicative.
Constructs containing the family IIIc CBD produce much more soluble product after filter paper hydrolysis than do those without this domain. This implies that E4 processivity requires this domain. After hydrolysis, the E4 residues shown to close over the substrate (22) move back to their open position, and the strand could directly dissociate from E4. The function of the type IIIc CBD, in part, must be to hold the enzyme onto the strand during this event and lessen the chance of dissociation. Yet, this CBD must also allow for enzyme migration along the strand, for the cellulose chain must travel the distance of four glucosyl residues to fully occupy the catalytic cleft before the next cleavage.
It is clear that CTet is readily cleaved further, since it does not accumulate. The CTet molecules would also compete with cellulose chains for the catalytic cleft. On the other hand, the cellulose chains would have many more potential (albeit weak) binding sites along the family IIIc CBD, making their binding in the cleft more favorable. CP is cleaved far more rapidly than CTet; in separate reactions with the oligosaccharides, after 15 min, there was no remaining CP in the reaction with E4, while after 2 h, about one-third of the CTet was not yet hydrolyzed (17).
The BMCC activities demonstrate that the family IIIc CBD plays a role in disrupting crystalline cellulose, as the activities increase 40- to 66-fold when this domain is present. BMCC, although crystalline, has a much more open structure than Avicel (3, 4), which would allow a relatively large protein such as E4 better access to this substrate. In contrast, hydrolysis of filter paper requires both binding domains. The liberation of a crystalline cellulose chain from its neighbors to make it available for hydrolysis requires time for a number of hydrophobic and hydrogen bonds to break and reform. It makes sense that having an anchor (the family II CBD) to keep the enzyme close to the solid substrate would be advantageous.
These experiments show the power of combining structural studies with enzymology and DNA techniques. They also illustrate that the more we learn about how enzymes function the more complicated and interesting the story becomes.
ACKNOWLEDGMENT
This work was supported by grant 95-37500-1823 from the NRI Competitive Grants Program/USDA.
REFERENCES
- 1.Barr B K, Hsieh Y-L, Ganem B, Wilson D B. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35:586–592. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., E. Morag, R. Lamed, S. Yaron, and Y. Shoham. Cellulosome structure: four-pronged attack using biochemistry, molecular biology, crystallography and bioinformatics. In Tricel 1997 Conference Proceedings, in press. Royal Society of Chemistry, Cambridge, United Kingdom.
- 3.Bothwell M, Daughhetee S, Chaua G, Wilson D B, Walker L. Binding capacities for Thermomonospora fusca E3, E4 and E5, the E3 binding domain, and Trichoderma reesei CBHI on avicel and bacterial microcrystalline cellulose. Bioresour Technol. 1997;60:169–178. [Google Scholar]
- 4.Bothwell M K. Binding kinetics of Thermomonospora fusca E3 and E5, and Trichoderma reesei CBHI. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1994. [Google Scholar]
- 5.Chirico W J, Brown R D. Separation of [1-3H] cellooligosaccharides by thin-layer chromatography: assay for cellulolytic enzymes. Anal Biochem. 1985;150:264–272. doi: 10.1016/0003-2697(85)90509-3. [DOI] [PubMed] [Google Scholar]
- 6.Chothia C, Lesk A M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doner L W, Irwin P L. Assay of reducing end-groups in oligosaccharide homologues with 2,2′ bicinchoninate. Anal Biochem. 1992;202:50–53. doi: 10.1016/0003-2697(92)90204-k. [DOI] [PubMed] [Google Scholar]
- 8.Ferchak J D, Hagerdal B, Pye E K. Saccharification of cellulose by the cellulolytic enzyme system of Thermomonospora sp. II. Hydrolysis of cellulosic substrates. Biotechnol Bioeng. 1980;22:1527–1542. [Google Scholar]
- 9.Gal L, Gaudin C, Belaich A, Pages S, Tardif C, Belaich J-P. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J Bacteriol. 1997;179:6595–6601. doi: 10.1128/jb.179.21.6595-6601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia E, Johnston D, Whitaker J R, Shoemaker S P. Assessment of endo-1,4-beta-d-glucanase activity by a rapid colorimetric assay using disodium 2,2′-bicinchoninate. J Food Biochem. 1993;17:135–145. [Google Scholar]
- 11.Gebler J, Gilkes N R, Claeyssens M, Wilson D B, Beguin P, Wakarchuk W W, Kilburn D G, Miller R C, Jr, Warren A J, Withers S G. Stereoselective hydrolysis catalyzed by related β-1,4-glucanases and β-1,4-xylanases. J Biol Chem. 1992;267:12559–12561. [PubMed] [Google Scholar]
- 12.Ghose T K. Measurement of cellulase activities. Pure Appl Chem. 1987;59:257–268. [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood D A, Bibb M J, Chater K F, Kiersen T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces—a laboratory manual. Norwich, England: The John Innes Foundation; 1985. [Google Scholar]
- 16.Irwin D, Walker L, Spezio M, Wilson D. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 17.Jung E D, Lao G, Irwin D, Barr B K, Benjamin A, Wilson D B. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl Environ Microbiol. 1993;59:3032–3043. doi: 10.1128/aem.59.9.3032-3043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juy M, Amit A G, Alzari P M, Poljak R J, Claeyssens M, Beguin P, Aubert J-P. Three-dimensional structure of a thermostable bacterial cellulase. Nature. 1992;357:89–91. [Google Scholar]
- 19.Lao G, Wilson D B. Cloning, sequencing, and expression of a Thermomonospora fusca protease gene in Streptomyces lividans. Appl Environ Microbiol. 1996;62:4256–4259. doi: 10.1128/aem.62.11.4256-4259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller G L, Blum R, Glennin W E, Benton A L. Measurement of carboxymethyl cellulase activity. Anal Biochem. 1960;21:127–132. [Google Scholar]
- 21.Pagès S, Gal L, Bèlaïch A, Gaudin C, Tardif C, Bèlaïch J-P. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. J Bacteriol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakon J, Irwin D, Wilson D B, Karplus P A. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat Struct Biol. 1997;4:810–817. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 25.Yaron, S., E. Morag, E. Bayer, R. Lamed, J. Tormo, and Y. Shoham. Identification and evaluation of ten functional residues of the cellulose-binding domain from the cellulosomal scaffoldin subunit of Clostridium thermocellum. Abstract. Tricel 1997 Conference. Ghent, Belgium.
- 26.Zhang S, Lao G, Wilson D B. Characterization of a Thermomonospora fusca exocellulase. Biochemistry. 1995;34:3386–3395. doi: 10.1021/bi00010a030. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Wilson D B. Surface residue mutations which change the substrate specificity of Thermomonospora fusca endoglucanase E2. J Biotechnology. 1997;57:101–113. doi: 10.1016/s0168-1656(97)00093-x. [DOI] [PubMed] [Google Scholar]