Abstract
Infection of Escherichia coli by the filamentous bacteriophage f1 is initiated by interaction of the end of the phage particle containing the gene III protein with the tip of the F conjugative pilus. This is followed by the translocation of the phage DNA into the cytoplasm and the insertion of the major phage capsid protein, pVIII, into the cytoplasmic membrane. DNA transfer requires the chromosomally encoded TolA, TolQ, and TolR cytoplasmic membrane proteins. By using radiolabeled phages, it can be shown that no pVIII is inserted into the cytoplasmic membrane when the bacteria contain null mutations in tolQ, -R and -A. The rate of infection can be varied by using bacteria expressing various mutant TolA proteins. Analysis of the infection process in these strains demonstrates a direct correlation between the rate of infection and the incorporation of infecting bacteriophage pVIII into the cytoplasmic membrane.
Infection of Escherichia coli by the Ff filamentous phages f1, fd, and M13 is initiated when the end of the particle containing the pIII protein interacts with the tip of the F conjugative pilus (22, 38). It is thought that the phage is then brought to the bacterial surface by retraction of the pilus (12). It is not known whether the retraction is a result of the normal polymerization-depolymerization cycles of the pilus or is triggered by the binding of the phage particle (8). Subsequent translocation of the phage DNA into the cytoplasm requires the products of the bacterial tolQRA genes. In the absence of any one of these gene products, no productive infection occurs (i.e., the bacteria are tolerant of the phages), even though the phages can bind to the pili and the bacteria are capable of producing progeny phages when transformed with phage DNA (27, 32). These three Tol proteins are also required for the uptake of the group A colicins (5, 15, 37) and are involved in maintaining the integrity of the outer membrane (7, 37). Bacteria containing mutations in any one of the tolQRA genes leak periplasmic proteins into the medium and are not killed by the group A colicins, even though these bacteriocins are able to bind to their respective outer membrane receptors.
TolQ, TolR, and TolA are integral cytoplasmic membrane proteins which appear to form a complex (6, 16), some of which is concentrated at contact sites between the cytoplasmic and outer membranes (10). TolQ contains three transmembrane helices, with the major portion of the protein located in the cytoplasm (13, 33, 35). TolR has a single transmembrane segment, with most of the protein exposed in the periplasm (13, 23). TolA is a three-domain protein anchored in the cytoplasmic membrane via its amino-terminal 47-residue domain I (18). The remaining 348 residues are exposed in the periplasm and are divided into the globular carboxyl-terminal domain III and the long, helical middle domain II. Presumably, the helical domain II is able to span the periplasm, positioning domain III to potentially interact with the outer membrane as well as with components of the periplasm.
TolA domain III appears to play an important role in the function of the TolQRA complex. The presence of a free form of domain III in the periplasm of wild-type bacteria results in the release of periplasmic components into the medium as well as an increased tolerance to the group A colicins, suggesting that domain III of TolA normally interacts with some periplasmic or outer membrane components (19). TolA domain III has also been shown to be essential for infection by the filamentous phages (4, 26), interacting with the amino terminal portion of the phage pIII (26). This interaction occurs only after initial interaction of the bacteriophage with the tip of the pilus (4). Thus, TolA domain III was recently designated the coreceptor of filamentous phage infection (26).
During infection, the DNA is translocated into the cytoplasm while the major capsid protein, pVIII, is inserted into the cytoplasmic membrane (29, 34). The pVIII from the infecting phage joins newly synthesized pVIII and is assembled into progeny phages (2, 29). Since TolA domain III appears to receive the phage from the retracting pilus, it is logical to assume that DNA translocation and pVIII membrane insertion occur after this step. However, a 1974 study suggested that pVIII could become associated with the inner membrane in a bacterium containing an undefined colicin-tolerant mutation in tolA (29). Since that time, the tolQRAB operon has been defined and its products have been characterized (36). In this paper, we reexamine the fate of pVIII from infecting f1 phages in bacteria by using defined tolQRA mutants. We show that insertion of the pVIII protein into the cytoplasmic membrane upon infection is clearly dependent upon functioning TolQ, TolR, and TolA proteins. Further, analysis of strains expressing mutant TolA proteins, which vary in their rates of infection, demonstrates a direct correlation between the rate of infection and the amount of pVIII from infecting phages incorporated into the cytoplasmic membrane.
MATERIALS AND METHODS
Bacteriophages, bacterial strains, and plasmids.
E. coli K91 (HfrC) and K17 (F−) were obtained from M. Russel (The Rockefeller University). GM1 (F′ lac pro) was obtained from D. Steege (Duke University). K17DE3 is K17 lysogenized with lambda DE3 carrying the inducible gene for T7 RNA polymerase (18). K17DE3/F+ contains the F′ lac pro from GM1, while K17DE3tolA/F+ and K91tolA each contain a mini Tn10 insertion in tolA (4). GM1-derived mutant strains TPS13 [tolQ(Am)], TPS66 (tolQ missense mutant), and TPS300 (tolR::Cm insertion mutant) have been previously described (32). Plasmid pSKL10 expresses wild-type TolA (18). Plasmids ptolAΔ1, ptolAΔ2, and ptolAΔ3 express TolA containing deletions of the first half of domain II (TolAΔIIn), the second half of domain II (TolAΔIIc), and the entire domain II (TolAΔII), respectively (4, 28). Plasmid pPGK101 expresses wild-type TolR. It was constructed by cloning the tolR gene from pTPS202 (32) by PCR and by subsequent insertion of the gene downstream of the ribosome binding site in pTrc99A (Pharmacia).
Media and chemicals.
Bacteria were grown in TY medium as described in Sun and Webster (32), supplemented with ampicillin (60 μg/ml) where appropriate. l-[4,5-3H(N)]lysine (108 Ci/mmol); Expre35S35S protein labeling mix (35S, >1,000 Ci/mmol), containing both [35S]cysteine and [35S]methionine; and [32P]orthophosphate (1 mCi/mmol) were purchased from DuPont, NEN. Subtilisin (bacterial protease type VIII) and phenylmethylsulfonyl fluoride were purchased from Sigma.
Infection with radiolabeled phages and removal of surface-bound phages.
Radiolabeled f1 phages were produced by infection of K91 in the presence of [35H]lysine, [35S]methionine-[35S]cysteine, or [32P]orthophosphate and purified by CsCl density centrifugation as previously described (20). Bacteria (100 ml) were grown to a density of 2 × 108 per ml and infected with the desired radioactive bacteriophage at a multiplicity of infection of 100. After 10 to 15 min, infection was stopped by rapid chilling to 0°C in the presence of 0.02% sodium azide, and the bacteria were harvested by centrifugation. The labeled bacteria were washed twice by resuspension in 100 ml of 10 mM HEPES, pH 7.8, containing 0.5 mM EDTA (HE) followed by centrifugation (washed bacteria). Surface-bound phages were removed from washed bacteria by two rounds of suspension in 100 ml of HE and shearing in a Sorvall omnimixer (sheared bacteria) as previously described by Lopez and Webster (21). For enzymatic removal of surface-bound phages, bacteria were suspended in 10 ml of 10 mM HEPES, pH 7.8, containing 2 mM CaCl2, divided into two aliquots, warmed to room temperature, and then incubated with or without 0.2 mg of subtilisin/ml for 15 min with occasional gentle mixing. Following rapid chilling in the presence of 0.8 mM phenylmethylsulfonyl fluoride, the bacteria were collected by centrifugation (protease-treated bacteria). Aliquots of suspended bacteria were boiled for 10 min in 2% sodium dodecyl sulfate (SDS), and radioactivity was determined in 10 ml of Hydrofluor with an Intertechnique scintillation spectrometer.
Cellular fractionation.
Bacteria, suspended in 10 ml of HE, were broken by passage through a prechilled French pressure cell at 18,000 lb/in2, and the total membrane fraction was isolated by density centrifugation onto a cushion of 55% sucrose (wt/wt) topped with 5% sucrose in HE buffer (25). The total membrane fraction was collected, adjusted to a volume of 1.5 ml with 30% sucrose in HE, and sheared three times through a 22-gauge needle. The sucrose concentration was raised to >55% by the addition of powdered sucrose (0.9 g per 1.5 ml). The sample, minus any undissolved sucrose crystals, was placed at the bottom of an ultracentrifugation tube and overlaid with a sucrose gradient in HE consisting of 50% sucrose (2.5 ml), 45% sucrose (2.5 ml), 40% sucrose (2.5 ml), and 35% sucrose (2.0 ml) and topped with 30% sucrose (approximately 0.8 ml) to fill the tube. After centrifugation for 72 h in a Beckman SW41 rotor at 4°C, fractions (0.5 ml) were collected from the bottom of each gradient and the radioactivity in aliquots (100 to 200 μl) was determined. NADH oxidase activity in each fraction was determined as previously described (17). Fractions containing the outer membrane, cytoplasmic membrane, and other regions of interest were pooled, diluted with HE, and pelleted by centrifugation for 3 h at 35,000 rpm in a TY 65 rotor. The pellets were dissolved in 4% SDS–0.25 M Tris, pH 6.8, and aliquots were subjected to analysis by SDS polyacrylamide gel electrophoresis followed by staining with Coomassie blue.
RESULTS
Fate of the major coat protein of the f1 bacteriophage following infection of E. coli.
The major coat protein, pVIII, of the bacteriophage f1 constitutes 98% (by weight) of the protein in the particle (22, 38). Upon infection, pVIII becomes associated with the membrane and later can be reutilized in the assembly of progeny phage particles (2, 29, 34). Therefore, the pVIII of the infecting phage probably assumes the same orientation in the membrane as newly synthesized pVIII. This would place the carboxyl-terminal 11 amino acids in the cytoplasm, the amino-terminal 20 amino acids in the periplasm, and the intervening 19 residues spanning the cytoplasmic membrane (24, 40).
Entry of the DNA into the cytoplasm appears to require the products of the bacterial tolQRA genes (4, 27, 33). Earlier studies suggested that pVIII from infecting phages became associated with the membrane in bacteria containing an undefined mutant of tolA (29). Our present knowledge about the topology of TolA (18) and its interactions with the phage pIII capsid protein (4, 26) would suggest that TolA must be required for the entry of pVIII into the membrane as well as for translocation of the DNA into the bacteria. To test this hypothesis, phages radiolabeled with either [3H]lysine or [35S]methionine-[35S]cysteine were used to infect both wild-type bacteria and bacteria containing a tolA null mutation. Based on the sequences of the mature capsid proteins (11) and the amount of each capsid protein per particle (22), approximately 99% of the [3H]lysine and 96.5% of the [35S]methionine-[35S]cysteine should be present in pVIII in these radiolabeled phages. Bacteria were infected at a multiplicity of infection of 100 for 10 min, conditions which result in at least 95% of the bacteria becoming infected (25). After the bacteria were washed, the percentage of the radioactive protein associated with the bacteria was determined (Table 1, experiment 1). Approximately 4% of the radioactivity remained with the F+ strain compared to 0.16% with the F− bacteria.
TABLE 1.
Fate of pVIII following infection of wild-type and tolA mutant E. coli
| Expt | Straina | Label in phagesb | % Input label remaining with bacteria afterc:
|
||
|---|---|---|---|---|---|
| Washing | Shearing | Subtilisin | |||
| 1 | K17/F− | [3H]Lys | 0.16 | ||
| K17/F+ | 3.98 | ||||
| K17tolA/F+ | 2.20 | ||||
| 2 | K91 | [3H]Lys | 4.18 | 3.62 | |
| K91tolA | 3.35 | 0.85 | |||
| 3 | K91 | 32P | 8.35 | 6.16 | |
| K91tolA | 7.97 | 1.68 | |||
| 4 | K17/F+ | [35S]Met-[35S]Cys | 3.84 | 2.65 | 2.57 |
| K17tolA/F+ | 2.61 | 0.47 | 0.35 | ||
| 5 | K17/F+ | [3H]Lys | 2.24 | 2.24d | |
| K17tolA/F+ | 3.21 | 1.08d | |||
All K17 strains are K17DE3, as described in Materials and Methods.
The specific activity of the [3H]lysine-labeled phages was 2.5 to 3.5 × 104 cpm/1010 PFU, that of the [35S]methionine-[35S]cysteine-labeled phages was 1.8 × 104 cpm/1010 PFU, and that of the 32P-labeled phages was 3.7 × 103 cpm/1010 PFU.
Bacteria were grown to 2 × 108 per ml, infected for 10 min with the appropriately labeled bacteriophage at a multiplicity of infection of 100, and harvested by centrifugation. The amount of radioactivity associated with the bacteria was measured after sequential washing, shearing, and treatment with subtilisin as described in Materials and Methods.
Washed bacteria which were treated with subtilisin without shearing.
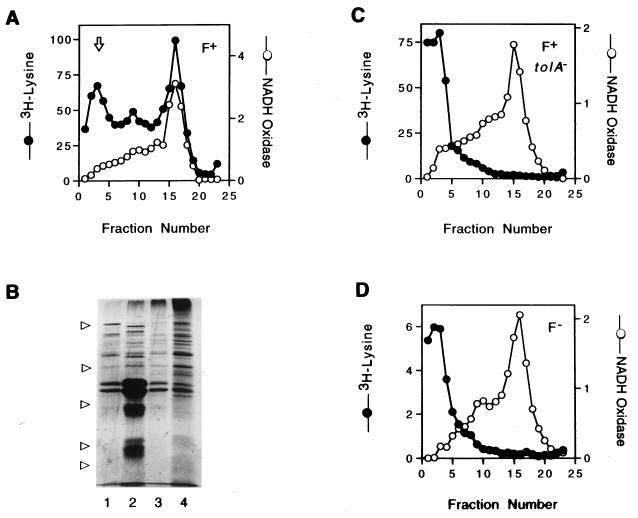
The infected F+ tolA null mutant strain (K17tolA/F+) contained approximately 2% of the input level of radioactivity, suggesting either that the pVIII had associated with the membrane or that intact phages were tightly attached to the bacteria. Membranes from the bacteria in this experiment were isolated on a sucrose flotation gradient. This procedure yields good separation of the cytoplasmic and outer membranes, as judged by the positions of the NADH oxidase activity (Fig. 1A) and the heavily stained outer membrane porins (Fig. 1B, lane 2), while leaving phages and phage fragments at the bottom of the gradient (Fig. 1A). A major portion of the radioactive label from the infected K17/F+ bacteria was associated with the cytoplasmic membrane (Fig. 1A). Some label also was associated with the fraction containing the outer membrane proteins. However, the membranes from infected K17tolA/F+ or K17/F− bacteria contained little if any radioactivity (Fig. 1C and D). This suggested that the radioactivity associated with the F+ tolA null bacteria was the result of phages attached to the pili and could be removed by subjecting the bacteria to shearing in an omnimixer or by treatment with subtilisin, a protease which has been shown to cleave the phage gene III protein (1, 9). Table 1 (experiments 2 and 3) shows that shearing removed radiolabeled phages from K91tolA mutant bacteria but not K91 wild-type bacteria, regardless of whether the phage contained the radioactive label in the protein or DNA. Shearing followed by treatment with subtilisin, or protease treatment alone, gave similar results (Table 1, experiments 4 and 5).
FIG. 1.
Phage coat protein pVIII from infecting phages is not found in the inner membranes of either F− or tolA mutant bacteria. (A, C, and D) Cultures of K17DE3 bacteria that were F+ (A), tolA/F+ (C), or F− (D) were infected with [3H]lysine-labeled phages (Table 1, experiment 1). The washed bacteria were broken in a French press, and the membrane fractions were separated by sucrose flotation gradient as described in Materials and Methods. The fractions, collected from the bottom of the gradient, were assayed for NADH oxidase activity, and radioactivity, which is expressed as counts per minute (in thousands), was determined. The arrow in panel A indicates the flotation position of both intact and broken phages. (B) Coomassie blue-stained SDS polyacrylamide gel of pooled fractions 1 to 5 (lane 1), 7 to 10 (lane 2), 11 to 13 (lane 3), and 14 to 18 (lane 4) from panel A. The arrows on the left indicate migration of protein standards with molecular masses (from the top) of 97, 45, 31, 21.5, and 14.4 kDa.
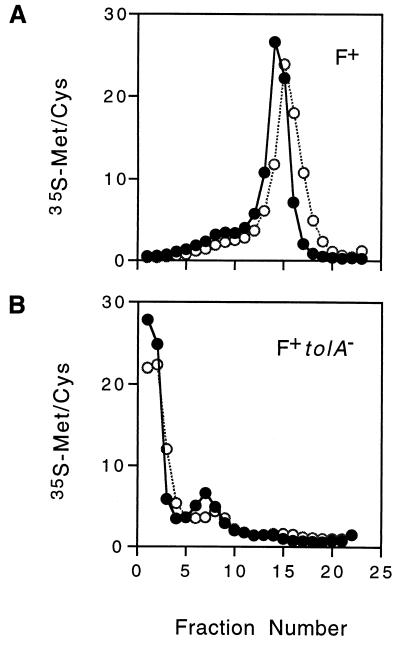
Presumably subtilisin would only cleave proteins located on the surfaces of the bacteria and therefore would not affect pVIII integrated into the cytoplasmic membrane. Even if subtilisin did have access to the periplasmic face of the cytoplasmic membrane under these experimental conditions, it should not affect the radiolabeled residues of pVIII, since the methionine is located in the transmembrane region and four of the five lysines are located in the cytoplasm. Membranes from infected K17/F+ bacteria which had been subjected to either shearing alone or shearing plus subtilisin (Table 1, experiment 4) were analyzed by sucrose gradient flotation centrifugation (Fig. 2A). The distribution of radioactivity was the same, regardless of protease treatment. When membranes from infected tolA mutant bacteria were examined, very little radioactivity was present in the cytoplasmic membrane (Fig. 2B). However, the protease treatment appeared to reduce the amount of radioactivity in the outer membrane portion of the gradient. The radioactivity present in the outer membrane portion of the gradient from infected tolA mutant bacteria might reflect phages attached to pili in a protein-rich portion of the membrane, such as an adhesion zone. The same experiments were repeated with lysine-labeled phages and yielded essentially the same results (Table 1 and data not shown).
FIG. 2.
Subtilisin treatment of sheared bacteria infected with [35S]methionine-[35S]cysteine-labeled phages. Cultures of K17DE3/F+ and K17DE3tolA/F+ bacteria infected with [35S]methionine-[35S]cysteine-labeled phages were washed, sheared, divided in half, and then incubated in the presence (○) or absence (•) of subtilisin (Table 1, experiment 4) as described in Materials and Methods. Radiolabeled membranes (approximately 2 × 105 cpm of K17DE3/F+ bacteria and 8 × 103 cpm of K17DE3tolA/F+ bacteria) were analyzed as described in the legend to Fig. 1. Radioactivity, measured by counting 20 to 40% of each fraction for 10 min, is expressed as a percentage of the total in the gradient.
Membrane insertion of pVIII correlates with the rate of infection.
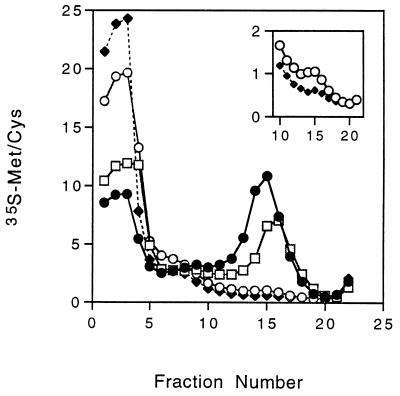
TolA has been shown to consist of three domains: an amino-terminal region anchored in the cytoplasmic membrane (domain I) and a periplasmic region consisting of a 232-residue-long helical region (domain II) attached to the carboxyl-terminal region (domain III) that is essential for activity (4, 18, 19). Click and Webster (4) showed that deletions of various regions of TolA slowed the rate of infection to varying degrees. Removal of the entire periplasmic helical domain of TolA (TolAΔII) resulted in a rate of infection approximately 10% that found for wild-type bacteria. Deletion of the amino-terminal half of domain II (TolAΔIIn) allowed a normal rate of infection, while deletion of the carboxyl-terminal half of domain II (TolAΔIIc) reduced the rate of infection by approximately 50%. If insertion of pVIII requires TolA, then the amount of pVIII inserted into the membrane in a 15-min infection period should correlate with the rate of infection.
Figure 3 shows the flotation gradient profile of membranes from K17tolA/F+ bacteria containing the TolA deletion proteins, which had been infected for 15 min with [35S]methionine-[35S]cysteine-labeled phages at a multiplicity of infection of 100. The membranes are from equal numbers of bacteria. The bacteria containing TolA lacking the carboxyl half of domain II (TolAΔIIC) incorporated only half as much pVIII as did bacteria lacking the amino-terminal half of domain II (TolAΔIIn). Membranes from bacteria containing TolA lacking the entire domain II (TolAΔII) had smaller, but detectable, amounts of pVIII in the inner membranes (Fig. 3, inset). These data give further evidence that TolA is required for insertion of the pVIII coat protein into the membrane during infection with the f1 filamentous phage.
FIG. 3.
Infection of strains expressing TolA deletion mutant proteins. Cultures of K17DE3tolA/F+ bacteria containing plasmids expressing either TolAΔIIn (•), TolAΔIIc (□), TolAΔII (○), or no TolA (⧫) were infected for 15 min with 35S-labeled phages, and the membrane fractions of the washed cultures (approximately 3 × 105 cpm) were analyzed as described in the legend to Fig. 1. Radioactivity is expressed as a percentage of the total in the gradient. The inset is an expanded scale comparing radioactivity in fractions 10 to 21 for bacteria with TolAΔII (○) and for bacteria with no TolA (♦). The washed cultures contained 6.4, 6.0, 5.2, and 5.2% of the input radioactivity, respectively.
TolQ and TolR are required for insertion of pVIII into the cytoplasmic membrane during infection.
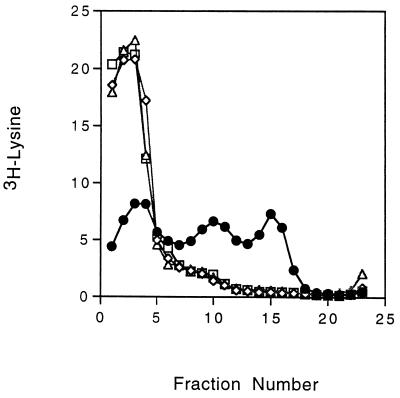
Both TolQ and TolR have been shown to be required for successful infection with the f1 phage (27, 33). Expression of these proteins is coupled, since translation of tolR is dependent on translation of the upstream tolQ region (36). Bacteria containing a missense mutation in tolQ (TPS66) or an insertion mutation in tolR (TPS300) were infected with [3H]lysine-labeled phages and analyzed for the presence of labeled pVIII in their cytoplasmic membranes. To test bacteria containing a null mutation in tolQ, a strain (TPS13) containing a polar amber mutation in tolQ was used and TolR was supplied from a plasmid (pPGK101). As shown in Fig. 4, no pVIII was detected in membranes from bacteria lacking either TolQ or TolR following infection with the radiolabeled phage.
FIG. 4.
Infection of tolQ and tolR mutant bacteria. Cultures of tol+ strain GM1 (•), tolR::Cm mutant TPS300 (◊), tolQ missense mutant TPS66 (▵), and tolQ amber mutant TPS13 expressing TolR from plasmid pPGK101 (□) were infected with [3H]lysine-labeled phages and analyzed as described in the legend to Fig. 1. Radioactivity is expressed as a percentage of the total in the gradient (approximately 1.5 × 105 cpm per strain).
DISCUSSION
Infection of E. coli by the Ff filamentous phages is initiated by the binding of one end of the particle to the tip of the F conjugative pilus. The phage capsid protein involved in this binding event is pIII, approximately five copies of which are located at one end of the phage particle. This minor capsid protein is composed of three domains (26, 31), with the carboxyl-terminal domain (pIII-D3) anchoring the protein to the phage particle, the amino-terminal domain (pIII-D1) involved in the translocation of the DNA into the cytoplasm, and the middle domain (pIII-D2) mediating the binding of the phage particle to the tip of the pilus. Retraction of the pilus (12) brings the bound phage to the bacterial surface, where the amino-terminal domain of pIII (pIII-D1) interacts with the carboxyl-terminal end of the TolA protein (TolA domain III), as described by Riechmann and Holliger (26). This interaction requires that pIII be associated with the tip of the pilus, since purified TolA domain III does not inhibit infection when added to phage particles but does inhibit infection when it is expressed in the periplasm (4). Since TolA domain III and pIII-D2 have been shown to compete for binding to pIII-D1 (26), it would appear that the binding of the phage to the pilus via pIII-D2 effectively removes pIII-D2 and makes pIII-D1 available for binding to domain III of TolA. This interpretation is consistent with the earlier observation by Boeke et al. that export into the periplasm of the amino-terminal 98 residues of pIII (pIII-D1) prevented infection by f1, presumably by interacting with domain III of TolA (3). However, these authors also showed that export of the amino-terminal 200 residues of pIII into the periplasm inhibited f1 phage infection. Since this region contains both domains 1 and 2 of pIII, perhaps pIII-D1 interacts with pIII-D2 only when the complete pIII molecule is present in the phage particle. In any event, the interaction of the phage pIII with TolA domain III is essential for subsequent steps in infection, since, in the absence of TolA, phage DNA is unable to enter the cytoplasm (4, 27) and the major coat protein, pVIII, is unable to enter the membrane (Table 1 and Fig. 1).
Following exposure to large numbers of radioactive phages, similar amounts of radioactive proteins remain associated with F+ and F+ tolA mutant bacteria after they are washed, in contrast to the smaller number associated with F− bacteria (Table 1). Membrane fractionation demonstrates that in the F+ bacteria, the radioactive proteins are associated with the cytoplasmic membrane fraction whereas the radioactivity associated with the F+ tolA bacteria appears to still be in phage particles. The radioactivity associated with the F+ tolA bacteria most likely reflects the ligand-receptor interaction of the phage with the tip of the F pilus. The presence of 2 to 3% of the radioactive phages (at a multiplicity of infection of 100) with washed F+ tolA bacteria (Table 1) suggests that two to three phages are associated with each bacterium, consistent with the average number of pili present per bacterium. These phages can be removed by shearing and washing, although further analysis by flotation gradient centrifugation showed that detectable radioactivity was still present in a dense fraction near the position of the outer membrane (Fig. 2B). Further treatment of the sheared bacteria with subtilisin removed some of this radioactivity, suggesting that it might be composed of pieces of sheared phages still attached to the withdrawn pilus tips. Therefore, the radioactivity associated with the outer membrane fractions of wild-type bacteria (Fig. 1A) may reflect phages attached to withdrawn pili in protein-dense portions of the membranes, such as adhesion zones. Further analysis is necessary to determine if such a structure is truly an intermediate in the infective process.
It is perhaps the tight binding of the phage to the pilus, making complete removal of phages from F+ bacteria difficult, that led to the earlier suggestion that pVIII is able to enter the membrane in a tolA mutant bacterium (29). We have found that phages, or fragments of phages produced by shearing in a French press, migrate with the inner membrane in tolA mutant bacteria when the membrane fractions are separated on sucrose step gradients after 18 to 24 h of centrifugation according to the procedures of Smilowitz et al. (30) or Levengood and Webster (17) (data not shown). The use of sucrose flotation gradients in this study demonstrates that pVIII is not inserted into the cytoplasmic membranes of the tolA mutant bacteria. An alternative explanation is that the mutant used in these earlier studies (29) may have been leaky to some extent.
TolA is required for the insertion of pVIII major capsid protein from an infecting phage into the cytoplasmic membrane, although the role that TolA plays in this process is not clear. It has been suggested that shortening the TolA molecule, by deleting domain II, might bring the phage closer to the periplasmic face of the cytoplasmic membrane (4). One might therefore expect the rate of entry of pVIII into the cytoplasmic membrane to be enhanced by this proximity. However, the efficiency of pVIII entry into the membrane remains proportional to the rate of infection in bacteria containing the TolA mutant proteins (Fig. 3), suggesting that it is not merely the proximity of the phage capsid proteins to the cytoplasmic membrane that allows the entrance of pVIII into the membrane. Rather it indicates that DNA translocation and membrane insertion of the capsid pVIII are closely coupled.
The pVIII major capsid protein from an infecting phage is inserted into the cytoplasmic membrane in such a manner that it can be assembled into a newly synthesized progeny phage particle (2, 29). Therefore, it presumably has the same topology in the membrane as newly synthesized pVIII, with the carboxyl-terminal 11 amino acids exposed in the cytoplasm. The process of insertion of pVIII from an infecting phage is certainly different from that for newly synthesized pVIII, which requires a 23-amino-acid signal sequence (14, 22). Insertion of pVIII from an infecting phage resembles the reverse of the assembly of mature pVIII around a newly synthesized phage DNA molecule. The minor capsid proteins pVII and pIX of an infecting phage may also be inserted into the membrane in a reusable manner. This prediction is based on the observation that infection of nonsuppressor strains of bacteria with gene VII or IX amber mutant phages gives rise to the production of between one and three infectious polyphages per bacterium, as if the input pVII and pIX proteins were able to direct the initiation of a phage particle (39). The fate of the input pIII or pVI minor capsid protein is not well understood at this time.
Ff bacteriophages are unable to infect bacteria containing mutations in any of the tolQ, -R, and -A genes (27, 32). The data presented here demonstrate the requirement for the TolQ, TolR, and TolA proteins for the insertion of pVIII into the membrane. Therefore, the translocation of the DNA into the cytoplasm may be coupled to the insertion of the capsid proteins into the membrane. After the formation of the TolA domain III-phage pIII complex, infection may proceed by subsequent interaction of this complex first with TolR, which has a large periplasmic domain (13, 23), and then with the entire TolQRA complex. The interacting transmembrane helices of TolQ, TolR, and TolA proteins (6, 16) could act as a channel for the phage DNA to cross the membrane (26) while pVIII, and perhaps the other capsid proteins, enters the cytoplasmic membrane. The TolQRA complex may be directly responsible for insertion of pVIII into the membrane in a manner similar to that for translocation of colicins (the TolQRA proteins are required for translocation of the group A colicins into or across the inner membrane [15]). If this is the case, the insertion of the coat protein into the cytoplasmic membrane may be the driving force for passage of the DNA across some channel in the membrane. Such a channel could be formed by pIII or by pIII plus the Tol proteins, as suggested by Riechmann and Holliger (26). Alternatively, the DNA channel may be solely a property of the three Tol proteins. Further experimentation is required to understand the interactions which occur at the cytoplasmic membrane during phage infection.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant GM 18306 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Armstrong J, Perham R N, Walker J E. Domain structure of bacteriophage fd adsorption protein. FEBS Lett. 1981;135:167–172. doi: 10.1016/0014-5793(81)80969-6. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J, Hewitt J A, Perham R N. Chemical modification of the coat protein in bacteriophage-fd and orientation of the virion during assembly and disassembly. EMBO J. 1983;2:1641–1646. doi: 10.1002/j.1460-2075.1983.tb01638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke J D, Model P, Zinder N D. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol Gen Genet. 1982;186:185–188. doi: 10.1007/BF00331849. [DOI] [PubMed] [Google Scholar]
- 4.Click E M, Webster R E. Filamentous phage infection: required interactions with the TolA protein. J Bacteriol. 1997;179:6464–6471. doi: 10.1128/jb.179.20.6464-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies J K, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol. 1975;123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deriouche R, Benedetti H, Lazzaroni J C, Lazdunski C, Lloubes R. Protein complex within Escherichia coli inner membrane-TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- 7.Fognini-Lefebvre N, Lazzaroni J C, Portalier R. tolA, tolB and excC, three cistrons involved in the control of pleiotropic release of periplasmic proteins by Escherichia coli K-12. Mol Gen Genet. 1987;209:391–395. doi: 10.1007/BF00329670. [DOI] [PubMed] [Google Scholar]
- 8.Frost L S. Conjugative pili and pilus-specific phages. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum; 1993. pp. 189–221. [Google Scholar]
- 9.Gray C W, Brown R S, Marvin D A. Adsorption complex of filamentous fd virus. J Mol Biol. 1981;146:621–627. doi: 10.1016/0022-2836(81)90050-4. [DOI] [PubMed] [Google Scholar]
- 10.Guihard G, Boulanger P, Benedetti H, Lloubes R, Besnard M, Letellier L. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J Biol Chem. 1994;269:5874–5880. [PubMed] [Google Scholar]
- 11.Hill D F, Petersen G B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982;44:32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson A. Role of F pili in the penetration of bacteriophage f1. J Virol. 1972;10:835–843. doi: 10.1128/jvi.10.4.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kampfenkel K, Braun V. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J Bacteriol. 1993;175:4485–4491. doi: 10.1128/jb.175.14.4485-4491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn A, Troschel D. Distinct steps in the insertion pathway of bacteriophage coat proteins. In: Newport W, Lills R, editors. Membrane biogenesis and protein targeting. New York, N.Y: Elsevier; 1992. pp. 33–47. [Google Scholar]
- 15.Lazdunski C. Colicin import and pore formation: a system for studying protein transport across membranes? Mol Microbiol. 1995;16:1059–1066. doi: 10.1111/j.1365-2958.1995.tb02331.x. [DOI] [PubMed] [Google Scholar]
- 16.Lazzaroni J C, Vianney A, Popot J L, Benedetti H, Samatey F, Lazdunski C, Portalier R, Geli V. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J Mol Biol. 1995;246:1–7. doi: 10.1006/jmbi.1994.0058. [DOI] [PubMed] [Google Scholar]
- 17.Levengood S K, Webster R E. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J Bacteriol. 1989;171:6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levengood S K, Beyer W F, Jr, Webster R E. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc Natl Acad Sci USA. 1991;88:5939–5943. doi: 10.1073/pnas.88.14.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levengood-Freyermuth S K, Click E M, Webster R E. Role of the carboxyl-terminal domain of TolA in protein import and integrity of the outer membrane. J Bacteriol. 1993;175:222–228. doi: 10.1128/jb.175.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin T C, Webster R E, Konigsberg W. Isolation and characterization of the C and D proteins coded by gene IX and gene VI in the filamentous bacteriophage f1 and fd. J Biol Chem. 1980;255:10331–10337. [PubMed] [Google Scholar]
- 21.Lopez J, Webster R E. Morphogenesis of filamentous bacteriophage f1: orientation of extrusion and production of polyphage. Virology. 1983;127:177–193. doi: 10.1016/0042-6822(83)90382-3. [DOI] [PubMed] [Google Scholar]
- 22.Model P, Russel M. Filamentous bacteriophage. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum; 1988. pp. 375–456. [Google Scholar]
- 23.Muller M M, Vianney A, Lazzaroni J C, Webster R E, Portalier R. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J Bacteriol. 1993;175:6059–6061. doi: 10.1128/jb.175.18.6059-6061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkawa I, Webster R E. The orientation of the major coat protein of bacteriophage f1 in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1981;256:9951–9958. [PubMed] [Google Scholar]
- 25.Rapoza M P, Webster R E. The products of gene I and the overlapping in-frame gene XI are required for filamentous phage assembly. J Mol Biol. 1995;248:627–638. doi: 10.1006/jmbi.1995.0247. [DOI] [PubMed] [Google Scholar]
- 26.Riechmann L, Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 27.Russel M, Whirlow H, Sun T-P, Webster R E. Low-frequency infection of F− bacteria by transducing particles of filamentous bacteriophages. J Bacteriol. 1988;170:5312–5316. doi: 10.1128/jb.170.11.5312-5316.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schendel S L, Click E M, Webster R E, Cramer W A. The TolA protein interacts with colicin E1 differently than with other group A colicins. J Bacteriol. 1997;179:3683–3690. doi: 10.1128/jb.179.11.3683-3690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smilowitz H. Bacteriophage f1 infection: fate of the parental major coat protein. J Virol. 1974;13:94–99. doi: 10.1128/jvi.13.1.94-99.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smilowitz H, Carson J, Robbins P W. Association of newly synthesized f1 major coat protein with infected host cell inner membrane. J Supramol Struct. 1972;1:8–18. doi: 10.1002/jss.400010103. [DOI] [PubMed] [Google Scholar]
- 31.Stengle I, Garces X, Giray J, Rasched I. Dissection of functional domains in phage fd adsorption protein: discrimination between attachment and penetration sites. J Mol Biol. 1990;212:143–149. doi: 10.1016/0022-2836(90)90311-9. [DOI] [PubMed] [Google Scholar]
- 32.Sun T-P, Webster R E. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986;165:107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun T P, Webster R E. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J Bacteriol. 1987;169:2667–2674. doi: 10.1128/jb.169.6.2667-2674.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trenkner E, Bonhoeffer F, Gierer A. The fate of the protein component of bacteriophage fd during infection. Biochem Biophys Res Commun. 1967;28:932–939. doi: 10.1016/0006-291x(67)90069-1. [DOI] [PubMed] [Google Scholar]
- 35.Vianney A, Lewin T M, Beyer W F, Jr, Lazzaroni J C, Portalier R, Webster R E. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J Bacteriol. 1994;176:822–829. doi: 10.1128/jb.176.3.822-829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vianney A, Muller M M, Clavel T, Lazzaroni J C, Portalier R, Webster R E. Characterization of the tol-pal region of Escherichia coli K-12: translational control of tolR expression by tolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J Bacteriol. 1996;178:4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webster R E. The tol gene products and the import of macromolecules into Escherichia coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
- 38.Webster R E. Biology of the filamentous bacteriophage. In: Kay B K, et al., editors. Phage display of peptides and proteins. San Diego, Calif: Academic Press Inc.; 1996. pp. 1–20. [Google Scholar]
- 39.Webster R E, Lopez J. Structure and assembly of the class I filamentous bacteriophage. In: Casjens S, editor. Virus structure and assembly. Boston, Mass: Jones and Bartlett; 1985. pp. 235–268. [Google Scholar]
- 40.Wickner W. Asymmetric orientation of a phage coat protein in cytoplasmic membrane of Escherichia coli. Proc Natl Acad Sci USA. 1975;72:4749–4753. doi: 10.1073/pnas.72.12.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]