Abstract
The rpoN region of Rhizobium etli was isolated by using the Bradyrhizobium japonicum rpoN1 gene as a probe. Nucleotide sequence analysis of a 5,600-bp DNA fragment of this region revealed the presence of four complete open reading frames (ORFs), ORF258, rpoN, ORF191, and ptsN, coding for proteins of 258, 520, 191, and 154 amino acids, respectively. The gene product of ORF258 is homologous to members of the ATP-binding cassette-type permeases. ORF191 and ptsN are homologous to conserved ORFs found downstream from rpoN genes in other bacterial species. Unlike in most other microorganisms, rpoN and ORF191 are separated by approximately 1.6 kb. The R. etli rpoN gene was shown to control in free-living conditions the production of melanin, the activation of nifH, and the metabolism of C4-dicarboxylic acids and several nitrogen sources (ammonium, nitrate, alanine, and serine). Expression of the rpoN gene was negatively autoregulated and occurred independently of the nitrogen source. Inactivation of the ptsN gene resulted in a decrease of melanin synthesis and nifH expression. In a search for additional genes controlling the synthesis of melanin, an R. etli mutant carrying a Tn5 insertion in ptsA, a gene homologous to the Escherichia coli gene coding for enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system, was obtained. The R. etli ptsA mutant also displayed reduced expression of nifH. The ptsN and ptsA mutants also displayed increased sensitivity to the toxic effects of malate and succinate. Growth of both mutants was inhibited by these C4-dicarboxylates at 20 mM at pH 7.0, while wild-type cells grow normally under these conditions. The effect of malate occurred independently of the nitrogen source used. Growth inhibition was decreased by lowering the pH of the growth medium. These results suggest that ptsN and ptsA are part of the same regulatory cascade, the inactivation of which renders the cells sensitive to toxic effects of elevated concentrations of malate or succinate.
Bacterial sigma (ς) factors confer promoter specificity to transcription initiated by the RNA polymerase holoenzyme (α2ββ′ς). On the basis of structural and functional criteria, ς factors fall into two major classes. Most ς factors are similar to the major vegetative ς factor of Escherichia coli, ς70 (18, 26). This sigma factor recognizes sequences similar to the canonical −35/−10 type of promoter and directs transcription of many housekeeping genes. Besides ς70, this family contains several alternative ς factors allowing cells to respond to many different environmental stimuli, each controlling a specific process such as sporulation, heat shock response, and flagellation (26). The second type of ς factor, ς54 (RpoN, NtrA, or GlnF), shows little sequence similarity to the first class and has been identified so far in many gram-negative and gram-positive bacterial species. Although it was originally recognized for its role in nitrogen metabolism, it is clear that ς54 also controls the expression of genes responding to many other physiological needs (24, 31). Promoters recognized by RpoN share conserved DNA sequences of which the consensus is 5′-CTGGCAC-N5-TTGCA-3′ (−24/−12 type of promoter; the invariant dinucleotides GG/GC are in boldface) (2).
Whereas the ς70-holoenzyme complex often initiates transcription in the absence of transcriptional activators, transcription from all known ς54-dependent promoters has an absolute dependence on the presence of an activator protein, such as NifA or NtrC (9, 24, 49). In the latter case, activators often bind to sequences located more than 100 bp upstream from the transcriptional start site (5, 9). This difference is probably correlated with the ability of ς54, but not ς70, to bind (in vitro) to the promoter in the absence of RNA polymerase core enzyme (6, 12). The strong interaction of ς54 with its promoter keeps the ς54 holoenzyme-promoter complex in a closed conformation which may then require activation by another protein to induce local DNA melting and initiate transcription (38, 46).
In most bacteria in which the rpoN region has been analyzed, a common organization of downstream open reading frames (ORFs) is found (31). The function of the conserved ORFs has been studied in only a few bacterial species, and their role has not been clearly defined yet. These genes are thought to modulate the activity of ς54 (30). In Klebsiella pneumoniae, the inactivation of the two ORFs, ORF95 and ptsN, located immediately downstream from rpoN increases the expression from several ς54-dependent promoters (30, 32), while a mutation in the fourth gene (ORF90) reduces the activity of these same promoters (32). For Pseudomonas aeruginosa, it was suggested that, together with rpoN, ORF2 functions as a coinducer of genes involved in the assimilation of glutamine (20). In Caulobacter crescentus, ORF159 modulates the expression from a ς54-dependent flagellar promoter.
Bacteria belonging to the genera Rhizobium, Bradyrhizobium, Azorhizobium, Mesorhizobium, and Sinorhizobium (collectively called rhizobia) elicit nitrogen-fixing nodules on the roots of their leguminous host plant. The rpoN genes in four rhizobial species, Rhizobium meliloti (43), Bradyrhizobium japonicum (23), Azorhizobium caulinodans (48), and Rhizobium sp. strain NGR234 (52), have been characterized. In these organisms, ς54 has been shown to control C4-dicarboxylate utilization (23, 43, 48), nitrate assimilation (23, 43, 48), and several symbiotic functions (23, 43, 48, 52). However, only partial DNA sequence information on the downstream regions of rhizobial rpoN genes is available (23, 43, 48). In addition, the role and regulation of the conserved ORFs located 3′ to rpoN have not been investigated yet.
Here we describe the identification and analysis of the rpoN region of Rhizobium etli, the nodulating symbiont of the common bean plant, Phaseolus vulgaris. In contrast to the case for most other bacterial species, the rpoN gene is separated by approximately 1.6 kb from the conserved ORFs, ORF191 and ptsN, which are normally found immediately downstream from rpoN. We analyzed the regulation of rpoN transcription and the phenotypes of rpoN and ptsN mutant strains. Moreover, we identified an R. etli Tn5 mutant strain displaying reduced melanin production. A further examination of this mutant revealed that the phenotype resulted from a mutation in ptsA, the gene coding for enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system (PTS). Our results indicate that in R. etli, ptsN and ptsA may coregulate several RpoN-dependent activities, including the activation of nifH, the production of the pigment melanin, and the assimilation of alanine. In addition, the ptsN and ptsA mutant strains display an increased sensitivity to the toxic effects of high concentrations of malate and succinate. Therefore, ptsN and ptsA are thought to be part of the same regulatory cascade which may be involved in the sensing of C4-dicarboxylates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Some are also diagrammed in Fig. 1. Plasmids used merely for sequencing are not shown. The Rhizobium isolate used in the present study is CNPAF512 (CNPAB/EMBRAPA culture collection), a Brazilian isolate from P. vulgaris nodules. Based on our nucleotide sequence analysis of a 260-bp fragment of the 16S rRNA gene, amplified by PCR with the primers Y1 (5′-TGGCTCAGAACGAACGCTGGCGGC-3′) and Y2 (5′-CCCACTGCTGCCTCCCGTAGGAGT-3′), and multilocus enzyme electrophoresis of this isolate (26a), CNPAF512 should be classified as R. etli. R. etli strains were routinely grown in liquid TY (0.5% tryptone, 0.3% yeast extract, 7 mM CaCl2) at 30°C and maintained on yeast-mannitol agar plates. E. coli was grown in Luria-Bertani medium at 37°C. Antibiotics supplied to the medium were at the following concentrations: nalidixic acid and neomycin, 40 μg/ml; kanamycin and gentamicin, 30 μg/ml; and ampicillin, 100 μg/ml. Tetracycline was added to a final concentration of 1 μg/ml (for R. etli) or 10 μg/ml (for E. coli).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | Gibco/BRL | |
| HB101 | Smr | 45 |
| R. etli | ||
| CNPAF512 | Nalr, wild type | This work |
| FAJ1154 | NmrrpoN::Ω-Km, opposite orientation | This work |
| FAJ1155 | NmrrpoN::Ω-Km, same orientation | This work |
| FAJ1156 | NmrrpoN::gusA-aphII | This work |
| FAJ1157 | NmrrpoN::aphII-gusA | This work |
| FAJ1164 | NmrptsN::Ω-Km, same orientation | This work |
| FAJ1165 | NmrptsN::Ω-Km, opposite orientation | This work |
| FAJ1166 | NmrptsA::Tn5-mob | This work |
| R. meliloti | ||
| 2011 | Wild type | |
| 1681 | Smr KmrrpoN::Tn5 | 43 |
| Plasmids | ||
| pUC18 | Apr, cloning vector | 35 |
| pJQ200-UC1 | GmrsacB | 40 |
| pHP45Ω-Km | Apr Kmr | 14 |
| pWM6 | Apr NmruidA2 | 33 |
| pLAFR1 | Tcr, broad-host-range vector | 15 |
| pFAJ21 | Tcr, A. brasilense pnifH-gusA | 51 |
| pFAJ1150 | Tcr, rpoN gene in pLAFR1 | This work |
| pFAJ1152 | Apr, pUC18, rpoN 6.5-kb HindIII fragment | This work |
| pFAJ302 | Tcr, fusion between A. brasilense ammonium transporter amtB and gusA | Anne Van Dommelen (this laboratory) |
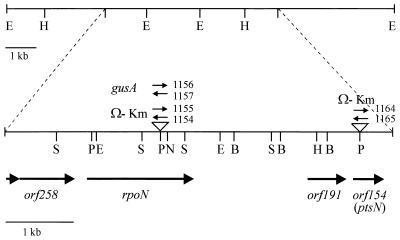
FIG. 1.
Physical map of the R. etli rpoN region. The 1.8-, 4.6-, and 6.1-kb EcoRI fragments hybridizing to the A. tumefaciens probe are shown above the physical map of the 5.6-kb region that was sequenced. Triangles represent insertions of the Ω-Km interposon or the uidA-aphII cassette. The positions and orientations of the identified ORFs are indicated below the restriction map. Restriction sites are abbreviated as follows: B, BstXI; H, HindIII; E, EcoRI; N, NotI; P, PstI.
Triparental conjugations and site-directed mutagenesis of R. etli were done as previously described (11).
Growth tests.
Tests of growth of R. etli in liquid medium were carried out in acid minimal salts (AMS) medium (37) containing 1 mM CaCl2. Cells were first grown overnight in TY, washed twice in 10 mM MgSO4, brought to an optical density (OD) at 600 nm of 0.02 in a Perkin-Elmer lambda 2 spectrometer, and diluted 100 times in AMS medium. Carbon and nitrogen sources were added to the appropriate concentrations with a sterile concentrated stock solution at pH 7.0. Cell growth was monitored in terms of turbidity at 600 nm in a microtiterplate reader. Carbon sources were d-(−)-mannitol, d-(+)-glucose, sucrose, l-(+)-arabinose, d-(+)-galactose, d-sorbitol, d-(−)-fructose, glycerol, trisodium citrate, cis-aconic acid, dl-isocitric acid, α-ketoglutaric acid, succinic acid, fumaric acid, dl-malic acid, oxaloacetic acid, and pyruvic acid. Nitrogen sources were NH4Cl, l-alanine, l-glutamine, and KNO3. For growth tests at various pH values, 3 (N-morpholino)propanesulfonic acid (pH 7.0) and 2(N-morpholino)ethanesulfonic acid (pH 6.5, 6.0, and 5.5) were used at a concentration of 30 mM.
For growth tests on plates, appropriate combinations of mannitol (0.2%, wt/vol), ammonium (10 mM), or amino acids (0.2%, wt/vol) were added to AMS agar (15 g/liter). Plates were supplied with (i) only the amino acid, (ii) the amino acid and ammonium, (iii) the amino acid and mannitol, or (iv) the amino acid, ammonium, and mannitol. Plates were incubated at 30°C, and colony size was monitored over a period of 3 to 7 days.
DNA methods.
General DNA manipulations were performed as described previously (3, 45). DNA fragments were recovered from agarose gels by using the Nucleotrap kit (Macherey-Nagel). Southern blotting and hybridizations were carried out as described previously (3, 34, 45). DNA probes were labeled with a digoxigenin labeling and detection kit (Boehringer). To generate blunt ends to incompatible DNA fragments, DNA was incubated with Klenow or T4 DNA polymerase in the presence of the four deoxynucleoside triphosphates. Automated DNA sequencing was performed on a Pharmacia A.L.F. sequencer with fluorescein-labeled universal and synthetic oligonucleotide primers. Both strands of overlapping pUC18 subclones covering the 5,600-bp DNA fragment were read with multiple sequencing.
PCR conditions.
Amplification of DNA fragments by PCR was performed in a TRIO-Thermoblock (Biometra). Twenty-five-microliter reaction mixes, containing 0.65 U of Taq DNA polymerase (Boehringer), each of the deoxynucleoside triphosphates at 200 μM, and each of the primers at 1 μM, were subjected to 30 cycles of incubation at 94°C for 60 s, 60°C (primers ojm065 and ojm072) for 60 s, and 72°C for 210 s.
Construction of mutants.
To mutagenize the R. etli rpoN gene, the 1.8-kb EcoRI fragment of pFAJ1150 was blunt-end ligated into the SmaI site of pJQ200-UC1. The resulting plasmid was digested with PstI. This plasmid was used in two separate reactions. First, it was blunt-end ligated to the 1.8-kb BamHI fragment from pHP45Ω-Km, resulting in two constructs in which the Ω-Km fragment is inserted in opposite directions. The Ω-Km interposon contains the aphII gene from Tn5 and confers resistance to kanamycin and neomycin. Second, to construct a transcriptional rpoN-gusA fusion, it was also blunt-end ligated to the 3.8-kb BamHI fragment from pWM6. Two plasmids carrying the gusA-Kmr cassette in opposite orientations were obtained. These four insertional mutations (Ω-Km and gusA) were finally recombined into the R. etli CNPAF512 chromosome. Insertion of these mutations was verified by Southern blot hybridization with the appropriate probes. In this way, the following mutants were constructed: FAJ1154 (orientations of Ω-Km and rpoN are opposed), FAJ1155 (same orientations), FAJ1156 (orientations of gusA and rpoN are the same), and FAJ1157 (opposite orientations).
For the inactivation of ptsN, a 1.7-kb fragment containing ptsN was amplified by PCR with pFAJ1150 as template DNA and the following two sequence-specific primers: ojm065 (5′-GAGCGCGGCCGCGTGGATCGGACTGATCTC-3′) and ojm072 (5′-ACTCGCGGCCGCGCTTCCGGGTCTCCGGTTCG-3′). The amplified fragment carries NotI recognition sites at both ends. Following digestion with NotI, this fragment was inserted at the corresponding site of pJQ200-UC1. Finally, the 2.2-kb BamHI fragment of pHP45Ω-Km was cloned into the PstI site of the 1.7-kb insert of pJQ200-UC1, after blunting of both fragments, thereby inactivating ptsN. The different constructs were used to generate site-directed mutants of R. etli CNPAF512. In the mutants FAJ1164 and FAJ1165, ptsN and Ω-Km read in the same and opposite directions, respectively.
Nucleotide sequence accession number.
The nucleotide sequence of the 5,600-bp DNA fragment containing R. etli rpoN and associated genes has been deposited with DDBJ-EMBL-GenBank under accession no. U23471.
RESULTS
Cloning of the R. etli rpoN gene.
To detect the rpoN gene, EcoRI-restricted genomic DNA from R. etli CNPAF512 was hybridized with, as a probe, the 1.7-kb EcoRI-HindIII fragment from pRJ7694 carrying the B. japonicum rpoN1 gene (23). One EcoRI fragment of 1.8 kb strongly hybridized to the probe. Also, only one hybridizing fragment was observed on HindIII- and SalI-restricted genomic DNA. The same probe was therefore used to screen a gene library of R. etli CNPAF512 maintained in E. coli HB101. This library was constructed in the EcoRI site of pLAFR1. One hybridizing cosmid, pFAJ1150, was identified. When pFAJ1150 was digested with EcoRI and hybridized with a 3.5-kb EcoRI fragment containing Agrobacterium tumefaciens rpoN and flanking DNA (53), three fragments of 1.8, 4.6, and 6.1 kb hybridized strongly. A physical map of the R. etli rpoN region is shown in Fig. 1.
To ascertain that pFAJ1150 contained a functional rpoN gene, this plasmid was transferred to the R. meliloti rpoN mutant 1680 (43). In contrast to the wild-type strain 1021, mutant 1680 cannot fix atmospheric nitrogen during symbiosis with alfalfa plants and is not able to grow on C4-dicarboxylates as carbon sources or to assimilate nitrate. Plasmid pFAJ1150 was shown to complement each of these defects in strain 1680 (data not shown).
Sequence analysis of the rpoN region.
To further characterize the R. etli rpoN gene, the nucleotide sequence of a 5,600-bp DNA fragment was determined. Examination of this nucleotide sequence revealed the presence of one partial and four complete (ORFs) (Fig. 1). All of the ORFs are transcribed in the same direction as rpoN. The initiation codons were assigned on the basis of sequence homology with homologous ORFs in other bacterial species.
The first complete ORF, ORF258, codes for a protein of 258 amino acids with a calculated molecular mass of 28,335 Da. No obvious E. coli-like Shine-Dalgarno sequence was detected upstream from the ORF258 initiation codon. A DNA sequence (GATTTCAGGCC-N5-GGCCTGAAATC) with the potential to form a hairpin loop secondary structure (ΔG [25°C] = −21.2 kcal) was detected 15 bp downstream from the termination codon. ORF258 shows homology with ORFs and partially sequenced ORFs located upstream from rpoN genes in other bacterial species (e.g., 81% amino acid identity with R. meliloti). The proteins encoded by these ORFs resemble members of the ATP-binding cassette-type permeases. ORF258 is preceded by the 3′ end of an incomplete ORF whose derived protein shows 50% amino acid identity with the corresponding R. meliloti protein.
The second ORF, named R. etli rpoN, is located 215 bp downstream from ORF258. The rpoN gene codes for an acidic (calculated isoelectric point, 4.32) protein of 520 amino acids with a predicted molecular mass of 57,477 Da. A putative ribosome-binding site (GGAG) is located 13 bp upstream from the rpoN initiation codon. Further analysis of the upstream region of rpoN revealed strong nucleotide sequence identity with the R. meliloti rpoN promoter. The R. meliloti rpoN start of transcription has been determined previously, and potential −35 and −10 regions have been identified (1). Sequences identical to the −35 and −10 regions of the R. meliloti rpoN promoter (CTTGAC-N17-CAATTT) as well as the transcriptional start site (CAATTTTTGGGCCAACT [the transcriptional start is underlined]) are conserved in the R. etli rpoN promoter region, suggesting that these sequences might also be operative in R. etli. Strong conservation of amino acid residues was found between R. etli RpoN and all known RpoN proteins of rhizobia (R. meliloti, 68% amino acid identity; Rhizobium sp. strain NGR234, 65%; A. caulinodans, 53%; and B. japonicum, 54% with RpoN1 and 50% with RpoN2). The three regions of the R. etli RpoN protein, as defined for other RpoN proteins (31, 52), are the amino-terminal region I (50 amino acids), region II (108 amino acids), and the carboxy-terminal region III (362 amino acids). A conserved helix-turn-helix motif, implicated in binding of the −24/−12 promoter (10), is found in the R. etli RpoN protein between amino acid positions 398 and 418 (helix [N-I]-turn [K-H]-helix [E-S]). A highly conserved sequence of 10 amino acids (ARRTVAKYRE), termed the RpoN box (52), is located near the carboxy terminus of R. etli RpoN between amino acids 488 and 497.
A third complete ORF was located 1.6 kb downstream from rpoN. This ORF is predicted to encode a protein of 191 amino acids with a calculated molecular mass of 21,172 Da. ORF191 is preceded by a putative ribosome-binding site, AGAAGG, located 8 bp upstream from the presumptive initiation codon. The protein sequence derived from ORF191 is homologous to sequences of proteins encoded by genes that are normally found immediately downstream from rpoN (e.g., E. coli ORF95). Eleven of these genes in various bacterial species have been sequenced (39); nine of them were completely sequenced. The number of amino acids encoded by these genes typically ranges from 95 to 130, with the exception of B. japonicum ORF203 and C. crescentus ORF208, which code for 203- and 208- amino-acid proteins, respectively. These proteins were shown to be homologous to an E. coli protein encoded upstream from pheA (SwissProt accession no. P11285 [31]).
The fourth ORF is located 76 bp downstream from ORF191 and codes for a protein of 154 amino acids (PtsN) with a predicted molecular mass of 16,675 Da. A putative ribosome-binding site (AGAAGG) is located 8 bp upstream from the initiation ATG. A DNA sequence (AAAAAAGGCGCCTG-N6-CAGGCGCCTTTTTT) with the potential to form a stable hairpin loop secondary structure (ΔG [25°C] = −28.2 kcal) was detected 19 bp downstream from the termination codon. This sequence may function as a rho-independent terminator (GC-rich stem loop followed by several Ts). This ORF is homologous to previously identified ptsN genes located at the same position in other bacterial species. This ORF encodes a protein that is similar to the enzyme IIA protein of the PTS, and in particular to enzyme IIA proteins specific for the transport of mannitol and fructose (39). No ORF with homology to E. coli ORF284 or ORF90 (encoding the Hpr-like protein Npr), located 3′ of the ptsN gene, was found downstream from ptsN (data not shown).
Expression of R. etli rpoN.
To monitor expression of R. etli rpoN, an rpoN-gusA (gusA in both orientations) fusion was inserted into the chromosome by site-directed mutagenesis, thereby inactivating the rpoN gene. The resulting strains are FAJ1156 (correct orientation of gusA) and FAJ1157 (opposite orientation). R. etli FAJ1156 was also complemented by using pFAJ1150. FAJ1156(pFAJ1150) did not differ from the wild type with respect to growth on nitrate or succinate, melanin production, and symbiotic properties (data not shown). Expression of rpoN was assayed in different media (Table 2). No difference between the expression levels in these media was observed, except that in FAJ1156 the expression level of rpoN was slightly lower when the strain was cultured in TY than when it was cultured in AMS medium containing ammonia and succinate. Also, expression was unaffected by the ammonia concentration (from 1 to 10 mM) (data not shown). However, under all free-living conditions tested, expression of rpoN-gusA was higher in FAJ1156 than in the complemented strain (Table 2). No β-glucuronidase activity was measured in the control strain FAJ1157. From these data it appears that the rpoN gene is expressed independently of the nitrogen concentration and is negatively autoregulated.
TABLE 2.
Expression of a chromosomally integrated R. etli rpoN-gusA fusion in wild-type [FAJ1156(pFAJ1150)] and rpoN mutant (FAJ1156) backgrounds
| Growth conditiona | β-Glucuronidase
activity (Miller units)b in:
|
|
|---|---|---|
| FAJ1156 | FAJ1156(pFAJ1150) | |
| Ammonium and mannitol | 82 (37) | 19 (13) |
| Nitrate and mannitol | 84 (37) | 11 (6) |
| Ammonium and succinate | 112 (20) | 19 (13) |
| TY, aerobic | 64 (12) | 20 (8) |
Cells were grown in TY or AMS medium containing the indicated combinations of ammonium, nitrate, and succinate (each at 10 mM) and mannitol (10 g/liter). Microaerobic inductions were carried out with 0.5% oxygen as detailed in Materials and Methods.
Data are the means for four replicates. Standard deviations are given in parentheses.
Growth of R. etli rpoN mutants.
To investigate the function of rpoN, the Ω-Km interposon was inserted in both orientations in the R. etli rpoN gene. These mutations were recombined into the chromosome by site-specific mutagenesis, yielding mutants FAJ1154 and FAJ1155 (Fig. 1 and Table 1). Insertion of the interposon at the correct site was confirmed by Southern hybridization. The rpoN mutants were tested for growth on defined media containing amino acids, ammonium, or KNO3 as a nitrogen source and mannitol or succinate as a carbon source.
First, growth of the rpoN mutant FAJ1154 was tested on different amino acids. Therefore, cells were grown on solid AMS minimal medium containing only the amino acid and compared with the parental strain CNPAF512. To evaluate possible toxic effects of the amino acid, mannitol (carbon source) or ammonium (nitrogen source) was additionally supplied to the plates (Table 3). No difference was observed for media containing lysine, glutamine, glutamate, proline, leucine, or isoleucine (Table 3). However, growth of the mutants was strongly inhibited on medium containing alanine or serine, in the presence or absence of ammonium or mannitol (Table 3). These results indicate that the rpoN gene is required for the metabolism of alanine and serine. In addition, since growth of the rpoN mutant is also restricted on media containing, in addition to alanine or serine, ammonium and mannitol, these amino acids are likely to be toxic to the cell or to inhibit the assimilation of ammonium or mannitol. To discriminate between these possibilities, glutamine was additionally supplied to the plates. Glutamine is a good nitrogen source but a poor carbon source in wild-type R. etli and the rpoN mutant (Table 3). Upon the addition of glutamine to these media, growth was restored to the wild-type level. Therefore, alanine and serine probably inhibit the assimilation of ammonia in the rpoN mutant.
TABLE 3.
Growth of R. etli wild-type strain CNPAF512 and rpoN mutant FAJ1154 on defined mediaa
| Amino acid(s) | Growthb with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Amino acid(s)
|
Amino acid(s) and
ammonium
|
Amino acid(s) and mannitol
|
Amino
acid(s), ammonium, and mannitol
|
|||||
| Wild type | rpoN mutant | Wild type | rpoN mutant | Wild type | rpoN mutant | Wild type | rpoN mutant | |
| Gln | + | + | + | + | +++ | +++ | +++ | +++ |
| Pro | ++ | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| Ala | + | − | + | − | +++ | − | +++ | − |
| Ser | + | +/− | + | +/− | +++ | +/− | +++ | +/− |
| Ala and Gln | + | + | + | + | +++ | +++ | +++ | +++ |
| Ser and Gln | + | + | + | + | +++ | ++ | +++ | ++ |
Growth tests were carried out on AMS plates containing the indicated combinations of amino acid(s) (0.2%), ammonium (10 mM), and mannitol (10 g/liter) (see Materials and Methods). Colony size was determined after 3 days of incubation at 30°C. Growth on Glu, Lys, Ile, and Leu was similar to that on Gln.
Symbols: −, no growth; +/−, colony size of <0.5 mm; +, colony size of 0.5 to 1 mm; ++, colony size of 1 to 2 mm; +++, colony size of >2 mm.
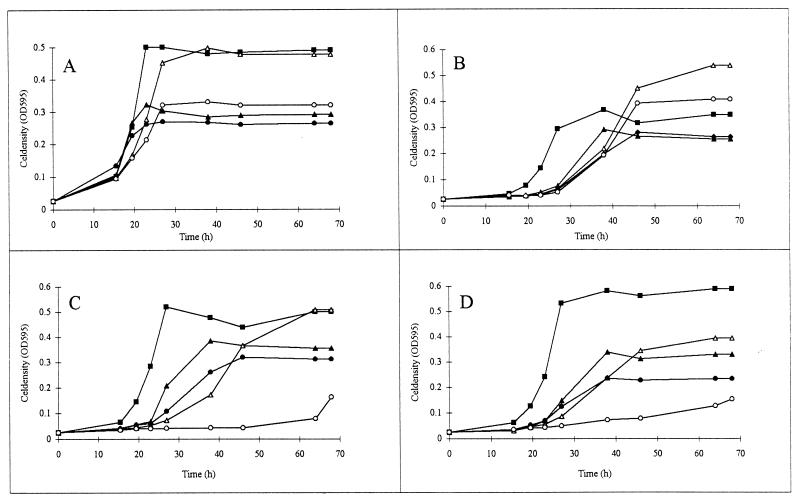
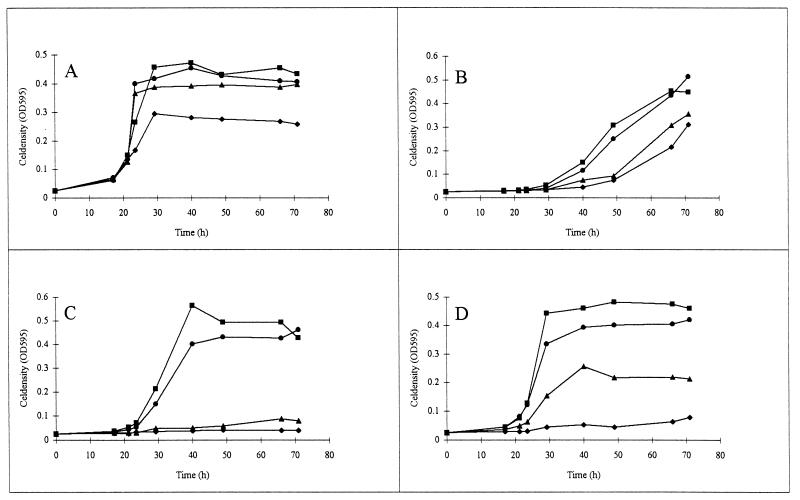
Second, growth of the rpoN mutants FAJ1154 and FAJ1155 in liquid AMS medium containing 10 mM NH4Cl or KNO3 (nitrogen source) and 10 mM mannitol, succinate, fumarate, or malate (carbon source) was monitored. When grown in the presence of ammonium and mannitol, both rpoN mutants have an extended lag phase which is approximately 10 h longer than that of the wild type (Fig. 2A and B). An additional delay of 10 h was observed when succinate, fumarate, or malate was used instead of mannitol (Fig. 2B). Finally, in comparison with that of the wild type, the lag phases of the rpoN mutants were extended for 30 h on mannitol and KNO3 and for 60 h on medium containing both succinate and KNO3 (data not shown).
FIG. 2.
Effects of malate and succinate concentrations on the growth curves of the R. etli wild-type strain CNPAF512 (A) and rpoN (B), ptsN (C), and ptsA (D) mutants. Cultures were grown in AMS medium containing NH4Cl (20 mM) as a nitrogen source and mannitol (20 mM) (▪), succinate (10 mM [▴] or 20 mM [▵]), or malate (10 mM [•] or 20 mM [○]) as a carbon source. OD595, OD at 595 nm.
Growth of R. etli ptsN mutants.
Insertion mutants of ptsN (FAJ1164 and FAJ1165) were constructed as detailed in Materials and Methods. The insertion of the Ω-Km interposon in both mutants was controlled by Southern hybridization with the appropriate probes.
In a preliminary experiment, growth of each of the ptsN mutants was assayed on AMS plates containing alanine. When compared with that of the parental strain, growth of the R. etli ptsN mutants was impaired on medium containing mannitol as a carbon source and alanine as a nitrogen source. This observation led us to analyze the effect of these mutations on the metabolism of two other carbon sources, glucose and succinate. Therefore, strains were grown on AMS plates containing alanine, serine, glutamine, or ammonium in combination with mannitol, glucose, or succinate. From Table 4 it can be seen that the inactivation of ptsN clearly reduces growth on medium containing succinate in the presence of alanine, serine, ammonium, or, to a lesser extent, glutamine. Such a strong effect was not observed with medium devoid of succinate but containing alanine or serine in the absence or presence of ammonium (data not shown), indicating that growth inhibition of the ptsN mutant is mediated primarily by the carbon source (see below).
TABLE 4.
Growth of wild-type R. etli and mutants on defined media containing Ala, Ser, Gln, or ammonium in combination with mannitol, glucose, or succinatea
| Strain | Growth with:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mannitol
and:
|
Glucose and:
|
Succinate and:
|
||||||||||
| Ala | Ser | Gln | Am | Ala | Ser | Gln | Am | Ala | Ser | Gln | Am | |
| CNPAF512 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| FAJ1154 | − | − | +++ | + | − | +/− | +++ | ++ | − | − | ++ | +/− |
| FAJ1165 | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +/− | + | ++ | + |
| FAJ1166 | + | ++ | + | + | + | ++ | + | + | +/− | +/− | +/− | +/− |
Growth tests were carried out on AMS plates containing the indicated combinations of amino acids (0.2%) or ammonium (Am) (10 mM) and sugars (0.2%). Growth conditions and symbols are as defined in the footnotes to Table 3.
In addition to tests with mannitol and glucose, colony size of the ptsN mutants was also evaluated on AMS plates containing NH4Cl, alanine, or glutamine as a nitrogen source and sucrose, arabinose, galactose, sorbitol, fructose, or glycerol as a carbon source. No difference or only a small decrease in colony size between the wild type and the ptsN mutants was observed (data not shown).
Production of melanin and expression of pnifH-gusA in R. etli rpoN and ptsN mutants.
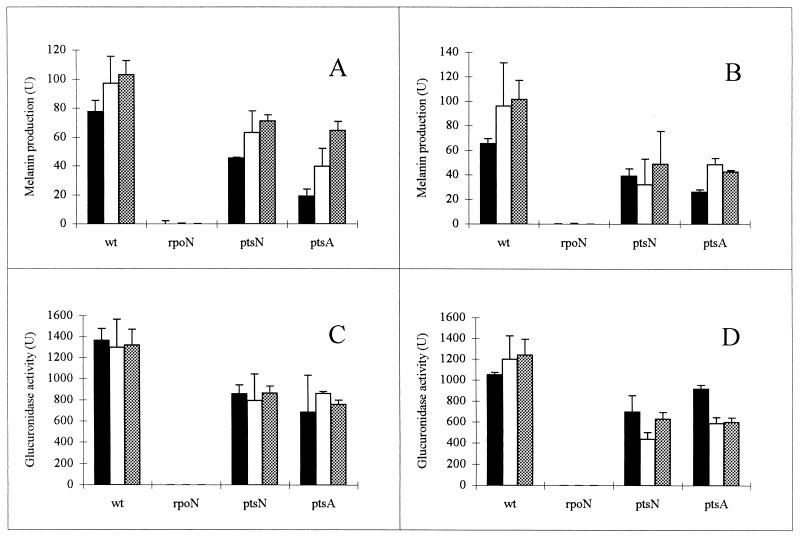
For R. etli CNPAF512, the production of the black pigment melanin was previously demonstrated to depend on the nifA gene (34). In addition, when reconstituted in E. coli, this phenotype was shown to be RpoN dependent (17). We therefore tested melanin synthesis in the rpoN mutant. These experiments were also performed with the ptsN mutant, since this gene was previously demonstrated to affect the expression of RpoN-dependent genes (30). Melanin production was first assayed on TY plates containing tyrosine and CuSO4 (17). As a control, we used the nifA mutant strain Rp1000, which is Mel− (34). Both rpoN mutant strains were unable to produce the black pigment melanin, while no difference between the wild-type strain and ptsN mutants was observed. However, since minor differences in the production of melanin cannot readily be detected by this method, melanin synthesis was also determined quantitatively on media containing different carbon sources (Fig. 3A and B). These quantified data clearly show a reduction of melanin synthesis in all mutants tested. Pigment production was abolished in the rpoN mutants and reduced two- to threefold in the ptsN mutant, depending on the composition of the growth medium.
FIG. 3.
Melanin production (A and B) and expression of nifH (C and D) in wild-type R. etli (wt) and rpoN (FAJ1154), ptsN (FAJ1165), and ptsA (FAJ1166) mutants. All data are the means from four independent replicates. Error bars denote the standard deviations. Precultures were grown overnight in TY medium at 30°C, diluted 20-fold in the different media, and incubated overnight with 0.5% oxygen (34). The nitrogen source used was alanine (20 mM). The carbon sources were mannitol (black bars), succinate (stippled bars), and malate (white bars) at 5 mM (A and C) and 20 mM (B and D). To quantify melanin production, cultures were lysed at 37°C in the presence of a solution containing sodium dodecyl sulfate (1%), CuSO4 (10 μg/ml), and tyrosine (30 μg/ml). The OD of the culture after lysis was measured at 340 nm in a microplate reader after 60 and 120 min of incubation. The difference between the ODs at 340 nm was used to calculate the units. Units are expressed as the ratio of the change in OD at 340 nm to the OD at 595 nm. β-Glucuronidase activities of the translational pnifH-gusA fusion plasmid pFAJ21 are expressed as Miller units.
Finally, we analyzed the induction of the pnifH-gusA fusion plasmid pFAJ21 (Fig. 3C and D). Similar to the case for the production of melanin, expression of pFAJ21 is abolished in both rpoN mutants and is reduced by 30 to 70% in the ptsN mutant compared to the wild type.
The decreased melanin synthesis or nifH expression is not caused by a reduction of nifA transcription. No difference in the expression level of a pnifA-gusA fusion was observed for the different mutant strains (data not shown). Also, expression of the pamtB-gusA fusion plasmid pFAJ302 was tested under the same conditions. The amtB gene is regulated by the RpoN and NtrC proteins. As expected, this fusion plasmid was expressed at a low level in the rpoN mutant strain. However, no reduction of the activity of this fusion was observed in the ptsN mutant compared to the wild type under the same conditions as tested in Fig. 3 (data not shown). The nifH and amtB genes, used to construct the fusions pFAJ21 and pFAJ302, were originally isolated from Azospirillum brasilense. Expression of pFAJ21 is RpoN/NifA dependent, while that of pFAJ302 is RpoN/NtrC dependent. These fusions have been clearly demonstrated to be regulated similarly in Azospirillum and Rhizobium and are therefore suitable for the expression studies detailed above.
Analysis of the R. etli Mel− strain FAJ1166.
In an attempt to isolate regulatory mutants affected in the production of melanin, a library of Tn5-mob mutants of R. etli CNPAF512 was previously screened on plates. Several mutants with a reduced level of melanin production were isolated. One of these mutants, FAJ1166 (which has the lowest level of melanin production among the isolated mutants), was further characterized. By the quantitative assay, FAJ1166 was shown to have a two- to fivefold reduction in melanin synthesis (Fig. 3A and B). Also, expression of pFAJ21 was reduced approximately twofold (Fig. 3C and D). These phenotypes were not caused by reduced expression of the nifA gene as demonstrated by the analysis of a pnifA-gusA fusion plasmid (data not shown). Also, under the same growth conditions, expression of pFAJ302 was not reduced in this mutant (data not shown). To further analyze this mutant, chromosomal DNA was partially digested with EcoRI and ligated to the cosmid vector pSUP205 (47). The ligation mixture was packaged into phage heads and used to infect E. coli HB101 cells. Cosmids containing the Tn5 insert were selected on kanamycin. Next, a fragment of approximately 14 kb carrying the Tn5 transposon was subcloned into pUC18. By using a primer derived from the sequence of the Tn5 inverted repeat (5′-GGTTCCGTTCAGGACGCT-3′), the DNA region of one site of the Tn5-flanking DNA was sequenced. This sequence coded for a peptide with homology to E. coli enzyme I of the PTS system (Fig. 4). Further analysis indicated that growth of mutant FAJ1166 was considerably impaired on all solid media tested, although the effect was strongest on medium containing succinate (Tables 3 and 4). However, growth of this mutant did not depend solely on the carbon source used. Higher growth rates were observed when serine instead of alanine, glutamine, or ammonia was used in combination with mannitol or glucose. Finally, in contrast to the wild type or the ptsN mutants, FAJ1166 had lost its mucoid morphology on all media tested.
FIG. 4.
DNA sequence flanking the Tn5-mob insertion of R. etli FAJ1166. The deduced amino acid sequence (RE) is compared to that of the E. coli (EC) phosphoenolpyruvate-protein phosphotransferase PtsA (enzyme I; accession no. P32670) from amino acid 111 to 226. The conserved phosphorylation site (H189) is underlined. Identical and similar (S-T-A, L-V-I-M, K-R, Q-N, and F-Y-W) residues are indicated below the amino acid sequences (cons). Stars denote similar residues.
Growth on TCA cycle acids.
As observed on solid medium, growth of ptsN and ptsA mutants is impaired in the presence of succinate (Table 4). Therefore growth of both mutants was also tested in liquid AMS medium containing mannitol or the C4-dicarboxylate succinate, fumarate, or malate as a carbon source and NH4Cl, alanine, or glutamine as a nitrogen source. Growth in the presence of NH4Cl is presented in Fig. 2. Growth of the ptsN and ptsA mutants was inhibited on malate and succinate. Inhibition was shown to depend on the concentration of the dicarboxylate. Complete growth inhibition of the ptsN and ptsA mutants, but not the wild-type strain, occurred in the presence of 20 mM malate (Fig. 2) or 30 mM succinate but not 30 mM fumarate (data not shown). Mannitol at concentrations of up to 30 mM or malate and succinate at a concentration of 5 mM did not interfere with growth (data not shown). Growth inhibition of these mutants decreased upon lowering of the concentration of malate or succinate. These observations were independent of the nitrogen source used.
Since cyclic AMP (cAMP) has been shown to play a central role in signalling of the PTS in E. coli, we tested whether this compound could modulate the inhibitory activity of malate. Therefore, cells of the wild-type, ptsN, ptsA, and rpoN strains were cultured in AMS medium containing alanine as the nitrogen source and mannitol, malate, succinate, or fumarate as a carbon source, either in the absence or in the presence of 0.5 mM cAMP. No effect of cAMP on the growth of these strains in the different media could be observed.
We also examined growth of the wild type and the ptsN or ptsA mutant on the other tricarboxylic acid (TCA) cycle intermediates, i.e., citrate, aconitate, isocitrate, α-ketoglutarate, oxaloacetate, and pyruvate. The organic acids were each at 20 mM, with the exception of oxaloacetate, which was used at 2 mM. Alanine was used as the nitrogen source. R. etli cells did not grow on citrate or isocitrate, while no obvious growth differences compared to the wild type were observed with the other acids.
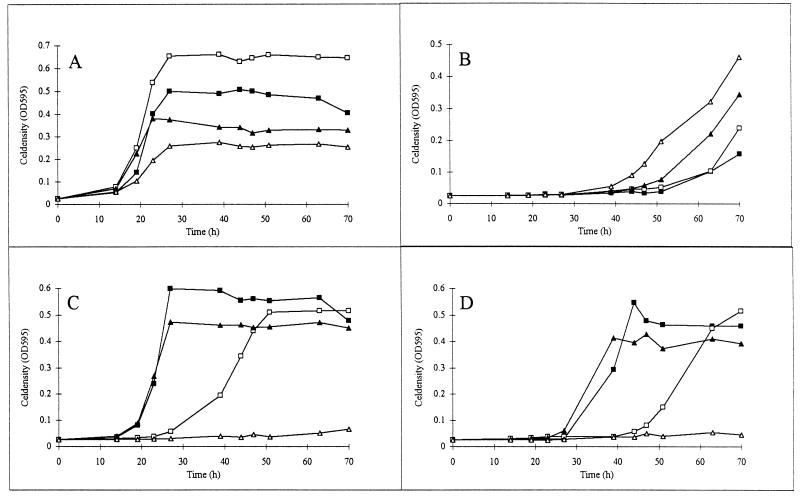
When malate was added to the growth medium in combination with mannitol or subinhibitory concentrations of succinate, growth of the ptsN and ptsA mutants was also strongly inhibited (Fig. 5). Inhibition was not observed in the presence of the same concentrations of fumarate, aspartate, or asparagine (data not shown). These data clearly indicate that, even in the presence of carbon sources that are normally metabolized by ptsN and ptsA mutants, malate inhibits growth and therefore probably affects an essential cellular function.
FIG. 5.
Toxic effects of malate on cell growth in the presence of mannitol and succinate. The strains tested are the R. etli wild-type strain CNPAF512 (A) and rpoN (B), ptsN (C), and ptsA (D) mutants. Cultures were grown in AMS medium containing alanine (20 mM) as a nitrogen source and mannitol (20 mM) (▪) or succinate (10 mM) (▴) alone or in combination with 20 mM malate (□, mannitol and malate; ▵, succinate and malate). OD595, OD at 595 nm.
The degree of ionization of dicarboxylic acids depends on the pH of the medium. In order to test whether the pH affects the toxicity of the C4-dicarboxylic acids, cell cultures were grown at various external pH values in the presence of either malate, succinate, or fumarate at 5 and 20 mM. The effect of malate is presented in Fig. 6. In contrast to the case for the wild type, the external pH affects the toxicity of malate and, to a lesser extent, succinate in the ptsN mutant at a concentration of 20 mM but not 5 mM (Fig. 6 and data not shown). Growth inhibition was highest at pH 7.0 and 6.5 but was absent at pH 6.0 or pH 5.5. Similar results were obtained with the ptsA mutant. No effect of pH on the assimilation of fumarate was observed (data not shown). Also, in the rpoN mutant at 20 mM malate, growth was inhibited by increasing the pH (Fig. 6). No effect of external pH values of between 5.5 and 7.0 on the assimilation of succinate in the rpoN mutant was found (data not shown).
FIG. 6.
Influence of pH on growth of R. etli wild-type strain CNPAF512 (A) and rpoN (B), ptsN (C), and ptsA (D) mutants. Alanine (20 mM) was used as a nitrogen source. Malate was at 20 mM. Symbols: ▪, pH 5.5; •, pH 6.0; ▴, pH 6.5; ⧫, pH 7.0. OD595, OD at 595 nm.
DISCUSSION
The rpoN locus of R. etli was isolated by using the B. japonicum rpoN1 gene as a probe. The nucleotide sequence of this region was determined, and four complete ORFs (ORF258, rpoN, ORF191, and ptsN) were identified. In most organisms analyzed so far, the organization of ORFs linked to the rpoN gene is remarkably well conserved (31). Exceptions are Rhodobacter capsulatus and Rhodobacter sphaeroides, in which rpoN is cotranscribed with nitrogen fixation genes (27, 28). The products of the conserved ORFs located upstream from rpoN genes, including R. etli ORF258, share features with the family of the ATP-binding cassette-type permeases. However, unlike those in other bacterial species, the two conserved downstream ORFs in R. etli (ORF191 and ptsN) are not adjacent to the rpoN gene but are separated by approximately 1.6 kb. Also, in C. crescentus homologs of these two genes are located 0.8 kb downstream from rpoN (19). The ptsN gene is homologous to genes coding for the enzyme IIA protein of the PTS (42). In bacteria, the PTS facilitates the uptake of many sugars (44) (see below). In E. coli and K. pneumoniae, sequence analysis of the DNA region downstream from ptsN has revealed the presence of two additional ORFs: ORF284 (also partially sequenced in Pseudomonas putida and P. aeruginosa [20]) and ORF90 (21, 32, 39). The product of the latter gene, NPr, is homologous to Hpr-like proteins of the PTS. No homologs of these two genes were detected in the DNA region downstream from R. etli ptsN. In addition, a rho-independent termination sequence was found 19 bp downstream from ptsN, suggesting that transcription did not extend further downstream. For E. coli, it was suggested that the NPr protein modulates the phosphorylation status of PtsN and as a result determines the activity of the protein (39) (see below). It is therefore possible that mechanisms of fine tuning of PtsN activity in R. etli are different from those in E. coli.
Expression of rpoN genes in E. coli and K. pneumoniae is at a low constitutive level (7, 29). In contrast, expression of the R. etli rpoN gene is increased in an rpoN mutant background. A similar situation exists in B. japonicum (23) and P. putida (22), where the rpoN gene (rpoN2 of B. japonicum) is also negatively autoregulated. Inspection of the R. etli rpoN promoter region revealed the presence of an RpoN consensus binding site, reading in the leftward direction (on the template strand). On the basis of sequence conservation between the R. meliloti and R. etli rpoN promoter regions (the R. etli rpoN gene can also functionally substitute for R. meliloti rpoN), this RpoN-binding site was shown to overlap the −10 promoter region and the transcription start site, as determined for R. meliloti (Fig. 7). Moreover, we observed that the promoter sequences and the oppositely oriented RpoN-binding sites were also conserved in the promoter regions of Rhizobium sp. strain NGR234 rpoN (one nucleotide missing) and B. japonicum rpoN2 (Fig. 7). We could not detect such a binding site in the 5′ region of the E. coli and K. pneumoniae rpoN genes, which are not autoregulated but are transcribed at a low constitutive level (7, 29). In P. putida, a putative RpoN-binding sequence also was identified three nucleotides downstream from the −10 region (22). However, this sequence was located on the noncoding strand. Also, in the Acinetobacter calcoaceticus rpoN gene, which is negatively autoregulated, a −24/−12 promoter consensus sequence is found on the noncoding strand near the transcriptional start site (13). The RpoN protein is able to bind in vitro to −24/−12-type promoters in the absence of the RNA polymerase core enzyme or of an activator protein (6, 11). It is therefore possible that negative autoregulation of R. etli and B. japonicum rpoN genes occurs by direct interference with Eς70 functioning through the binding of ς54 (or Eς54) to the −10 to +1 promoter region of the rpoN gene, thereby inhibiting transcription initiation. Negative regulation of ς70 promoters by binding of the repressor to the −10 to +1 region of the promoter is well documented for E. coli (16).
FIG. 7.
Alignment of promoter regions from rhizobial rpoN genes. The aligned rpoN sequences are from R. meliloti (Rm), R. etli (Re), Rhizobium sp. strain NGR234, and B. japonicum rpoN2 (BjrpoN2). The −10 and −35 regions from the R. meliloli rpoN promoter are overlined, and the transcription start site is indicated by an arrow (43). The inverse/complement of the consensus (Cons) sequence of a −24/−12 promoter is indicated; conserved GC and CC residues are in boldface. Nucleotides identical to the consensus −24/−12 sequence are underlined. Ec, E. coli.
R. etli rpoN mutants were constructed by insertion of the Ω-Km interposon in both directions in the gene. These mutations are polar and may thus affect the functioning of the genes located downstream from and belonging to the same operon as rpoN. However, the phenotypes of an R. etli strain carrying a mutation immediately downstream from the rpoN gene (data not shown) and of the ptsN mutants were different from that of the rpoN mutants. In addition, insertion of a gusA-aphII cassette into the rpoN gene, giving rise to nonpolar mutations (33), produced the same phenotypic defects as in the interposon mutants. Therefore, the phenotype of the rpoN mutants is not caused by a polar effect on downstream genes but can be attributed to inactivation of rpoN.
Growth defects of the R. etli rpoN mutants on different C and N sources were observed. When growth was tested on plates containing different amino acids as carbon and/or nitrogen sources, growth of the mutant, but not that of the wild-type strain, was inhibited on plates containing either serine or alanine as the sole C and N source. Growth defects may be attributed to the inability of the rpoN mutant strain to catabolize serine and alanine or to assimilate its degradation products. In E. coli, the metabolism of serine and that of alanine involve different genes (41). Both amino acids are degraded to pyruvate and ammonium. The ammonia produced is used to synthesize glutamate and glutamine. The corresponding R. etli genes may also be subject to an RpoN-type regulation. However, serine and alanine also inhibit growth of the R. etli rpoN mutants on medium containing ammonium and mannitol. The supplementary addition of glutamine to these media relieved growth inhibition. In E. coli, alanine and serine inhibit the activity of glutamine synthetase. A similar inhibitory action on the glutamine synthetase enzyme in R. etli might explain the observed phenotypes. The analysis of ammonia assimilation in R. etli is complicated by the presence of three different glutamine synthetase genes, glnA (coding for GSI), glnII (GSII), and glnT (GSIII). Rhizobium GSI enzymes are similar to the GSI of E. coli and might therefore also be inhibited by alanine and serine. GSII is similar to eukaryotic glutamine synthetases. In R. etli, transcription of glnII is RpoN dependent (36), while that of the glnA gene is not (8). Possibly, the wild-type strain synthesizes GSI and/or GSII, while the rpoN mutant produces only GSI. The observed phenotypes may therefore be attributed to the inability of the rpoN mutant to assimilate ammonia produced by the degradation of alanine and serine. A defect in the assimilation of ammonia by the rpoN mutant strain was also observed in liquid medium containing ammonium as the sole nitrogen source. The presence of a second glutamine synthetase, encoded by glnII, may thus confer a selective advantage under those conditions where GSI activity is inhibited. It is not known whether Rhizobium is subject to such conditions during its life cycle.
Growth of the rpoN mutants on succinate, fumarate, and malate was delayed compared to that of the parental strain. Therefore, in R. etli, an RpoN-dependent system for the uptake of C4-dicarboxylates is likely to exist. However, after an extended lag phase, the mutants grew as fast as the wild type, indicating that in the absence of RpoN, the activation of alternative control mechanisms of the RpoN-dependent C4-dicarboxylate transport (dct) system or the existence of a second, inducible, dct system may account for the observed growth kinetics. This is in contrast to the case for R. meliloti, Rhizobium sp. strain NGR234, and A. caulinodans (43, 48, 52). rpoN mutants of these bacteria cannot grow on succinate as the sole carbon source. Alternatively, a B. japonicum rpoN double mutant is not impaired in the assimilation of dicarboxylates. For this species, the existence of an RpoN-independent dicarboxylate uptake system has been suggested (23).
In R. etli, interposon mutagenesis of ptsN results in reduced expression of pnifH and decreased production of the pigment melanin. Due to the presence of a putative rho-independent terminator, the ptsN gene is probably not cotranscribed with downstream genes, and the observed phenotype is likely due to its inactivation. Growth of the ptsN mutant in media containing mannitol, glucose, and dicarboxylic acids in combination with different nitrogen sources (alanine, serine, glutamine, and ammonium) was tested. Although growth inhibition of the mutant on plates containing alanine and serine in the presence of mannitol and glucose was observed, the strongest inhibition resulted from the presence of the C4-dicarboxylates. Also, in liquid medium, growth of the ptsN mutant was impeded in the presence of malate or succinate but not fumarate or mannitol, independently of the nitrogen source (alanine, ammonia, or glutamine) used. This growth inhibition was dependent on the concentration of the dicarboxylic acid. Malate was inhibitory at lower concentrations than succinate. Also, a clear effect of pH on the toxicity of malate was observed; toxicity was greatly reduced upon lowering of the pH of the growth medium. Finally, the inhibitory activity of malate is dominant over the assimilation of additional carbon sources that are normally metabolized (mannitol or low concentrations of succinate). These observations indicate that malate inhibits an essential cellular function or metabolic activity.
Independently of the analysis of the rpoN locus, we identified a Tn5-induced R. etli mutant with a reduced production of the black pigment melanin. Partial sequence analysis indicated that the mutated gene corresponds to a homolog of the E. coli gene coding for enzyme I of the PTS. Interestingly, this mutant also displayed reduced expression of pnifH. In addition, growth of this mutant was inhibited in the presence of malate or succinate, similar to what was observed with the ptsN mutant. Presently, we cannot exclude the possibility that the phenotype of FAJ1166 may be due to polarity of the Tn5 transposon on a thus-far-unidentified downstream gene.
From these observations, two conclusions can be drawn. First, the rpoN, ptsN, and ptsA mutants display several similar phenotypes. The expression of nifH or the production of melanin is abolished in the rpoN mutant and reduced in the ptsN and ptsA mutant strains. Also, depending on the carbon source used, growth of the ptsN mutant is impaired in the presence of alanine and serine, as observed in the rpoN mutant. These data indicate that PtsN and PtsA may act as coregulators of RpoN-dependent genes. However, no effect of these mutations on the activation of the RpoN-dependent amtB promoter was observed, indicating that not all of the RpoN-regulated genes are also controlled by PtsN and PtsA. Analysis of the conserved ORFs located downstream from rpoN genes has been carried out with only a few bacterial species. In K. pneumoniae, mutations in the two ORFs immediately downstream from rpoN (ORF95 and ptsN) increase the expression from the ς54-dependent pnifH, pnifL, and pglnAp2 (30). However, inactivation of the fourth gene (encoding an HPr-like product) results in decreased expression from these same promoters (32). In P. aeruginosa, ptsN positively controls genes involved in the assimilation of glutamine and does not influence pilin or flagellin genes (20). In E. coli, induction of the glnAp2 promoter is not dependent on ptsN, although ptsN is required for maximal growth on alanine or adenosine as the sole nitrogen source. In addition, ptsN facilitates the use of these compounds as organic nitrogen sources in the presence of additional carbon sources, especially TCA cycle intermediates. Finally, ptsN suppresses mutations in the GTPase Era (39). In C. crescentus, ptsN is involved in fine tuning the expression from the fljK promoter but not from those of other ς54-dependent flagellar genes (19). Clearly, the phenotypes of mutations in the rpoN-linked genes largely differ between different species and between the experimental systems used. Merrick and Coppard (30) proposed that the ORF95 and PtsN proteins control the stability of either the Eς54 complex or the Eς54-DNA complex. Alternatively, it was recently suggested that these downstream ORFs may also control the level of reinitiation of transcription by affecting the rate of release or conformational change of the ς54 protein bound at the −24/−12 promoter after transcription has initiated (50). In both models, since the primary sequence of the promoter affects the stability of ς54 binding, this sequence may also account for the differential effects of mutations in the downstream ORFs on the expression of these genes.
Second, it is striking that both the rpoN mutant and the ptsN (or ptsA) mutant have growth defects on C4-dicarboxylates. However, these defects clearly result from two different mechanisms. The ptsN and ptsA mutant strains have wild-type growth on low concentrations of malate and succinate, while these concentrations are strongly inhibitory for the rpoN mutant. In addition, increasing concentrations of these dicarboxylates stimulate growth of the rpoN mutant strain (Fig. 2 and 5) but repress growth of the ptsN and ptsA mutants. The defects of the rpoN mutant are probably due to the absence of a high-affinity dct system (see above). Growth inhibition of the ptsN and ptsA mutants is relieved by low pH. Assuming that the deprotonation of C4-dicarboxylates increases their permease-mediated uptake, it is likely that at pH 7.0, the toxic effects of both dicarboxylates are caused by their increased cellular concentrations. Possibly, the input of larger amounts of dicarboxylates can affect the TCA cycle activities, resulting in growth inhibition. In R. etli, the catabolism of glucose is arrested in the presence of dicarboxylates (25). Therefore, a sensing system for dicarboxylates is likely to be operative. This system might signal to the cell the concentration of C4-dicarboxylates in the growth medium. Depending on the concentration of these compounds, cells may, for example, have to decrease their uptake. Given the well-described role of the PTS in signalling in other microorganisms and since toxicity of malate and succinate is strongly increased in ptsN and ptsA mutants, this system might involve ptsN and ptsA. In other microorganisms, the PTS consists of two general proteins, enzyme I and Hpr, and the sugar-specific permease enzyme II. Enzyme II comprises up to four different polypeptides or domains (IIA, IIB, IIC, and IID). A phosphoryl group is sequentially transferred from phosphoenolpyruvate to enzyme I, Hpr, enzyme II, and, concomitant with its uptake, the sugar (44). PtsN, which is homologous to the enzyme IIA subunit, from E. coli is phosphorylated in vitro by the standard PTS phosphorelay involving phosphoenolpyruvate, enzyme I, and Hpr (39). Similar results were obtained with the K. pneumoniae ORF162 product PtsN (4, 32). Since ptsA and ptsN mutants have similar phenotypes, the phosphotransfer cascade of the PTS system in R. etli possibly controls the phosphorylation of the PtsN protein, thereby modulating its activity.
ACKNOWLEDGMENTS
We thank E. Martínez-Romero for characterizing Rhizobium strain CNPAF512 and H.-M. Fischer for providing plasmid pRJ7694.
J.M. is a postdoctoral fellow of the Fund for Scientific Research—Flanders.
REFERENCES
- 1.Albright L M, Ronson C W, Nixon B T, Ausubel F M. Identification of a gene linked to Rhizobium meliloti ntrAwhose product is homologous to a family of ATP-binding proteins. J Bacteriol. 1989;171:1932–1941. doi: 10.1128/jb.171.4.1932-1941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M. Regulation of nitrogen fixation genes. Cell. 1984;37:5–6. doi: 10.1016/0092-8674(84)90294-0. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 4.Begley G S, Jacobson G R. Overexpression, phosphorylation, and growth effects of ORF162, a Klebsiella pneumoniae protein that is encoded by a gene linked to rpoN, the gene encoding ς54. FEMS Microbiol Lett. 1994;119:389–394. doi: 10.1111/j.1574-6968.1994.tb06918.x. [DOI] [PubMed] [Google Scholar]
- 5.Buck M, Miller S, Drummond M, Dixon R. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature. 1986;320:374–378. [Google Scholar]
- 6.Buck M, Cannon W. Specific binding of the transcription factor ς54to promoter DNA. Nature. 1992;358:422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- 7.Castano I, Bastarrachea F. glnF::lacZ fusions in Escherichia coli: studies on glnFexpression and its chromosomal orientation. Mol Gen Genet. 1984;195:228–233. doi: 10.1007/BF00332751. [DOI] [PubMed] [Google Scholar]
- 8.Chiurazzi M, Iaccarino M. Transcriptional analysis of the glnB-glnA region of Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1990;4:1727–1735. doi: 10.1111/j.1365-2958.1990.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 9.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppard J R, Merrick M J. Cassette mutagenesis implicates a helix-turn-helix motif in promoter recognition by the novel RNA polymerase sigma factor ς54. Mol Microbiol. 1991;5:1309–1317. doi: 10.1111/j.1365-2958.1991.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 11.D’hooghe I, Michiels J, Vlassak K, Verreth C, Waelkens F, Vanderleyden J. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoliCNPAF512. Mol Gen Genet. 1995;249:117–126. doi: 10.1007/BF00290243. [DOI] [PubMed] [Google Scholar]
- 12.Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 13.Ehrt S, Ornston L N, Hillen W. RpoN (ς54) is required for conversion of phenol to catechol in Acinetobacter calcoaceticus. J Bacteriol. 1994;176:3493–3499. doi: 10.1128/jb.176.12.3493-3499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitroinsertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 15.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobiummutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 16.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 17.Hawkins F K L, Johnston A W B. Transcription of a Rhizobium leguminosarum biovar phaseoli gene needed for melanin synthesis is activated by nifA of Rhizobium and Klebsiella pneumoniae. Mol Microbiol. 1988;2:331–337. doi: 10.1111/j.1365-2958.1988.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 18.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 19.Janakiraman R S, Brun Y V. Transcriptional and mutational analyses of the rpoN operon in Caulobacter crescentus. J Bacteriol. 1997;179:5138–5147. doi: 10.1128/jb.179.16.5138-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S, Ishimoto K, Lory S. Nucleotide sequence of the rpoN gene and characterization of two downstream open reading frames in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1316–1322. doi: 10.1128/jb.176.5.1316-1322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones D H A, Christopher F, Franklin H, Thomas C M. Molecular analysis of the operon which encodes the RNA polymerase sigma factor ς54 of Escherichia coli. Microbiology. 1994;140:1035–1043. doi: 10.1099/13500872-140-5-1035. [DOI] [PubMed] [Google Scholar]
- 22.Köhler T, Alvarez J F, Harayama S. Regulation of the rpoN, ORF102 and ORF154 genes in Pseudomonas putida. FEMS Microbiol Lett. 1994;115:177–184. doi: 10.1111/j.1574-6968.1994.tb06634.x. [DOI] [PubMed] [Google Scholar]
- 23.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H-M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lafontaine P J, Lafrenière C, Antoun H. Some properties of carbohydrate and C4-dicarboxylic acid utilization negative mutants of Rhizobium leguminosarum biovar phaseolistrain P121. Plant Soil. 1989;120:195–201. [Google Scholar]
- 26.Lonetto M, Gribskov M, Gross C A. The ς70family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Martínez-Romero, E. Personal communication.
- 27.Masepohl B, Klipp W, Pühler A. Genetic characterization and sequence analysis of the duplicated nifA/nifB gene region of Rhodobacter capsulatus. Mol Gen Genet. 1988;212:27–37. doi: 10.1007/BF00322441. [DOI] [PubMed] [Google Scholar]
- 28.Meijer W G, Tabita F R. Isolation and characterization of the nifUSVW-rpoN gene cluster from Rhodobacter sphaeroides. J Bacteriol. 1992;174:3855–3866. doi: 10.1128/jb.174.12.3855-3866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrick M J, Stewart W D P. Studies on the regulation and function of the Klebsiella pneumoniae ntrAgene. Gene. 1985;35:297–303. doi: 10.1016/0378-1119(85)90008-3. [DOI] [PubMed] [Google Scholar]
- 30.Merrick M J, Coppard J R. Mutations in genes downstream of the rpoN gene (encoding ς54) of Klebsiella pneumoniae affect expression from ς54-dependent promoters. Mol Microbiol. 1989;3:1765–1775. doi: 10.1111/j.1365-2958.1989.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 31.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 32.Merrick M J, Taylor M, Saier M H, Jr, Reizer J. The role of the genes downstream of the ςN structural gene rpoN in Klebsiella pneumoniae. In: Tikhonovich I A, Provorov N A, Romanov V I, Newton W E, editors. Nitrogen fixation: fundamentals and applications. London, United Kingdom: Kluwer Academic Publishers; 1995. pp. 189–194. [Google Scholar]
- 33.Metcalf W W, Wanner B L. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- 34.Michiels J, D’hooghe I, Verreth C, Pelemans H, Vanderleyden J. Characterization of the Rhizobium leguminosarum biovar phaseoli nifA gene, a positive regulator of nifgene expression. Arch Microbiol. 1994;161:404–408. doi: 10.1007/BF00288950. [DOI] [PubMed] [Google Scholar]
- 35.Norrander K, Kempe T, Messing K. Construction of improved M13 vectors using oligonucleotide directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 36.Patriarca E J, Chiurazzi M, Manco G, Riccio A, Lamberti A, De Paolis A, Rossi M, Defez R, Iaccarino M. Activation of the Rhizobium leguminosarum glnIIgene by NtrC is dependent on upstream DNA sequences. Mol Gen Genet. 1992;234:337–345. doi: 10.1007/BF00538692. [DOI] [PubMed] [Google Scholar]
- 37.Poole P S, Schofield N A, Reid C J, Drew E M, Walshaw D L. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 38.Popham D, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 39.Powell B S, Court D L, Inada T, Nakamura Y, Michotey V, Cui X, Reizer A, Saier M H, Reizer J. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. J Biol Chem. 1995;270:4822–4839. doi: 10.1074/jbc.270.9.4822. [DOI] [PubMed] [Google Scholar]
- 40.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 41.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 42.Reizer J, Reizer A, Saier M H, Jacobsen G R. A proposed link between nitrogen and carbon metabolism involving protein phosphorylation in bacteria. Protein Sci. 1992;1:722–726. doi: 10.1002/pro.5560010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronson C W, Nixon B T, Albright L M, Ausubel F M. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol. 1987;169:2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saier M H, Jr, Reizer J. Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1992;174:1433–1438. doi: 10.1128/jb.174.5.1433-1438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Sasse-Dwight S, Gralla J D. Probing the Escherichia coli glnALGupstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon R U, Priefer U, Pühler A. A broad host range mobilization system for in vivogenetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 48.Stigter J, Schneider M, de Bruijn F J. Azorhizobium caulinodans nitrogen fixation (nif/fix) gene regulation: mutagenesis of the nifA −24/−12 promoter element, characterization of a ntrA (rpoN) gene, and derivation of a model. Mol Plant-Microbe Interact. 1993;6:238–252. doi: 10.1094/mpmi-6-238. [DOI] [PubMed] [Google Scholar]
- 49.Thöny B, Hennecke H. The −24/−12 promoter comes of age. FEMS Microbiol Rev. 1989;63:341–358. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 50.Tintut Y, Jonathan T W, Gralla Y D. A novel bacterial transcription cycle involving ς54. Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 51.Vande Broek A, Michiels J, de Faria S M, Milcamps A, Vanderleyden J. Transcription of the Azospirillum brasilense nifHgene is positively regulated by NifA and NtrA and is negatively controlled by the cellular nitrogen status. Mol Gen Genet. 1992;232:279–283. doi: 10.1007/BF00280007. [DOI] [PubMed] [Google Scholar]
- 52.van Slooten J C, Cervantes E, Broughton W J, Wong C H, Stanley J. Sequence and analysis of the rpoN sigma factor gene of Rhizobiumsp. strain NGR234, a primary coregulator of symbiosis. J Bacteriol. 1990;172:5563–5574. doi: 10.1128/jb.172.10.5563-5574.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Z, Charles T C, Wang H, Nester E W. The ntrA gene of Agrobacterium tumefaciens: identification, cloning, and phenotype of a site-directed mutant. J Bacteriol. 1992;174:2720–2733. doi: 10.1128/jb.174.8.2720-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]